Abstract
Exposure to cumulative SES risk in childhood may interact with variability at the serotonin transporter linked polymorphic region (5HTTLPR) to alter DNA methylation across interacting sets of proteins. DNA was obtained from 388 African Americans at age 19. Genotype at the 5HTTLPR was determined, and methylation ratios for C-phosphate-G (CpG) residues were assessed. Exposure to cumulative SES risk was determined using repeated parental reports at ages 11–13. At high SES risk, CpG methylation patterns indicated altered cellular stress response in women, but not men, who carried a short allele at the 5HTTLPR. These changes in methylation patterns may lead to increases in mental and physical health risks. No genotype effect emerged for either women or men at low SES risk. Methylation patterns provide guidance in identifying pathways by which genetic susceptibility is transformed into adverse outcomes years later.
Keywords: 5HTTLPR, Gene × environment interaction, Health, Resources, Susceptibility
Cumulative socio-economic status (SES) risk in childhood exerts effects on a range of behavioral and health consequences through mechanisms that are currently poorly understood. Effects on those growing up in low SES conditions include having a shorter life expectancy (Braveman, Cubbin, Egerter, Williams, & Pamuk, 2010) and an increased likelihood of a range of interrelated negative health outcomes across the lifespan (Starfield, Robertson, & Riley, 2002). One potential mechanism that may link cumulative SES risk in childhood with later outcomes is epigenetic change, a process that may be initiated by effects of chronic stress on cellular level processes. In particular, chronic stressors during key developmental periods, such as exposure to high cumulative SES related risk, appears to have the potential to stimulate sustained Hypothalamic-Pituitary-Adrenal (HPA) axis activation and thereby to alter systemic functioning, in some cases producing long-lasting change (McQuillan, Pan, & Porter, 2006). The accumulation of such long-term health risks as a function of repeated stress over time is often referred to as the development of allostatic load (McEwen & Gianaros, 2010). Because these changes have the potential to alter cellular environments in both the brain and the body, they may set the stage for epigenetic change in response to changed intracellular environments across many systems, and so precipitate changes in DNA methylation in the central nervous system and the periphery.
African American youth living in the southeastern coastal plain are exposed to substantial SES related risk at developmentally sensitive stages. Indeed, in large areas of the southeast, a sizable percentage of the population lives at or below the official poverty level; the families participating in the current study resided in nine rural counties in which poverty rates are among the highest in the nation and unemployment rates are above the national average (Proctor & Dalaker, 2003). Although all youth exposed to these circumstances may be adversely affected to some degree, there is considerable basic research indicating that sequence variation at the 5HTTLPR, which is a functional variable nucleotide repeat found ~1400 bp upstream of the transcription start site of the serotonin transporter gene (SLC6A4), has potential to moderate environmental stress (e.g., Caspi, Hariri, Holmes, Uher, & Moffitt, 2010). Numerous studies have documented the functionality of this polymorphism, with estimates indicating that cells bearing two copies of the short or “s” allele (14 copies of a 22 bp repeat element) have an approximate 40% reduction in the amount of SLC6A4 mRNA as compared to those having two copies of the long or “l” allele (16 copies of a 22 bp repeat element) (Philibert et al., 2008).
In several high profile publications, investigators have shown that exposure to environmental stressors among carriers of at least one copy of the “s” allele confers increased risk for the subsequent development of adult depression as compared to those with two copies of the “l” allele (Caspi et al., 2003; Hammen, Brennan, Keenan-Miller, Hazel, & Najman, 2010). If true, the exact mechanism of increased vulnerability to later life depression and other adverse outcomes remains unexplained. One possible mechanism leading to differential effect of environmental stress on delayed outcomes is differential methylation of the promoter region of SLC6A4, an effect which has previously been shown to influence expression of transcripts of SLC6A4 (Brenet et al., 2011; Vijayendran, Beach, Plume, Brody, & Philibert, 2012). A broader effect is also possible, with a recent investigation demonstrating that variation in the promoter of the serotonin transporter gene (SLC6A4) influenced the extent to which cumulative SES risk was associated with level of methylation across a number of depression associated genes (Beach et al., in press). This study compared genetic vulnerability and genetic susceptibility models. A vulnerability model posits that carriers of the “risk” allele are less resilient to high levels of stress, placing them at increased risk of adverse outcomes in the context of elevated early cumulative SES stress. A susceptibility model also predicts differences in the presence of elevated early cumulative SES stress, but also predicts differences in the opposite direction in response to positive early environments, leading to a “for better or worse” pattern of outcomes (Belsky & Pluess, 2009). Beach et al. (in press) found that across a number of aggregated indices of methylation there were stronger correlations between level of methylation and cumulative stress for “s” allele carriers than for those not carrying an “s” allele, and that regression lines showed the cross-over pattern indicative of 5HTT operating as a “susceptibility” gene rather than as a “vulnerability” gene (See Belsky and Pluess, 2009 for an in depth discussion of genetic susceptibility vs. vulnerability effects). Greater correlations between environmental stress and level of methylation were found for s-allele carriers relative to non-carriers at 93.2% of differentially methylated CpG sites. Consistent with a susceptibility perspective, differences in methylation at both higher and lower levels of cumulative stress were attributable to genotype, albeit in opposite directions under high vs. low risk conditions.
