Abstract
Variation of metabolic profiles in Cordyceps pruinosa mycelia cultivated under various media and light conditions was investigated using 1H nuclear magnetic resonance (NMR) analysis and gas chromatography mass spectrometry (GC-MS) with multivariate statistical analysis. A total of 71 metabolites were identified (5 alcohols, 21 amino acids, 15 organic acids, 4 purines, 3 pyrimidines, 7 sugars, 11 fatty acids, and 5 other metabolites) by NMR and GC-MS analysis. The mycelia grown in nitrogen media and under dark conditions showed the lowest growth and ergosterol levels, essential to a functional fungal cell membrane; these mycelia, however, had the highest levels of putrescine, which is involved in abiotic stress tolerance. In contrast, mycelia cultivated in sabouraud dextrose agar with yeast extract (SDAY) media and under light conditions contained relatively higher levels of fatty acids, including valeric acid, stearic acid, lignoceric acid, myristic acid, oleic acid, palmitoleic acid, hepadecenoic acid, and linoleic acid. These mycelia also had the highest phenolic content and antioxidant activity, and did not exhibit growth retardation due to enhanced asexual development caused by higher levels of linoleic acid. Therefore, we suggested that a light-enriched environment with SDAY media was more optimal than dark condition for cultivation of C. pruinosa mycelia as biopharmaceutical or nutraceutical resources.
Introduction
The anamorph of Cordyceps pruinosa is the fungus Mariannaea pruinosa. C. pruinosa, which belongs to the Ascomycota phylum, is a well known entomogenous fungus that parasitizes the larvae of Lepidopter [1], [2]. Many beneficial genetic and biological characteristics of C. pruinosa mycelia have been identified. Among these are the many secondary metabolic compounds such as polysaccharides, N-6-(2-hydroxyethyl) adenosine, adenosine, and anti-ultraviolet radiation constituents [3]–[5].
Previous studies have shown that the methanol extract of the fruit body of C. pruinosa is able to curb NF-κB–dependent inflammatory gene expression, and that the mycelium of C. pruinosa improves cellular immune functions [6], [7]. Furthermore, the butanol fraction of C. pruinosa might play an effective role as an anti-proliferative agent for cancer therapy [8].
However, global metabolite profiling of C. pruinosa has not yet been performed. Metabolic profiling has the potential to help improve our understanding of the variety of physiological properties associated with diverse metabolites of C. pruinosa. However, it is known that metabolite production is affected by environmental factors such as light exposure and culture medium components. Many previous studies have investigated the effects of environmental changes on the production of secondary metabolites in Cordyceps species [9]–[12]. In addition, there have been many studies of antioxidants from other Cordyceps species, including C. militaris and C. sinensis [13], [14]. Until recently, metabolic profiles and antioxidative activity in C. pruinosa mycelium had not been investigated.
We hypothesized that the metabolic profile and antioxidative activity of C. pruinosa mycelia could be modulated by controlling cultivation conditions. The main objective of this study was to perform metabolic profiling in mycelia of C. pruinosa cultivated under various culture environments and determine the optimal culture conditions for the production of the mycelia with the highest antioxidant activity.
Materials and Methods
Solvents and chemicals
First grade methanol, hexane, water, D2O [99.%, containing 0.05% 3-(trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt (TSP) as internal standard for NMR], methoxylamine hydrochloride, pyridine, L-ascorbic acid, 1,1-diphenyl-2-picrylhydrzyl (DPPH), Folin-Ciocalteu's phenol reagent, gallic acid, and sodium carbonate were purchased from Sigma (St. Louis, MO, USA). NaOD were obtained from Cortec (Paris, France). BSTFA [N,O-bis (trimethylsilyl) trifluoroacetamide containing 1% TMCS (trimethyl chlorosilane)] were purchased from Alfa Aesar (Ward Hill, MA, USA) and 2-Chloronaphtahalene as internal standard for GC-MS were purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan).
Culture conditions for C. pruinosa mycelia
C. pruinosa ascospores discharged from fresh stroma were purchased from Mushtech Co. (Chuncheon, Korea). Culture materials including dextrose, agar, peptone, and yeast extract (Difco, Detroit, MI, USA) were purchased from Sigma (Sigma, St. Louis, MO, USA), and iron was purchased from Junsei Chemical (Junsei Chemical Co., Ltd, Japan). The ascospores were inoculated with SDY medium (cultivation conditions and media components are shown in Table 1) and incubated at 25°C. After storage for 2 days, 300 µL of the discharged ascospore were suspended in sterilized water and cultivated in the following media: sabouraud dextrose agar with yeast extract (SDAY) medium without light (S+D), SDAY medium with light (S+L), nut medium without light (L+D), nut with SDAY medium without light (SL+D), iron-supplemented SDAY medium without light (SF+D), and nitrogen-supplement medium without light (N+D).
Table 1. Composition of growth medium and light conditions for cultivation of C. pruinosa mycelia.
| Medium | Medium composition (g/L) | Light condition | Abbreviation | ||||||||||||
| Dextrose | Peptone | Yeast extract | Iron- EDTA | Walnut powder | Peanut powder | Pine nut powder | Macro- elements* | Micro- elements** | FeCl3 •6H2O | amino acid*** | yeast nitrogen | Agar | Dark /light | ||
| SDAY | 20 | 5 | 5 | – | – | – | – | – | – | – | – | – | 15 | Dark | S+D |
| SDAY | 20 | 5 | 5 | – | – | – | – | – | – | – | – | – | 15 | Light | S+L |
| Nut | – | – | – | – | 3.4 | 3.4 | 3.4 | – | – | – | – | – | 15 | Dark | L+D |
| SDAY- Nut | 20 | 5 | 5 | – | 3.4 | 3.4 | 3.4 | – | – | – | – | – | 15 | Dark | SL+D |
| SDAY -Fe | 20 | 5 | 5 | 0.04 | – | – | – | – | – | – | – | – | 30 | Dark | SF+D |
| Nitrogen | – | – | – | – | – | – | – | 100 (ml) | 100 (ml) | 1 (ml) | 10 (ml) | 6.7 | 15 | Dark | N+D |
*KH2PO4 10 g/L, MgSO4 7H2O 5 g/L, CaCl2 2H2O 1 g/L. **CuSO4 5H2O 0.04 mg/100 mL, BO3H3 0.5 mg/100 mL, ZnSO4 7H2O 0.4 mg/100 mL, MnSO4 H2O 0.4 mg/100 mL, NaMoO4 2H2O 0.2 mg/100 mL. ***0.005% amino acid (L-glutamic acid, L-methionine, L-lysine, L-leucine, L-isoleucine).
To clearly separate cultivated mycelia from media at the harvest period, we used sterilized cellophane (583 Gel Dryer, Bio-SDAY medium Rad, Hercules, CA, USA) on the individual medium. All cultivation plates were incubated for 3 days at 25°C under either light (3,000 lux) or dark conditions, after which mycelia were collected from the cellophane. The mycelia were then lyophilized (FDU-1200, EYELA, Miyagi, Japan) for 24 h and frozen at −80°C until further analysis.
NMR analysis
KH2PO4 (1.232 g) was added to 100 mL of D2O for NMR measurements as a buffering agent; 0.05% 3-(trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt (TSP) was used as the internal standard for D2O. The pH of D2O was adjusted to 6.0 with the addition of 520 µL NaOD. The lyophilized powders of the mycelia from individual C. pruinosa samples (30 mg from each culture condition) were then extracted with 1 mL of D2O to obtain metabolic profiles of water-soluble metabolites. After sonication for 40 min, the samples were centrifuged at 2,000 rpm for 10 min (1730MR; Gyrogen Ltd., Daejeon, Korea), and then the supernatant was collected from each sample and filtered using a 0.45 µm filter (0.45 µm PTFE syringe filter for D2O). Each filtered sample (600 µL) was then transferred to a 5 mm NMR tube for analysis.
