Abstract
The neuroscience research landscape has changed dramatically over the past decade. An impressive array of neuroscience tools and technologies have been generated, including brain gene expression atlases, genetically encoded proteins to monitor and manipulate neuronal activity and function, cost effective genome sequencing, new technologies enabling genome manipulation, new imaging methods and new tools for mapping neuronal circuits. However, despite these technological advances, several significant scientific challenges must be overcome in the coming decade to enable a better understanding of brain function and to develop next generation cell type-targeted therapeutics to treat brain disorders. For example, we do not have an inventory of the different types of cells that exist in the brain, nor do we know how to molecularly phenotype them. We also lack robust technologies to map connections between cells. This review will provide an overview of some of the tools and technologies neuroscientists are currently using to move the field of molecular neuroanatomy forward and also discuss emerging technologies that may enable neuroscientists to address these critical scientific challenges over the coming decade.
Introduction
Progress in neuroscience over the past decade has relied heavily on gene-centric strategies such as the genetic or pharmacological manipulation of gene function affecting many cell types and tissues in the nervous system. While progress in the gene-centric realm has been substantial, the fundamental organizing principle of the nervous system is the cell and not the gene. Transmission of information and the generation of behavior are directly determined by cell type and connectivity among various cell types. Improved cell-centric strategies, such as the ability to functionally manipulate specific neuronal cell types and circuits, are critical for understanding the nervous system and may be essential for a full mechanistic understanding of important brain disorders and the eventual development of next generation cell type-targeted therapeutics to treat important brain disorders. Furthermore the development of nanoparticles that are targetable to specific cell types (as defined by molecular phenotype and neuronal circuit) could enable the non-invasive mapping, monitoring and manipulation of the activity of millions of neurons at the single cell and millisecond resolution as conceived of by projects such as the Brain Activity Map (BAM). 1-3
This review will describe several of the most important gene-centric technologies and resources that have been developed and describe the ways in which they provide a firm foundation for the further development of new and improved cell-centric strategies for analysis of the nervous system in the coming decade. These technologies and resources include gene expression atlases of the brain, gene expression profiling, knockouts and transgenic animals, CRE driver lines, viral vectors, connectivity maps, and genetically-encoded biosensors and modulators, and molecular phenotype datasets4. Cell-specific genetic manipulation has been inhibited by: 1) The limited number of cell type-specific promoters; 2) the very few genes that are selectively expressed in a given cell type and; 3) our still limited knowledge of the mechanisms that specify cell type. Emerging single cell technologies used to profile cell types and synapses in heterogeneous tissues, such as cell-specific barcoding strategies, provides a means to overcome this barrier.
Brain Atlases
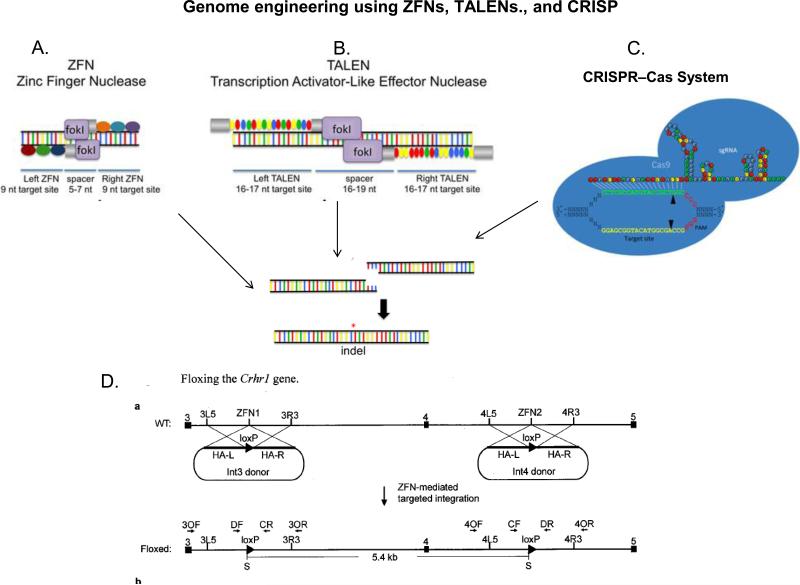
Eighty percent of 20,000 genes in the mammalian genome are expressed in the central nervous system5. These distinct patterns of gene expression underlie neuronal identity, anatomical boundaries, and the specification of neuronal circuits. Characterization of changes in neuronal gene expression has provided key insights into the development, and the response of the nervous system to the environment and drugs of abuse. The Allen Brain Gene Expression Atlas and the GENSAT atlas were developed with the expectation that gene expression arrayed in either 2-D or 3-D would identify cell type-specific molecular markers. The identification of cell type-specific molecular markers would provide targets that would facilitate the delivery of genes and gene products to these cell types for the analysis of cellular development, connectivity, and function as well as the principles by which genes organize the nervous system. Early advances in mouse genome sequencing and in manipulating the mouse genome through transgenesis and homologous recombination led to a strong preference for the mouse over other organisms such as the rat. Targeted mutation in the rat can now be achieved using Transcription Activator-Like Effector Nucleases (TALEN), Zinc Finger Nucleases (ZFN), and CRISPR. Fig 1.
Fig 1. Creating CRE/LoxP rat using Zinc Finger Nuclease, TALEN, and CRISPR technologies for inducible gene knock-in or knock-out.
A. Zinc Finger Nuclease (ZFN) links engineered DNA binding zinc finger (ZF) domain with a DNA-cutting nuclease domain, which contains a Fok1 restriction enzyme site. Each zinc finger recognizes and binds to three targeted nucleotides. The pairing of left ZFN and right ZFN acts like a pair of genomic scissors to produce a double-strand DNA incision in the spacer region. The ZF domains are often extended, doubled or tripled, for longer sequence recognition and increased specificity on each side. ZFNs induced chromosomal breaks are then randomly reconnected by endogenous cellular DNA repair mechanisms called non-homologous end joining, leading to gene knockout. Nonetheless, if there are manipulated DNA strands ending with nucleotides on each side that can form homologous bindings to the DNA break at the time of ZFN incision, a homology dependent repair can occur, resulting in gene knock-in. Adapted with permission151
B. TALENs bind DNA using TAL effector repeat domains derived from Xanthomonas that recognize individual nucleotides. These TALE repeats are ligated together to create binding arrays that recognize extended DNA sequences. The coupled nuclease domain of TALEN cleaves the DNA in the same fashion as ZFN within the intervening spacer region. Adapted with permission151
C. CRISPRs (clusters of regularly interspaced short palindromic repeats) were first found in the Escherichia coli genome likely involved in its immune defense against foreign DNA. CRISPR loci are surrounded by a cohort of conserved CRISPR-associated genes (Cas genes) adjacent to the cluster repeats. The processed CRISPR RNA (crRNAs) serve as sequence-specific guides for the Cas proteins/nucleases to target and generating double strand break (DSB) in the DNAs. In CRISPR genome editing, the crRNA and the trans-activating crRNA (tracrRNA) forms a double strand RNA structure that directs the Cas9 to generate DSBs in the target DNA. At the genomic site where is complementary to the crRNA, the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like domain cleaves the non-complementary strand. Reprinted with permission from152.
D. Floxing the Crhr1 gene. In conditional knockin rats, a plasmid DNA that encodes the CRE or LoxP proteins is used together with the genomic editing methods. The plasmid DNA contains homologous binding sequences to bind to the DSB. Insertion of the plasmid DNA is achieved through a mechanism called homologous end joining during DNA repair. This allows CRE-LoxP to be inserted at desired genomic sites to created inducible mutations and generate CRE/LoxP rat for specific neural circuitries in the brain. Figure Courtesy of Dr. Xiaoxia Cui, Sage Labs, St. Louis MO..
The Allen Mouse Brain Atlas
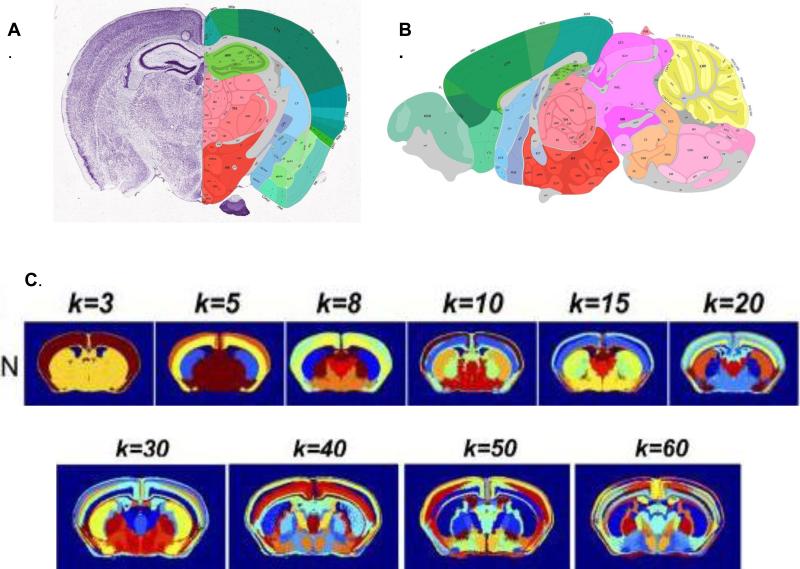
The Allen Mouse Brain Atlas, is a high resolution 2-D and 3-D digital atlas of the C57BL/J mouse brain populated by 20,000 transcripts5-7. This effort led to the development of high-throughput methods for doing in situ hybridization, a standardized coordinate system for displaying mouse brain gene expression and an informatics framework for data integration and analysis. The standardized coordinate system is particularly important for correlating expression of multiple genes to infer the number of cells types in a given brain region, delineate anatomical boundaries and ultimately correlate gene expression patterns with neuronal connectivity8. Using data from the Allen Mouse Brain Atlas, Wolf et al9 suggest that regional gene expression predicts neuronal connectivity. Recent work by Ko et al10 suggests that the anatomical boundaries within a mouse brain can be defined by the clustering of just 170 neuron-specific genes (Fig. 2). Work by Pascal et al11 also suggests that anatomical boundaries for cortex, hippocampus, striatum, ventral midbrain, medulla, and cerebellum can be delineated based on the density of cell types and gene expression profiling of 64 cell types from these different regions. Experiments in which these gene expression patterns are subtracted from each voxel suggest that many more mouse brain cell types may exist. Expression profiling of many additional neuronal cell types is needed to provide combinatorial gene expression data that can be used to better characterize brain anatomical boundaries and cell-type specific organization.
Fig 2.
Spatial clustering of neuron-specific transcripts delineates anatomical boundaries in the brain, revealing more than 50 brain regions. The spatial resolution and delineation of brain-structures increases as then number of k-mean clusters of neuron-specific transcripts used in the analysis increases. Further refinement of c ell types and their projection may increase anatomical resolution. A. Coronal section from Allen Brain Reference Atlas. B. Sagital section from Allen Brain Atlas. C. Spatial cluster analysis of neuron-specific transcripts. Adapted with permission9.
