Abstract
Stem cell based-therapies are novel therapeutic strategies that hold key for developing new treatments for diseases conditions with very few or no cures. Although there has been an increase in the number of clinical trials involving stem cell-based therapies in the last few years, the long-term risks and benefits of these therapies are still unknown. Detailed in vivo studies are needed to monitor the fate of transplanted cells, including their distribution, differentiation, and longevity over time. Advancements in non-invasive cellular imaging techniques to track engrafted cells in real-time present a powerful tool for determining the efficacy of stem cell-based therapies. In this review, we describe the latest approaches to stem cell labeling and tracking using different imaging modalities.
Keywords: Stem cells, Cell labeling, Cellular imaging, Reporter genes, Nanoparticles
Introduction
Unlike the hydrozoans and planarians, characterized by their ability to remodel tissues and organs after a wound, or amphibians who can repair a damaged limb in just 70 days, the human body is extremely poor at repair and regeneration of damaged cells. A possible solution to this problem, stem cell transplantation, was introduced in 1945, the year of the first atomic bomb explosion in World War II. Leon Jacobson at the University of Chicago demonstrated that protecting mice spleens with a lead shield could protect them from the lethal effects of ionizing radiation on bone marrow. His team further reported a similar effect with the mice femur. Jacobson and colleagues eventually found that protection against ionizing radiation could also be achieved by intravenous bone marrow infusion [1–3]. Initially, it was believed that this protection was due to some unknown humoral factor. However, in the 1950s, Barnes and Loutit [4] and Main and Prehn [5] showed evidence that the protection from radiation was the result of bone marrow stem cells.
Stem cell-based therapies hold great promises in regenerative medicine because of their inherent biological properties of plasticity, self-renewal, and migration. Edmund B. Wilson first mentioned the term “stem cell” in his book, The Cell in Development and Inheritance, in 1896. Stem cells can be classified as (i) pluripotent embryonic stem cells derived from the inner mass of the blastocyst, (ii) multipotent adult stem cells found throughout the body after development, (iii) oligopotent progenitor cells, the “mid-point” between stem cells and fully differentiated cells, and (iv) induced pluripotent stem cells, somatic cells that have been genetically reprogrammed to an embryonic stem cell-like state. After systemic or local transplantation, stem cells may be able to migrate and differentiate at pathologic sites in the body to produce a therapeutic effect. In 1959, the first human bone marrow transplantation by Edward Thomas operated on the concept that infusing bone marrow could provide hematological reconstitution in lethally irradiated patients with acute leukemia [6]. Since then, several advancements have been made in the field of regenerative medicine, with some very promising outcomes. There has been a rise in clinical trials involving stem cell therapies in the last 3 years. The public clinical trials database, http://www.clinicaltrials.gov, shows 290 trials using mesenchymal stem cells (MSCs) in different disease conditions (accessed 06/18/2013, unknown status excluded).
CartiCel, a cell therapy product to repair articular cartilage injuries in the knee of adults, was launched by Genzyme, USA and approved by the Food and Drug Administration (FDA) in 2010 (www.fda.gov; Submission Tracking Number: BL 103661). In Europe, ChondroCelect (TiGenix, Leuven, Belgium) was licensed in 2009 by the European Medicines Agency as advanced therapeutic medicinal product for cartilage regeneration in the knee (www.emea.europa.eu). In the USA, the Cardiovascular Cell Therapy Research Network (CCTRN), established by the National Heart, Lung, and Blood Institute (NHLBI), develops, coordinates, and conducts multiple collaborative protocols to test the effects of stem cell therapy in cardiovascular diseases. The main goal of the CCTRN is to determine whether stem cell therapies are safe for use in humans and will not lead to any adverse events. The success of any stem cell-based therapy depends on several critical issues, such as type of cell, route and accuracy of cell transplant, functionality of transplanted cells (long-term assessment), and, above all, how cells interact with the microenvironment after transplantation (short-term assessment). Long-term assessment of transplanted cells may not be relevant if the cells do not initially home and engraft. To address these questions, it is imperative to monitor transplanted cells in real time.
While safety has been consistently demonstrated, sustained therapeutic benefits may not always be obtained with stem cell-based therapies. Several risks may arise following these therapies, including teratoma formation, the potential for malignant transformation, graft-versus-host disease, graft failure, and organ damage [7, 8]. According to the FDA guidelines on stem cell-based therapies, although in vitro assays may indicate how the stem cell-based product is likely to function in vivo, pre-clinical animal studies are needed to monitor the function of the cells after transplantation. Such observations are particularly important for a stem cell-based product that contains cells that are not terminally differentiated at the time of transplantation. These studies should mimic the route and method of administration to be used in subsequent clinical studies (http://www.fda.gov). Therefore, in vivo tracking of transplanted cells over time is a crucial step in determining the safety and efficacy of cell therapy.
Traditionally, histological investigation has been required to determine the fate of transplanted cells. However, it is highly invasive, requires multiple tissue biopsies, and also limits our ability to monitor transplanted cells in real time. In recent years, the development of in vivo cell imaging techniques has significantly improved our ability to noninvasively track transplanted cells in real time. We can now monitor distribution, differentiation, and viability of these cells at the site of injury or elsewhere in the body. Along with these benefits, other important information about the action of engrafted stem cells can also be gained by non-invasively visualizing the changes in the target tissue. For example, the efficacy of transplanted neural stem cells for new myelination in a dysmyelinated animal model can be detected by diffusion tensor imaging (DTI) [9]. Similarly, the therapeutic outcome of stem cell therapy for treatment of myocardial infarct can be evaluated by magnetic resonance imaging (MRI) [10], without labeling and visualizing cells. Positron emission tomography (PET) imaging can be used for monitoring myelin repair in the spinal cord [11].
Cell tracking can be useful for the preclinical assessment of new cell therapies, the design of clinical trials, and for monitoring these therapies in clinical practice. In this review, we provide an overview of different methods of cell labeling, as well as discuss the use of different imaging modalities for cell tracking and their key strengths and limitations.
Cell Labeling
Labeling or tagging of cells is necessary to track the migration and differentiation of cells by imaging after transplantation. A cell can be labeled by direct or indirect methods. Direct methods are simple and do not involve genetic modification of the cell. Indirect cell labeling methods require genetic alteration of the cell to introduce a reporter gene into it.
Direct Labeling Methods
Direct labeling is a commonly used cell labeling method. In this method, a labeling agent is introduced into the cells prior to transplantation. The labeling agent is then imaged as a surrogate for the transplanted cells. Depending on the imaging modality to be used, cells can be labeled with fluorescent semiconductor nanocrystals or fluorochromes for optical fluorescence imaging, paramagnetic and superparamagnetic iron oxide particles (SPIO) for MRI, and radionuclides for single photon emission computed tomography (SPECT) or PET.
Direct Labeling for Optical Imaging
In this method, cells are labeled with optical fluorescent probes (fluorochromes). After the transplantation of labeled cells in the body, the fluorescent probes are excited with light of a defined wavelength and the emitted photons are registered with a highly sensitive charge-coupled device (CCD) camera. Organic fluorochromes are affordable, easy to use, and have a favorable toxicity profile. Past approaches have investigated several fluorochromes, including the most commonly used fluorochromes, cyanine dyes. Organic fluorochromes are prone to photobleaching, an obstacle when longer observation periods are required. Another major limitation of these fluorochromes is the low tissue penetration of light. The visible spectrum emitted by fluorescence has only a limited tissue penetration of a few hundred micrometers; thus, fluorochromes accumulated a few centimeters from the skin may not be detected. An alternative to conventional, traditional fluorochrome probes is semi-conductive, light-emitting inorganic nanocrystals, also called quantum dots (QDs). These are a new group of agents for cell labeling whose emission spectra can be tuned to any desired wavelength [12]. Contrary to traditional fluorochromes, the emission wavelength of QDs does not depend on their chemical structure, but rather, on the size, shape, and chemical composition. These nanoparticles are highly photostable and preferred over organic fluorochromes for multicolor cell labeling because of their high quantum yield, high molar absorption coefficient, broad excitation spectra, long fluorescence lifetime, and narrow emission spectra. One of the early applications of QDs for cell tracking was demonstrated by Lei and colleagues in which MSCs labeled with QDs were systemically injected into immuodeficient NOD/SCID beta2 M null mice [13]. The fluorescence PEG-encapsulated CdSe/ZnS QDs were used to quantify the tissue distribution of MSCs in different organs. Ohyabu et al. further showed that QDs have no adverse effects on the differentiation of MSCs in vitro or in vivo [14]. In the above-mentioned studies, histological evaluations were performed to track the cells labeled with QDs. A noninvasive tracking of cells is possible with the use of near-infrared QDs. Sugiyama and colleagues showed that MSCs labeled with near-infrared QDs (QD800) could be tracked noninvasively after transplantation into the ipsilateral striatum of rats with cerebral stroke [15]. Another study discovered that QDs do not affect growth, differentiation, or proliferation of primary nerve cells, mouse embryonic stem cells, or kidney stem cells, and are suitable for short-term cell tracking [8, 16]. Recently developed organic far-red/near-infrared dots with aggregation-induced emission have also been demonstrated to be useful as long-term non-invasive cell tracers [17].
Despite the promise of this labeling technique, the main limitation with the use of QDs is heavy metal toxicity. QDs are often composed of atoms from groups II–VI or III–V elements in the periodic table, such as CdSe, CdTe, InP, and InAs. This raises concerns about metal toxicity, and limits the use of these QDs in humans.
Alternatively, another near-infrared fluorescent lipophilic dye 1,1-dioctadecyl-3,3,3,3-tetramethylindotricarbocyanine iodide (DiR), can be used for direct cell labeling and optical imaging. DiR has been primarily used for macrophage labeling [18] and also for embryonic stem cell labeling [19]. DiR is safe and typically provides a rapid and stronger labeling [20]. The two long 18-carbon chains of DiR dye can enter through the cell membrane, resulting in a stable labeling with negligible dye transfer between cells, which has been a significant problem when obtaining cell specificity [21].
Direct Labeling for Magnetic Resonance Imaging
In this method, cells are labeled with a suitable contrast agent. MR contrast agents contain metal ions that define their relaxation enhancement properties. These agents alter the relaxation rate of nearby tissue water protons, making them discernible on post-contrast MRI. Contrast agents can be introduced to cells by binding to the external surface of the cell membrane by immuno-magnetic linking to an antibody [22, 23] or by internalization into the cytoplasm through facilitated transmembrane uptake [24]. In immunomagnetic labeling, contrast agents in general do not enter the cells, and thus, do not affect cell viability. However, there is the risk of contrast agent detachment from the cell membrane or the transfer of contrast agent to other cells with this method.
The following types of MR contrast agents are generally used for cell labeling and tracking: (i) paramagnetic agents; (ii) superparamagnetic agents; and (iii) contrast agents that contain nuclei other than hydrogen.
Labeling with Paramagnetic Agents
Gadolinium (III)-Based Contrast Agents
Paramagnetic agents are the most widely used contrast agents in the clinical setting. Gadolinium (III) [Gd (III)] chelates, such as gadopentetate dimeglumine pentaacetic acid (Gd-DTPA), are effective paramagnetic contrast agents owing to their seven unpaired electrons. These unpaired electrons create a magnetic moment that increases the relaxation rates of the surrounding water proton spins and shortens the longitudinal relaxation rate (T1). This increases the signal by creating “positive” contrast on a T1-weighted scan. The clinical safety of paramagnetic contrast agents is largely dependent on the stability of the chelate in vivo. Major factors that determine safety include thermodynamics, solubility, selectivity, and kinetics. Gadolinium is a metal of the lanthanide series, which are rare earth elements. The safety of this class of agents is based on the use of chelates, firmly holding the gadolinium ion and ensuring rapid and complete excretion. If dechelated, the gadolinium ion can be deposited in organs. Gadolinium has been used to label different types of stem cells, such as hematopoietic progenitor cells, monocytic cells, endothelial progenitor cells, and MSCs for cell tracking [25–27]. However the long-term presence of gadolinium in engrafted cells may affect cell function and hence the therapeutic benefit of cell therapy. For instance, Modo and colleagues found that in rats with middle cerebral artery occlusion (MCAO), transplantation of the neural stem cell line MHP36 labeled with Gadolinium-Rhodamine Dextran (GRID) had an actual negative therapeutic effect. Over a period of 1 year, administration of GRID-labeled cells resulted in a slight increase in lesion size compared to untreated MCAO rats [28], a highly undesirable outcome.
The low cellular uptake of paramagnetic contrast agents is the main obstacle in cellular imaging. Different methods, e.g., transfection using transfection agents (lipofectin and lipofectamine) [29], or coupling of the contrast agent to a membrane translocation peptide (13-mer HIV-tat peptide) [30], can be used to increase the uptake of Gd (III) chelates. Several advancements have been made in the last several years to improve the efficacy of paramagnetic contrast agents. For example, Tseng et al. developed a paramagnetic contrast agent, gadolinium hexanedione nanoparticles (GdH-NP), to track transplanted stem cells [31]. They found that GdH-NP was nontoxic to human MSCs and had the ability to spontaneously internalize in the cells. A liposomal-based Gd nanoparticle as a MR contrast agent has also been developed. This dual-Gd liposomal contrast agent consists of a core-encapsulated Gd liposome and a surface-conjugated Gd liposome. This provides the contrast agent with an even greater ability to deliver a higher concentration of Gd to the cell, resulting in greater signal enhancement [32]. Furthermore, the use of PEGylated ultra-small (mean diameter 5 nm) gadolinium oxide (Gd2O3) nanoparticles has also been reported to enhance signal. Very small particles provide the optimal surface-to-volume ratios necessary to reach high relaxivities and generate strong positive contrast enhancement on T1-weighted MRI [33, 34].
Paramagnetic Chemical Exchange Saturation Transfer (PARACEST) Contrast Agents
A new class of paramagnetic contrast agents is the PARACEST (paramagnetic chemical exchange saturation transfer) agents. Contrary to conventional Gd-based contrast agents that affect T1 relaxation, these agents enhance image contrast only at a specific radiofrequencies [35]. Therefore, this technique may make it possible to track two different types of cell populations in the same experiment [36]. Ferrauto and colleagues used two PARACEST agents, Yb- and Eu-HPDO3A, to track two cell populations: J774.A1 murine macrophages and B16-F10 murine melanoma cells that co-localize in tumor phenotypes. They found that tracking of the two cell types is possible using these two agents. Both PARACEST agents have potential for clinical translation as they share the same stability and in vivo pharmacokinetic properties of clinically approved Gd-HPDO3A (ProHance) [37].
Manganese (Mn)-Based Contract Agents
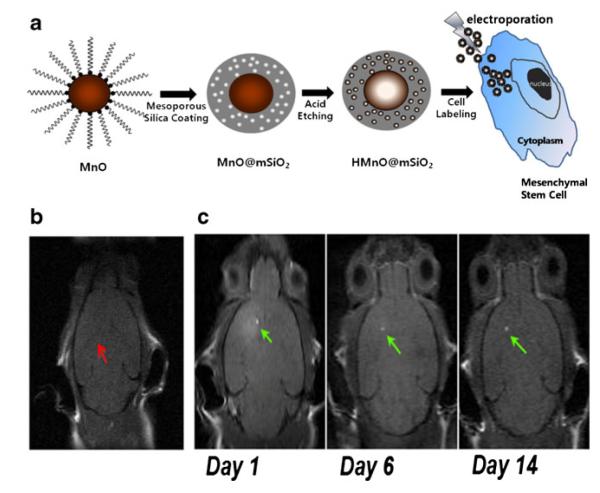
After Gd, manganese (Mn) has been the most studied T1 contrast agent. Mn-enhanced MRI (MEMRI) has been used to study neuronal activity, and monitor the neuronal tract and neuronal connectivity in different animal models [38]. MnO nanoparticles are a novel T1 contrast agent for cell labeling and tracking. They can be used to detect cells with positive contrast in vivo, and can be combined with iron oxide to detect two cell populations simultaneously with opposite cell contrast [39]. