Abstract
Precise control over activation of the apoptotic machinery is critical for development, tissue homeostasis and disease. In Drosophila, the decision to trigger apoptosis--whether in response to developmental cues or to DNA damage--converges on transcription of inhibitor of apoptosis protein (IAP) antagonists reaper, hid and grim. Here we describe a parallel process that regulates the sensitivity to, rather than the execution of, apoptosis. This process establishes developmental windows that are permissive or restrictive for triggering apoptosis, where the status of cells determines their capacity to die. We characterize one switch in the sensitivity to apoptotic triggers, from restrictive to permissive, that occurs during third-instar larval (L3) development. Early L3 animals are highly resistant to induction of apoptosis by expression of IAP-antagonists, DNA-damaging agents and even knockdown of the IAP diap1. This resistance to apoptosis, however, is lost in wandering L3 animals after acquiring a heightened sensitivity to apoptotic triggers. This switch in sensitivity to death activators is mediated by a change in mechanisms available for activating endogenous caspases, from an apoptosome-independent to an apoptosome-dependent pathway. This switch in apoptotic pathways is regulated in a cell-autonomous manner by the steroid hormone ecdysone, through changes in expression of critical pro-, but not anti-, apoptotic genes. This steroid-controlled switch defines a novel, physiologically-regulated, mechanism for controlling sensitivity to apoptosis and provides new insights into the control of apoptosis during development.
Keywords: apoptosis, apoptosome-dependent, apoptosome-independent, ecdysone, mid-L3 transition, sensitivity to apoptosis
INTRODUCTION
The ability to commit suicide by apoptosis is a fundamental, and irreversible, cellular process in multicellular organisms. Accordingly, normal development and homeostasis depend on precise control over when and where apoptosis occurs. Apoptosis is regulated by a signaling cascade of cysteine proteases called caspases (Fuentes-Prior and Salvesen, 2004). Initiator caspases sit at the top of the signaling cascade; they exist as monomers and are activated by dimerization on specialized signaling platforms (Bratton and Salvesen, 2010). One such signaling platform, the apoptosome, is composed of oligomers of the caspase adaptor Apaf-1 and the initiator caspase Caspase-9 (Bratton and Salvesen, 2010). Unlike initiator caspases, effector caspases exist as inactive dimers and are activated by caspase-dependent proteolytic cleavage (Riedl and Shi, 2004). Although caspases are expressed in all cells, their inappropriate activation is prevented by Inhibitor of Apoptosis Proteins (IAPs) (Orme and Meier, 2009; Salvesen and Duckett, 2002).
Activation of apoptosis during development is regulated by relieving the IAP-dependent inhibition of caspases. This mechanism is best elucidated in Drosophila where the activation of caspases converges on transcription of the IAP-antagonists reaper, hid and/or grim (Steller, 2008). These IAP-antagonists bind to Diap1 (Drosophila IAP 1), disrupting its interaction with caspases and initiating caspase activation and apoptosis (Ryoo et al., 2002; Yoo et al., 2002). Elimination of all three IAP-antagonists blocks apoptosis, while ectopic expression triggers apoptosis (Chen et al., 1996; Grether et al., 1995; White et al., 1994; 1996). Similarly, loss of diap1 has been shown to result in immediate activation of apoptosis in embryos (Goyal et al., 2000; Lisi et al., 2000; Wang et al., 1999). The mammalian death activator Smac/Diablo acts as an IAP-antagonist, inhibiting IAPs such as Survivin and XIAP, demonstrating that this pathway has been conserved through evolution (LaCasse et al., 2008).
Apoptosis is executed when the rapidly expanding cascade of caspase activation crosses a critical threshold; cells that do not cross this apoptotic threshold do not initiate apoptosis (Thompson, 1995). In turn, the ability to achieve this apoptotic threshold is predetermined by the endogenous expression levels of critical pro- and anti-apoptotic factors (Florentin and Arama, 2012; Lowe et al., 2004). This threshold model for activation of apoptosis was formulated primarily through characterization of oncogenic mutations and changes in gene expression observed in tumor cells, changes that disable the ability to trigger apoptosis in malignant cells (Adams and Cory, 2007; Lowe et al., 2004). It is now well established that the ability to evade apoptosis is a hallmark of cancer (Hanahan and Weinberg, 2011). However, physiological contexts for changes in the ability to trigger apoptosis and the mechanisms that regulate them remain poorly understood. Here, we characterize a dramatic and global switch in the sensitivity to apoptosis during Drosophila development at the onset of metamorphosis and demonstrate that this switch, mediated by changes in expression of critical pro-apoptotic genes, is regulated by the steroid hormone 20-hydroxyecdysone (hereafter referred to as ecdysone).
MATERIALS AND METHODS
Stocks
The following stocks were obtained from the Bloomington Drosophila Stock Center: hs-Gal4, UAS-rpr, en-Gal4,UAS-RFP, Sgs3-Gal4, Sgs3-GFP, nub-Gal4, UAS-GFP, UAS-EcRF645A and Nc51. The following stocks were kindly provided by colleagues in the fly community: hs-rpr (White et al., 1996), hs-hid (Grether et al., 1995), hs-diap1 RNAi (Yin and Thummel, 2004), Ark82 (Akdemir et al., 2006) and driceΔ1 (Muro et al., 2006).
Developmental staging
Early and wandering third instar larvae (eL3 and wL3, respectively) were identified by developmental age, wandering behavior and expression of a mid-L3-specific reporter (Sgs3-GFP)(Biyasheva et al., 2001). To collect eL3 animals, embryos were aged at 25°C about 76–88 hours after egg lay (AEL) and Sgs3-GFP-expressing larvae, if any, were removed. The wL3 animals were collected as rapidly wandering larvae from the sides of un-crowded bottles with robust expression of Sgs3-GFP (these animals are within 10 hours from puparium formation). Sugar feeding experiments were performed at 25°C on agar plates with 20% sucrose and without yeast.