GENSAT
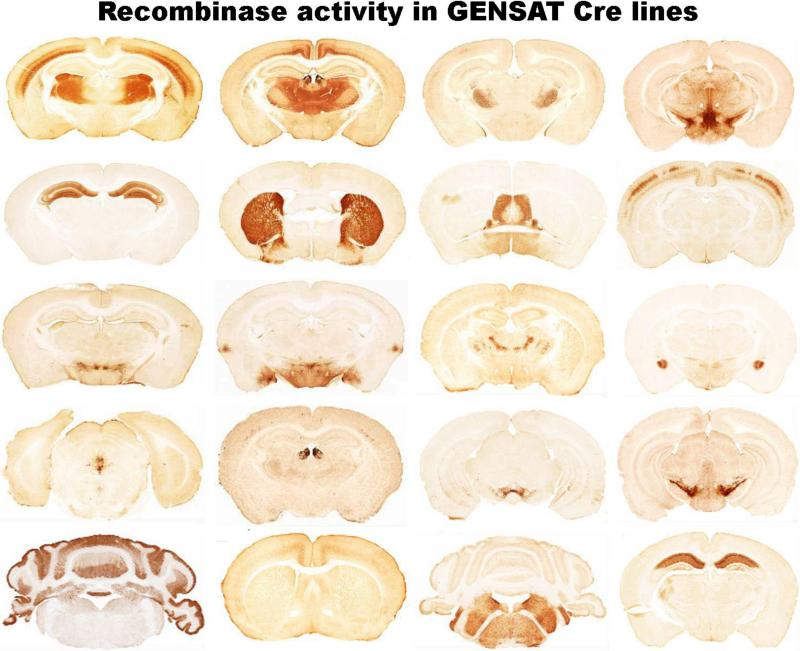
GENSAT provides images of gene expression in the adult and developing nervous system of the FVB/N mouse as demonstrated by transgenic bacterial artificial chromosome (BAC)-expressing green fluorescent protein (GFP) or cre-recombinases12-14. In BAC recombineering, the eGFP reporter or cre-recombinase is placed in frame in the first exon with an inframe stop codon to prevent overexpression of the protein. Since BACs are usually of large size, they typically include control elements that recapitulate in vivo temporal and spatial expression of that gene. This increases the likelihood that the BAC transgene expression pattern will be congruent with the wild-type expression pattern but positional effects are still possible. GENSAT BAC mice are widely used by the scientific community since they enable biochemical and electrophysiological analysis of individual eGFP-expressing neurons and glia. The cre-driver lines provide a method for temporal and spatial control of gene activity or reporter genes15-17. GENSAT has characterized over 10,000 BAC transgenic founder lines and 1347 eGFP and Cre founder lines (Fig. 3) have been made available to the scientific community through Mutant Mouse Regional Resource Centers (MMRRC) (see Table I and Table II for links to digital atlases and resources for molecular neuroanatomy).
Fig 3.
Micrograph of anti-EGFP stained coronal section through the brains of a subset of Cre recombinase BAC transgenic mouse line generated by the GENSAT project. All Cre lines are tested for recombinase activity by breeding with a standard Rosa26-loxP-Stop-loxP-EGFP reporter. Examples of over two hundred other Cre recombinase lines can be viewed on the Gensat website. Mice are available to investigators via the Mutant Mouse Regional Resource Center (MMRRC),. Image courtesy of Nathaniel Heintz, Rockefeller University and Charles Gerfen, NIMH.
Table I.
| Resource | Summary | References or Website |
|---|---|---|
| General Resource to Brain Atlases | ||
| Neuroscience Information Framework | Provides links to many brain atlases | https://www.neuinfo.org/mynif/search.php?q=atlas&t=registry |
| Mouse Brain/CNS Atlases | ||
| Allen Mouse Brain Atlas | Reference Atlas of C57BL/6J adult mouse brain and high resolution gene expression atlas | http://mouse.brain-map.org/ |
| Gene Expression Nervous System Atlas (GENSAT) | An NIH-funded, publicly available, gene expression atlas of the developing and adult CNS in mice. | http://www.gensat.org/index.html |
| Whole Brain Catalog | An open source, downloadable, multi-scale, virtual catalog of the mouse brain and its cellular constituents. | http://wholebraincatalog.org/ |
| Rodent Brain Workbench | A consortium website and portal to atlases, databases, and tools. | http://www.rbwb.org/ |
| Waxholm Standard Space | A coordinate-based reference space for the mapping and registration of neuroanatomical data. | http://incf.org/programs/atlasing/projects/waxholm-space |
| GenePaint | A digital atlas of gene expression patterns in the mouse. | http://www.genepaint.org/Frameset.html |
| Mousebrain Gene Expression Map | A database containing gene expression patterns assembled from mouse nervous tissues at 4 time points throughout brain development, including embryonic (e) day 11.5, e15.5, postnatal (p) day 7 and adult p42. | http://www.stjudebgem.org/web/mainPage/mainPage.ph |
| Mouse Spinal Cord Atlas | A gene expression atlas of mouse spinal cord at P4 and P56 | http://mousespinal.brain-map.org/ |
| The Mouse Brain Library | High-resolution images and databases of brains from many genetically characterized strains of mice. | http://www.mbl.org/ |
| Atlases of the Developing Mouse | ||
| Allen Brain Developing Mouse Brain Atlas | In situ hybridization of developing mouse brain at E11.5, E13.5, E15.5, E18.5, P4, P14, P56 | http://developingmouse.brain-map.org/ |
| Molecular Anatomy of Mouse Embryo Project (MAMEP) | A comprehensive information resource for the functional analysis of pattern formation, tissue development, and organogenesis in mice. Whole-mount in situ hybridizations on mid-gestation mouse embryos are used to assign genes to cell types and organs. | http://mamep.molgen.mpg.de/ |
| EMBRYS | A database of gene expression patterns mapped in the whole-mount mouse embryo (ICR strain) of mid-gestational stages (Embryonic Day 9.5, 10.5, 11.5), in which the most striking dynamics in pattern formation and organogenesis is observed. | http://embrys.jp/embrys/html/MainMenu.html |
| Eurexpress | A transcriptome Atlas database for mouse embryo. | http://www.eurexpress.org/ee/ |
| e-mouse Atlas (EMA and EMAGE) | EMA is a 3D anatomical atlas of mouse embryo development with histology. EMAGE is a database of mouse gene expression which can be mapped into EMA 3D space. | http://www.emouseatlas.org/emap/home.html |
| Gene Expression Database | A collection of gene expression information during mouse development | http://www.informatics.jax.org/expression.shtml |
| Mouse Phenome Database (MPD) | A NIH-funded database with characterizations of hundreds of strains of laboratory mice to facilitate translational discoveries and to assist in selection of strains for experimental studies. | http://phenome.jax.org/db/q?rtn=docs/home |
| Non-Human Primate Brain Atlases | ||
| NIH Blueprint NHP Atlas | A suite of gene expression data, neuroanatomical data, and informatics tools for exploring the cellular and molecular architecture of the developing postnatal rhesus macaque brain. | http://www.blueprintnhpatlas.org/ |
| Rhesus Macaque Brain Atlases | These atlases enable alignment of individual scans to improve localization and statistical power of the results, and allow comparison of results between studies and institutions. A set of multi-subject atlas templates is constructed specifically for functional and structural imaging studies of rhesus macaque. | http://brainmap.wisc.edu/monkey.html |
| Human Brain Atlases | ||
| Big Brain: An Ultrahigh Resolution 3D Human Brain Model | An Ultrahigh Resolution 3D Human Brain Model | https://bigbrain.loris.ca/main.php |
| Allen Human Brain Atlas | A multi-modal, multi-resolution atlas detailing gene expression across the adult human brain | http://human.brain-map.org/ |
| BrainSpan (Atlas of the Developing Brain) | An atlas describing the transcriptional mechanism of human brain development, which includes RNA sequencing and exon microarray data profiling up to 16 cortical and subcortical structures across the full course of human brain development. Such information is referenced to MRI and diffusion weighted imaging (DWI) data. | http://www.brainspan.org |
| Multiple Species Atlases | ||
| Brain Maps | Interactive multi-resolution brain atlases based on over 20 million megapixels of sub-micron resolution, annotated, scanned images of serial sections of human, non-human primate, cat, rat, mouse, barn owl, opossum, red jungle fowl, spiny anteaters, platypus and goldfish brains. Brain structures and functions can be quickly queried and retrieved over the internet. | http://brainmaps.org/ |
| The Scalable Brain Atlas | A web based display engine for different atlases created for mouse, macaque, and human | http://scalablebrainatlas.incf.org/main/index.php |
| Laboratory of Neuroimaging Atlas | Website contains brain atlases for fetal and adult human, Alzheimer's, monkey, vervet, mouse, and rat | http://www.loni.ucla.edu/Atlases/ |
| Adult Rat Brain Atlas | ||
| Adult Wistar Rat Brain | Atlas combines the Paxino Watson atlas with magnetic resonance histology and diffusion tensor imaging. | http://www.civm.duhs.duke.edu/neuro2012ratatlas/ |
| Rodent Brain Workbench | A consortium website and portal to atlases, databases, and tools. | http://www.rbwb.org/ |
| Other species | ||
| Atlas of the Drosophila Nervous System Armstrong HD et al 1995 | Atlas of the adult and developing Drosophila central nervous system | http://flybrain.neurobio.arizona.edu/ |
| Zebrafish Brain Atlas | Curated information and images of neuroanatomical structures in the developing zebrafish | http://zebrafishbrain.org/ |
| Wormatlas | A description of anatomical location and function of 302 neurons in the adult C. elegans | http://www.wormatlas.org/hermaphrodite/nervous/Neuroframeset.html |
| Zebra Finch Expression Brain Atlas | A NIH-funded online public repository of in situ hybridization images from a large collection of genes expressed in the brain of adult male zebra finches. | http://www.zebrafinchatlas.org/ |
| Human Connectome | ||
| Human Connectome | A five-year project sponsored by NIH including tween two consortia of research institutions to build a “network map” in order to better understand the anatomical and functional connectivity in healthy subjects. |
http://www.humanconnectome.org/
http://www.humanconnectomeproject.org/ |
| Human Brain Connectivity Database | Curated by the Brain Architecture Project, this is a preliminary database of neuroanatomical connectivity reports specifically for the human brain. | http://mitraweb1.cshl.edu:8080/BrainArchitecture/pages/publications.faces |
| Mouse Connectome | ||
| Mouse Connectome | An NIH-funded online database of neuroanatomical connectivity in the mouse brain. |
http://www.mouseconnectome.org/
http://brainarchitecture.org/ http://connectivity.brain-map.org/ |
| Allen Mouse Brain Connectivity Atlas | A cell-based connectivity map of the C57BL/6J mouse brain | http://connectivity.brain-map.org/ |
| Mouse Brain Architecture Project | A connectivity map of the C57BL/6J mouse brain | http://brainarchitecture.org/mouse |
Table II.