Unfortunately, this prior research was focused on broad patterns of change only and so leaves unaddressed the characterization of potential functional effects of methylation changes as well as characterization of the functional gene networks that might be implicated by a differential epigenetic response to stress as a function of genotype. For this reason, prior work identifying change in level of methylation at particular CpG sites is unsatisfying for the purpose of understanding likely impact and functional significance of these changes at the cellular level. Although methylation of specific CpG sites may have implications for the production of particular proteins by physically impeding the binding of transcriptional proteins to the gene, or by facilitating the binding of DNA by Methyl-CpG-binding domain proteins (MBDs), the link to actual gene transcription is often modest (Plume, Beach, Brody, & Philibert, 2012). More importantly, because proteins do not act alone, but rather work together to promote biological activities at the cellular level, it is necessary to place findings regarding methylation change in the context of broader maps of protein–protein interactions (Cusick, Klitgord, Vidal, & Hill, 2005) in order to identify the type of potentially coordinated changes that might signal a cellular level stress response and provide the basis for potential long-term change in cellular functioning.
Emerging work on protein–protein interactions (e.g., iRefIndex data base, Razick, Magklaras, & Donaldson, 2008) identifies possible ways proteins interact or make physical contact with each other in various cells of the human body. This places important constraints on plausible effects of shifts in epigenetic regulation of gene activity. By combining currently available information about protein–protein interactions with information about changes in level and patterning of methylation, it is possible to further explore the likely functional significance of methylation pattern associated with gene × environment interaction effects, and to better characterize the likely impact on functional networks of genes. Specifically, utilizing protein–protein interaction information allows us to ask how particular observed changes in methylation are likely to influence cellular level biological interactions, constraining attention to those combinations of proteins with physiologically relevant protein–protein interactions. In turn, this refined characterization of effects allows exploration of specific hypotheses regarding impact of carrying the s-allele on stress response at the cellular level. This approach therefore compliments information available from popular algorithms such as “GoMiner” that do not take advantage of experimentally derived systematic information on the functional connectivity of the proteome. The importance of the approach and its validity for incorporating a systems perspective into examination of disease processes has recently been presented in a very accessible manner by Jordan, Nguyen, and Lui (2012). As they note, the investigation of changes in protein–protein interaction networks can provide novel inferences regarding mechanisms of disease progression. A more technical presentation, also demonstrating the potential utility and validity of approach for identifying interacting gene networks can be found in Li, Huang, Liu, Cai, & Chou (2012). As they note, analysis of protein–protein interaction networks can extend the identification of functional gene networks beyond what is possible using genomewide profiling alone. In the current analysis, we focus on changes in methylation of CpG sites in promoter regions because, given their stronger association with transcriptional control (Suzuki & Bird, 2008), they are particularly appropriate in the context of studies of protein–protein interactions.
1. Specific hypotheses
In the current investigation, we examine changes in methylation patterns in promoter regions genome-wide that are associated with the experience of cumulative stress in a population best described as “rural, African American, working poor” to see if there is evidence of an effect of cumulative exposure to SES risk in childhood on shifts in epigenetic patterns in gene promoters and whether this effect is moderated by genotype at SLC6A4. We also examine the potential effect on broader coordinated sets of changes that might suggest a functional epigenetic signature of cellular level stress. We hypothesize that such effects are most likely to emerge among those who are genetically susceptible to environmental stressors, i.e., carriers of the “s” allele of SLC6A4.
2. Method
2.1. Participants
Participants were primary caregivers and a 19-year-old target youth selected from each of 388 African American families who participated in annual data collections. Participants were selected randomly from a larger sample (see Brody, Yu, Chen, Miller, Kogan, and Beach, 2013, for a full description) that had been followed since the youth were in the 5th grade, providing prospective reports of socio-economic conditions. Each caregiver assessed the family’s economic circumstances. Target youth provided blood for genetic and epigenetic assessments at age 19. All youth who were successfully genotyped were included in the study (n = 388; 213 females and 175 males). The target youth mean age was 11.2 years at the first assessment of socio-economic circumstances and 19.3 years at the epigenetic assessment. Of the youth in the sample, 55% were female.
All methods were approved by the University of Georgia Institutional Review Board. Data for waves 1–3 were collected from August 2001 to July 2004; blood draws and young adult depression data were collected from June 2009 to August 2011.
2.2. Procedure
At each wave of data collection, all data were collected in participants’ residences using a standardized protocol. Two African American field researchers worked separately with the primary caregiver and the target youth. Interviews were conducted privately, with no other family members present or able to overhear the conversation. Primary caregivers consented to their own and to their minor youths’ participation in the study, and minor youth assented to their own participation. At the age 19 assessment, youth consented to their own participation.
2.3. Measures
2.3.1. Preadolescent cumulative SES-related risk
Three waves of preadolescent data were collected when the target youths were 11, 12, and 13 years of age, and these assessments were used to establish levels of cumulative SES-related risk in childhood using primary caregiver report. Six standard indicators of risk were assessed, with each risk factor scored dichotomously (0 = absent, 1 = present; see, Evans, 2003; Kim & Brody, 2005). Cumulative risk was defined as the average of the risk factors across the three assessments, yielding an index with a theoretical range of 0–6 (M = 2.33, SD = 1.35). The six risk indicators were: (a) family poverty (see http://www.census.gov/hhes/www/poverty/data/threshld/) using a threshold that incorporates both family income and household size, (b) primary caregiver non-completion of high school or an equivalent, (c) primary caregiver unemployment, (d) single-parent family structure, (e) family receipt of Temporary Assistance for Needy Families, and (f) income rated by the primary caregiver as not adequate to meet all needs. For the current study, families were characterized as “at risk”(i.e., High SES risk) or “not at risk” (i.e., Low SES risk) based on a median split on the SES index. All indicators were successful in separating high and low risk subsamples. Because of the high level of poverty in the sample, the median provided a convenient point at which to break the sample and corresponded roughly to those living below vs. above the poverty level. Family poverty was the strongest single predictor of subgroup membership (t = 16.836, p < .001), but all indicators differentiated the subgroups at p < .001. Correlation among SES risk indicators in the full sample were all positive and significant, ranging from a low of r = .126, p = .013, between completion of high school and reported adequacy of family income to a high of r = 455, p < .001, between family poverty and single parent household. Supplemental Table A provides descriptive statistics for indicator values for the full sample and subsamples and provides subgroup t-tests for each indicator and the total score.