Delivery of apoptotic activators and survival assays
To trigger apoptosis in a temporally-controlled manner, we used transgenic lines with death activators directly fused to the hsp70 heat-shock promoter (e.g., hs-rpr and hs-diap1-RNAi) or exposure to DNA-damaging UV-C light. Appropriately staged animals were heat-shocked for 30 minutes (unless otherwise noted) by submerging Parafilm-sealed grape agar plates in a water bath at the desired temperature; after heat-shock, animals were transferred to 25°C on grape agar plates with a thin layer of yeast. Exposure to UV-C light was done inside a Stratalinker 1800 (Stratagene) equipped with a UV-C bulb. Larvae were washed with distilled water, transferred as a single layer onto an empty petri dish and exposed to the appropriate dose of UV-C light. The larvae were then moved to grape agar plates with a thin layer of yeast and allowed to recover at 25°C. Death was assessed by touch response in larvae, heartbeat in prepupae, ability to head evert properly in pupae and by eclosion into adult flies.
Immunofluorescence
Tissues dissected from appropriately staged larvae were fixed and immunostained using standard methods (Yin et al., 2007). Staining for caspase activity was performed using an antibody raised against cleaved human Caspase-3 (rabbit α-cleaved caspase-3, Cell Signaling; used at 1:200 dilution), that detects activity of the Drosophila Caspase-9 homolog, Nc (Fan and Bergmann, 2010). Other primary antibodies used were mouse α-elav (1:50, Developmental Studies Hybridoma Bank), mouse α-Diap1 (1:200, gift from B. Hay), rabbit α-Nc and α-Drice (1:200, gift from P. Friesen). Secondary antibodies used were Cy3 α-rabbit (1:200, Jackson Immuno-Research Labs), AlexaFluor 488 α-mouse and α-rabbit (1:200, Invitrogen) and AlexaFluor 633 α-mouse and α-rabbit (1:200, Invitrogen). Images were taken on an Olympus FluoView FV1000 confocal microscope and optimized with the FV10-ASW software.
Quantitative RT-PCR
To measure mRNA expression levels of target genes, we used quantitative real time PCR (qPCR) with standard methods as previously described (Ihry et al., 2012). qPCR was performed using a Roche 480 LightCycler with the LightCycler 480 DNA SYBR Green I Master kit (Roche). Samples on the same graph were run simultaneously with three independent biological samples for each target gene (primer sequences shown in Table S2) and rp49 was used as the reference gene. Relative Expression Software Tool (REST) (Pfaffl et al., 2002) was used to calculate changes in relative expression.
Western Blotting
To measure protein expression levels, we performed western blots using standard methods as previously described (Ihry et al., 2012). For whole animal lysates, eL3 or wL3 animals were homogenized in 75 µl of hi-salt lysis buffer. Primary antibodies used were rabbit α-DIAP1 (1:1,000, gift from B. Hay), rabbit α-Drice (1:5,000, gift from P. Friesen), guinea pig α-Nc (1:1,000, gift from H. Steller), α-β-tubulin (1:1,000, Millipore) and α-β-actin (1:1,000, Cell Signaling). Secondary antibodies used were alkaline phosphatase conjugated goat α-rabbit IgG (1:30,000, GE Healthcare), α-guinea pig and α-mouse IgG (1:30,000, Sigma). Membranes were developed for imaging with ECF substrate (GE Healthcare) and were imaged using a Storm 840 Scanner (Amersham Bioscience) with ImageQuant TL software version 7.0 (GE Healthcare).
RESULTS AND DISCUSSION
Global changes in sensitivity to death activators define permissive and restrictive windows for apoptosis during development
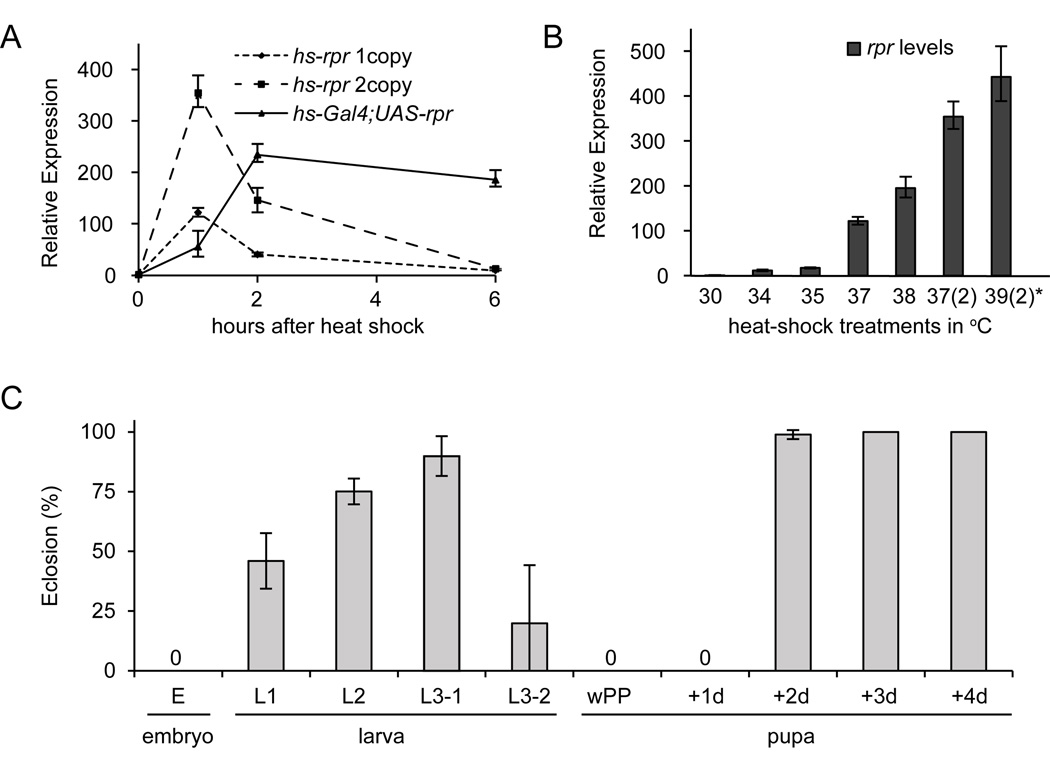
To measure changes in the sensitivity to apoptotic triggers during development, we calibrated a heat-controlled promoter expressing the IAP-antagonist reaper (hs-rpr)(White et al., 1996). This hs-rpr transgene delivered ubiquitous and highly-reproducible bursts of reaper expression that lasted about two hours (Fig. 1A). The dose of reaper was controlled by adjusting the temperature of the heat treatment and the number of transgenes used (Fig. 1B). Our standard protocol, unless otherwise stated, is a 30 minute heat-shock at 37°C with one copy of hs-rpr; this treatment delivers ~120-fold induction of reaper and represents ten times the minimal lethal dose (Table S1). By comparison, an endogenous death response in larval tissues during metamorphosis generates a ~50-fold induction of reaper (Ihry et al., 2012).