Enabling Technologies and Resources
| General Resource for Data and Resources | ||
| Neuroscience Information Framework | A web portal and search engine that links to data and research resources and has the capability of searching data and resources in the hidden web. | http://neuinfo.org |
| Genetic Resources for Rat and Mouse | ||
| Mouse Genome Informatics Database | International database of biological resources for the mouse. | www.informatics.jax.org |
| Genenetwork | GeneNetwork is a database and open source bioinformatics software resource for systems genetics | www.genenetwork.org |
| Geneweaver | Gene Weaver allows users to integrate phenotype centered gene sets across species, tissue and experimental platform | http://geneweaver.org/ |
| Mouse Phenome Database | Collaborative collection of baseline phenotypic data on inbred mouse strains | http://phenome.jax.org |
| International Mouse Phenotyping Consortium (IMPC) | Phenotype and characterize the function of 20,000 known and predicted mouse genes | http://www.mousephenotype.org/ |
| The Rat Genome Database | Genetic resources for rat | http://rgd.mcw.edu/ |
| Rat Resource and Research Center | A NIH-funded Resource Center to supply biomedical investigators with the rat models, embryonic stem cells, related reagents, and protocols they require for their research. | http://www.rrrc.us/ |
| Resources for Knockout Mice | ||
| International Gene Trap Consortium (IGTC) | IGTC represents all publicly available gene trap cell lines, which are available on a non-collaborative basis for nominal handling fees. | http://www.genetrap.org/ |
| International Knockout Mouse Consortium | Knockout Mouse Project (KMP, funded by NIH) European Conditional Mouse Mutagenesis Project (EUCOMM, funded by European Union) North American Conditional Mouse Mutagenesis Project (NorCOMM, funded by Genome Canada) Texas A&M Institute for Genomic Medicine (TIGM) |
www.knockoutmouse.org |
| Mutant Mouse Regional Resource Centers (MMRRCs) | Distributes and cryopreserves scientifically valuable, genetically engineered mouse strains and mouse ES cell lines with potential value for the genetics and biomedical research community. | http://www.mmrrc.org/about/generalInfo.php |
| International Mouse Strain Resource (IMSR) | A searchable database of mouse strains, stocks, and mutant ES cells that are available in public repositories | www.findmice.org |
| Cre-Driver and FLP-Driver Lines | ||
| Cre-driver Network | Database and Expression profile of Cre driver lines supported by the NIH Blueprint | www.credrivermice.org |
| The Jackson Laboratory Cre Repository | Provides the scientific community information about a comprehensive set of well characterized Cre driver lines and their availability | http://cre.jax.org/index |
| The Jackson Laboratory FLPe repository | Provides information about available FLP-FRT recombinases in mice and constructs | http://www.jax.org/search/Main.jsp?qt=Research+tools+FLP-FRT+System&sg=0 |
| Cre-X-Mice: A database of Cre Transgenic Lines | A database of Cre-transgenic lines maintained by Andras Nagy at Mt Sinai Hospital, Toronto, Canada | http://nagy.mshri.on.ca/cre_new/index.php |
| Allen Brain Atlas expression map of Cre and other drivers | Expression maps of transgenic Cre and other driver lines in mice | http://connectivity.brain-map.org/transgenic/search/basic/ |
| Gensat Cre-Mice | Provides tissue specific expression of Cre recombinases expressed in Bacterial Artificial Chromosomes | http://www.gensat.org/cre.jsp |
| Full Length cDNAs | ||
| Mammalian Gene Collection (MGC) | Full length cDNAs for human, mouse, rat, and cow | http://mgc.nci.nih.gov/ |
| The Xenopus Gene Collection | Full length cDNAs for Xenopus | http://xgc.nci.nih.gov/ |
| The Zebrafish Gene Collection | Full length cDNAs for Zebrafish | http://zgc.nci.nih.gov/ |
| Viral Vectors | ||
| National Gene Vector Biorepository (NGVB) | A NIH-funded Center to address the increasing needs of the research community for access to the use of viral transneuronal tracing tracers. This approach is the most widely used method to gain a circuit perspective on the functional architecture of the nervous system at a cellular level. | https://www.ngvbcc.org/Home.action |
| Tissue-Specific Enhancers and Promoters | ||
| Pleiades Promoter Project: | This resource provides mini-promoters to drive tissue specific expression. | http://www.pleiades.org/ |
| The ENCODE Project | Then ENCyclopedia Of DNA Elements provides information about the functional elements in the genome | http://www.genome.gov/10005107 |
| Genome Browsers | ||
| University of California at Santa Cruz Genome Browser | Provides reference sequences and working draft assemblies for large collection of genomes and access to the ENCODE and Neanderthal projects. | http://genome.cse.ucsc.edu/ |
| Ensembl Genome Browser | Produces genome database for the eukaryotic and vertebrate organisms. | http://www.ensembl.org/index.html |
| NCBI Genome | This resource organizes information on genomes including sequences, maps, chromosomes, assemblies, and annotations. | http://www.ncbi.nlm.nih.gov/genome |
| Epigenomes | ||
| NIH Roadmap Epigenomics Mapping Consortium | A consortium leverages experimental pipelines built around next-generation sequencing technologies to map DNA methylation, histone modifications, chromatin accessibility, and small RNA transcripts in stem cells and primary ex vivo tissues selected to represent the normal counterparts of tissues and organ systems frequently involved in human disease. | http://www.roadmapepigenomics.org/ |
| The ENCODE Project | The ENCyclopedia Of DNA Elements provides information about the functional elements in the genome | http://www.genome.gov/10005107 |
| Epigenomics | Genome wide maps of DNA and chromatin modifications | http://www.ncbi.nlm.nih.gov/epigenomics/ |
| MicroRNA | ||
| microRNA.org | A website service to predict microRNA targets & target downregulation scores. Experimentally observed expression patterns. |
http://www.microrna.org/microrna/home.do http://www.rnainterference.org/(for background) |
| Proteins | ||
| NIF Antibody Registry | Provides a stable, traceable, permanent identifier to all antibody products created by large commercial vendors and individual | http://www.neuinfo.org/products/antibodyregistry/ |
| Protein Database | Databases of protein sequences and 3dimensional structures of proteins | http://www.ncbi.nlm.nih.gov/protein |
| Other Important Research Resources | ||
| Drosophila Genomics Resource Center (DGRC) | A NIH-funded Center which serves the Drosophila research community to provide services on DNA clones, cell lines, microarrays and the emerging genomics technologies. | https://dgrc.cgb.indiana.edu/ |
| Zebrafish International Resource Center | A NIH-funded Resource Center to provide a central repository for wild-type and mutant strains of zebrafish (Danio rerio) and for materials and information about zebrafish research. | http://zebrafish.org/zirc/home/guide.php |
| Model Organisms for Biomedical Research | Provides information about resources for model organisms | http://www.nih.gov/science/models/ |
Connectome
The detailed description of neuronal circuitry of the mammalian brain remains highly incomplete, a knowledge gap that was emphasized in an article by Francis Crick and Ted Jones in 199318. This knowledge gap has led to the development of projects to systematically map the connections of the nervous system at the macroscopic, mesoscopic, and microscopic levels. Maps at all three of these levels provide a critical foundation for understanding brain function and dysfunction 19-22. To meet this need, the human connectome and mouse projectome projects have begun to characterize neuronal connectivity at the macroscopic and mesoscopic level. Cell-based connectivity maps based on molecular phenotypes are just beginning to be developed.
Cell-based connectivity is important because the location of cell types and the connectivity between types of cell in the nervous system creates structural constraints on the functional properties of the nervous system by limiting and directing the flow of information23. In support of this hypothesis, Varshney, et al24, Sohn et al 25, Jerrell et al 26, have been able to make testable predictions about functional connectivity underlying behavior in C. elegans based on EM reconstruction of the wiring diagram of the C. elegans and computational methods that weight synaptic strength instead of synaptic number. Their analysis is limited by the inability to discern from electron micrographs whether a synapse is excitatory or inhibitory and the effect of neuromodulators on these synapses when released from distant sites.27
The Human Connectome Project
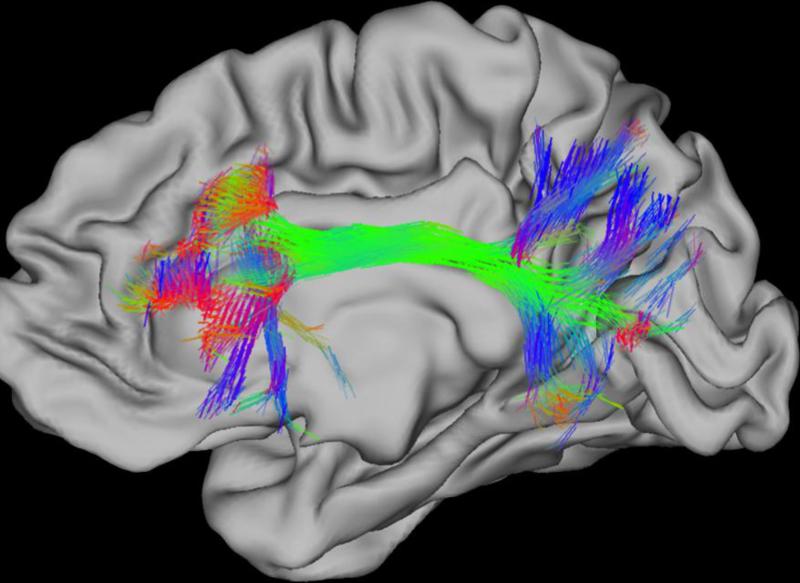
The NIH Neuroscience Blueprint Human Connectome Project is a five year project designed to map the brain connectivity of 1200 healthy adults, including 300 twin pairs and their non-twin siblings, to correlate connectivity with genetic and behavioral data (Fig. 4). The four methods that will be used for mapping structural and functional connections in the human brain include diffusion MRI (d-MRI), task functional MRI (T-fMRI), resting-state fMRI, T1- and T2-weighted MRI. To compensate for the limited temporal resolution of the MRI, EEG and magneto-electroencephalography will also be used. The first phase of the project (2010 to mid-2012) has performed optimization of hardware and imaging protocols, while the second phase (mid-2012 to 2015) will acquire data from 400 research participants each year. The ultimate goal of the project is to make the data freely available to the scientific community.28, 29
Fig 4. Human Connectome.
The diffusion of water in the brain is greater along the lengths of axons than it is across axons. Diffusion MRI (dMRI) reveals the directions along which water diffusion is greatest. The probabilistic trajectories from a frontal cortex starting point that end at other locations on the cortical surface are shown. 3D orientation vectors shown as RGB colored sticks (red: left-right, green: anterior-posterior, blue:inferior- superior) show the estimated directions of white matter fibers through each 1.25mm cubic volume of brain (voxel), the resolution of the diffusion MRI data. Intersection of color tracts suggests connectivity. Adapted with permission153
The Mouse Connectome
Three complementary projects, The Brain Architecture Project, the iConnectome and the Allen Mouse Brain Connectivity Atlas30, are aligned with the scientific recommendations of meetings held at the Banbury Conference Center at the Cold Spring Harbor Laboratory from 2006 to 2008. The published recommendations 31 advocated the brain-wide mapping of mesoscopic level neuronal connectivity across species. The decision to initially map the whole male C57BL/6J mouse connectome at the mesoscopic level will enable the connectome data to be integrated with the Allen Reference Brain Atlas and exploit the genetic resources that exist for the mouse. (Fig 5.)
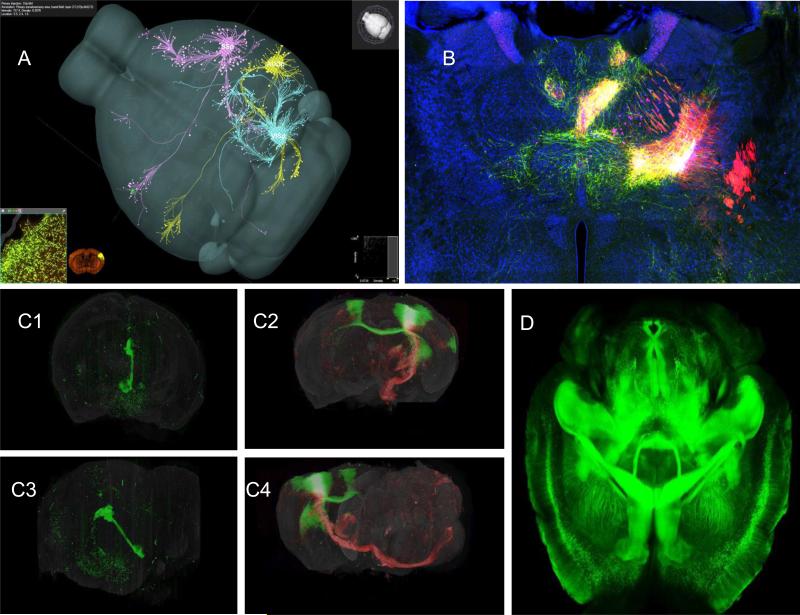
Fig 5. Mouse connectome.