1H-NMR spectra were recorded at a temperature of 300 K on a 600.13-MHz Bruker Avance spectrometer (Bruker Analytische GmbH, Rheinstetten, Germany) using a cryoprobe. To suppress the residual water signal, we applied a zgpr pulse sequence. In total, 128 transients were gathered into 32 K data points with a relaxation delay of 2 s. An acquisition time per scan of 1.70 s and a spectral width of 9615.4 Hz were used. Before Fourier transformation, an exponential line broadening function of 0.30 Hz was applied to the free induction decay. In addition, 2D 1H-13C heteronuclear single quantum correlation (HSQC) experiments were additionally performed to confirm the assignment of metabolites. The HSQC spectra were obtained with a 2.0 s relaxation delay, 32 scans, and 5,896.2 Hz spectral width in F2 and 30, 864.2 Hz in F1.
MestReNova (version 6.0.4; Mestrelab Research SL, Santiago de Compostela, Spain) was used to obtain the NMR spectra, which were automatically encoded in ASCII files using AMIX (version 3.7; Bruker BioSpin, Billerica, MA, USA) software. The spectral 1H-NMR region from d = 0.56 to d = 10.00 was separated into regions with widths of 0.04 ppm, rendering 236 integrated regions in each NMR spectrum. 1H-NMR signal assignments were obtained by comparing their chemical shift with splitting patterns using the Chenomx NMR suite software (version 5.1; Chenomx, Edmonton, Canada). Each spectral intensity dataset was normalized to the total sum of the spectral regions.
GC-MS analysis
To obtain separate extractions of non-polar and polar metabolites, both methanol and n-hexane were used as extraction solvents. Each sample (20 mg) cultivated under different conditions was put into a glass eppendorf tube (Axygen, Union City, CA, USA), and then extracted with 1 mL of 70% methanol and 100% n-hexane. Following sonication, the tube containing the extract was centrifuged at 2,000 rpm for 10 min. The supernatant was collected from each sample and filtered through a 0.45 µm filter (Acrodisc Syringe Filters, Pall Corporation, NY, USA). After extracting the samples, 100 µL of each sample solution was transferred into a GC vial to conduct derivatization and GC-MS analysis.
The solutions in GC vials were dried with a nitrogen gas flow for 5 min at 60°C, and then 30 µL of 20,000 µg/mL methoxylamine hydrochloride in pyridine for oximation, 50 µL of BSTFA (N,O-Bis (trimethylsilyl) trifluoroacetamide; Alfa Aesar, Ward Hill, MA, USA) containing 1% TMCS (trimethyl chlorosilane) for trimethylsilylation (TMS) derivatization, and 10 µL of 2-chloronaphthalene (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan; 250 µg/mL in pyridine as an internal standard) were separately added to the dried samples. The derivatized samples were stored at 60°C for 60 min, and then subjected to GC-MS analysis.
To analyze the samples, a 7890A Agilent GC (Agilent Technologies, CA, USA) model equipped with a 5975C MSD detector (Agilent Technologies), autosampler (7683 B series, Agilent Technologies), split/splitless injector, injection module, and Chemstation software were used. Before analyzing the sample, the GC inlet temperature was set to 250°C with an injection volume of 1.0 µL and a split ratio of 1∶10, using helium as a carrier gas in constant-flow mode of 1.0 mL/min. The column used for analysis was a fused silica capillary column of 5% phenyl methylpolysiloxane phase (DB-5, Agilent Technologies) with dimensions of 30 m×0.25 mm i.d. ×0.25 µm film thickness.
Electron impact (EI) ionization mode was used for ionization. The detector voltage was 1400 V. The aux temperature, MS source temperature, and MS quad temperature were set to 280°C, 230°C, and 150°C, respectively. The mass range was 50–700 Da, and data were gathered in full scan mode.
For the polar metabolite analysis, The oven temperature was 80°C and was programmed to increase to 130°C (at 3°C/min) and then to 240°C (at 5°C/min) and then to 320°C (at 10°C/min; hold 3 min). For the non-polar metabolite analysis, The oven temperature was 80°C and was programmed to increase to 260°C (at 5°C/min) and then to 300°C (at 10°C/min; hold 3 min).
The mass spectra of the compounds were accepted when the match quality was more than 70% compared to the NIST-Wiley Mass Spectra Library for identification. It is important to obtain the raw GC-MS data to quantitatively compare metabolic profiles among all samples as described previously [15]. We used the Automated Mass Spectral Deconvolution and Identification System (AMDIS; http://chemdata.nist.gov/mass-spc/amdis/) for mass spectral deconvolution, which distinguishes peaks from noise and overlapping peaks. The setting values were as follows: component width = 12, adjacent peak subtraction = 1, medium resolution, medium sensitivity, and medium shape requirement. After finishing the program, ELU and FIN files were obtained as the output files of deconvolution and peak picking. However, the ELU files needed to be further analyzed with an online peak-filtering algorithm (SpectConnect, http://spectconnect.mit.edu) due to scatter and a tendency of the AMDIS data in multiple datasets to include false positives. The identification was performed using the spectra of each component, which were transferred to the NIST mass spectral search program MS Search 2.0, where they were matched to entries in the NIST MS library. The criterion for peak assignment was adopted when a match quality value was higher than 70%. Normalization to an internal standard peak area was used before multivariate statistical analysis. The relative intensities of assigned metabolites using GC-MS analysis were obtained.
Statistical analysis
The results were evaluated by SIMCA-P software (version 13.0, Umetrics, Umeå, Sweden) for principal component analysis (PCA) and partial least squares regression (PLSR) was performed using mean-centered and unit variance scaled data. Clear differences in the content of the metabolites were detected by one-way analysis of variance (ANOVA) using IBM SPSS Statistics 19 software (IBM, Somers, NY), followed by the Tukey's significant-difference test. The level of statistical significance was set at p<0.05.
DPPH assay
The latent antioxidant activity of each extract was evaluated based on the scavenging ability of the stable 1,1-diphenyl-2-picrylhydrazyl (DPPH) free radical, as described in a previous publication [16]. With a test tube, all sample solutions were added to separate wells (0.2 mL per sample), and then 3.8 mL of 100 mM DPPH was added to each tube. The tube was kept at room temperature in the dark for 30 min, and then the solution was transferred to a 96-well plate; the absorbance of the sample was measured at 520 nm. Blank solutions were prepared with 0.2 µL methanol and 3.8 µL DPPH. The antioxidant ability of each sample was calculated using the following formula: (inhibition rate, %) = (blank OD – sample OD)/blank OD ×100. Ascorbic acid was used as the positive control.
Total phenolic content
The total phenolic content (TPC) in C. pruinosa cultivated under various conditions was measured using a modified version of the Folin-Ciocalteu method with gallic acid as the standard [17]. Each extract (0.2 mL) was mixed with 4.8 mL of distilled water, followed by addition of 0.5 ml of Folin-Ciocalteu's phenol reagent (Sigma). After incubating the mixture for 2 min at room temperature, 1.5 mL of 20% (w/v) sodium carbonate solution was added. The mixture was allowed to react for 2 h at room temperature, and its absorbance was then measured at 765 nm using a microplate spectrophotometer. The results were estimated using a calibration curve of gallic acid in the concentration range of 100–500 ppm; the results are expressed as mg of gallic acid equivalents (GAE) per dried extract.