Fig. 1. Sensitivity to apoptosis is dynamically regulated during Drosophila development.
(A) Expression of reaper, measured by qPCR in eL3 animals, peaks an hour after heat-shock and is back to basal levels within 6 hours. With the same heat-shock treatment (at 37°C for 30 minutes), animals carrying two copies of the hs-rpr transgene reach a higher peak of maximal reaper expression but maintain a similar temporally-restricted expression profile. On the other hand, the use of the Gal4/UAS binary expression system (hs-Gal4;UAS-rpr) results in a perduring and thus disproportionately higher cumulative dose of reaper. (B) The scalable nature of heat-induced expression was exploited by varying the temperature of heat-shock and the number of copies of the hs-rpr transgene (indicated in parenthesis). Shown are peak expressions, one hour after heat-shock. All heat-shock treatments were 30 minutes long, except one (marked by an asterisk) which was 60 minutes long. (C) Percent of animals eclosing as adults after temporally-restricted expression of reaper at representative stages during the life cycle. Embryos (6–18 hours after egg lay or AEL), first-instar (L1: 28–40 AEL), second-instar (L2: 52–64 AEL), third instars (L3-1: 76–88 AEL and L3-2: 100–112 AEL), newly pupariated animals (white prepupa or wPP), and various stages during metamorphosis (1–4 days after wPP). Partial lethality in L1 and L3-2 reflects additional developmental heterogeneity within these stages. Each experiment was done with three replicates of at least 20 animals each.
The original description of the hs-rpr transgene reported that two developmental stages--the late embryo and the late pupae--were not sensitive to killing by reaper (White et al., 1996). We have extended that analysis to the entire fly life cycle and demonstrate that resistance to reaper is the norm, not the exception. Most stages during larval and pupal development appear to be highly resistant to reaper (Fig. 1C). In fact, sensitivity to killing by reaper appears to be primarily restricted to embryogenesis and to the larval-to-pupal transition when wandering larvae transform into immature pupae. These two developmental windows of sensitivity to reaper show a striking overlap with stages where most, if not all, programmed cell death occurs during the fly life cycle (Abrams et al., 1993; Wolff and Ready, 1991). Thus, the sensitivity to apoptosis appears to be regulated globally and may play an important role in restricting programmed cell death to discrete stages during development.
Animals within restrictive windows for apoptosis are 50 times more resistant to death activators and can resist knockdown of diap1
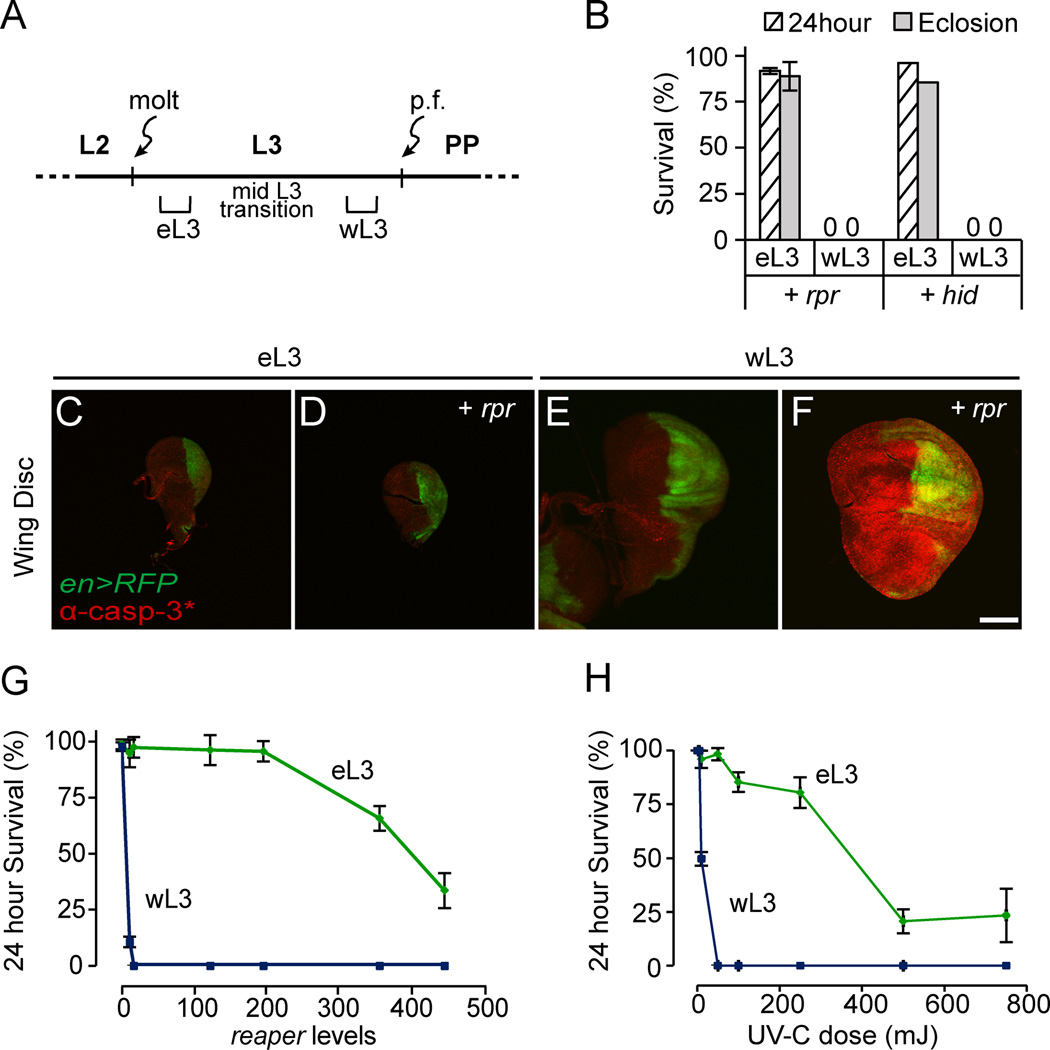
To understand the changes in sensitivity to apoptosis, we focused on the responses to reaper during L3 development. L3 development starts with the L2-L3 molt and ends about two days later with puparium formation and the onset of metamorphosis. The changes in sensitivity to apoptosis start half-way through L3 development and coincide with glue protein synthesis in salivary glands, a molecular marker of the mid-L3 transition (Andres and P. Cherbas, 1992; Andres et al., 1993). The mid-L3 transition, however, is strongly influenced by environmental conditions like crowding and nutrition, generating significant developmental asynchrony among individuals of the same age. To avoid this heterogeneity, we selected L3 animals well before and well after the mid-L3 transition: “early L3” (eL3) and “wandering L3” (wL3), respectively (Fig. 2A; see methods for details). These two populations of larvae have dramatically different sensitivities to reaper. Expression of reaper in wL3 kills animals within hours (Fig. S1A). In contrast, most reaper-treated eL3 animals, despite a comparable dose of reaper expression (Fig. S1B), eclose as adult flies (Fig. 2B). Heat-induced expression of hid gave similar results (Fig. 2B). The hs-hid transgene was not used extensively in this study because leaky expression of hid complicated phenotypic analysis (Fig. S1C). Consistent with the strong resistance to apoptosis in whole animals, expression of reaper does not initiate caspase activation in eL3 tissues. Staining wing imaginal discs with antibodies that detect caspase activity showed that reaper triggers caspase activation in wL3, but not in eL3, animals (Fig. 2C–F).