A. Three AAV-GFP tracer injections into the mouse primary somatosensory (SSp), the primary auditory (AUDp) or the primary visual (VISp) cortical area, respectively, are co-displayed in a common 3D mouse brain model in the Brain Explorer program. Courtesy of Hongkui Zeng, Allen Brain Institute B. A representative digital image of the iConnectome project (www.MouseConnectome.org) showing multiple fluorescent labeled neuronal pathways through one coronal thalamic level of the mouse brain, after multiple fluorescent tracers were injected into the infralimbic (PHAL and CTb) and prelimbic (AAV and CTb) cortical areas. Green axons are labeled with PHAL; red axons are labeled with AAV; magenta neurons are labeled with CTb. The yellow regions also contain fluorogold labeled neurons, which are mostly intermixed with AAV axons. This approach effectively reveals two sets of afferents and two sets of efferent pathways associated with two anterograde/retrograde tracer coinjection sites within the same brain and simultaneously exposes reciprocal connections . Courtesy of Hongwei Dong, Univ. Southern California. C. The 3-D reconstruction of two whole mouse brains injected in the left hemisphere with fluorescent virus tracers to visualize specific sets of connectivity. Top, frontal view; bottom, left lateral view. (C1) Left, GFP labeled modified ‘retrograde’ rabies virus injected in the medial nucleus of the habenula (mH) showing retrogradely labeled cell bodies in the hypothalamus and frontal cortex, as well as the passively filled output projection from the mH (Fasciculus Retroflexus) . (2) Right, double injection of primary motor cortex (MOp) with modified ‘anterograde’ adeno-associated virus (AAV2/1 CAG) injection to highlight axons of efferent projections by supragranular neurons (injection with GFP-AAV), and infragranular neurons (injection with RFP-AAV). Courtesy of Partha Mitra, Cold Spring Harbor Laboratory. D. CLARITY is a newly developed technology to transform intact and opaque biological tissue into a hybrid form of formaldehyde-fixed tissue and hydrogel monomers which is physically stable and permeable to exogenous macromolecules. Because the hybrid is permeable to exogenous macromolecules, the whole tissue can be uniformly stained with antibodies to reveal specific cell types and allows multi-round molecular phenotyping of an intact brain. Adapted with permission from Chung and Deisseroth154 .
In the Brain Architecture and the iConnectome projects, anterograde and retrograde tracers are systematically injected together at 200 to 250 different sites in a grid dependent fashion and micrographs are obtained using automated wide field fluorescent microscopy. The inputs and outputs of each brain region are mapped to a grid based on the Allen Reference Atlas. Both projects provide a detailed map of axonal projections and are likely to reveal a distributed network map with overlapping circuitry instead of the classic labeled lines. Convergent and divergent projections will be visualized with this approach. This method does not distinguish cell types within a region from which a projection arises. These regions are often composed of heterogeneous cell types with each cell type possibly having its own unique projection. This method also does not permit connections to be defined directly as excitatory or inhibitory.
To address the issue of cell heterogeneity giving rise to different projections within a given region, the Allen Mouse Brain Connectivity Atlas further characterizes projections from 300 anatomically defined structures by using genetically defined populations of neurons within these regions that express Cre-recombinases. Injection of Cre-dependent recombinant AAV (rAAV) virus expressing a fluorescent protein only labels the anterograde projections of neurons expressing CRE in a region. Fluorescent images of the rAAV tracer are acquired with serial two-photon (STP) tomography. The limitation of this gene-based method is that cell type is being defined by a single gene and its projection. Furthermore, only a limited number of CRE transgenic mice can be generated with cell-specific or region-specific expression. Thus, many cell type-specific projections not defined by a single gene are missed.
The Cre-dependent recombinant AAV (rAAV) virus used by the Allen Brain Mouse Connectivity Atlas is not transynaptically transported and thus cellular connectivity cannot be established. This problem can be surmounted by exploiting other viruses. Rabies and Pseudo Rabies Virus (PRV) are used as retrograde trans-synaptic tracers, while the herpes simplex virus type 1 strain H129 virus has been used as an anterograde trans-synaptic tracer32, 33, 34,35. The direction of transport appears to be determined by the viral glycoprotein 36. This directionality is an advantage over classic transynaptic tracers such as wheat germ agglutinin (WGA), which are transported in both anterograde and retrograde directions35. Since neurotropic viruses replicate within neurons, the problem of serial dilution faced by conventional transynaptic tracers is minimized.
Wickersham, et al37, 38 used a complementation strategy to restrict the transsynaptic spread of rabies virus to monosynaptic connections in genetically defined populations of neurons. This method uses a PRV in which the G glycoprotein needed for replication is replaced with EGFP linked to the glycoprotein, EnvA from ASLV-A. This PRV can only infect mammalian neurons if its receptor TVA, a protein found in birds but not mammals, is expressed on the cell surface. Expression of the TVA receptor can be restricted to a subpopulation of neurons by placing TVA under the control of an inducible promoter. Thus, cells expressing TVA replicate the virus and presynaptically infect a neighboring cell. The presynaptic cell not expressing TVA renders the virus replication incompetent preventing further spread to adjacent neurons, thereby labeling only monosynaptic connections.
Integration of the connectivity datasets described above with super resolution array tomography39, 40 (TEXT BOX 1) and other data in the Whole Brain Catalog will enhance the mesoscopic connectivity data in these atlases by combining cellular connectivity data and receptor types, thereby enabling the determination of whether major projections are excitatory or inhibitory. The Whole Brain Catalog utilizes Waxholm Space41 that allows translation across the different coordinate systems used by different atlases such that the datasets are interoperable. The Whole Brain Catalog is an open source 3-D atlas of the mouse brain that allows linkage and integration of information and data from a number of sources, including the Cell Centered Database (CCDB) 42 and other sources cataloged in the Neuroscience Information framework43.
Reconstruction of Microcircuits
Reconstruction of microcircuits has been based on automated serial block-face electron microscopy with semi-automated image reconstruction44, serial section electron tomography45, and super-resolution array tomography39, 46. EM techniques recognize cell types based on morphology; super-resolution array tomography uses protein expression based on antibody detection and morphology. The development of a genetically encoded tag based on the Arabidopsis phototropic2 protein, a fluorescent protein, called miniSOG (mini Singlet Oxygen Generator) permits long distance serial EM reconstruction of neural connections of cells expressing this genetic tag47. Exposure of miniSOG to blue light generates singlet oxygen that polymerizes diaminobenzidine, permitting tissues to be stained with osmium for electron microscopic visualization. Improved correlation of images obtained with fluorescent microscopy and EM can be obtained because the same tag is used enabling direct association of structure with cellular physiology using genetically encoded sensors in the same cell.
Genetically Encoded Biosensors and Activators
Electrophysiological recording from identified neurons in simple model organism such as Aplysia has contributed greatly to the functional analysis of neural circuits and to the understanding of the cellular basis of behavior. Systems with identified neurons can permit assignment of function to a given cell type within a neuronal circuit and can enable analysis of structure-function relationships. Tissues tagged with genetically encoded biosensors (GEBs) and genetically encoded activators (GEAs) are beginning to allow functional dissection of specific tissues and brain regions comprised of highly heterogeneous cell types. Neuronal tissues can be tagged via transgenes, through infection with viral vectors, and/or by electroporation of embryos. In the case of transgenes, cellular specificity is dependent on the cis regulatory elements driving expression of the GEB and GEA or the recombinase used to activate the GEB/GEA. Attempts to use split recombinases (e.g.split CRE)48, 49 may increase cellular selectivity, but the expression of two genes may not be sufficient to uniquely tag a cell type. Selective expression of GEBs/GEAs using viral vectors and plasmids is critically dependent upon tissue specific promoters.
GEBs encode a fluorescent or bioluminescent protein linked with a functional sensing or target binding sensor. The fluorescent proteins used are often modified GFP derivatives or analogous proteins from marine organisms, while the bioluminescent proteins (e.g. aequorin or luciferase) are typically enzymes that emits light upon oxidizing substrates such as coelenterazines50. Fluorescent protein GEBs depend either on a single fluorescent protein, on a biomolecular fluorescent complementation strategy (BiFC), on a double dimer fluorescent strategy (ddFP)51 or on fluorescent energy transfer (FRET). The functional or target detecting component of the GEB is a linked protein that may bind to ions (e.g., Ca2+, Zn2+, Cl−, and H+); small molecules (e.g., cAMP or hydrogen peroxide; or enzymes, (e.g., myosin light chain kinase)52. Thus, biochemical, metabolic, and physiological changes occurring within cells can now be visualized.
Genetically encoded calcium indicators (GECIs)
The most widely used sensors of neuronal activity are genetically encoded calcium indicators based on GFP. GCaMP5 and GCaMP6, which can sense calcium influx, are currently the most sensitive of these detectors53, 54. New genetically encoded calcium indicators with different emission wavelengths have been created enabling multi-color imaging of different cell types and organelles. Overcoming the problem of spectral overlap between genetically encoded calcium indicators (GECI) and optogenetic activators allows simultaneous use of blue-activated channel rhodhopsin to stimulate neuronal activity with a red-shifted GECI to record neuronal activity55 . GECIs have been used to map the sequential activation of neurons in the parietal cortex during a memory task56. GECIs, combined with high speed light-sheet microscopy, have permitted whole brain functional imaging in larval zebrafish with the activity determined for 80% of neurons at single cell resolution57, 58 .
Genetically Encoded Voltage Sensors
Imaging of calcium transients is an indirect measure of neuronal activity and does not provide information about hyperpolarization and sub-threshold events. A number of versions of genetically encoded voltage sensors have been developed59-61 . These include Flash, SPARC, FLARE, VSFP, and Arch3 (Arch) 62,63-65. These voltage sensors, except for Arch, have time constant greater than 1ms, making it difficult to temporal resolve action potentials. However Arch suffers from low brightness. A major advance for the field will be the development of voltage sensors that have time constants of less than 1 millisecond, a large change in fluorescence in relationship to baseline (ΔF/F) with signal to noise ratio (SNR) greater than 2, some detectable fluorescence at the resting potential, a peak molecular brightness equivalent to eGFP, limited bleaching, a capacitance change of <1%, ability to target to the soma or dendrites, and are non-toxic as well as being genetically encodable.60, 61
Genetically Encoded Sensors of Neurotransmitter Release
pHflourins are pH sensitive fluorescent proteins used as genetically encoded reporters to measure synaptic release. pHfluorins can be targeted to synaptic vesicles by fusing pHflourin to synaptic vesicle proteins such as SNARE, synaptophysin, and the vesicular glutamate transporter. The acidic environments inside synaptic vesicles quench fluorescence while exocytosis exposes the lumen of the vesicle to neutral pH,which reduces quenching and increasing fluorescence 66. The development of mOrange2-pHluorins tagged to VGLUT1 together with a green calcium reporter GCaMP3 fused with synaptophysin-GaMP3 (SyGCaMP3) permits analysis of vesicle release with calcium concentration at the same synapse 67. In addition, glutamate-sensitive fluorescent reporters have also been developed to visualize the release and diffusion of glutamate at the synapse 68.
Genetically Encoded Activators and Inhibitors Manipulate Neuronal Activity
Optogenetic Methods
Over the past ten years, a large number of genetically encoded proteins that affect the electrophysiological characteristics of neurons and that are activated by light or controlled by small molecules have been developed to manipulate the activity of neurons. Lasers, mercury arc lamps, and light emitting diodes are used to activate optogenetic probes69. Three different optogenetic methods have been used to control membrane potential70. When exposed to bluelight, neuronally-expressed channelrhodopsin depolarizes neurons by activating a non-selective cation ion channel. In response to green-yellow light, neurons expressing halorhodopsin hyperpolarize by eliciting an inward chloride current. Suppression of neuronal activity can also be achieved with archaerhodopsin which,when stimulated with green-yellow light, generates an outward proton current produces hyperpolarization. (Fig, 6)
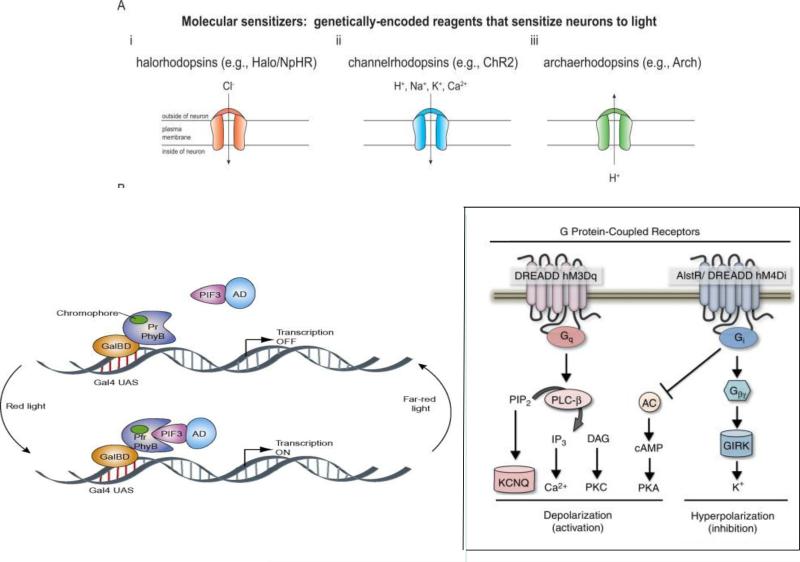
Fig. 6.