2.3.2. Genotyping
Genotype at the 5-HTTLPR was determined for each sample as described previously (Bradley, Dodelzon, Sandhu, & Philibert, 2005) using the primers F-GGCGTTGCCGCTCTGAATGC and R-GAGGGACTGAGCTGGACAACCAC, standard Taq polymerase and buffer, standard dNTPs with the addition of 100 μM 7-deaza GTP, and 5% DMSO. The resulting polymerase chain reaction products were electrophoresed on a 6% nondenaturing polyacrylamide gel, and products were visualized using silver staining. Two individuals blind to the study hypotheses and other information about the participants called the genotypes. Genotypes were in Hardy-Weinberg equilibrium, p = .6151, with 21 participants homozygous for the “s” allele, 131 heterozygous, and 236 with no “s” alleles.
2.3.3. Characterization of methylation
After bisulfite conversion, methylation status for each sample was assessed using the Illumina (San Diego, CA) HumanMethylation450 Beadchip at the University of Minnesota Genome Center (Minneapolis, MN) following the protocol specified by the manufacturer. The Illumina array has been validated against other measurement strategies (Das, Tan, & Easteal, 2011; Vijayendran et al., 2012, June 13) and has shown good convergence with independent assays. The chip contains 485,577 probes. Male and female subjects were randomly assigned to 12 sample “slides.” Eight replicates of the same DNA were also included to monitor slide to slide and batch-related bisulfite conversion variability. Data were inspected for outliers or confounds by plate and chip variables. The resulting microarray data were inspected for complete bisulfite conversion of the DNA. The average beta value (i.e. average methylation) for each CpG residue was determined using the GenomeStudio V2009.2; Methylation module Version 1.5.5, Version 3.2 (Illumina, San Diego). Beta values for each probe were calculated as the ratio of methylated probes to the sum of methylated and unmethylated probes, ranging from 0 = entirely unmethylated to 1 = fully methylated. With respect to this sample, >99.76% of the 485,577 probes yielded statistically reliable data. Any probe with detection >.05 was removed to insure that only high confidence probes were included. Beta values were exported into Microsoft Excel, and initial analyses were conducted in R.
Because the analyses focused on promoter methylation, CpG sites that map to promoter regions were extracted with respect to each gene annotated in the data set. A total of 22,609 gene-associated methylation CpG sets were generated. To characterize changes in methylation of promoter regions across the 22,609 genes associated with variation at the 5HTTLPR, two sets of promoter methylation vectors were formed; one for “s” allele carriers and the other for non-“s” allele carriers. For each gene examined, one vector summarized methylation of promoter associated loci for those with one or two “s” alleles whereas the other vector summarized methylation for those with no “s” alleles. An unpaired t-test was then performed using the statistical programming language, R (R Development Core Team, 2009) on the vector pair for each of the 22,609 genes, within level of cumulative stress, for males and females separately (Ns = 38, 31, 41, 68 for low SES risk, and Ns = 38, 45, 58, and 69 for high SES risk male s-allele, female s-allele, male non-s-allele, female non-s-allele carriers, respectively). Resulting p-values were adjusted for multiple comparisons using the Benjamini and Hochberg (BH) method (Benjamini & Hochberg, 1995).
Human protein–protein interactions (PPI) data were downloaded from the comprehensive iRefIndex data base (Razick et al., 2008). The methylation probes that mapped to the promoter regions of the 22,609 genes assessed were compared to the PPI data. A subset of 29,912 possible protein–protein interactions mapping to the 22,609 genes were obtained. We then proceeded to calculate PPI edge weights for interactions using the adjusted p-values in order to construct an edge-weighted network.
Edge weight between two genes of each interaction was calculated based on the following equation:
where wij is the edge weight between genes i and j, padj,i is the BH adjusted p-value of the t-test between “s” allele carriers and non-“s” allele carriers for gene i, padj,j is the corresponding BH adjusted p-value for gene j, and padj,min is the minimum BH adjusted p-value among all adjusted p-values calculated for the respective 22,609 genes. The resulting edge weight therefore identifies gene pairs showing significant and coordinated up or down regulation.
The Module Inference by Parametric Local Modularity (miPALM) algorithm (Kessler, Mickelson, & Williams, 1999) was used to identify significantly differentially methylated protein sub-networks using the PPI data alongside the computed edge weights at the .05 significance level. This greedy search algorithm implemented in JAVA, identifies highly connected (i.e., up or down regulated together) sets of genes that comprise networks with the potential to interact inside cells as physically interacting protein complexes. Significantly differentially methylated sub-networks identified by the algorithm were visualized using Cytoscape (Smoot, Ono, Ruscheinski, Wang, & Ideker, 2011). To delineate enriched Gene Ontology (GO) categories of statistically overrepresented genes in each sub-network, the Cytoscape plugin BiNGO (Maere, Heymans, & Kuiper, 2005) was used. Node and edge attributes were also visualized. The node attribute was the BH adjusted p-values that were negative log transformed and visualized as a color gradient, going from green at the smallest p-values (most significant) to blue for less highly significant values. The edge attribute was the edge weight of an interaction between two nodes, with more strongly connected nodes having a thicker line between them. Accordingly, results depict alterations in functional gene networks with potential implications for cellular level activity and outcomes (Jordan, Nguyen, & Lui, 2012; Li, Huang, Liu, Cai, & Chou, 2012).
The single locus examination (at SLC6A4) of the relationship of methylation and genotype data was conducted using general linear model algorithms contained within SPSS 21 (Armonk, New York). Correction for multiple comparisons at the SLC6A4 locus was accomplished by dividing the observed values by the number of effective independent variables as calculated by the method of Cheverud and colleagues (Chen, Miller, Lachman, Gruenewald, & Seeman, 2012). Specifically, we identified 5 independent dimensions among the CpG residues associated with 5HTT and so we considered values as significant if they were p < .01 (i.e. p < .05/5).
3. Results
The clinical and demographic characteristics of the 388 participating African American youth whose data are included in the study are given in Table 1 by gender. As can be seen, there were no significant differences as a function of gender. To test the hypothesis that the effects of stress on SLC6A4 would depend on genotype, potentially leading to differential changes in function, we extracted the beta values for each of the 16 probes mapping to the SLC6A4 locus. We then used two-way ANOVAs to examine whether stress had direct or genetically contextual effects on DNA methylation at each of the 16 CpG residues. The results of these analyses are shown in Table 2. As the Table illustrates, there were no main effects of genotype on methylation at any of the 16 residues for either women or men. However, cumulative SES risk had significant main effects at 2 residues for women (cg14692377 and cg24984698) and there were significant interactive effects of stress with genotype at 3 residues (cg05016953, cg14692377 and cg26126367). Both cg05016953 and cg14692377 map to the first exon portion of the center of the CpG island which has been shown to be critical for transcriptional silencing (Bradley et al., 2005). Only one residue, cg14692377 demonstrated both main and genetically contextual effects of cumulative SES risk for women. The interaction effect at cg14692377 was attributable to relatively greater methylation among “s” allele carriers experiencing SES risk conditions (M = .133) than among “non-s” allele carriers experiencing risk (M = .122) or among those in lower SES risk conditions regardless of genetic context (M = .110 and M = .123 for s-carriers and non-s carriers, respectively). For men, there were two CpG sites that demonstrated genetically contextual effects (cg26741280 and cg10901968), and the latter demonstrated a marginally significant main effect of cumulative stress as well. Accordingly, there was not conservation of effects across gender and effects were stronger, in general, for women.
Table 1.
ANOVA and descriptive statistics of components of the cumulative risk index for carriers of the s or l allele at the 5-HTTLPR, by gender.
| Variables | Males (n = 175) |
Females (n = 213) |
ANOVA |
|||
|---|---|---|---|---|---|---|
| ss/sl (76) Mean (SD) |
ll (99) Mean (SD) |
ss/sl (76) Mean (SD) |
ll (137) Mean (SD) |
F-test | p-Value | |
| Age at the first assessment | 11.623 (.363) |
11.647 (.368) |
11.639 (.361) |
11.695 (.328) |
.879 | .452 |
| Hours of work per week | 39.28 (12.049) |
39.19 (10.057) |
40.72 (10.148) |
40.40 (11.483) |
.350 | .789 |
| Family incomes | 2129.957 (1287.168) |
2231.787 (1660.089) |
2012.244 (1820.411) |
1902.079 (1165.585) |
.982 | .401 |
| Family poverty (150% below line) | .648 (.481) |
.637 (.483) |
.771 (.423) |
.685 (.466) |
1.268 | .285 |
| Parent unemployment | .145 (.354) |
.232 (.424) |
.211 (.410) |
.256 (.438) |
1.216 | .304 |
| Single-parent family | .560 (.500) |
.660 (.477) |
.630 (.487) |
.540 (.500) |
1.229 | .299 |
| TANF receipt | .040 (.196) |
.090 (.289) |
.110 (.309) |
.070 (.261) |
.880 | .451 |
| Parent education (<high school) | .540 (.502) |
.459 (.501) |
.526 (.503) |
.555 (.499) |
.742 | .527 |
| Inadequate income for needs | .408 (.495) |
.367 (.485) |
.329 (.473) |
.229 (.460) |
.965 | .409 |
TANF, Temporary Assistance for Needy Families.
Table 2.
The significance of the association between methylation at CpG sites on SLC6A4 and cumulative stress level, genotype, and their interaction, by gender.
| Probe | CpG Site | Position | Island status | CS | G | CS×G |
|---|---|---|---|---|---|---|
| I | cg12074493 | TSS1500 | South shore | 0.588 (0.838) |
1.000 (0.626) |
0.656 (0.229) |
| II | cg06841846 | TSS1500 | South shore | 0.198 (0.474) |
0.635 (0.127) |
0.920 (0.787) |
| III | cg18584905 | TSS1500 | South shore | 0.494 (0.077) |
0.598 (0.931) |
0.054 (0.023) |
| IV | cg27569822 | TSS200 | Island | 0.816 (0.290) |
0.516 (0.627) |
0.061 (0.244) |
| V | cg10901968 | TSS200 | Island | 0.241 (0.010) |
0.241 (0.188) |
0.515 (0.006*) |
| VI | cg26741280 | TSS200 | Island | 0.138 (0.799) |
0.319 (0.784) |
0.242 (0.006*) |
| VII | cg25725890 | TSS200 | Island | 0.500 (0.472) |
0.875 (0.777) |
0.019 (0.176) |
| VIII | cg05016953 | 1st Exon/5′UTR | Island | 0.922 (0.837) |
0.804 (0.249) |
0.005* (0.790) |
| IX | cg14692377 | 1st Exon/5′UTR | Island | 0.001* (0.682) |
0.608 (0.705) |
0.000* (0.136) |
| X | cg03363743 | 5′UTR | Island | 0.322 (0.942) |
0.393 (0.227) |
0.219 (0.709) |
| XI | cg22584138 | 5′UTR | North shore | 0.502 (0.085) |
0.406 (0.480) |
0.478 (0.390) |
| XII | cg05951817 | 5′UTR | North shore | 0.555 (0.123) |
0.105 (0.208) |
0.027 (0.842) |
| XIII | cg26126367 | 5′UTR | North shore | 0.139 (0.342) |
0.917 (0.853) |
0.001* (0.118) |
| XIV | cg01330016 | 5′UTR | 0.032 (0.435) |
0.837 (0.575) |
0.011 (0.055) |
|
| XV | cg24984698 | Body | 0.006* (0.687) |
0.508 (0.261) |
0.013 (0.018) |
|
| XVI | cg20592995 | 3′UTR | 0.640 (0.172) |
0.481 (0.331) |
0.159 (0.840) |
Note: CS, cumulative stress; G, genotype. CS, G, and CS × G values not in parentheses refer to females; values in parentheses refer to males.