Fig. 2. Early third-instar larvae are highly resistant to expression of IAP-antagonists.
(A) Developmental staging of our “early” and “wandering” third-instar larvae (“eL3” and “wL3,” respectively). p.f.: puparium formation. PP: prepupae. (B) Most eL3 animals complete development after expression of reaper (hs-rpr/+) or hid (hs-hid/+). Each condition was tested in triplicate with at least 25 animals each. (C–F) Wing imaginal discs from control and hs-rpr-treated eL3 and wL3 animals dissected 30 minutes after heat-shock, stained with antibodies that detect caspase activity (in red; α-casp-3*: α-cleaved caspase-3); en-Gal4, UAS-RFP used as a morphological marker (RFP pseudo-colored green). Scale bars indicate 100 µM. (G–H) Dose response curves for 24 hour survival after expression of reaper (G) or exposure to DNA-damaging UV-C light (H) in eL3 (green) or wL3 (blue) animals.
To measure the difference in sensitivity to apoptotic activators, we tested increasing doses of death activators for their ability to kill eL3 animals. The resulting dose response curves demonstrate that while wL3 animals are very sensitive to reaper, eL3 animals require 50 times higher doses to trigger lethality (eL3 LD50=400 and wL3 LD50=7; Fig. 2G). Surprisingly, similar dose response curves were observed with UV-C light (Fig. 2H), a known DNA-damaging agent (Zhou and Steller, 2003), demonstrating that the switch in sensitivity to apoptosis is not limited to expression of IAP-antagonists.
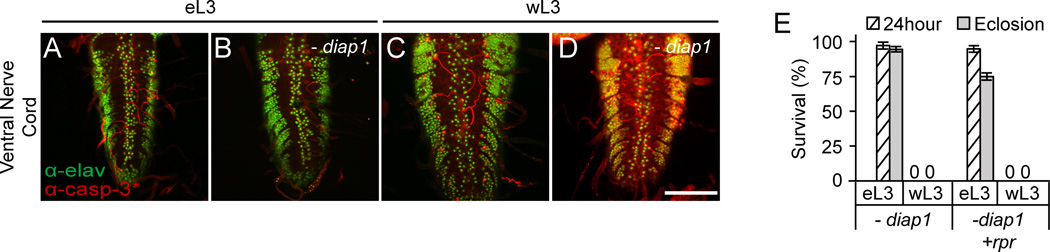
Given that diap1 is thought to be essential for preventing activation of apoptosis, we tested if eL3 animals could resist knockdown of diap1. Expression of diap1-RNAi triggered caspase activation in wL3 tissues, shown by staining for activated caspases in ventral nerve cords dissected from RNAi-treated wL3 animals (Fig. 3C,D). However, caspase activation was not detected in ventral nerve cords dissected from RNAi-treated eL3 animals (Fig. 3A,B). In fact, knockdown of diap1 in wL3 kills every animal within 24 hours while a similar treatment has little effect on eL3, with most animals eclosing as adults (Fig. 3E). Although the trace amounts of Diap1 protein remaining in RNAi-treated animals may be sufficient to block apoptosis in eL3 tissues (Fig. S1D–H), simultaneous expression of reaper does not dramatically alter the observed resistance (Fig. 3E). Thus, our data suggests that the resistance to apoptotic triggers in eL3 animals is mediated downstream of the diap1-dependent inhibition of apoptosis.
Fig. 3. Early third instar larvae are highly resistant to knockdown of diap1.
(A–D) Staining with antibodies that detect caspase activity (in red; α-casp-3*: α-cleaved caspase-3) showing resistance to apoptotic triggers in eL3 tissues. Ventral nerve cords from control and hs-diap1-RNAi-treated eL3 and wL3 animals dissected two hours after heat-shock (staining with antibodies directed against elav used as a morphological marker). Scale bars indicate 100 µM. (E) Most eL3 animals complete development after knockdown of diap1 (hs-diap1-RNAi), even with co-expresssion of reaper. Each condition was tested in triplicate with at least 25 animals each.
These results define a whole animal developmental context for understanding the mechanisms that control global changes in sensitivity to apoptosis. The unexpected resistance to a strong knockdown of Diap1 protein levels further demonstrate that expression of IAP-antagonists during development (i.e., “execution”) is not always sufficient to trigger apoptosis. Thus, our data suggests that apoptosis during development is regulated by two parallel processes whereby cells need to be “primed,” by increasing their sensitivity to apoptosis, before they can be “executed.”