Genetically encoded biosensors A. Some genetically encoded biosensors have been used as molecular sensitizers to manipulate neuronal activities. Neurons express them can be activated or silenced by light. a. Halorhodopsins, such as N. pharaonis halorhodopsin, pump chloride ions inward and hyperpolarize the neuron upon yellow light. b. Channel rhodopsins are inward nonspecific cation channels for H+, Na+, K+, and Ca2+. They respond to brief, millisecond-timescale pulses of blue light, transporting cations inward and depolarizing neuron. c. Stimulation of Archaerhodopsins (H. sodomense opsin) with yellow or green lights shuts down neuronal activity by hyperpolarizing neurons. Adapted with permission70.
B. Biosensors can be designed to control protein-protein interactions. Here red light triggers PhyB binding of PIF3. Since PhyB is fused to a Gal4-binding domain (GalBD) and PIF3 fused to a Gal4 activation domain (Gal4AD), binding of PhyB to PIF3 in turn dimerizes GalBD with Gal4AD, and initiates gene expression. Remarkably, in the dark or upon far-red light PhyB reverts to the non-binding conformation and releases PIF3, shutting down transcription. In this way, light can be used to either induce or shut down transcription. Adapted with permission from155.
C. Biosensors are effective tools to rapidly manipulate neuronal activity. DREADDs are engineered from muscarinic acetylcholine receptors (mAChRs). It is potently activated by the pharmacologically inert molecule clozapine-N-oxide (CNO). When it is coupled to Gq, Gi or Gs the GPCR signaling pathways can be manipulated without obvious interference with endogenous GPCR signaling. In this figure the presence of CNO selectively activates (Gq-coupled hM3Dq) or inhibits (Gi-coupled hM4Di) G-protein signaling. The response is robust and reversible for the study of neuronal activity. Activation of Gq signaling leads to neuronal depolarization and increased activity. In contrast, CNO activation of Gi signaling triggers inward rectifying potassium channels (GIRKs), resulting in hyperpolarization and inhibition of the neuron.
The utility of optogenetic methods and genetically encoded reporters to monitor and control brain activity in vivo is limited by the need to use invasive methods to stimulate the optogenetic tool or visualize light being emitted by a genetically encoded reporter because the opaque nature of nervous tissue scatters light. The depth of fluorescent imaging by two photon microscopy, which images to a maximum depth of 1000 μM, exceeds laser confocal microscopy71. This depth can be extended by penetrating tissue with a fluorescent microendoscope with gradient refractive index (GRIN) microlenses72 that also permit recording in alive and awake animals. Activation of neurons in awake behaving animal via light stimulation of genetically encoded proteins in deep brain structures requires fiber optic probes with light emitting diode arrays to be inserted deeply in the brain to reduce light scattering73, 74. An advantage of the optogenetic method is that it permits the brain-wide mapping of activity generated by stimulating neurons (potentially of identified type) in the region near the stimulating optical fiber. This has permitted the development of optogenetic fMRI 75, 76 which promises to help disentangle neuronal circuitry involved in Deep Brain Stimulation therapy of neurological and neuropsychiatric disease. Furthermore recent advances may enable locus specific and/or temporal manipulation of gene expression levels in neurons through targeted alteration of transcription factor binding or chromatin state via opto-epigenetic or chemo-epigenetic methods77-79.
Chemogenetic Methods
An alternative to the optogenetic approach is to express genetically engineered receptors that respond to a ligand without activating endogenous receptors. These approaches include the expression of TRPV1 and P2X activated by photocaged ligands80, activation of a trp channel coupled to transferrin by a pulsing magnetic field81, C. elegans glutamate-gated chloride channel activated by ivermectin82, the Drosophila allatostatin receptor83, and designer receptors exclusively activated by designer drugs (DREADDs) 84, 85. The discovery of Lynx, a family of endogenous peptides with homology to nicotinic acetylcholine receptor blocking snake toxins, has led to development of tethered toxins to permit long-term activation or inactivation of a neural circuit 86.
Molecular Phenotyping of Brain Cell and Tissue Types
The transgenic technologies described have significant therapeutic potential but their translational utility will remain limited unless transgenes can be systemically delivered and expressed in specific cell types and circuits to correct circuit abnormalities that underlie brain diseases. Tools and technologies enabling researchers to restrict expression of a transgene to any specific cell type of interest would greatly enhance our ability to investigate nervous system circuitry and mechanisms. Yet it has become apparent that no single gene expressed in the nervous system defines a cellular phenotype. For example, work by the Rubin lab suggests that multiple enhancers are required to drive cell specific expression of a transcriptional activator such as GAL4 in Drosophila80, 81.
To begin to understand the genome-wide molecular characteristics and enhancer elements for specific cell types, scientific consortia such as the NIH Roadmap Epigenomics Program and the NHGRI-funded Encyclopedia of DNA elements (ENCODE) project have been generating molecular phenotype data sets for a variety of Drosophila, C. elegans, mouse and human cell and tissue types 87-91. These data sets can be used to identify molecular signatures and enhancer elements specific for particular cell types. In particular, identification of unique cell-specific enhancer elements may facilitate our ability to spatially and temporally express transgenes with much greater specificity.
The NIH Roadmap Epigenomics Program has been generating comprehensive maps of chromatin from a wide variety of “normal” human cell and tissue types including components of the nervous system from post-mortem adult and fetal tissue92. These maps typically include DNA methylation information, chromatin immunoprecipitation data for several informative activating or silencing histone modifications, chromatin accessibility information using the DNAse1 hypersensitivity assay, and gene expression information. A major discovery made using data generated by the NIH Roadmap Epigenomics Program is the finding that GWAS variants are highly enriched in regulatory DNA regions and often have activity during fetal development93.
While the transcriptional profile of a number of cell types or tissue types have been characterized and published94-103, no systematic analysis of the transcription profiles of nervous system cell types has yet been reported. Several different approaches have been used to transcriptionally profile individual cell types in the nervous system. These methods include immunopanning (PAN), Laser Capture Microscopy (LCM), Fluorescent Activated Cell Sorting (FACS), manual sorting of fluorescently labeled cell (manual) and Translating Ribosome Affinity Purification, and Ribo-tag. LCM and manual sorting of fluorescently labeled cells are limited by throughput 104, 105. FACS for isolating cells106, 107 and PAN can introduce artifactual gene expression changes due to cellular stress108. These problems quantitating translated mRNAs are overcome with genetic methods of tagging of ribosomes using BACs/ TRAP 109 or the induction of the floxed ribosomal tag (Ribo-Tag) with a “cell specific Cre”110.
The BAC-TRAP (Bacterial Artificial Chromosome- Translating Ribosome Affinity Purification) method dynamically measures translated mRNAs in genetically identified cells, providing molecular phenotype information for cell types in the nervous system109, 111 . In this method, the L10 ribosomal subunit is inserted into the first exon of a gene tagged with eGFP into a BAC. Polysomes are affinity purified using an antibody against eGFP and the mRNAs are profiled using microarrays or RNAseq. Background transcriptional contamination from other cell types is removed by subtracting the unbound RNA isolated from the tissue from the RNA isolated from the bound polysomes or from a BAC-TRAP that is ubiquitously expressed. This method provides a quantitative measure of gene expression unlike the qualitative measurement made with eGFP BACs or in situ data, which is a static representation of gene expression. Already this method has identified and characterized a number of specific cell types on the basis of molecular phenotype112-115.
Ribo-tag uses a gene targeting approach in which the ribosomal gene, Rpl22, is floxed with hemagglutinin (HA) epitope tag inserted before the stop codon. Crossing the Ribo-Tag mouse with a Cre tissue-specific driver induces the expression of the epitope tagged Rpl22. The epitope-tagged Rpl22 incorporated ribosomes is used to immunoprecipitate ribosome-specific transcripts from specific tissues.110
For profiling miRNAs, floxed Ago2 tagged with GFP and myc (miRAP) has also been used to identify miRNAs in glutamatergic and GABAergic cells in the cortex and cerebellum116. Tissue-specific CRE drives expression of the Ago2 trangene in a specific neuronal region and the myc tag permits immunoprecipitation of the miRNA-Ago2 complex, followed by deep sequencing.
The determination of cell specific expression by TRAP, Ribo-Tag, and miRAP is limited by either BACs that tag specific cell types or knowledge of cell specific promoters. Microfluidic and lab on chip technology may offer a solution to an unbiased analysis of gene expression, epigenetic marks, protein expression, and metabolomics of single cells in a heterogeneous population of cell types117, 118. However, dissociation of nervous tissue disrupts the ability to correlate cell type and cellular physiology with cell type-connectivity as well as introducing artifacts. The ability to analyze changes in cellular connectivity with cellular epigenetic changes in individual neurons on network scale is essential for understanding the mechanisms of synaptic plasticity and learning, a major goal of neuroscience, as well as for determining the importance of structural connectivity in influencing function.
Using Barcodes to Inventory Neuronal Cell Types and Connections
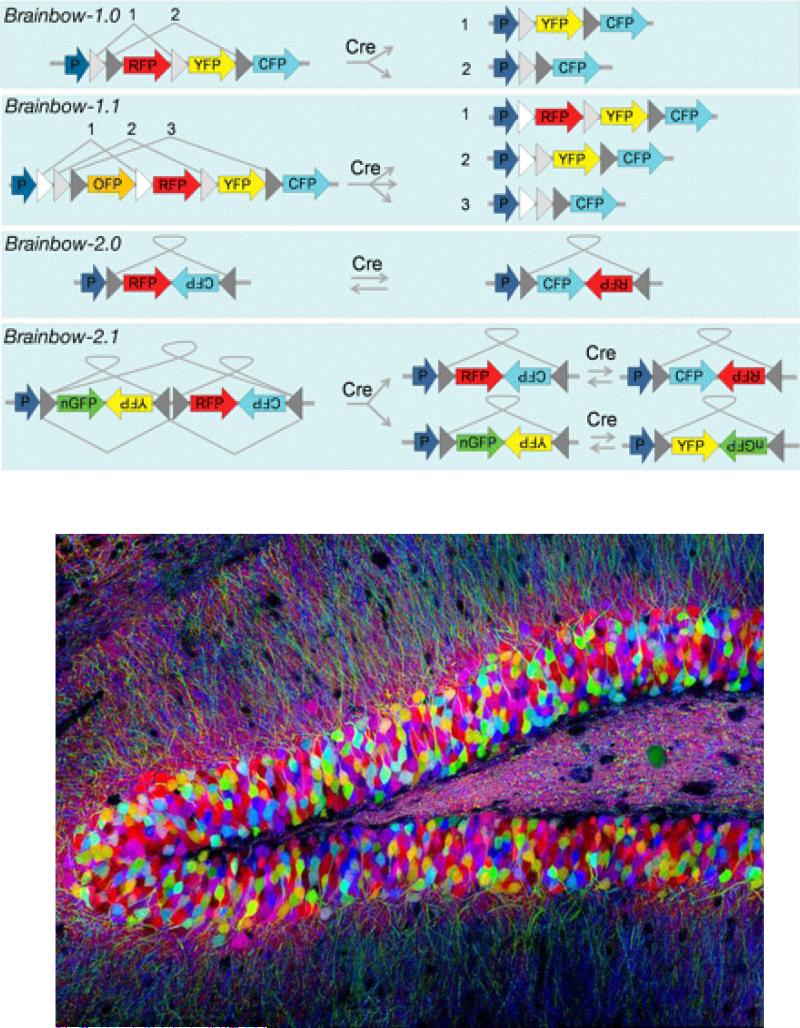
Correlation of cell type with cell-type specific connectivity in the nervous system is ultimately an inventory problem. In commerce, optical barcodes have been used for decades to handle inventory problems. Recently cellular barcodes based on DNA sequence have been used to track hematopoietic cell lineages119-124. The development of robust cell-specific DNA barcode technology that uniquely labels ribosomes could provide a means to compare the gene expression profiles of a specific cell with other cells. This unique tag could also label proteins at the synapse, making possible the identification of connections between cells. Brainbow provides a partial solution to this inventory problem 125, 126. In Brainbow, different genes encoding different fluorescent proteins are placed in between loxp sites (Fig. 7). A palette of different color neurons are generated when CRE acting on the loxp sites causes random rearrangements and deletions of the fluorophores. However, the number fluorophores and their dynamic range given the number of cells and synapses limit the utility of using Brainbow.
Fig 7.
Brainbow. A. Strategy for creating Brainbow mice. In Brainbow 1.0 and Brainbow 1.1 excision of different fluorophores from incompatible lox variants generates 2 to 3 different color combinations of XFP. In 2.0 the XFP genes are placed head to head between two asymmetric loxP sites. Activation of recombination by CRE causes inversion and change in the expression of the fluorophore from red to cyan. Place the inverted fluorophore genes between asymmetric loxP sites in tandom increases the number of combinations to four. B. Fluorescent fluorophore expression in mouse hippocampus. Adapted with permission from 128.
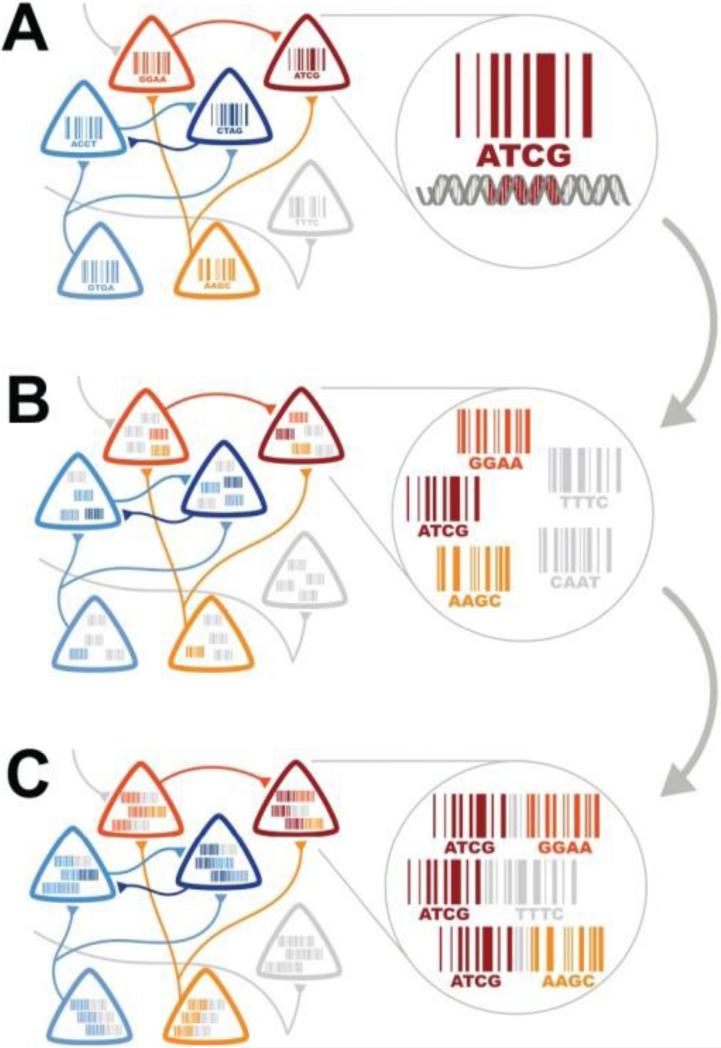
As suggested by Zador et al127 a solution to the connectivity problem could be to tag cells with viruses expressing DNA barcodes and profile the DNA barcodes with NextGen Sequencing. In this scheme each neuron is infected with a transynaptic virus with a unique sequence generated by a recombinase (Fig. 8). Invader viruses would be ligated together with host neuronal viral sequence by phi31 integrase and sequenced to identify synaptically connected cells. In the future, neurons tagged with DNA barcodes may enable molecular profiling of single cells with RNA-seq as well as identification of synaptic connections.
Fig 8.
DNA Barcoding of Individual cells and neuronal connectivity converts connectivity problem into a sequencing problem. A. Each neuron is assigned a unique identity by labeling the cell with PRV virus that expresses a unique DNA barcode. B. PRV virus expressing a unique DNA Barcode is transynaptically transported (the invader virus) across a synapse. C. The DNA Barcodes from host PRV virus and the Invader PRV virus are joined by PhiC31 integrase and high throughput sequencing is performed. Reprinted from Zador119.
The use of DNA barcodes overcomes the lack of dynamic range of fluorophores used in Brainbow since there are far more barcodes available than there are fluorophores128. For example, 1015 barcode combinations can be generated with 25 nucleotides 425 more than enough to label either 6.3 × 107 neurons and 5 × 107 non-neuronal cells in the mouse brain129, 130 or the 8.6 × 1010 neurons and 8.5 × 1010 non neuronal cell in the human brain, and sufficient to label 8.8 ×1011 synapse in the mouse brain 131. The problem of inventorying neuronal identity and connectivity is then essentially transformed into a more tractable high-throughput DNA sequencing problem. Our current ability to generate and analyze large amounts of DNA sequence at low cost is extremely robust.
The barcode invader method described by Zador may be limited by the problem of infection of host neuron with multiple viruses, although statistical methods may help overcome this problem. Secondly, this method does not provide a way to correlate cell type (as defined by transcription profile) with connectivity. Nor does it allow quantification of changes in gene expression or epigenetic changes with changes in synaptic strength.
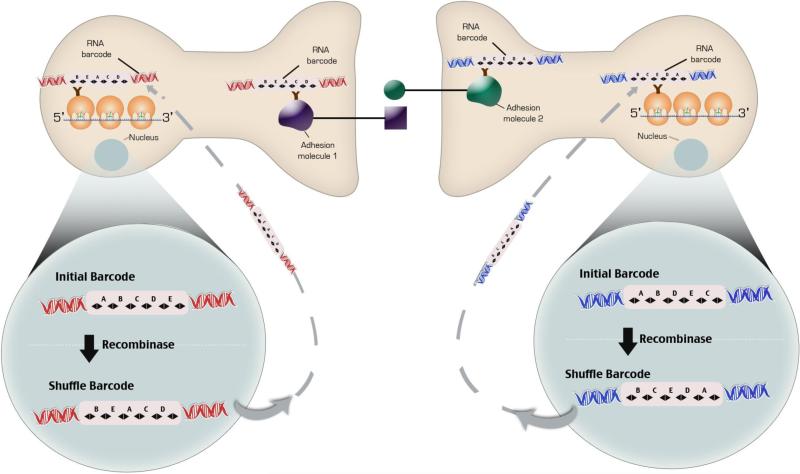
The development of new methods to tag ribosomal subunits and synaptic proteins with cell-specific nucleic acid barcodes using zinc finger DNA or RNA binding proteins132 may provide a method to correlate cell type with connectivity and unambiguously label specific cells (Fig 9). Isolation of synaptic adhesion molecules from a pre-and post-synaptic neurons that bind one another tagged with a cell-specific barcode would show two different barcodes of two mono-synaptically connected neurons. Any increase in the number of sequenced correlated barcodes would indicate a change in synaptic strength. Transcriptional profiles obtained from ribosomes tagged with the same cell-specific barcode can then be associated with the barcode connectivity data and changes in synaptic plasticity. Cell-specific barcodes are generated by combining a unique nucleic acid sequence with 20 to 30 DNA nucleotides inserted between LoxP sites in a transgene that is expressed throughout the nervous system. By crossing transgene with a CRE mouse a unique barcode identifier is created.
Fig 9.
Schema for correlating cell-type gene expression with connectivity. A Lentiviral vector carrying a unique DNA barcode infects neurons so that each neuron expresses a unique cellular barcode. The DNA element in the viral genome are transcribed and translated such that an RNA barcode is generated. B. In the second schema DNA Barcodes inserted into the Rosa26 locus are placed between loxp sites or another site specific recombinase. Recombination of the barcode sequence is induced by the activation of the recombinase and RNA barcode is generated. The RNA barcode binds to an RNA binding protein inserted in into a ribosomal subunit with EGFP of the cells ribosomes. A RNA binding protein is also inserted into pre- and post-synaptic adhesion molecules that bind to each other. The ribosomes are purified. Complementary DNA sequences attached to a longer DNA sequence are hybridized to the RNA Barcode on the ribosome. PCR primers to the 5' sequence of the complementary DNA sequences attached to a longer DNA and to polyA tail are made such that they join upon the PCR reaction. PCR and sequencing performed. Only mRNAs from that cell will contain the unique barcode sequence. At the same time synaptic adhesion complexes are isolated from synaptomsomes with immunoprecipitation. The DNA barcodes are hybridized to beads containing the complementary sequence and sequenced in a microfluidic chamber.
Wouldn't sequencing barcodes to establish connectivity destroy useful 3-D information needed for reconstruction of circuits? The problem of loss of spatial resolution might be partly overcome by voxelating the brain into small cubes followed by RNA seq profiling of the DNA barcoded neurons. Alternatively, strategies to image cellular DNA barcodes in serial sections of nervous tissue may help to overcome this obstacle that include in situ sequencing133-138.
The use of cellular barcodes will not lead directly to the ultimate goal of unambiguously identified neurons as in Aplysia because the barcodes labeling each cell occur randomly in each animal and are not specified by a developmental program. The use of barcodes to identify cell types and cellular connectivity may help to identify a molecular combinatorial code of cell surface molecules that defines cell types and connectivity as has been suggested for cadherins139 that could ultimately lead to the discovery of a complete catalog of cell types in the nervous system. The generalization of this code across species is testable.
Association of a unique set of specific cell surface makers with a particular neuronal cell type would enable the targeting of nanovesicles containing shRNAs, viral vectors, and nanoparticles to specific cells with a unique set of surface markers. Even if such a cell surface marker code is not discovered, DNA barcoded Zinc finger proteins expressed on cell surface may facilitate the targeted delivery of viruses and nanoparticles and may provide unique identifiers140, 141.
The transcriptional profiling of cell types using barcodes and the tagging of nuclei with cell-specific barcodes could also assist in the identification of enhancer combinations that specify neuronal cell types. The discovery of an enhancer combinatorial code that specifies gene expression in specific cell types would aid the development of viral vectors that are uniquely expressed in specific cells. The development of such vectors would be not only useful for gene therapy approaches but could greatly aid in mapping functional connectivity of the primate nervous system where the opportunities and capability for doing transgenic delivery of gene through the germline is limited.