Significant after correction for multiple comparisons.
To determine whether genetically contextual changes in DNA methylation at SLC6A4 were reflected in broader patterns of functionally significant change elsewhere in the genome, we used the miPALM algorithm to detect significant genetically contextual changes in protein–protein interaction networks in the presence or absence of stress. There were no significant loci or networks identified when examining the effects of genotype for men at either level of cumulative SES stress, or under low cumulative stress conditions for women. However, in the presence of high cumulative stress, a number of differentially impacted gene networks were generated for women, indicating differential outcomes for “s” vs. “non-s” carriers. The network with the highest genome-wide significance and its corresponding gene pathway enrichment analysis is shown in Fig. 1. Fig. 1 illustrates that methylation status of the gene promoter of CD3D was significantly greater in the context of exposure to cumulative SES risk in childhood among females carrying an s-allele. In turn, methylation at CD3D, which is a T-cell receptor subunit critical for innate immune responses, is highly correlated with level of methylation of the promoter regions of a number of other genes, with whose protein product the CD3D protein subunit physically interacts. As can be seen in Table 3, gene pathway analysis conducted using the BiNGO algorithm demonstrated a significant enrichment of gene pathways involved in cellular response to unfolded proteins and to stress. Accordingly, the pathway portrayed in Fig. 1 for female s-allele carriers exposed to high cumulative SES risk can be characterized as reflecting an altered cellular level stress response.
Fig. 1.
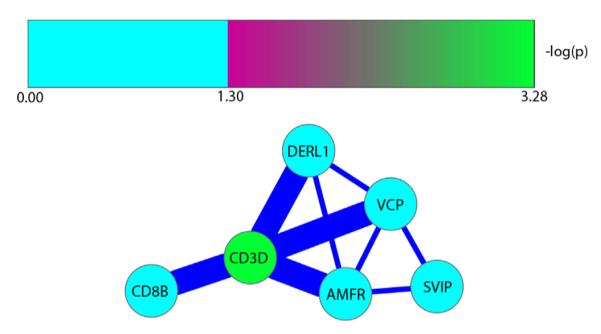
The most significant protein sub-network (p-value approximately 0) consists of 6 proteins. The central node is CD3D. Node colors represent the negative log p-value from the t-test between female s-allele carriers and non-s-allele carriers in the presence of cumulative stress after correction for multiple comparisons. Line thickness between nodes represents strength of interaction between proteins. Subnetwork enriched for pathway involved with cellular response to unfolded proteins and stress. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Enriched GoMiner categories for pathway 1 (centered on CD3D) among female s allele carriers experiencing high cumulative stress.
| GO category | Category name | Genes |
Corrected p-value | |
|---|---|---|---|---|
| Total | Changed | |||
| GO:0034620 | Cellular response to unfolded protein | 21 | 3 | 2.58E–6 |
| GO:0030968 | Endoplasmic reticulum unfolded protein | 21 | 3 | 2.58E–6 |
| GO:0071445 | Cellular response to protein stimulus | 27 | 3 | 2.84E–6 |
| GO:0030433 | ER-associated protein catabolic process | 27 | 3 | 2.84E–6 |
| GO:0034976 | Response to endoplasmic reticulum stress | 34 | 3 | 4.62E–6 |
| GO:0006984 | ER-nucleus signaling pathway | 36 | 3 | 4.62E–6 |
| GO:0071216 | Cellular response to biotic stimulus | 49 | 3 | 1.02E–5 |
| GO:0006986 | Response to unfolded protein | 66 | 3 | 2.06E–5 |
| GO:0006986 | Retrograde protein transport, ER to cytosol | 5 | 2 | 2.06E–5 |
| GO:0051789 | Response to protein stimulus | 117 | 3 | 1.00E–4 |
Depictions of the remaining 15 protein sub-networks that differed significantly between “s” and “non-s” carrying females exposed to high levels of cumulative SES risk in childhood, and their corresponding pathway enrichment analyses, are available in Supplementary Figs. 1–4 and Supplementary Tables 1–15, respectively. The size of the protein networks identified range from a minimum of 6 proteins to a maximum of 27 proteins. Recurrent themes in the functional significance of the protein networks as indicated by their pathway analyses include immune response and apoptosis. Pathways involved in immune response include those associated with viral replication, chemo taxis, NK-Kappa B activity and interleukin 1 production. With respect to apoptosis, large networks centered on P TEN and TP53 (a.k.a. P53), both of which are well known oncogenes, were identified. Finally, one fairly large network involved in the regulation of coagulation was also identified. Accordingly, all but one of the identified pathways are related to cellular level stress response, suggesting that they capture a range of long-term adaptations to chronic stress with the potential to influence a range of physical and psychiatric outcomes.