The change in the sensitivity to death activators reflects a switch from apoptosome-independent to apoptosome-dependent execution of apoptosis
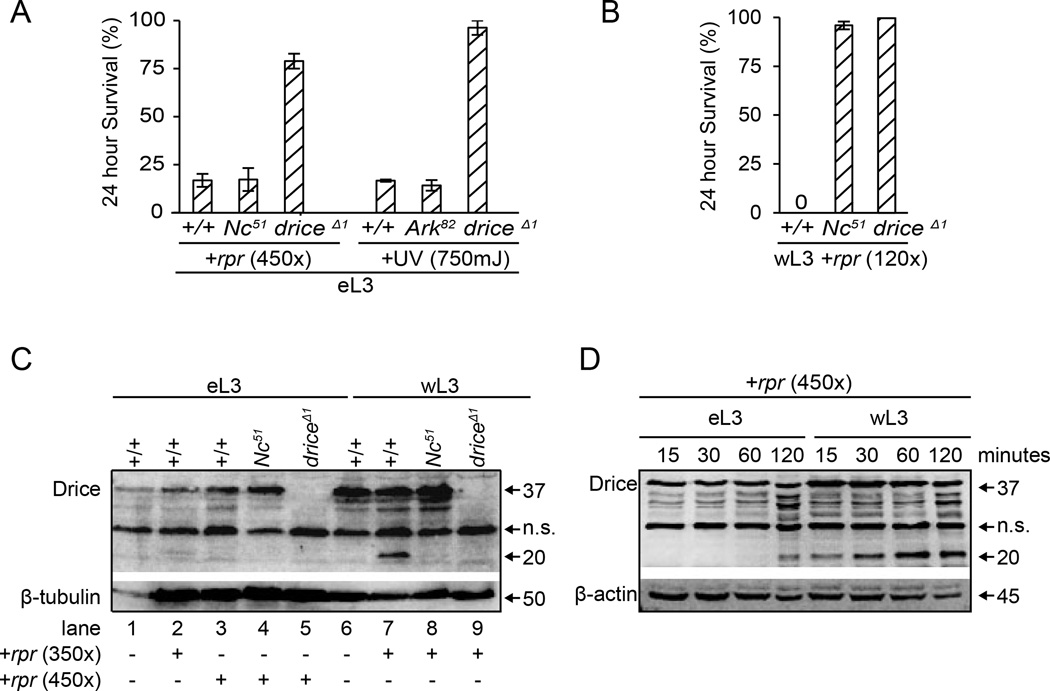
The striking difference in sensitivity to apoptosis between eL3 and wL3 animals prompted us to ask whether these two stages used different mechanisms to execute apoptosis. To our surprise, at the very high doses of reaper and UV-C light required to kill eL3 animals, the ensuing lethality occurs equally well in eL3 animals without functional Ark or Nc (orthologs of mammalian Apaf-1 and caspase-9, respectively; Fig. 4A). Importantly, this Ark- and Nc-independent death response requires the effector caspase drice (ortholog of mammalian caspase-3; Fig. 4A), suggesting that drice is activated in an apoptosome-independent manner in eL3 animals. In wL3 animals, however, the death response requires drice and apoptosome components like Nc (Fig. 4B), highlighting a switch in the apoptotic pathways that execute apoptosis at these two stages.
Fig. 4. eL3 and wL3 animals use different mechanisms for executing apoptosis.
(A) Lethality after extreme doses of apoptotic triggers in eL3 animals is independent of the apoptosome. Control, Ark82 and Nc51 mutant eL3 animals show similar rates of lethality; however, most driceΔ1 mutant animals survive the same treatment. (B) Lethality after expression of reaper in wL3 animals depends on the apoptosome. Most Nc51 and driceΔ1 mutant wL3 animals are still alive 24 hours after our standard dose of reaper. Each condition was tested in triplicate with at least 20 animals each. (C) Western blot with antibodies directed to the large subunit of Drice was used to detect full-length (p37) and cleaved (p20) protein products in whole animal extracts. Extracts from eL3 or wL3 animals one hour after 350- or 450-fold expression of reaper (summarized below the blot) show that Drice is cleaved after reaper expression in wL3 (cf. lanes 6 and 7) but not in eL3 animals (cf. lane 1 with 2 and 3). This activation of Drice requires Nc activity (lane 8). (D) A time course of Drice products after 450-fold expression of reaper in eL3 and wL3 animals. Drice is activated within 15 minutes of reaper expression in wL3 animals but requires 120 minutes in eL3 animals. Antibodies to β-tubulin and β-actin used as controls. n.s.= non-specific band, present in all lanes including those from drice mutant animals (lanes 5 and 9 in C).
To characterize the biochemical effect of this switch in apoptotic pathways, we examined the proteolytic activation of the Drice zymogen (p37). Cleavage of effector caspases like Drice define the point-of-no-return for apoptosis (Riedl and Salvesen, 2007). In wL3 animals, the large cleaved Drice product (p20) is observed within an hour after reaper treatment on western blots and this processing is Nc-dependent (lanes 6–8, Fig. 4C). In contrast, only negligible amounts of the activated Drice product is observed under the same conditions in eL3 animals (lanes 2–3; Fig. 4C). This difference in the processing of Drice results from a change in the kinetics of proteolytic activation. In response to the same extreme apoptotic stimuli, wL3 animals activate Drice within 15 minutes whereas it takes 120 minutes to process Drice in eL3 animals (Fig. 4D). In addition to approximately an order of magnitude increase in the rate of Drice cleavage, the fraction of pro-Drice that is proteolytically activated is also considerably higher in wL3 animals (Fig. S2). Thus, the switch to an apoptosome-dependent pathway in wL3 animals confers a faster (increased rate) and higher capacity (increased efficacy) activation of Drice. In turn, these changes in kinetics of caspase activation translate into dramatic differences in the sensitivity to apoptotic triggers.