Results and Discussion
Morphological characteristics of C. pruinosa mycelia
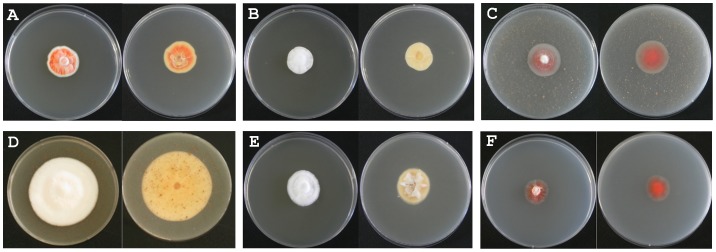
The nutritional and cultural requirements of Cordyceps species have been studied for many years with the aim of cultivating them under artificial conditions, from which nutritionally rich SDAY medium has been determined to be the most optimal medium [18]. C. pruinosa mycelia cultivated in SDAY medium are shown in Figure 1.
Figure 1. Morphological characteristics of C. pruinosa mycelia cultivated under various conditions.
Left photo in each condition: front of plate. Right photo in each condition: back of plate. (A) S+L condition; (B) S+D condition; (C) L+D condition; (D) SL+D condition; (E) SF+D condition, (F) N+D condition.
C. pruinosa is deep or light red due to a red pigment present in its cells. We grew the mycelia of C. pruinosa under light and dark conditions, adding lipid, iron, and nitrogen individually to each mycelium under dark conditions. The following 6 culture conditions were used: dextrose agar supplemented with 0.5% SDAY medium without light (S+D), which were usually used as traditional cultivation conditions;, SDAY medium with light (S+L) as a control group; nut medium without light (L+D), which was used to measure lipid metabolism of the mycelium by adding unsaturated fatty acid to the medium; nut with SDAY medium without light (SL+D) to investigate the effect of complemented lipid nutrients on mycelial metabolism; iron-supplemented SDAY medium without light (SF+D) to analyze the effects of iron, known to be important in growth of Cordyceps specie [19]; and nitrogen medium without light (N+D) because of its role in the biosynthesis of alkaloids, including cordycepin and adenosine (Table 1).
Growth characteristics such as mycelia density, colony diameter, and pigmentation were similar in samples grown in the S+L and L+D conditions, which were different from the characteristics observed for the S+D and SF+D samples. In particular, mycelia density and colony diameter in the SL+D group were the highest compared to mycelia grown in the other conditions. Mycelia cultivated in the N+D condition grew more slowly than those cultivated under other conditions.
Mycelia grown under all conditions appeared white at the top of the colonies, whereas those at the bottom of the colonies exhibited the following differences. Mycelia cultivated under S+L, L+D, and N+D conditions were red, while those of S+D, SL+D, and SF+D appeared white. Morphological differences were also observed between mycelia cultivated under various conditions, implying that there were likely different metabolic profiles induced by different culture conditions.
Metabolite assignment by NMR and GC-MS analysis
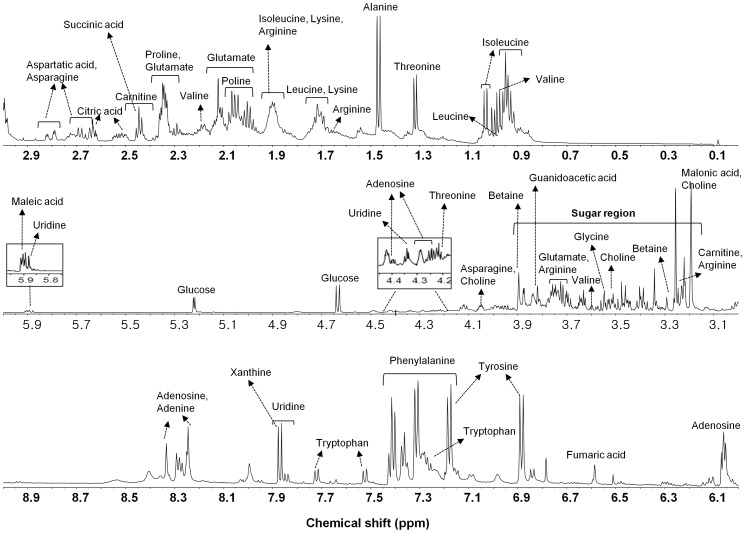
The D2O extracts of the C. pruinosa mycelia cultivated under various conditions were analyzed using NMR spectroscopy to characterize global metabolic profiles. A representative 1H NMR spectrum of the extracts obtained from C. pruinosa mycelia is shown in Figure 2. The metabolites were identified by comparing the chemical shifts of standard compounds using the NMR Suite software (version 5.1; Chenomx). As shown in Table 2, thirty-three metabolites were detected. 1H-13C HSQC analysis was performed to confirm the assignment, and fourteen compounds, such as isoleucine, leucine, valine, threonine, alanine, lysine, proline, asparagine, choline, glucose, betaine, glycerol, uridine and adenosine were confirmed by 1H-13C HSQC (Fig. S1A and Fig. S1B).
Figure 2. Representative 1H NMR spectra (600 MHz) of C. pruinosa mycelia extracts.
Table 2. Assignment of 1H NMR spectral peaks for C. pruinosa mycelia using D2O extracts.
| No | Compound | Chemical shift (multiplicity, J value) |
| 1 | Isoleucine | 0.93(t, J = 7.20), 1.03(d, J = 7.03), 1.21–1.29(m) |
| 2 | Leucine | 0.95(t, J = 6.00), 1.64–1.76(m) |
| 3 | Valine | 0.98(d, J = 6.96), 1.03(d, J = 6.97), 2.15–2.22(m), 3.60 (d, J = 426) |
| 4 | Threonine | 1.32(d, J = 6.6), 3.60(d, J = 4.98), 4.22–4.28(m) |
| 5 | Alanine | 1.47(d, J = 7.2), 3.74–3.80(m) |
| 6 | Arginine | 1.65–1.77(m), 1.85–1.96(m), 3.21–3.25(m), 3.70(t, J = 6.18) |
| 7 | Lysine | 1.67–1.76(m), 1.83–1.95(m), 3.01(t, J = 7.68) |
| 8 | Proline | 1.94–2.09(m), 2.30–2.37(m), 4.10–4.14(m) |
| 9 | Glutamate | 2.00–2.10(m), 2.10–2.16(m), 2.29–2.40(m), 3.76(dd, J 1 = 6.2, J 2 = 3.8) |
| 10 | Carnitine | 2.41–2.48(m), 3.22(s) |
| 11 | Succinic acid | 2.44(s) |
| 12 | Citric acid | 2.53(d, J = 16.2), 2.64 (d, J = 16.56) |
| 13 | Asparagine | 2.67(dd, J 1 = 12, J 2 = 7.74), 2.80(dd, J 1 = 18, J 2 = 4.32) |
| 14 | Aspartatic acid | 2.68(dd, J 1 = 18.3, J 2 = 9.5), 2.82(dd, J 1 = 17.5, J 2 = 3.6), |
| 15 | Choline | 3.19(s), 3.49–3.53(m), 4.03–4.08(m), |
| 16 | Malonic acid | 3.20(s) |
| 17 | Glucose | 3.21–3.25(m), 3.37–3.42(m), 3.43–3.48(m), 3.50–3.55(m), 3.67–3.78(m), 3.80–3.85(m), 3.88(dd, J 1 = 12.18, J 2 = 2.10), 4.63(d, J = 7.95), 5.22(d, J = 3.74) |
| 18 | Betaine | 3.26(s), 3.90(s) |
| 19 | Glycerol | 3.52–3.57(m), 3.66(dd, J 1 = 12, J 2 = 4.32) |
| 20 | Glycine | 3.55(s) |
| 21 | Glucitol | 3.61–3.67(m), 3.72–3.79(m), 3.80–3.86(m) |
| 22 | Guanidoacetic acid | 3.82(s) |
| 23 | Uridine | 4.21(t, J = 5.52), 4.34(t, J = 5.1), 5.87–5.92(m), 7.87(d, J = 8.13) |
| 24 | Adenosine | 4.27–4.30(m), 4.38–4.40(m), 6.06(d, J = 6.12), 8.24(s), 8.33(s) |
| 25 | Tartric acid | 4.34(s) |
| 26 | Maleic acid | 5.90(s) |
| 27 | Fumaric acid | 6.59(s) |
| 28 | Tyrosine | 6.89(d, J = 8.52), 7.16–7.20(m) |
| 29 | Tryptophan | 7.19(t, J = 7.23), 7.25–7.30(m), 7.53(d, J = 8.24), 7.72(d, J = 9.53) |
| 30 | Phenylalanine | 7.30–7.33(m), 7.34–7.38(m), 7.39–7.44(m) |
| 31 | Phenylacetic acid | 7.25–7.33(m), 7.34–7.39(m) |
| 32 | Xanthine | 7.87(s) |
| 33 | Adenine | 8.24(s), 8.33(s) |
To maximize the number of metabolites that could be investigated, GC-MS was employed to perform global metabolite profiling. There were 44 identified compounds in the 70% methanol extracts and 14 identified compounds in the 100% n-hexane extracts. The detected metabolites could be divided into several classes, including alcohols, amino acids, organic acids, purines, sugars, fatty acids, sterols, and pyrimidines (Table 3 and Table 4). The relative levels of various metabolites in the 70% methanol extracts and 100% n-hexane extracts are presented in Table S1 and Table S2.