4. Discussion
African American youth in the rural South confront sustained socio-economic stress and other related difficulties during sensitive developmental periods. This experience appears to confer greater risk for several problematic health conditions many years later (Braveman et al., 2010; Starfield et al., 2002) and may set the stage for co-occurrence of both psychiatric and physical health problems in adulthood. Though the exact genetic architecture moderating differential susceptibility to these childhood stressors is likely to be extensive, a number of investigators have suggested that variability in the 5HTTLPR may be important in setting the stage for changes relevant to mental and physical health outcomes (Caspi et al., 2010). Accordingly, mapping impact of this region in interaction with a robust environmental stressor has the potential to illuminate the source of some mental and physical heath disparities. In this cohort of 19-year-old African-American young adults who are at high risk for the future onset of a variety of complex medical and behavioral disorders including type II diabetes (T2DM), coronary artery disease, depression, and obesity, we provide an initial molecular explication of potential pathways through which differential genotype at 5HTTLPR may exert its effects by amplifying the impact of environmental stressors. This effect was evidenced by differential DNA methylation in the critical 1st exon region of SLC6A4 and at the genome wide level as shown by gene networks indicative of changes in level of cellular stress response.
The effects we observed were more robust in females than males. Part of the reason for the more robust effects observed in our current findings may be the greater power afforded by having a larger number of female subjects. Alternatively, females may be more reactive to the stressors examined in this study. Suggesting the latter, the rate of depression and obesity, both of which are correlated with exposure to adolescent stress, are significantly higher in young adult African-American females than in males. Conceivably, differences in the rates could rise from X-chromosome influences or gender specific cultural effects.
Our current finding that genetic variability at the 5HTTLPR has significant impact only in the presence of high SES risk contrasts with previous findings that the 5HTTPR functions as a susceptibility gene with regard to its impact on methylation (Beach et al., in press). Whereas the prior findings suggested a significant impact of genotype at both low and high cumulative SES risk, the current focus on plausible cellular level effects on functional protein–protein networks suggests differences only under high risk. By restricting attention to possible protein–protein interactions, and to CpG changes with genomewide significance, we highlight the potential for impact on cellular level functioning to be greater at high levels of stress. Likewise, when we restricted our focus to only the SLC6A4 we found significant main effects of stress and G × E interaction effects mainly mapping to the critical 1st exon region of SLC6A4. This is consistent with our global understanding of the preeminence of this 5′ region in regulating gene expression (Brenet et al., 2011). Accordingly, it appears that, among women, gene expression controlled by the 5HTTLPR may be differentially affected by experience of economic stress during childhood depending on “s” vs. “l” genotype. This provides an interesting and plausible mechanism for G × E effects on later behavioral tendencies and so helps address the controversy over G × E effects related to the serotonin transporter (Caspi et al., 2003; Risch et al., 2009).
The finding of genetically contextual effects of economic stress during childhood on likely longer-term functioning of the 5HTTLPR also makes plausible the potential for additional accumulation of differential impacts on methylation genome-wide. Although the specific developmental sequence leading to physical and psychiatric difficulties is not illuminated by the current analyses, the use of protein–protein interaction mapping provides evidence of broad impact at the cellular level. At the genome wide level, in the absence of cumulative SES risk, we did not observe any significant effects of 5HTTLPR genotype on alteration of coherent patterns of promoter methylation that would result in changes in protein–protein networks. However, in the presence of high cumulative SES risk, a number of significantly affected gene promoter networks were observed for women. Hence, the interactive effects of stress on SLC6A4 methylation in a critical regulatory region and the interactive effects on gene pathways indicating heightened stress response are consistent with current conceptualizations that SLC6A4 variation exerts its effect by increasing reaction to elevated psychosocial stress.
The finding that the most highly differentially regulated network maps to a T-Cell immune network is consistent with existing biology and suggests a molecular framework for existing phenomenological studies of the genetically contextual effect of adverse childhood environments on later life outcomes such as premature aging. Although the reasons for its existence outside the CNS are not clear, the serotonin transporter is selectively functionally expressed in lymphocytes, which constitute ~20–30% of all peripheral white blood cells (WBC). But with the possible exception of some production in monocytes which constitute 1–2% of peripheral white blood cells (WBC), it seems to be largely absent from the rest of the other WBC’s (neutrophils, basophils, eosinophils, etc.) that constitute the majority of the WBC population. Studies of SLC6A4 on lymphocytes have consistently shown that it is both functional and that the level of expression is regulated by the 5HTTLPR (Lesch et al., 1996, November 29; Philibert et al., 2008). In contrast to its role in the CNS where its predominant role is to clear serotonin from the region of the synaptic cleft, in vitro studies of lymphocytes show a powerful role for this transporter in moderating the role of the production of cytokines such as tumor necrosis factors alpha (TNFα) (Coe, Irwin, Lippner, & McCabe, 2011). Pertinent to this line of reasoning is that fact that the gene that produces TNFα (TNF) and TNF receptor superfamily, member 1A (TNFSRF1A) are hub genes in one of the gene networks listed in the Supplementary Figure section. Second, exposure to childhood stress is known to stimulate the expression of cytokines in T lymphocytes (Leggate, Nowell, Jones, & Nimmo, 2010; Mensah, Mokdad, Ford, Greenlund, & Croft, 2005). Third, although the conclusions are controversial (Risch et al., 2009), there is a wealth of evidence to support a critical role for the 5HTTLPR in moderating the effects of childhood stressors on adverse adult outcomes, many of which are inflammatory in nature (Caspi et al., 2003; Sen, Burmeister, & Ghosh, 2004). Accordingly, the results are encouraging of future efforts to examine particular inflammatory pathways as mechanisms linking cumulative SES risk in childhood to later adverse health and mental health outcomes. Given recent evidence suggesting roles for altered TNF signaling in hypothalamic neurons and microglia that results in premature aging (Gabuzda & Yanker, 2013) and evidence suggesting that peripheral DNA methylation can predict CNS methylation (Davies et al., 2012), these findings additionally suggest that studies of peripheral DNA methylation may shed insight into the complex mind–brain interplay underlying a diverse set of processes such as premature aging. Unfortunately, understanding the systems biology of the exact mechanisms through which these gene pathways are modulated may be difficult and may necessitate coordinated prospective clinical and in vitro studies.
Several potential limitations of the current investigation should also be noted. First, it is possible that observed effects may be due to unmeasured variation within SLC6A4 (cf. Murdoch, Speed, Pakstis, Heffelfinger, & Kidd, 2013) or due to combinations of genes that were not examined in the current investigation. However, Murdoch et al. (2013) found strong decay of LD between the 5HTTLPR and the downstream elements of the SLC6A4 gene, limiting these potential concerns. In addition, although the study is longitudinal, it is correlational in design, precluding causal inference, and raising the possibility of third variable explanations of the apparent impact of cumulative SES risk. Likewise, the sample size is small for genomewide examination, increasing the likelihood of type II errors due to low power to detect differences. In addition, the method used to examine edge weights was based on dichotomized scores, further limiting power to detect real differences and potentially underestimating genetic effects at low cumulative SES risk that might have been observable with a larger sample. This is particularly important in the context of prior observations of an impact of variation at SLC6A4 at both high and low risk. Accordingly, the findings should be viewed as preliminary and suggestive until they are replicated.
In sum, the current investigation advances understanding of possible SLC6A4 interactions with the environment by highlighting allele specific influences on methylation, and by clarifying the nature of potentially affected gene pathway with the potential to affect cellular level functioning. In this manner the current research helps connect investigation of methylation with the broader investigation of allostasis. At the same time, considerable work remains to establish transcriptional effects of observed methylation changes within each of the identified protein–protein networks. Likewise, it will be important to establish links between transcriptional effects and changes in key cellular level processes. Doing so has the potential to help more rapidly advance the study of health disparities.
Supplementary Material
Footnotes
This research was supported by Award Number 5R01HD030588-16A1 from the National Institute of Child Health and Human Development and Award Number 1P30DA027827 from the National Institute on Drug Abuse. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Child Health and Human Development, the National Institute on Drug Abuse, or the National Institutes of Health.
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.biopsycho.2013.10.006.
References
- Beach SRH, Brody GH, Lei MK, Kim S, Cui J, Philibert RA. Is Serotonin Transporter Genotype Associated with Epigenetic Susceptibility or Vulnerability? Examination of the Impact of SES Risk on Among African American Youth. Development and Psychopathology. doi: 10.1017/S0954579413000990. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky J, Pluess M. Beyond diathesis-stress: differential susceptibility to environmental influences. Psychological Bulletin. 2009;135(6):885–908. doi: 10.1037/a0017376. http://dx.doi.org/10.1037/a0017376. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57(1):289–300. [Google Scholar]
- Bradley SL, Dodelzon K, Sandhu HK, Philibert RA. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2005;136(1):58–61. doi: 10.1002/ajmg.b.30185. http://dx.doi.org/10.1002/ajmg.b.30185. [DOI] [PubMed] [Google Scholar]
- Braveman PA, Cubbin C, Egerter S, Williams DR, Pamuk E. Socioeconomic disparities in health in the United States: What the patterns tell us. American Journal of Public Health. 2010;100(S1):S186–S196. doi: 10.2105/AJPH.2009.166082. http://dx.doi.org/10.2105/AJPH.2009.166082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PloS ONE. 2011;6(1):e14524. doi: 10.1371/journal.pone.0014524. http://dx.doi.org/10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody GH, Yu T, Chen E, Miller GE, Kogan SM, Beach SRH. Is resilience only skin deep?: Rural African Americans’ socioeconomic status-related risk and competence in preadolescence and psychological adjustment and allostatic load at age 19. Psychological Science. 2013;24(7):1285–1293. doi: 10.1177/0956797612471954. http://dx.doi.org/10.1177/0956797612471954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. http://dx.doi.org/10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: The case of the serotonin transporter gene and its implications for studying complex diseases and traits. American Journal of Psychiatry. 2010;167(5):509–527. doi: 10.1176/appi.ajp.2010.09101452. http://dx.doi.org/10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Miller GE, Lachman ME, Gruenewald TL, Seeman TE. Protective factors for adults from low-childhood socioeconomic circumstances: The benefits of shift-and-persist for allostatic load. Psychosomatic Medicine. 2012;74(2):178–186. doi: 10.1097/PSY.0b013e31824206fd. http://dx.doi.org/10.1097/PSY.0b013e31824206fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe LM, Irwin R, Lippner D, McCabe LR. The bone marrow microenvironment contributes to type I diabetes induced osteoblast death. Journal of Cellular Physiology. 2011;226(2):477–483. doi: 10.1002/jcp.22357. http://dx.doi.org/10.1002/jcp.22357. [DOI] [PubMed] [Google Scholar]
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: Gateway into systems biology. Human Molecular Genetics. 2005;14(Suppl. 2):R171–R181. doi: 10.1093/hmg/ddi335. http://dx.doi.org/10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- Das D, Tan X, Easteal S. Effect of model choice in genetic association studies: DRD4 exon III VNTR and cigarette use in young adults. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156(3):346–351. doi: 10.1002/ajmg.b.31169. http://dx.doi.org/10.1002/ajmg.b.31169. [DOI] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biology. 2012;13(6) doi: 10.1186/gb-2012-13-6-r43. http://dx.doi.org/10.1186/gb-2012-13-6-r43. Article R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GW. A multimethodological analysis of cumulative risk and allo-static load among rural children. Developmental Psychology. 2003;39(5):924–933. doi: 10.1037/0012-1649.39.5.924. http://dx.doi.org/10.1037/0012-1649.39.5.924. [DOI] [PubMed] [Google Scholar]
- Gabuzda D, Yanker BA. Physiology: Inflammation links ageing to the brain. Nature. 2013;497(7448):197–198. doi: 10.1038/nature12100. http://dx.doi.org/10.1038/nature12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammen C, Brennan PA, Keenan-Miller D, Hazel N, Najman J. Chronic and acute stress, gender, and serotonin transporter gene–environment interactions predicting depression in youth. Journal of Child Psychology and Psychiatry. 2010;51(2):180–187. doi: 10.1111/j.1469-7610.2009.02177.x. http://dx.doi.org/10.1111/j.1469-7610.2009.02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan F, Nguyen T-P, Liu W-C. Studying protein-protein interaction networks: a systems view on diseases. Briefings in Functional Genomics. 2012;11(6):497–504. doi: 10.1093/bfgp/els035. http://dx.doi.org/10.1093/bfgp/els035. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Mickelson KD, Williams DR. The prevalence, distribution, and mental health correlates of perceived discrimination in the United States. Journal of Health and Social Behavior. 1999;40(3):208–230. http://dx.doi.org/10.2307/2676349. [PubMed] [Google Scholar]
- Kim S, Brody GH. Longitudinal pathways to psychological adjustment among Black youth living in single-parent households. Journal of Family Psychology. 2005;19(2):305–313. doi: 10.1037/0893-3200.19.2.305. http://dx.doi.org/10.1037/0893-3200.19.2.305. [DOI] [PubMed] [Google Scholar]
- Leggate M, Nowell MA, Jones SA, Nimmo MA. The response of interleukin-6 and soluble interleukin-6 receptor isoforms following intermittent high intensity and continuous moderate intensity cycling. Cell Stress and Chaperones. 2010;15(6):827–833. doi: 10.1007/s12192-010-0192-z. http://dx.doi.org/10.1007/s12192-010-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- Li BQ, Huang T, Liu L, Cai YD, Chou KC. Identification of colorectal cancer related genes with mRMR and shortest path in protein-protein interaction network. PLoS ONE. 2012;7:e33393. doi: 10.1371/journal.pone.0033393. http://dx.doi.org/10.1371/journal.pone.0033393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. http://dx.doi.org/10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Gianaros PJ. Central role of the brain in stress and adaptation: Links to socioeconomic status, health, and disease. Annals of the New York Academy of Sciences. 2010;1186:190–222. doi: 10.1111/j.1749-6632.2009.05331.x. http://dx.doi.org/10.1111/j.1749-6632.2009.05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan GM, Pan Q, Porter KS. Consent for genetic research in a general population: An update on the National Health and Nutrition Examination Survey experience. Genetics in Medicine. 2006;8(6):354–360. doi: 10.1097/01.gim.0000223552.70393.08. http://dx.doi.org/10.1097/01.gim.0000223552.70393.08. [DOI] [PubMed] [Google Scholar]
- Mensah GA, Mokdad AH, Ford ES, Greenlund KJ, Croft JB. State of disparities in cardiovascular health in the United States. Circulation. 2005;111(10):1233–1241. doi: 10.1161/01.CIR.0000158136.76824.04. http://dx.doi.org/10.1161/01.CIR.0000158136.76824.04. [DOI] [PubMed] [Google Scholar]
- Murdoch JD, Speed WC, Pakstis AJ, Heffelfinger CE, Kidd KK. Worldwide population variation and haplotype analysis at the serotonin transporter gene SLC6A4 and implications for association studies. Biological Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.02.006. http://dx.doi.org/10.1016/j.biopsych.2013.02.006. Advance online publication. [DOI] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B(5):543–549. doi: 10.1002/ajmg.b.30657. http://dx.doi.org/10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plume JM, Beach SRH, Brody GH, Philibert RA. A cross-platform genome-wide comparison of the relationship of promoter DNA methylation to gene expression. Frontiers in Genetics: Epigenomics. 2012;3 doi: 10.3389/fgene.2012.00012. http://dx.doi.org/10.3389/fgene.2012.00012. Article 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor BD, Dalaker J. Poverty in the United States: 2002 (Current Population Reports, P60-222) US Census Bureau; Washington, DC: 2003. [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. iRefIndex: A consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-405. http://dx.doi.org/10.1186/1471-2105-9-405. Article 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang K-Y, Eaves LJ, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: A meta-analysis. Journal of the American Medical Association. 2009;301(23):2462–2471. doi: 10.1001/jama.2009.878. http://dx.doi.org/10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Burmeister M, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;127B(1):85–89. doi: 10.1002/ajmg.b.20158. http://dx.doi.org/10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. http://dx.doi.org/10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starfield B, Robertson J, Riley AW. Social class gradients and health in childhood. Ambulatory Pediatrics. 2002;2(4):238–246. doi: 10.1367/1539-4409(2002)002<0238:scgahi>2.0.co;2. http://dx.doi.org/10.1367/1539-4409. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nature Reviews Genetics. 2008;9(6):465–476. doi: 10.1038/nrg2341. http://dx.doi.org/10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Vijayendran M, Beach SRH, Plume JM, Brody GH, Philibert RA. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Frontiers in Psychiatry: Child and Neurodevelopmental Psychiatry. 2012;3 doi: 10.3389/fpsyt.2012.00055. http://dx.doi.org/10.3389/fpsyt.2012.00055. Article 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