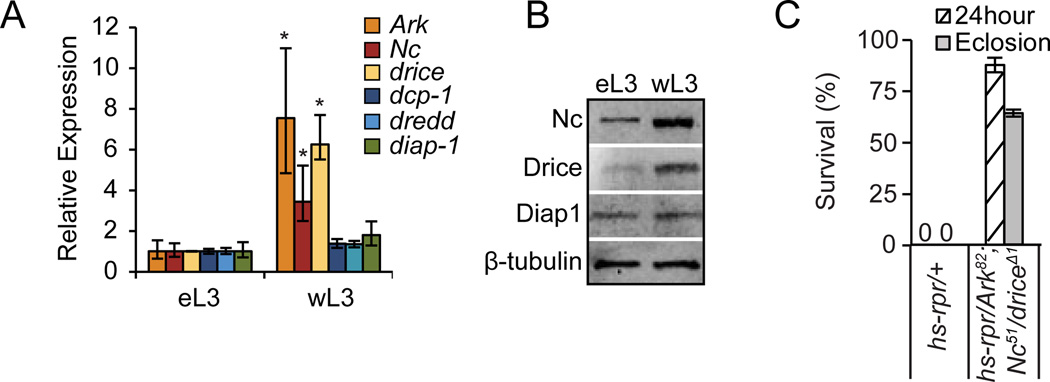
To understand how the switch in the ability to execute apoptosis through the apoptosome is regulated, we measured the expression levels of key pro- and anti-apoptotic genes at these stages. Our results indicate that endogenous mRNA levels, as measured by qPCR, of at least three critical regulators of apoptosis--the apoptosome components Ark and Nc and the effector caspase drice--are several fold higher in wL3 animals (Fig. 5A). On the other hand, endogenous mRNA levels of diap1 and the caspases dredd and dcp-1 do not change (Fig. 5A). Consistently, western blots of total protein extracted from eL3 and wL3 animals showed higher levels of Nc and Drice proteins in wL3, while Diap1 protein levels appear not to change (Fig. 5B). To demonstrate that changes in expression levels of three pro-apoptotic genes are sufficient to explain the dramatic switch in sensitivity to apoptosis, we examined the response to reaper in wL3 animals heterozygous for null mutations in Ark, Nc and drice. Animals carrying a single allele of any one of these mutations did not survive expression of reaper. Triple heterozygous wL3 animals, however, showed a striking resistance to reaper: 90% of reaper-treated animals survived 24 hours and nearly 65% eclosed as adults (Fig. 5C). Thus, the changes in endogenous levels of these three pro-apoptotic genes are sufficient to explain the switch in sensitivity to apoptosis.
Fig. 5. Changes in expression levels of pro-apoptotic genes is sufficient to predetermine apoptotic outcome.
(A) qPCR expression analysis shows that mRNA levels of Ark, Nc, and drice (but not dcp-1, dredd and diap1) significantly increase in wL3 animals. qPCR results reflect triplicate biological samples; asterisk indicates p-values <0.01. (B) Western blots show changes in protein expression of Nc and Drice; levels of Diap1 protein, however, are unchanged. β-tubulin was used as loading control. Total mRNA or protein was extracted from appropriately staged eL3 and wL3 whole animals. (C) Effects of reaper expression on animals heterozygous for mutations in Ark82, Nc51 and driceΔ1. All control animals die within hours after hs-rpr (120×) treatment while most triple heterozygous animals survive. Each condition tested in triplicate with at least 20 animals each.
Taken together, our results demonstrate that stage-specific, coordinately-regulated changes in gene expression of key pro-apoptotic genes, determine the ability to execute apoptosis through the apoptosome, thereby controlling the rate and efficacy of caspase activation and, as a result, establish windows during development that are either permissive or restrictive for apoptosis.
Single-cell quantitative studies of apoptosis have suggested that relatively minor changes in the endogenous levels of apoptotic regulators, even among cells within a clonal population, can change the outcome to uniform apoptotic stimuli (Spencer et al., 2009). Moreover, reduced expression of individual pro-apoptotic genes like Apaf-1 has been described in many cancers (Dai et al., 2004; Fu et al., 2003; Soengas et al., 2001; Zlobec et al., 2007); although a causal relationship between changes in expression of pro-apoptotic genes and reduced sensitivity to apoptosis are difficult to establish from these studies because tumor cells often carry many mutations. Our results, however, demonstrate that a two-fold reduction in levels of three pro-apoptotic genes can effectively neutralize apoptotic outcomes in response to death activators. Our results also place more importance on the canonical apoptosome-dependent pathway for execution of cell death during development. Consistent with these results, most, but not all, developmentally-triggered apoptosis are disrupted in Ark and Nc mutant embryos (Mills et al., 2006; Xu et al., 2005). The mechanisms that regulate the apoptosome-independent activation of apoptosis, however, remain poorly characterized.
The switch in sensitivity to apoptosis is regulated by ecdysone in a cell-autonomous manner
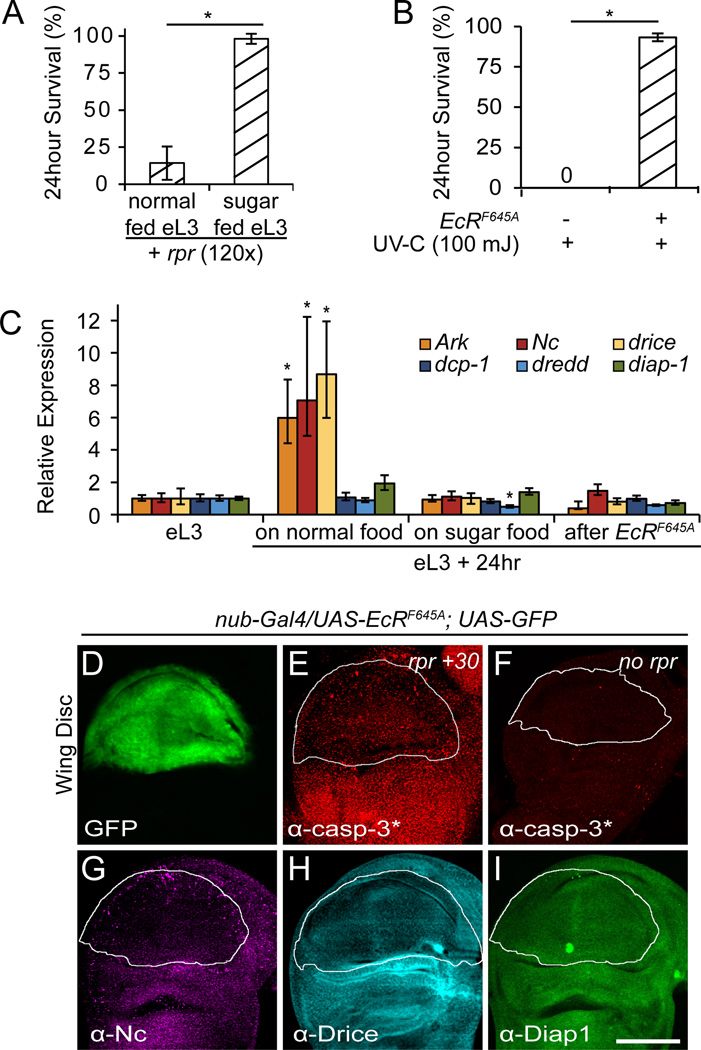
To identify regulators of the switch in sensitivity to apoptosis, we examined the role of ecdysone and the mid-L3 transition. The mid-L3 transition can be disrupted by transferring eL3 animals to sugar-only food (Britton and Edgar, 1998), arresting the progression of L3 development until a complete food source is re-supplied. These sugar-fed eL3-arrested animals remained resistant to apoptosis while their normal-fed counterparts switched their sensitivity to apoptosis (Fig. 6A). Accordingly, expression levels of core death genes increased in normal-fed but not in sugar-fed eL3 animals (Fig. 6C). These results demonstrate that the switch in sensitivity to apoptosis during L3 development occurs during the mid-L3 transition. In turn, the mid-L3 transition coincides with a low titer systemic pulse of ecdysone (Warren et al., 2006). Although the role of ecdysone in the mid-L3 transition is not well understood, ecdysone is known to regulate expression of Nc and drice at other stages during development (Dorstyn et al., 1999; Kilpatrick et al., 2005). To examine the role of ecdysone in the switch in sensitivity to apoptosis during the mid-L3 transition, we used a dominant negative form of the ecdysone receptor EcR (EcRF645A)(L. Cherbas et al., 2003). eL3 animals treated with ubiquitous expression of EcRF645A and aged for 24 hours in normal food remained highly resistant to apoptotic triggers (Fig. 6B). Moreover, this continued resistance to apoptosis was accompanied by a failure to increase expression of core death genes Ark, Nc and drice (Fig. 6C).
Fig. 6. The switch in sensitivity to apoptosis during L3 development is regulated by ecdysone in a cell-autonomous manner during the mid-L3 transition.
(A) Preventing the mid-L3 transition by transferring eL3 animals to sugar food for 24 hours, blocks the switch in sensitivity to apoptosis. Sensitivity to apoptosis measured by 24 hour survival after exposure to ubiquitous reaper. (B) Ubiquitous expression of a dominant negative ecdysone receptor (EcRF645A) in eL3 animals also blocks the switch in sensitivity to apoptosis. Tested by measuring 24 hour survival after exposure to UV-C light. Survival examined with three replicates of at least 20 animals for each condition. (C) Sugar-fed and EcRF645A-treated eL3 animals also block the increase in expression of Ark, Nc and drice observed in similarly aged control siblings. All qPCR results are in triplicate; asterisks indicate p-values <0.01. (D–I) Wing imaginal discs, dissected from wL3 animals, expressing the dominant negative ecdysone receptor in the pouch region using nub-Gal4 (nub-Gal4, UAS-EcRF645A, UAS-GFP). (D) Expression of GFP used to outline the pouch region. (E–F) Wing discs before and 30 minutes after ubiquitous expression of reaper (from hs-rpr), stained with antibodies that detect caspase activity (in red; α-casp-3*: α-cleaved caspase-3). Expression of reaper does not induce caspase activation in EcRF645A-expressing cells. Staining with antibodies directed to Nc (E), Drice (F) and Diap1 (G) proteins showing that EcRF645A-expressing cells have lower levels of Nc and Drice but not Diap1.
Strikingly, the effects of ecdysone on the switch in sensitivity to apoptosis were cell-autonomous and therefore independent of the global mid-L3 transition. Expression of EcRF645A in the pouch region of the wing imaginal disc with the nub-Gal4 driver, allowed animals to proceed through the mid-L3 transition. Importantly, however, EcRF645A-expressing cells in the wing pouch did not activate caspases in response to ubiquitous reaper, while the region around the wing pouch did (Fig. 6D–F). Moreover, the resistance to caspase activation was accompanied by a reduction in levels of Nc and Drice but not of Diap1 proteins, as observed by staining with antibodies directed to these core death regulators (Fig. 6G–I). The cell-autonomous changes in expression of pro-apoptotic genes, parallel the changes observed in whole animals and provide a unique molecular signature to the ecdysone-regulated switch in sensitivity to apoptosis during L3 development.
In Drosophila, genetic manipulations of the hippo and Jak/Stat signaling pathways can alter the sensitivity to apoptosis through changes in the expression levels of critical apoptotic regulators. In fact, both of these pathways regulate transcription of diap1 (Betz et al., 2008; Halder and Johnson, 2011; Staley and Irvine, 2012). The IKK-related kinase also regulates the sensitivity to apoptosis through its effects on diap1, controlling stability of Diap1 protein (Kuranaga et al., 2006). In contrast to these pathways, the ecdysone-controlled switch in sensitivity to apoptosis described here does not involve changes in levels of diap1 transcripts or proteins. Thus, this ecdysone-regulated process represents a novel mechanism for regulating sensitivity to apoptosis during normal development. The effects of ecdysone are likely mediated through binding of the ecdysone nuclear receptor complex and recruitment of a unique subset of coregulators and chromatin-modifying complexes to the promoters of critical apoptotic genes. Although nuclear receptor coregulators remain poorly characterized in Drosophila, inappropriate function or expression of coregulators are implicated in a large number of human pathologies including endocrine-based diseases like breast, prostate and ovarian cancers (Lonard and O'Malley, 2012). Thus, our results may provide new insights into the role of steroid hormones in human disease.
Conclusions
Programmed cell death was originally described in the context of the hormonal regulation of insect and frog metamorphosis (Lockshin and Williams, 1964; Tata, 1966). Since then, the role steroid hormones have been shown to control apoptosis in many tissues during mammalian development (Evans-Storms and Cidlowski, 1995; Kiess and Gallaher, 1998). In Drosophila, the steroid hormone ecdysone controls the execution of apoptosis during metamorphosis by inducing expression of IAP-antagonists (Jiang et al., 2000; Yin and Thummel, 2005). The results described here define a parallel ecdysone-dependent process that regulates the sensitivity to, rather than the execution of, apoptosis. This independently controlled sensitivity to apoptosis generates permissive and restrictive windows for apoptosis during development. Control of sensitivity to apoptosis may be used to protect essential cells or to allow IAP-antagonist-dependent initiation of non-apoptotic functions of caspases. These latter non-apoptotic roles include a growing list of functions like proliferation, differentiation, dendritic pruning and migration (Kuranaga and Miura, 2007; Lamkanfi et al., 2007; Rudrapatna et al., 2013; Yi and Yuan, 2009). Many of these non-apoptotic roles of caspases may be normally restricted to developmental stages with reduced sensitivity to apoptosis. Finally, our results highlight the importance of considering global changes in sensitivity to apoptosis when studying the regulation of cell death during development.
Supplementary Material
Highlights.
-
*
we define a process that controls sensitivity to, rather than execution of, apoptosis
-
*
this generates permissive and restrictive windows for apoptosis during development
-
*
during restrictive windows, cells fail to execute apoptosis through the apoptosome
-
*
available pathways for apoptosis determined by levels of key pro-apoptotic genes
-
*
this process is controlled, cell-autonomously, by the steroid hormone ecdysone
Acknowledgements
We are grateful to the Bloomington Stock Center (Bloomington, IN, USA) and colleagues in the community for Drosophila stocks; DSHB (Iowa City, IA, USA), P. Friesen (Madison, WI, USA), B. Hay (Pasadena, CA, USA) and H. Steller (New York, NY, USA) for antibodies; Gina Castelvecchi for technical assistance; Kate O’Connor-Giles for critical reading of the manuscript. This work was supported by an NIGMS/NIH grant (R01GM095944) to A.B..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved'ová L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Cherbas P. Tissue-specific ecdysone responses: regulation of the Drosophila genes Eip28/29 and Eip40 during larval development. Development. 1992;116:865–876. doi: 10.1242/dev.116.4.865. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev Biol. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- Betz A, Ryoo HD, Steller H, Darnell JE. STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc Natl Acad Sci USA. 2008;105:13805–13810. doi: 10.1073/pnas.0806291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyasheva A, Do TV, Lu Y, Vaskova M, Andres AJ. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev Biol. 2001;231:234–251. doi: 10.1006/dbio.2000.0126. [DOI] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. Journal of Cell Science. 2010;123:3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- Dai DL, Martinka M, Bush JA, Li G. Reduced Apaf-1 expression in human cutaneous melanomas. Br J Cancer. 2004;91:1089–1095. doi: 10.1038/sj.bjc.6602092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc Natl Acad Sci USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Storms RB, Cidlowski JA. Regulation of apoptosis by steroid hormones. The Journal of Steroid Biochemistry and Molecular Biology. 1995;53:1–8. doi: 10.1016/0960-0760(95)00034-w. [DOI] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentin A, Arama E. Caspase levels and execution efficiencies determine the apoptotic potential of the cell. J Cell Biol. 2012;196:513–527. doi: 10.1083/jcb.201107133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W-N, Bertoni F, Kelsey SM, Mcelwaine SM, Cotter FE, Newland AC, Jia L. Role of DNA methylation in the suppression of Apaf-1 protein in human leukaemia. Oncogene. 2003;22:451–455. doi: 10.1038/sj.onc.1206147. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L, Mccall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Ihry RJ, Sapiro AL, Bashirullah A. Translational Control by the DEAD Box RNA Helicase belle Regulates Ecdysone-Triggered Transcriptional Cascades. PLoS Genet. 2012;8:e1003085. doi: 10.1371/journal.pgen.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Molecular Cell. 2000;5:445–455. doi: 10.1016/s1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- Kiess W, Gallaher B. Hormonal control of programmed cell death/apoptosis. Eur. J. Endocrinol. 1998;138:482–491. doi: 10.1530/eje.0.1380482. [DOI] [PubMed] [Google Scholar]
- Kilpatrick ZE, Cakouros D, Kumar S. Ecdysone-mediated up-regulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J Biol Chem. 2005;280:11981–11986. doi: 10.1074/jbc.M413971200. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, Miura M. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 2006;126:583–596. doi: 10.1016/j.cell.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Miura M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends in Cell Biology. 2007;17:135–144. doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin RA, Williams CM. Programmed cell death—II. Endocrine potentiation of the breakdown of the intersegmental muscles of silkmoths. Journal of Insect Physiology. 1964;10:643–649. [Google Scholar]
- Lonard DM, O'Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Mills K, Daish T, Kieran Harvey, Pfleger C, Hariharan I, Kumar S. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J Cell Biol. 2006;172:809. doi: 10.1083/jcb.200512126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, Guo M, Baehrecke EH, Hay BA. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development. 2006;133:3305–3315. doi: 10.1242/dev.02495. [DOI] [PubMed] [Google Scholar]
- Orme M, Meier P. Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis. 2009;14:950–960. doi: 10.1007/s10495-009-0358-2. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rudrapatna VA, Bangi E, Cagan RL. Caspase signalling in the absence of apoptosis drives Jnk-dependent invasion. EMBO Rep. 2013;14:172–177. doi: 10.1038/embor.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. Apoptosis: IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, Cordón-Cardó C, Lowe SW. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Dev Dyn. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- Tata JR. Requirement for RNA and protein synthesis for induced regression of the tadpole tail in organ culture. Dev Biol. 1966;13:77–94. doi: 10.1016/0012-1606(66)90050-9. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Müller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Warren JT, Yerushalmi Y, Shimell MJ, O'connor MB, Restifo LL, Gilbert LI. Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Dev Dyn. 2006;235:315–326. doi: 10.1002/dvdy.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16:21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thummel CS. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc Natl Acad Sci USA. 2004;101:8022–8027. doi: 10.1073/pnas.0402647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thummel CS. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin Cell Dev Biol. 2005;16:237–243. doi: 10.1016/j.semcdb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thummel CS, Bashirullah A. Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J Cell Biol. 2007;178:85–92. doi: 10.1083/jcb.200703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RMR, Clem RJ, Müller H-AJ, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zhou L, Steller H. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev Cell. 2003;4:599–605. doi: 10.1016/s1534-5807(03)00085-6. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Minoo P, Baker K, Haegert D, Khetani K, Tornillo L, Terracciano L, Jass JR, Lugli A. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur. J. Cancer. 2007;43:1101–1107. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.