Table 3. Chromatographic data of the identified compounds from the 70% MeOH extract of C. pruinosa mycelia analyzed using GC-MS.
| Identified compounds | RT | MS fragment ion (m/z) | TMS |
| Alcohols | |||
| Glucitol | 39.14 | 387, 357, 299, 217, 147, 73 | 6TMS |
| Glycerol | 13.14 | 205, 147, 117, 103, 73 | 3TMS |
| Myo- inositol | 32.03 | 318, 217, 191, 147, 73 | 6TMS |
| 33.46 | 318, 217, 191, 147, 73 | 6TMS | |
| 34.61 | 318, 217, 191, 147, 73 | 6TMS | |
| 34.75 | 318, 217, 191, 147, 73 | 6TMS | |
| 40.05 | 318, 217, 191, 147, 73 | 6TMS | |
| 31.53 | 319, 205, 147, 103, 73 | 6TMS | |
| Ribo-hexitol | 29.56 | 231, 205, 147, 129, 73 | 6TMS |
| Xylitol | 26.71 | 307, 217, 205, 147, 103, 73 | 5TMS |
| 27.05 | 307, 217, 205, 147, 103, 73 | 5TMS | |
| Amine | |||
| Putrescine | 27.35 | 361, 214, 200, 174, 86 | 4TMS |
| Amino acids | |||
| Alanine | 18.93 | 290, 248, 174, 147, 73 | 3TMS |
| Asparagine | 25.86 | 231, 188, 147, 132, 116, 73 | 3TMS |
| Aspartic acid | 21.95 | 232, 218, 147, 100, 73 | 3TMS |
| Cystathionine | 36.86 | 278, 218, 128, 73 | 4TMS |
| Glutamine | 24.69 | 348, 246, 147, 128, 73 | TMS |
| 28.32 | 347, 245, 156, 73 | 3TMS | |
| Glycine | 14.25 | 276, 248, 174, 147, 73 | TMS |
| 20.31 | 320, 218, 174, 147, 73 | 3TMS | |
| Histidine | 31.33 | 254, 182, 154, 73 | 3TMS |
| Homoserine | 19.72 | 320, 218, 147, 128, 103, 73 | 3TMS |
| Isoleucine | 13.82 | 218, 158, 147, 73 | 2TMS |
| Lysine | 26.80 | 362, 156, 102, 84, 73 | 3TMS |
| 31.47 | 317, 230, 174, 156, 128, 73 | 4TMS | |
| Ornithine | 29.22 | 420, 200, 174, 142, 73 | 4TMS |
| Proline | 13.96 | 216, 142, 73 | 2TMS |
| 21.79 | 258, 230, 156, 147, 73 | 2TMS | |
| 22.04 | 230, 140, 73 | 3TMS | |
| Serine | 12.44 | 219, 147, 132, 116, 103, 73 | 2TMS |
| 16.49 | 278, 218, 204, 147, 100, 73 | 3TMS | |
| Threonine | 17.46 | 291, 218, 147, 117, 101, 73 | 3TMS |
| Tyrosine | 31.74 | 382, 280, 218, 179, 147, 73 | 3TMS |
| Valine | 10.91 | 218, 144, 73 | 2TMS |
| Organic acids | |||
| Acetic acid | 19.21 | 263, 204, 177, 147, 117, 73 | 2TMS |
| Citric acid | 29.35 | 465, 363, 347, 273, 147, 73 | 4TMS |
| Fumaric acid | 16.18 | 245, 147, 73 | 2TMS |
| Galactonic acid | 32.82 | 433, 319, 292, 205, 147, 73 | 6TMS |
| Gluconic acid | 32.90 | 333, 319, 292, 205, 147, 73 | 6TMS |
| 40.16 | 387, 357, 333, 299, 147, 73 | 6TMS | |
| Glucuronic acid | 39.58 | 449, 305, 204, 217, 147, 73 | 6TMS |
| Glutaric acid | 23.53 | 304, 288, 198, 147, 73 | 2TMS |
| Glyceric acid | 15.29 | 292, 205, 189, 147, 133, 73 | 3TMS |
| Succinic acid | 14.71 | 247, 147, 73 | 2TMS |
| 20.99 | 304, 174, 147, 86, 73 | 3TMS | |
| γ-aminobutyric acid | 9.41 | 204, 147, 130, 73 | 2TMS |
| 22.11 | 304, 174, 147, 86, 73 | 3TMS | |
| Purines | |||
| Adenine | 30.22 | 279, 264, 192 84, 73 | 2TMS |
| Adenosine | 42.65 | 540, 245, 236, 230, 217, 192 | 4TMS |
| 44.06 | 498, 236, 188, 175, 148, 73 | 4TMS | |
| 46.15 | 692, 315, 258, 230, 169, 73 | 5TMS | |
| Uric acid | 34.87 | 456, 444, 384, 369, 147, 73 | 4TMS |
| Xanthine | 33.31 | 368, 353, 294, 279, 147, 73 | 3TMS |
| Pyrimidine | |||
| Cytidine | 40.54 | 245, 217, 168, 151, 73 | 3TMS |
| Sugars | |||
| Arabinose | 29.13 | 217, 204, 191, 147,73 | 4TMS |
| Erythrose | 21.57 | 307, 217, 205, 147, 103, 73 | 3TMS |
| Fructose | 26.07 | 307, 217, 147, 103, 73 | 6TMS |
| 30.58 | 307, 217, 147, 103, 73 | ||
| Galactose | 29.82 | 319, 217, 147, 133, 73 | 5TMS |
| 30.66 | 319, 217, 205, 160, 147, 73 | 6TMS | |
| 31.00 | 217, 204, 191, 147, 73 | 6TMS | |
| 39.44 | 217, 204, 191, 147, 73 | 6TMS | |
| 41.19 | 319, 217, 205, 160, 147, 73 | 6TMS | |
| Glucose | 29.00 | 217, 204, 191, 147, 73 | 6TMS |
| 31.24 | 319, 205, 160, 147, 103, 73 | 6TMS | |
| 32.57 | 435, 217, 204, 191, 147, 73 | 6TMS | |
| 34.74 | 494, 259, 173, 147, 131, 73 | 5TMS | |
| 36.80 | 364, 319, 217, 205, 160 | 5TMS | |
| 39.74 | 387, 299, 204, 147, 129, 73 | 6TMS | |
| 43.69 | 437, 361, 217, 73 | 8TMS | |
| 44.78 | 387, 299, 204, 147, 129, 73 | 6TMS | |
| Mannose | 30.83 | 435, 217, 204, 191, 147, 73 | 6TMS |
| N-acetylglucosamine | 33.86 | 245, 217, 191, 147, 73 | 6TMS |
| 35.10 | 245, 217, 191, 147, 73 | 6TMS |
Table 4. Chromatographic data of the identified compounds from the 100% n-hexane extract of C. pruinosa mycelia analyzed using GC-MS.
| Identified compounds | RT | MS fragment ion (m/z) | TMS |
| Pyrimidine | |||
| Uracil | 11.70 | 256, 241, 147, 99, 73, 45 | 2TMS |
| Saturated fatty acids | |||
| Myristic acid | 23.45 | 285, 145, 132, 117, 73, 55 | TMS |
| Valeric acid | 25.42 | 299, 145, 129, 117, 73, 55 | TMS |
| Arachidic acid | 33.72 | 367, 129, 117, 73, 55, 45 | TMS |
| Lignoceric acid | 39.30 | 425, 145, 132, 117, 73, 57 | TMS |
| Margaric acid | 29.12 | 327, 145, 132, 117, 73, 55 | TMS |
| Palmitic acid | 36.41 | 459, 371, 239, 203, 147, 73 | 2TMS |
| Stearic acid | 30.89 | 341, 145, 132, 117, 73 | TMS |
| Unsaturated fatty acids | |||
| Heptadecenoic acid | 28.67 | 325, 145, 129, 117, 73, 55 | TMS |
| Linoleic acid | 29.42 | 308, 263, 109, 87, 67, 55 | TMS |
| 29.92 | 335, 129, 108, 95, 73, 55 | TMS | |
| 30.30 | 337, 262, 150, 129, 95, 75 | TMS | |
| 31.22 | 335, 129, 108, 95, 73, 55 | TMS | |
| 31.91 | 337, 262, 150, 129, 95, 75 | TMS | |
| Oleic acid | 29.54 | 264, 222, 111, 97, 88, 55 | TMS |
| 30.42 | 339, 145, 129, 117, 73, 55 | TMS | |
| 30.50 | 339, 145, 129, 117, 73, 55 | TMS | |
| 30.60 | 339, 145, 129, 117, 73, 55 | TMS | |
| 38.45 | 485, 397, 147, 129, 103, 73 | TMS | |
| Palmitoleic acid | 26.91 | 311, 145, 129, 117, 73, 55 | 2TMS |
| Sterols | |||
| Dehydroergosterol | 43.66 | 470, 343, 255, 147, 107, 69 | TMS |
| Ergosterol | 43.41 | 468, 363, 337, 253, 143, 69 | TMS |
Significantly higher levels of γ-aminobutyric acid (GABA) were observed in mycelia from the SF+D group (Table S1). It has been reported that, compared to other mushrooms, C. mycelia are good sources of GABA [20]. As a result of C. mycelia grown under usual conditions in prior study, it seems that iron supplementary medium could stimulate and be optimal production of GABA. Compared to the S+D group, significant changes in lipid metabolism were not observed in mycelia grown under L+D and SL+D conditions (Table S2).
Compounds identified by NMR and GC-MS are presented in Table 5. The following 19 metabolites were observed from both NMR and GC-MS analysis: isoleucine, valine, threonine, alanine, lysine, proline, succinic acid, citric acid, asparagine, aspartic acid, glucose, glycerol, glycine, glucitol, adenosine, fumaric acid, tyrosine, xanthine, and adenine. 14 metabolites, such as phenylalanine, tryptophan, betaine, choline, and leucine were only observed using NMR. 39 metabolites, including inositol, putrescine, ornithine, γ-aminobutyric acid, uric acid, uracil, fatty acids, and ergosterol were only observed in the GC-MS analysis.
Table 5. The list of metabolites identified by 1H NMR and GC-MS analysis.
| Analytical platform | Identified compounds |
| 1H NMR | Leucine, arginine, carnitine, choline, malonic acid, betaine, uridine, guanidoacetic acid, tartaric acid, maleic acid, tryptophan, phenylalanine, phenylacetic acid |
| GC-MS | Inositol, ribo-hexitol, xylitol, putrescine, cystathionine, histidine, homoserine, ornithine, serine, acetic acid, galactonic acid, gluconic acid, glucuronic acid, glutaric acid, glyceric acid, γ-aminobutyric acid, uric acid, cytidine, arabinose, erythrose, fructose, galactose, mannose, N-acetylglucosamine, uracil, myristic acid, valeric acid, arachidic acid, lignoceric acid, margaric acid, palmitic acid, stearic acid, heptadecenoic acid, linoleic acid, oleic acid, palmitoleic acid, dehydroergosterol, ergosterol |
| 1H NMR and GC-MS | Isoleucine, valine, threonine, alanine, lysine, proline, glutamic acid, succinic acid, citric acid, asparagine, aspartic acid, glucose, glycerol, glycine, glucitol, adenosine, fumaric acid, tyrosine, xanthine, adenine |
PCA based on NMR and GC-MS datasets
To provide comparative interpretations and visualization of metabolic changes under various culture conditions, PCA was applied to the NMR and GC-MS spectral datasets. PCA is a powerful tool to selectively identify the major controlling factors contributing to differences between samples without a priori knowledge of what those factors might be. PCA can allow for the identification of not only how a specific sample differs from other samples, but also what variables contributed to those differences [21].
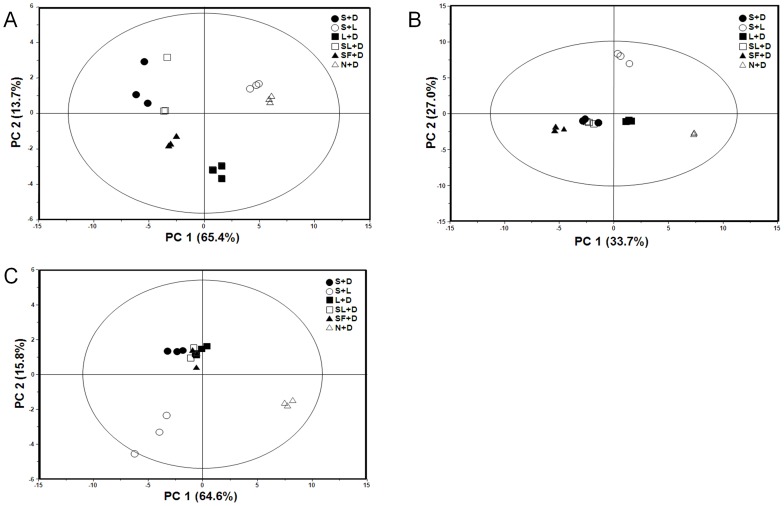
The PCA score plot and loading plot of the D2O extracts analyzed by NMR are presented in Figure 3A and S2, respectively. In the score plots, PC 1 explained 65.4% of the variation and PC 2 explained 13.7% of the variation. S+D, SF+D, and SL+D were clearly distinct from the other conditions, S+L, N+D, and L+D along PC 1, while SF+D and L+D were separated from the others along PC 2. It was shown that relatively higher levels of glutamine, proline, alanine, arginine, and glucose were contained in S+D, SF+D, and SL+D, while those of isoleucine, leucine, tryptophan, tyrosine, adenosine, and phenylalanine were higher in S+L, N+D, and L+D from the loading plot analysis of D2O extracts (Fig. S2).
Figure 3. PCA score plots of C. pruinosa mycelia cultivated under various conditions.
(A) D2O extracts characterized using NMR analysis. (B) 70% methanol extracts and (C) 100% n-hexane extracts characterized using GC-MS analysis. PCA, Principal component analysis.
In Figure 3B and S3, the PCA score plot and loading plot for 70% methanol extracts determined using GC-MS are shown; PC 1 and PC 2 represented 33.7% and 27.0% of the variance across samples, respectively. The trend of the clusters was similar to that observed for the D2O extracts (Fig. 3A). S+D, SL+D, and SF+D were clearly distinguished from S+L, L+D, and N+D along PC 1, while S+L was completely separated from the others. From the loading plot analysis of 70% methanol extracts, relatively higher levels of aspartic acid, mannose, glucose, and γ-aminobutyric acid were observed ,in S+D, SF+D, and SL+D, whereas those of myo-inositol, putrescine, histidine, cystathionine, and adenine were observed in S+L, L+D, and N+D (Fig. S3).
The PCA score plot and loading plot for 100% n-hexane extracts analyzed by GC-MS are shown in Figure 3C and S4. PC 1 and PC 2 represented 64.6% and 15.8% of the sample variance, respectively. Unlike Figure 3A and 3B, S+D, SL+D, L+D, and SF+D were closely aggregated, whereas S+L and N+D were separated from the other samples. The loading plot analysis of 100% n-hexane extracts showed the lignoceric acid, linoleic acid, palmitoleic acid, oleic acid, and heptadecenoic acid were relatively abundant in S+L, and dehydroergosterol, margaric acid, and arachidic acid were relatively higher in N+D (Fig. S4).
Interestingly, there was a similar clustering tendency observed for both the NMR D2O extracts (Fig. 3A) and GC-MS 70% methanol extracts (Fig. 3B) based on the color of the bottom portions of the collected mycelia. The S+L, N+D, and L+D conditions, which had mycelia that were red in color, clustered in the PCA plots along PC 1, whereas the S+D, SL+D, and SF+D, which were white in color, were also clustered together in the score plots (Fig. 3A and 3B).
Antioxidant activity
The free radical scavenging activities (FRSAs) of the 70% methanol extracts of C. pruinosa mycelia grown under various cultivation conditions are summarized in Table 6. The highest FRSAs were obtained from samples grown under the SL+D condition. The FRSA of the SL+D sample (64.08%) was similar to that of 0.2 mg/mL ascorbic acid (62.65%). The FRSAs from samples of the S+L and N+D conditions were similar to each other. Relatively lower FRSAs were observed in the samples from the S+D, L+D, and SF+D conditions.
Table 6. Antioxidant ability and total phenolic content of the 70% methanol extracts of C. pruinosa mycelium grown in different cultivation conditions.
| Sample | Free radical scavenging activity (%) (50,000 mg/L) | TPC (GAE mg/g extract) (50,000 mg/L) |
| S+D | 23.48±2.43a | 2.95±0.26a |
| S+L | 59.57±2.20bc | 8.77±0.57b |
| L+D | 38.21±0.72d | 3.99±0.04c |
| SL+D | 64.08±2.93c | 5.87±0.27d |
| SF+D | 11.74±0.95e | 2.06±0.11e |
| N+D | 57.56±1.58b | 6.94±0.30f |
| Ascorbic acid (200 mg/L) | 62.65±1.93bc |
GAE: gallic acid equivalents.
Cytotoxic effect of the mycelia was investigated by MTT assay and the cell viabilities in all extracts were over 80% in all conditions, indicating that there was no significant cytotoxicity of C. pruinosa mycelia extracts (data not shown).
Polyphenolic compounds are important constituents of mushrooms. Mushrooms contain various polyphenolic compounds that serve as excellent antioxidants due to their ability to scavenge free radicals [22]–[24]. The TPCs of C. pruinosa samples cultivated under various conditions are listed in Table 6. Samples from the S+L condition had the highest TPC, followed by samples from the N+D, SL+D, L+D, S+D, and SF+D conditions.
There was a strong positive linear relationship (correlation coefficient of 0.8058) between antioxidant activity and TPC for C. pruinosa mycelia extracts. This result suggests that the TPC contributed significantly to the antioxidant ability in C. pruinosa mycelia cultivated under various conditions.
Partial least squares regression (PLSR) was performed to evaluate the correlation between FRSAs and metabolites analyzed by NMR and GC-MS. As shown in Figure S5, the correlation efficient was R2 = 0.69. However, phenolic compounds known to contribute to antioxidant activity were not fully covered by NMR and GC-MS analysis in this study. Thus, LC-MS analysis will be employed to analyze metabolites including various phenolic compounds having FRSAs in further studies.
Comparison of metabolic profiles
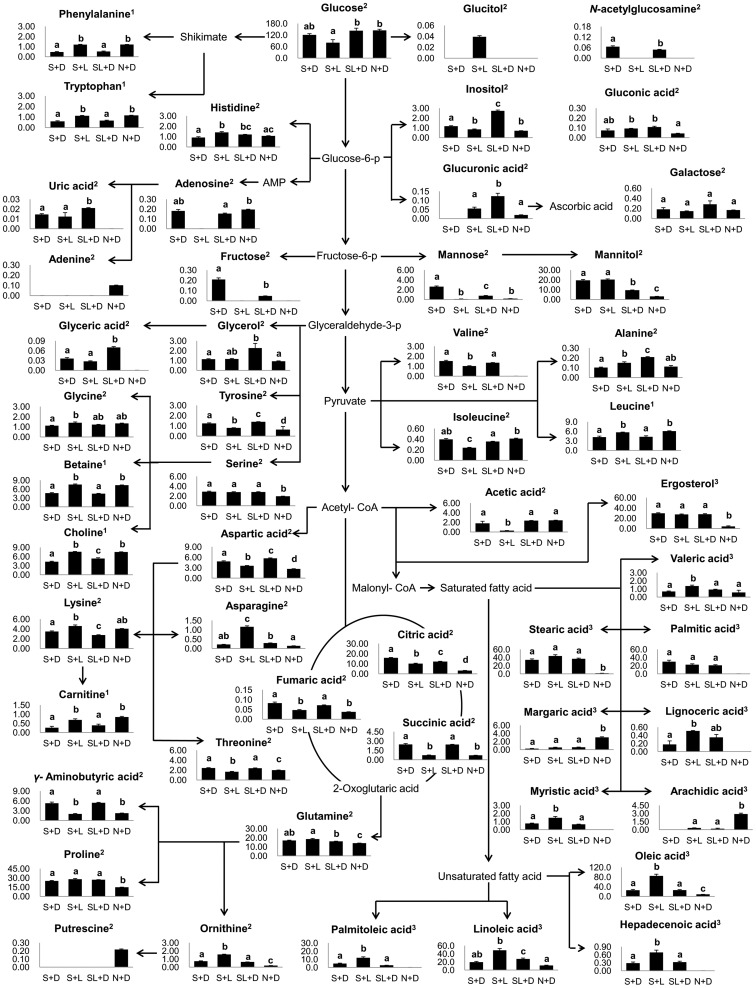
Our results showed that there were distinct differences in morphological, metabolic, and FRSA characteristics between C. pruinosa mycelia grown under different cultivation conditions. Metabolic profiles of S+L, N+D, and SL+D samples showing relatively higher antioxidant activities were further investigated and compared to S+D samples. This comparison is illustrated in Figure 4. The metabolic pathways were prepared by combining data from 5 metabolites identified using NMR and 47 metabolites identified by the GC-MS analysis.
Figure 4. Schematic diagram of the metabolic pathway and relative levels of the major compounds detected in C. pruinosa extracted with 70% MeOH and 100% n-hexane plus D2O.
This was modified from pathways presented in KEGG database (http://www.genome.jp/kegg/). ANOVA was performed to assess the statistical significance of differences between samples (p<0.05). Data are mean values with error bars representing standard deviation values. Different letters in bars represent the difference of statistical significance of metabolites levels. The superscript numbers in compounds represent analysis methods. 1: NMR analysis of D2O extracts, 2: GC-MS analysis of 70% methanol extracts, 3: GC-MS analysis of n-hexane extracts.
The highest levels of adenosine, adenine, and carnitine were obtained in the N+D samples, but the mycelial growth was retarded in these samples. Putrescine, which was only detected in the N+D samples, is known to be one of the major polyamines involved in cell division and tolerance against abiotic stress in mushrooms [25]. The existence of putrescine only in the N+D samples may be due to an adaptation to an adverse environment for mycelia formation. Putrescine is synthesized from ornithine by ornithine decarboxylase [26]. We found that ornithine levels were the lowest in the N+D condition. The low ornithine levels in N+D samples might also be correlated with an increase in putrescine synthesis.
In addition, ergosterol is a specific component of the fungal cell membrane and is a precursor of vitamin D2 [27]. Similar levels of ergosterol were detected in samples grown under the S+L and SL+D conditions, but not under N+D condition. Given that ergosterol in C. pruinosa plays an important role in cell maintenance, the relatively lower ergosterol content in N+D samples indicates that this is an unsuitable condition for cell growth.
Levels of fatty acids, such as valeric acid (C5:0), myristic acid (C14:0), stearic acid (C18:0), lignoceric acid (C24:0), heptadecenoic acid (C17:1), linoleic acid (C18:2), oleic acid (C18:1), and palmitoleic acid (C16:1), were highest in samples grown under the S+L condition. However, sugar levels, including those of glucose, galactose, and mannose were lowest in S+L samples. Moreover, fructose and N-acetylglucosamine were not detected in the S+L condition.
Contrary to our results, it was previously reported that Aspergillus ornatus and Blastocladiella emersonii exposed to light showed growth retardation due to limited uptake of essential nutrients [28], [29]. Aspergillus species are known to develop either asexually in the light or sexually in the dark [30], [31]. Asexual spore development was also shown to be enhanced by linoleic acid [32], [33]. We suggest that enhanced linoleic acid in response to light might stimulate conidia formation in the context of asexual development in C. pruinosa. Thus, growth in samples cultivated under the S+L condition might not be retarded compared to that observed for S+D samples. The mycelia of C. cardinalis, C. bassiana, and C. militaris are commonly cultivated in SDAY agar medium in the dark to produce fruiting bodies [34]–[36]. However, our data suggest that the light condition is more optimal than dark condition for antioxidative activity, linoleic acid content, and mycelial growth in cultivation of C. pruinosa mycelia with SDAY medium.
Conclusion
In this study, C. pruinosa mycelia cultivated under various media and light conditions showed differences in growth, metabolic profiles, and FRSAs. Higher levels of FRSAs were achieved in samples cultivated under SL+D, S+L, and N+D conditions. The mycelia grown under the S+L condition contained relatively higher levels of fatty acids, including linoleic acid, which is known for stimulating fungal conidia formation. Therefore, growth retardation was not observed in S+L samples because of enhanced asexual development caused by linoleic acid. Our finding revealed that C. pruinosa mycelia cultivated under the S+L (SDAY media and light condition) condition had higher antioxidative activity, linoleic acid content, and mycelial growth. To the best of our knowledge, this is the first report on effect of light on mycelial growth and metabolic profiling of C. pruinosa. Light was more beneficial condition than dark for antioxidative activity, linoleic acid content, and mycelial growth in cultivation of C. pruinosa mycelia with SDAY medium. This result can be applied to C. pruinosa fruiting body cultivation as a means of producing biopharmaceutical or natural medicinal resources.
Supporting Information
Representative 1H-13C HSQC NMR spectra of Cordyceps pruinosa mycelia by D2O extraction. (a) 1H chemical shift of 0–3 ppm, (b) 1H chemical shift of 3–6 ppm (1: isoleucine, 2: leucine, 3: valine, 4: threonine, 5: alanine, 7: lysine, 8: proline, 13: asparagine, 15: choline, 17: glucose, 18: betaine, 19: glycerol, 23: uridine, 24: adenosine).
(TIF)
PCA loading plots derived from NMR analysis of D2O extracts of C. pruinosa mycelia cultivated under various conditions.
(TIF)
PCA loading plots derived from GC-MS analysis of 70% methanol extracts of C. pruinosa mycelia cultivated under various conditions.
(TIF)
PCA loading plots derived from GC-MS analysis of 100% n -hexane extracts of C. pruinosa mycelia cultivated under various conditions.
(TIF)
PLS t1/u1 score plots representing relationship between the free radical scavenging activities (u1) and metabolic profiles (t1) obtained by NMR and GC-MS.
(TIF)
A GC-MS-based metabolic profile of 70% methanol extracts of C. pruinosa mycelia. The relative levels of each metabolite were obtained by dividing the percentage area of each metabolite by the percentage area of the internal standard. Different letters in the same row indicate a significant difference. Mean±SD values for triplicate measurements are shown. ‘ND’ means ‘not detected’.
(DOCX)
A GC-MS-based metabolic profile of 100% n-hexane extracts of C. pruinosa mycelia. The relative levels of each metabolite were obtained by dividing the percentage area of each metabolite by the percentage area of the internal standard. Different letters in the same row indicate a significant difference. Mean±SD values for triplicate measurements are shown. ‘ND’ means ‘not detected’.
(DOCX)
Funding Statement
This work was supported by a grant from the Next-Generation Biogreen 21 Program (No. PJ008154), Rural Development Administration, and by Mid-career Researcher Program (NRF-2012R1A2A2A02011748) through NRF grant funded by the MSIP, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Liang Z (1991) Verification and identification of the anamorph of Cordyceps pruinosa Petch. Jun Wu Xue Bao 10: 104–107. [Google Scholar]
- 2. Liu Z, Liang Z, Liu A, Yao Y, Hyde KD, et al. (2002) Molecular evidence for teleomorph-anamorph connections in Cordycepsbased on ITS-5.8 S rDNA sequences. Mycol Res 106: 1100–1108. [Google Scholar]
- 3. Furuya T, Hirotani M, Matsuzawa M (1983) N-6-(2-hydroxyethyl) adenosine, a biologically active compound from cultured mycelia of Cordyceps and Isaria species. Phytochemistry 22: 2509–2512. [Google Scholar]
- 4. Huang J, Shi Q, Wu S, Liang Z, Liu A, et al. (1998) Isolation and Identification and Characterization of Polysaccharide ps1 and ps2 from Mycelia of Cordyceps pruinosa . J Fujian Teach Univ (Nat Sci Ed) 14: 82–85. [Google Scholar]
- 5. Hao Y, Liu A, Liang Z (1999) Studies on high N-6-(2-hydroxythyl) adenosine yielding strain of Cordyceps pruinosa Petch induced by UV irradiation of protoplast. Jun Wu Xue Bao 18: 307–310. [Google Scholar]
- 6. Kim K, Kwon Y, Chung H, Yun Y, Pae H, et al. (2003) Methanol extract of Cordyceps pruinosa inhibits in vitro and in vivo inflammatory mediators by suppressing NF-κB activation. Toxicol Appl Pharmacol 190: 1–8. [DOI] [PubMed] [Google Scholar]
- 7. Liu J, Fei Y (2001) Enhancement of Cordyceps taii polysaccharide and Cordyceps pruinosa polysaccharide on cellular immune function in vitro. Immunol J 17: 189–191. [Google Scholar]
- 8. Kim HG, Song H, Yoon DH, Song B, Park SM, et al. (2010) Cordyceps pruinosa extracts induce apoptosis of HeLa cells by a caspase dependent .pathway. J Ethnopharmacol 128: 342–351. [DOI] [PubMed] [Google Scholar]
- 9. Dong J, Liu M, Lei C, Zheng X, Wang Y, et al. (2012) Effects of selenium and light wavelengths on liquid culture of Cordyceps militaris link. Appl Biochem Biotechnol 166: 2030–2036. [DOI] [PubMed] [Google Scholar]
- 10. Fan D, Wang W, Zhong J (2012) Enhancement of cordycepin production in submerged cultures of Cordyceps militaris by addition of ferrous sulfate. Biochem Eng J 60: 30–35. [Google Scholar]
- 11. Prathumpai W, Kocharin K, Phimmakong K, Wongsa P (2007) Effects of different carbon and nitrogen sources on naphthoquinone production of Cordyceps unilateralis BCC 1869. Appl Biochem Biotechnol 136: 223–232. [DOI] [PubMed] [Google Scholar]
- 12. Xiao JH, Chen DX, Xiao Y, Liu JW, Liu ZL, et al. (2004) Optimization of submerged culture conditions for mycelial polysaccharide production in Cordyceps pruinosa . Process Biochem 39: 2241–2247. [Google Scholar]
- 13. Dong C, Yao Y (2008) In vitro evaluation of antioxidant activities of aqueous extracts from natural and cultured mycelia of Cordyceps sinensis . Lebensm Wiss Technol 41: 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Won S, Park E (2005) Anti-inflammatory and related pharmacological activities of cultured mycelia and fruiting bodies of Cordyceps militaris . J Ethnopharmacol 96: 555–561. [DOI] [PubMed] [Google Scholar]
- 15. Styczynski MP, Moxley JF, Tong LV, Walther JL, Jensen KL, et al. (2007) Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal Chem 79: 966–973. [DOI] [PubMed] [Google Scholar]
- 16. Ou Z, Schmierer DM, Rades T, Larsen L, McDowell A, et al. (2013) Application of an online post-column derivatization HPLC-DPPH assay to detect compounds responsible for antioxidant activity in Sonchus oleraceus L. leaf extracts. J Pharm Pharmacol 65: 271–279. [DOI] [PubMed] [Google Scholar]
- 17. De Oliveira AC, Valentim IB, Silva CA, Bechara EJH, Barros MPd, et al. (2009) Total phenolic content and free radical scavenging activities of methanolic extract powders of tropical fruit residues. Food Chem 115: 469–475. [Google Scholar]
- 18. Shrestha B, Lee W, Han S, Sung J (2006) Observations on some of the mycelial growth and pigmentation characteristics of Cordyceps militaris isolates. Mycobiology 34: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson L (2008) Iron and siderophores in fungal–host interactions. Mycol Res 112: 170–183. [DOI] [PubMed] [Google Scholar]
- 20. Chen S, Ho K, Hsieh Y, Wang L, Mau J, et al. (2012) Contents of lovastatin, γ-aminobutyric acid and ergothioneine in mushroom fruiting bodies and mycelia. Lebensm Wiss Technol 47: 274–278. [Google Scholar]
- 21. Ramadan Z, Jacobs D, Grigorov M, Kochhar S (2006) Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta 68: 1683–1691. [DOI] [PubMed] [Google Scholar]
- 22. Kalogeropoulos N, Yanni AE, Koutrotsios G, Aloupi M (2013) Bioactive microconstituents and antioxidant properties of wild edible mushrooms from the island of Lesvos, Greece. Food Chem Toxicol 55: 378–385. [DOI] [PubMed] [Google Scholar]
- 23. Kim M, Seguin P, Ahn J, Kim J, Chun S, et al. (2008) Phenolic compound concentration and antioxidant activities of edible and medicinal mushrooms from Korea. J Agric Food Chem 56: 7265–7270. [DOI] [PubMed] [Google Scholar]
- 24. Palacios I, Lozano M, Moro C, D'arrigo M, Rostagno M, et al. (2011) Antioxidant properties of phenolic compounds occurring in edible mushrooms. Food Chem 128: 674–678. [Google Scholar]
- 25. Dadáková E, Pelikánová T, Kalač P (2009) Content of biogenic amines and polyamines in some species of European wild-growing edible mushrooms. Eur Food Res Technol 230: 163–171. [Google Scholar]
- 26. Gill SS, Tuteja N (2010) Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 5: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vanegas JM, Contreras MF, Faller R, Longo ML (2012) Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys J 102: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hill E (1976) Effect of light on growth and sporulation of Aspergillus ornatus. J Gen Microbiol 95: 39–44. [DOI] [PubMed] [Google Scholar]
- 29. Goldstein A, Cantino E (1962) Light-stimulated polysaccharide and protein synthesis by synchronized, single generations of Blastocladiella emersonii. J Gen Microbiol 28: 689–699. [DOI] [PubMed] [Google Scholar]
- 30. Park H, Yu J (2012) Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol 15: 669–677. [DOI] [PubMed] [Google Scholar]
- 31. Dyer PS, O'Gorman CM (2012) Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev 36: 165–192. [DOI] [PubMed] [Google Scholar]
- 32. Calvo AM, Hinze LL, Gardner HW, Keller NP (1999) Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl Environ Microbiol 65: 3668–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Calvo AM, Gardner HW, Keller NP (2001) Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans . J Biol Chem 276: 25766–25774. [DOI] [PubMed] [Google Scholar]
- 34. Sung J, Park Y, Lee J, Han S, Lee W, et al. (2006) Selection of superior strains of Cordyceps militaris with enhanced fruiting body productivity. Mycobiology 34: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee J, Shrestha B, Kim T, Sung G, Sung J, et al. (2007) Stable formation of fruiting body in Cordyceps bassiana . Mycobiology 35: 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim S, Shrestha B, Sung G, Han S, Sung J, et al. (2010) Optimum conditions for artificial fruiting body formation of Cordyceps cardinalis . Mycobiology 38: 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative 1H-13C HSQC NMR spectra of Cordyceps pruinosa mycelia by D2O extraction. (a) 1H chemical shift of 0–3 ppm, (b) 1H chemical shift of 3–6 ppm (1: isoleucine, 2: leucine, 3: valine, 4: threonine, 5: alanine, 7: lysine, 8: proline, 13: asparagine, 15: choline, 17: glucose, 18: betaine, 19: glycerol, 23: uridine, 24: adenosine).
(TIF)
PCA loading plots derived from NMR analysis of D2O extracts of C. pruinosa mycelia cultivated under various conditions.
(TIF)
PCA loading plots derived from GC-MS analysis of 70% methanol extracts of C. pruinosa mycelia cultivated under various conditions.
(TIF)
PCA loading plots derived from GC-MS analysis of 100% n -hexane extracts of C. pruinosa mycelia cultivated under various conditions.
(TIF)
PLS t1/u1 score plots representing relationship between the free radical scavenging activities (u1) and metabolic profiles (t1) obtained by NMR and GC-MS.
(TIF)
A GC-MS-based metabolic profile of 70% methanol extracts of C. pruinosa mycelia. The relative levels of each metabolite were obtained by dividing the percentage area of each metabolite by the percentage area of the internal standard. Different letters in the same row indicate a significant difference. Mean±SD values for triplicate measurements are shown. ‘ND’ means ‘not detected’.
(DOCX)
A GC-MS-based metabolic profile of 100% n-hexane extracts of C. pruinosa mycelia. The relative levels of each metabolite were obtained by dividing the percentage area of each metabolite by the percentage area of the internal standard. Different letters in the same row indicate a significant difference. Mean±SD values for triplicate measurements are shown. ‘ND’ means ‘not detected’.
(DOCX)