Conclusion
While significant progress has been made in the gene-based analysis of the nervous system, improved cell-centric strategies based on molecular phenotype being developed are necessary for a full understanding of brain function. The organizational principle of nervous system is the cell; cell types and the connectivity pattern among cell types control the organization of behavior and the processing of information. The development of new technologies to define cell types, connectivity, improved voltage sensors, and new methods that non-invasively image the activity of thousands of single neurons at the depth of centimeters opens new horizons for a cell-centric understanding of brain function. The identification of cell types and their connectivity based on molecular phenotype provide not only a means to observe nervous system function, but to experimentally manipulate specific cell types and connectivity. The ability to manipulate specific cell type is fundamental to the development of the next generation of cell-type targeted therapeutics to treat brain disorders.
Text Box 1. High Resolution Microscopic Methods for Mapping Neuronal Circuits.
Super Resolution Microscopy
Advances in light microscopy and high throughput electron microscopy have significantly improved our ability to analyze the structure of the nervous system at the microscopic level. The limit of light microscopic resolution for subcellular structures and organelles when diffraction reaches half the wavelength of light (150-300nm) is now overcome with super-resolution fluorescent microscopy that provides 2-10 times the resolution of conventional light microscopes142, 143. These methods include stochastic optical reconstruction microscopy (STORM), stimulated emission depletion microscopy (STED), saturated-illumination microscopy (SSIM), photoactivated localization microscopy (PALM), and fluorescent photoactivation localization microscopy (FPALM).
Array Tomography combined with Super Resolution Microscopy
Dani and colleagues144, using STORM to image proteins at the synapse, were able to show the 3-D relationship among many synaptic proteins using immunolabeling with three spectrally distinct activated dye pairs. STORM only produces high resolution 3D images at tissue surfaces because antibody penetration and optical restrictions in thick tissue limit the resolution at deeper depths. By integrating array tomography, an automated serial section/volume reconstruction imaging with super-resolution fluorescent microscopy like STORM, the problems with variable antibody penetration and optical aberrations can be resolved because ultrathin sections can be used. In the technique of array tomography39,40, tissue is imbedded in acrylic resin and serially sliced into a stack of ultrathin sections of 50 to 200nm in width. These sections can be repeatedly stained with, and stripped of, by 20 or more immunofluorescent antibodies for repeated imaging of 20 or more antigens before the final staining with heavy metals for imaging with electron microscopy is performed. The stack of images is placed in register to produce high resolution large volumetric 3D images. When super-resolution imaging is used in array tomography throughput is decreased. Thus, decisions in experimental design require evaluations of the tradeoffs between resolution and throughput needed to resolve the cellular and molecular structures of synapses, neurons, and circuits.
Automated Sectioning, Image Acquisition, and 3-D Reconstruction
Despite automated methods to collect images145, 146 months to years are now required to reconstruct the synaptic connections at the microscopic level in a cubic millimeter of nervous tissue147. Development of improved algorithms for segmentation of images and new computational methods will automate image analysis and increase throughput148, 149. Toward this goal Helmstaedter150 has developed KNOSSOS software for visualization and annotation of imaged sections that is 50 times faster than volume labeling. Instead of standard volume labeling, point-to-point lines along the length of a neurite or skeletons are created. Multiple human annotators create skeletons for the same neurite. Redundant-skeleton consensus procedure (RESCOUP), a statistical method, is used to resolve discrepancies among annotators.
Text Box 2: Unanswered Questions.
How many cell types are defined by their molecular profile and connectivity in the Mammalian Brain?
Dot invariant gene expression profiles define a cell type? What are they?
Are there species-specific brain cell types?
How do cell types impact the functioning of specific genes and their variants?
Can barcode technologies be exploited to provide high throughput methods to identify cell types and connectivity in the brain?
Can improved voltage sensors be developed with the fidelity of microelectrodes with temporal resolution less than 500 microseconds and are highly sensitive to changes in voltage?
Do any laws of physics prevent the non-invasive recording of single neurons at millisecond resolution through centimeters of tissue?
Highlights.
Compelling technologies and resources for neuroscience are highlighted.
Atlases, the connectome, genetically encoded sensors and activators are reviewed.
Methods for molecular phenotyping of brain cell and tissue types are described.
The use DNA barcoding to catalog neuronal cell types and connections is presented.
The argument for new technologies to define cell types and connectivity is given.
Acknowledgment
JDP wishes to thank Drs. Anthony Zador and Ed Boyden for helpful discussions regarding neuronal barcoding. We also thank Dr. Christina Liu for helpful discussions in preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Alivisatos AP, Andrews AM, Boyden ES, et al. Nanotools for neuroscience and brain activity mapping. ACS Nano. 2013;7(3):1850–1866. doi: 10.1021/nn4012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alivisatos AP, Chun M, Church GM, et al. Neuroscience. The brain activity map. Science. 2013;339(6125):1284–1285. doi: 10.1126/science.1236939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, Yuste R. The brain activity map project and the challenge of functional connectomics. Neuron. 2012;74(6):970–974. doi: 10.1016/j.neuron.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 5.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 6.Hawrylycz M, Baldock RA, Burger A, et al. Digital atlasing and standardization in the mouse brain. PLoS Comput Biol. 2011;7(2):e1001065. doi: 10.1371/journal.pcbi.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawrylycz M, Ng L, Page D, et al. Multi-scale correlation structure of gene expression in the brain. Neural Netw. 2011;24(9):933–942. doi: 10.1016/j.neunet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Wolf L, Goldberg C, Manor N, Sharan R, Ruppin E. Gene expression in the rodent brain is associated with its regional connectivity. PLoS Comput Biol. 2011;7(5):e1002040. doi: 10.1371/journal.pcbi.1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko Y, Ament SA, Eddy JA, et al. Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain. Proc Natl Acad Sci U S A. 2013;110(8):3095–3100. doi: 10.1073/pnas.1222897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grange P, Bohland JW, Okaty B, et al. A cell-based model explaining co-expression patterns of genes in the brain. Proc.Natl.Acad.Sci.U.S.A. 2013 doi: 10.1073/pnas.1312098111. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heintz N. Gene expression nervous system atlas (GENSAT). Nat Neurosci. 2004;7(5):483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 13.Gong S, Kus L, Heintz N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nat Protoc. 2010;5(10):1678–1696. doi: 10.1038/nprot.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong S, Zheng C, Doughty ML, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 15.Madisen L, Mao T, Koch H, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15(5):793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi H, He M, Wu P, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crick F, Jones E. Backwardness of human neuroanatomy. Nature. 1993;361(6408):109–110. doi: 10.1038/361109a0. [DOI] [PubMed] [Google Scholar]
- 19.Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1(4):e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson LW, Bota M. Foundational model of structural connectivity in the nervous system with a schema for wiring diagrams, connectome, and basic plan architecture. Proc Natl Acad Sci U S A. 2010;107(48):20610–20617. doi: 10.1073/pnas.1015128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson LW. Quest for the basic plan of nervous system circuitry. Brain Res Rev. 2007;55(2):356–372. doi: 10.1016/j.brainresrev.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denk W, Briggman KL, Helmstaedter M. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat Rev Neurosci. 2012;13(5):351–358. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- 23.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314(1165):1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 24.Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol. 2011;7(2):e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohn Y, Choi MK, Ahn YY, Lee J, Jeong J. Topological cluster analysis reveals the systemic organization of the Caenorhabditis elegans connectome. PLoS Comput Biol. 2011;7(5):e1001139. doi: 10.1371/journal.pcbi.1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarrell TA, Wang Y, Bloniarz AE, et al. The connectome of a decision-making neural network. Science. 2012;337(6093):437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- 27.Bargmann CI. Beyond the connectome: how neuromodulators shape neural circuits. Bioessays. 2012;34(6):458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- 28.Van Essen DC, Ugurbil K, Auerbach E, et al. The Human Connectome Project: A data acquisition perspective. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Essen DC, Ugurbil K. The future of the human connectome. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josh HZ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 31.Bohland JW, Wu C, Barbas H, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput Biol. 2009;5(3):e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ugolini G. Rabies virus as a transneuronal tracer of neuronal connections. Adv Virus Res. 2011;79:165–202. doi: 10.1016/B978-0-12-387040-7.00010-X. [DOI] [PubMed] [Google Scholar]
- 33.Ekstrand MI, Enquist LW, Pomeranz LE. The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends Mol Med. 2008;14(3):134–140. doi: 10.1016/j.molmed.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18(6):617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72(6):938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beier KT, Saunders A, Oldenburg IA, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci U S A. 2011;108(37):15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickersham IR, Lyon DC, Barnard RJ, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53(5):639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4(1):47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55(1):25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Micheva KD, O'Rourke N, Busse B, Smith SJ. Array tomography: semiautomated image alignment. Cold Spring Harb Protoc. 2010;2010(11):db. doi: 10.1101/pdb.prot5527. [DOI] [PubMed] [Google Scholar]
- 41.Johnson GA, Badea A, Brandenburg J, et al. Waxholm space: an image-based reference for coordinating mouse brain research. Neuroimage. 2010;53(2):365–372. doi: 10.1016/j.neuroimage.2010.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martone ME, Zhang S, Gupta A, et al. The cell-centered database: a database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics. 2003;1(4):379–395. doi: 10.1385/NI:1:4:379. [DOI] [PubMed] [Google Scholar]
- 43.Gardner D, Akil H, Ascoli GA, et al. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6(3):149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471(7337):183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 45.Bock DD, Lee WC, Kerlin AM, et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471(7337):177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleinfeld D, Bharioke A, Blinder P, et al. Large-scale automated histology in the pursuit of connectomes. J Neurosci. 2011;31(45):16125–16138. doi: 10.1523/JNEUROSCI.4077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu X, Lev-Ram V, Deerinck TJ, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9(4):e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 49.Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35(1):70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- 50.Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29(3):144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alford SC, Wu J, Zhao Y, Campbell RE, Knopfel T. Optogenetic reporters. Biol Cell. 2013;105(1):14–29. doi: 10.1111/boc.201200054. [DOI] [PubMed] [Google Scholar]
- 52.Tantama M, Hung YP, Yellen G. Optogenetic reporters: Fluorescent protein-based genetically encoded indicators of signaling and metabolism in the brain. Prog Brain Res. 2012;196:235–263. doi: 10.1016/B978-0-444-59426-6.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akerboom J, Chen TW, Wardill TJ, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32(40):13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen TW, Wardill TJ, Sun Y, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akerboom J, Carreras CN, Tian L, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual- navigation decision task. Nature. 2012;484(7392):62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10(5):413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 58.Panier T, Romano SA, Olive R, et al. Fast functional imaging of multiple brain regions in intact zebrafish larvae using Selective Plane Illumination Microscopy. Front Neural Circuits. 2013;7:65. doi: 10.3389/fncir.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mutoh H, Akemann W, Knopfel T. Genetically engineered fluorescent voltage reporters. ACS Chem Neurosci. 2012;3(8):585–592. doi: 10.1021/cn300041b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mutoh H, Knopfel T. Probing neuronal activities with genetically encoded optical indicators: from a historical to a forward-looking perspective. Pflugers Arch. 2013;465(3):361–371. doi: 10.1007/s00424-012-1202-z. [DOI] [PubMed] [Google Scholar]
- 61.Peterka DS, Takahashi H, Yuste R. Imaging voltage in neurons. Neuron. 2011;69(1):9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9(1):90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker BJ, Lee H, Pieribone VA, et al. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods. 2007;161(1):32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Guerrero G, Siegel MS, Roska B, Loots E, Isacoff EY. Tuning FlaSh: redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys J. 2002;83(6):3607–3618. doi: 10.1016/S0006-3495(02)75361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perron A, Mutoh H, Akemann W, et al. Second and third generation voltage-sensitive fluorescent proteins for monitoring membrane potential. Front Mol Neurosci. 2009;2:5. doi: 10.3389/neuro.02.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miesenbock G. Synapto-pHluorins: genetically encoded reporters of synaptic transmission. Cold Spring Harb Protoc. 2012;2012(2):213–217. doi: 10.1101/pdb.ip067827. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Foss SM, Dobryy YL, et al. Concurrent imaging of synaptic vesicle recycling and calcium dynamics. Front Mol Neurosci. 2011;4:34. doi: 10.3389/fnmol.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hires SA, Zhu Y, Tsien RY. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc Natl Acad Sci U S A. 2008;105(11):4411–4416. doi: 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prakash R, Yizhar O, Grewe B, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9(12):1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernstein JG, Garrity PA, Boyden ES. Optogenetics and thermogenetics: technologies for controlling the activity of targeted cells within intact neural circuits. Curr Opin Neurobiol. 2012;22(1):61–71. doi: 10.1016/j.conb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2(12):932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 72.Wilt BA, Burns LD, Wei Ho ET, Ghosh KK, Mukamel EA, Schnitzer MJ. Advances in light microscopy for neuroscience. Annu Rev Neurosci. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de LL. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450(7168):420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zorzos AN, Scholvin J, Boyden ES, Fonstad CG. Three-dimensional multiwaveguide probe array for light delivery to distributed brain circuits. Opt Lett. 2012;37(23):4841–4843. doi: 10.1364/OL.37.004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JH, Durand R, Gradinaru V, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465(7299):788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Desai M, Kahn I, Knoblich U, et al. Mapping brain networks in awake mice using combined optical neural control and fMRI. J Neurophysiol. 2011;105(3):1393–1405. doi: 10.1152/jn.00828.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Konermann S, Brigham MD, Trevino AE, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gilbert LA, Larson MH, Morsut L, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013 doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121(1):141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 81.Huang H, Delikanli S, Zeng H, Ferkey DM, Pralle A. Remote control of ion channels and neurons through magnetic-field heating of nanoparticles. Nat Nanotechnol. 2010;5(8):602–606. doi: 10.1038/nnano.2010.125. [DOI] [PubMed] [Google Scholar]
- 82.Lerchner W, Xiao C, Nashmi R, et al. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron. 2007;54(1):35–49. doi: 10.1016/j.neuron.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 83.Tan EM, Yamaguchi Y, Horwitz GD, et al. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51(2):157–170. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 84.Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol Rev. 2011;63(2):291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wulff P, Arenkiel BR. Chemical genetics: receptor-ligand pairs for rapid manipulation of neuronal activity. Curr Opin Neurobiol. 2012;22(1):54–60. doi: 10.1016/j.conb.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ibanez-Tallon I, Nitabach MN. Tethering toxins and peptide ligands for modulation of neuronal function. Curr Opin Neurobiol. 2012;22(1):72–78. doi: 10.1016/j.conb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roy S, Ernst J, Kharchenko PV, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330(6012):1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerstein MB, Lu ZJ, Van Nostrand EL, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Okaty BW, Sugino K, Nelson SB. Cell type-specific transcriptomics in the brain. J Neurosci. 2011;31(19):6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nelson SB, Hempel C, Sugino K. Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol. 2006;16(5):571–576. doi: 10.1016/j.conb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 96.Nelson SB, Sugino K, Hempel CM. The problem of neuronal cell types: a physiological genomics approach. Trends Neurosci. 2006;29(6):339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 97.Sugino K, Hempel CM, Miller MN, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9(1):99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 98.Rossner MJ, Hirrlinger J, Wichert SP, et al. Global transcriptome analysis of genetically identified neurons in the adult cortex. J Neurosci. 2006;26(39):9956–9966. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paul A, Cai Y, Atwal GS, Huang ZJ. Developmental Coordination of Gene Expression between Synaptic Partners During GABAergic Circuit Assembly in Cerebellar Cortex. Front Neural Circuits. 2012;6:37. doi: 10.3389/fncir.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29(39):12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45(2):207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 102.Cahoy JD, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J Neurosci. 2009;29(21):7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meguro R, Lu J, Gavrilovici C, Poulter MO. Static, transient and permanent organization of GABA receptor expression in calbindin-positive interneurons in response to amygdala kindled seizures. J Neurochem. 2004;91(1):144–154. doi: 10.1111/j.1471-4159.2004.02701.x. [DOI] [PubMed] [Google Scholar]
- 105.Rossner MJ, Hirrlinger J, Wichert SP, et al. Global transcriptome analysis of genetically identified neurons in the adult cortex. J Neurosci. 2006;26(39):9956–9966. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tomomura M, Rice DS, Morgan JI, Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur J Neurosci. 2001;14(1):57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- 107.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 108.Okaty BW, Sugino K, Nelson SB. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS One. 2011;6(1):e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heiman M, Schaefer A, Gong S, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Doyle JP, Dougherty JD, Heiman M, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dalal J, Roh JH, Maloney SE, et al. Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 2013;27(5):565–578. doi: 10.1101/gad.207654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dougherty JD, Maloney SE, Wozniak DF, et al. The disruption of Celf6, a gene identified by translational profiling of serotonergic neurons, results in autism-related behaviors. J Neurosci. 2013;33(7):2732–2753. doi: 10.1523/JNEUROSCI.4762-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Warner-Schmidt JL, Schmidt EF, Marshall JJ, et al. Cholinergic interneurons in the nucleus accumbens regulate depression-like behavior. Proc Natl Acad Sci U S A. 2012;109(28):11360–11365. doi: 10.1073/pnas.1209293109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schmidt EF, Warner-Schmidt JL, Otopalik BG, Pickett SB, Greengard P, Heintz N. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149(5):1152–1163. doi: 10.1016/j.cell.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73(1):35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.White AK, Heyries KA, Doolin C, Vaninsberghe M, Hansen CL. High Throughput Microfluidic Single Cell Digital PCR. Anal Chem. 2013 doi: 10.1021/ac400896j. [DOI] [PubMed] [Google Scholar]
- 118.Yin H, Marshall D. Microfluidics for single cell analysis. Curr Opin Biotechnol. 2012;23(1):110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 119.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29(10):928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schepers K, Swart E, van Heijst JW, et al. Dissecting T cell lineage relationships by cellular barcoding. J Exp Med. 2008;205(10):2309–2318. doi: 10.1084/jem.20072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gerrits A, Dykstra B, Kalmykowa OJ, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115(13):2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 122.Grosselin J, Sii-Felice K, Payen E, Chretien S, Roux DT, Leboulch P. Arrayed lentiviral barcoding for quantification analysis of hematopoietic dynamics. Stem Cells. 2013 doi: 10.1002/stem.1383. [DOI] [PubMed] [Google Scholar]
- 123.Naik SH, Perie L, Swart E, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496(7444):229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 124.Verovskaya E, Broekhuis MJ, Zwart E, et al. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 2013 doi: 10.1182/blood-2013-01-481135. [DOI] [PubMed] [Google Scholar]
- 125.Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9(6):417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jefferis GS, Livet J. Sparse and combinatorial neuron labelling. Curr Opin Neurobiol. 2012;22(1):101–110. doi: 10.1016/j.conb.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 127.Zador AM, Dubnau J, Oyibo HK, Zhan H, Cao G, Peikon ID. Sequencing the connectome. PLoS Biol. 2012;10(10):e1001411. doi: 10.1371/journal.pbio.1001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weissman TA, Sanes JR, Lichtman JW, Livet J. Generating and imaging multicolor Brainbow mice. Cold Spring Harb Protoc. 2011;2011(7):763–769. doi: 10.1101/pdb.top114. [DOI] [PubMed] [Google Scholar]
- 129.Fu Y, Rusznak Z, Herculano-Houzel S, Watson C, Paxinos G. Cellular composition characterizing postnatal development and maturation of the mouse brain and spinal cord. Brain Struct Funct. 2012 doi: 10.1007/s00429-012-0462-x. [DOI] [PubMed] [Google Scholar]
- 130.Herculano-Houzel S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc Natl Acad Sci U S A. 2012;109(Suppl 1):10661–10668. doi: 10.1073/pnas.1201895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schuz A, Palm G. Density of neurons and synapses in the cerebral cortex of the mouse. J Comp Neurol. 1989;286(4):442–455. doi: 10.1002/cne.902860404. [DOI] [PubMed] [Google Scholar]
- 132.Mackay JP, Font J, Segal DJ. The prospects for designer single-stranded RNA-binding proteins. Nat Struct Mol Biol. 2011;18(3):256–261. doi: 10.1038/nsmb.2005. [DOI] [PubMed] [Google Scholar]
- 133.Pregibon DC, Toner M, Doyle PS. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 2007;315(5817):1393–1396. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 134.Banholzer MJ, Qin L, Millstone JE, Osberg KD, Mirkin CA. On-wire lithography: synthesis, encoding and biological applications. Nat Protoc. 2009;4(6):838–848. doi: 10.1038/nprot.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fernandez-Rosas E, Gomez R, Ibanez E, et al. Intracellular polysilicon barcodes for cell tracking. Small. 2009;5(21):2433–2439. doi: 10.1002/smll.200900733. [DOI] [PubMed] [Google Scholar]
- 136.Cai L. Turning single cells into microarrays by super-resolution barcoding. Brief Funct Genomics. 2013;12(2):75–80. doi: 10.1093/bfgp/els054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lin C, Jungmann R, Leifer AM, et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat Chem. 2012;4(10):832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mitra RD, Shendure J, Olejnik J, Edyta KO, Church GM. Fluorescent in situ sequencing on polymerase colonies. Anal Biochem. 2003;320(1):55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 139.Krishna K, Hertel N, Redies C. Cadherin expression in the somatosensory cortex: evidence for a combinatorial molecular code at the single-cell level. Neuroscience. 2011;175:37–48. doi: 10.1016/j.neuroscience.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 140.Mali P, Aach J, Lee JH, Levner D, Nip L, Church GM. Barcoding cells using cell-surface programmable DNA-binding domains. Nat Methods. 2013;10(5):403–406. doi: 10.1038/nmeth.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nolan G. Inner-outer beauty: DNA-binding surface tags as cellular barcodes. Nat Methods. 2013;10(5):399–401. doi: 10.1038/nmeth.2452. [DOI] [PubMed] [Google Scholar]
- 142.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143(7):1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sigrist SJ, Sabatini BL. Optical super-resolution microscopy in neurobiology. Curr Opin Neurobiol. 2012;22(1):86–93. doi: 10.1016/j.conb.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 144.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68(5):843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2(11):e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Knott G, Rosset S, Cantoni M. Focussed ion beam milling and scanning electron microscopy of brain tissue. J Vis Exp. 2011;(53):e2588. doi: 10.3791/2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lichtman JW, Denk W. The big and the small: challenges of imaging the brain's circuits. Science. 2011;334(6056):618–623. doi: 10.1126/science.1209168. [DOI] [PubMed] [Google Scholar]
- 148.Jain V, Seung HS, Turaga SC. Machines that learn to segment images: a crucial technology for connectomics. Curr Opin Neurobiol. 2010;20(5):653–666. doi: 10.1016/j.conb.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cardona A, Saalfeld S, Preibisch S, et al. An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol. 2010;8(10) doi: 10.1371/journal.pbio.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Helmstaedter M, Briggman KL, Denk W. High-accuracy neurite reconstruction for high- throughput neuroanatomy. Nat Neurosci. 2011;14(8):1081–1088. doi: 10.1038/nn.2868. [DOI] [PubMed] [Google Scholar]
- 151.Moore FE, Reyon D, Sander JD, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). PLoS One. 2012;7(5):e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hwang WY, Fu Y, Reyon D, et al. Heritable and Precise Zebrafish Genome Editing Using a CRISPR-Cas System. PLoS One. 2013;8(7):e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10(6):508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 155.Tucker CL. Manipulating cellular processes using optical control of protein-protein interactions. Prog Brain Res. 2012;196:95–117. doi: 10.1016/B978-0-444-59426-6.00006-9. [DOI] [PubMed] [Google Scholar]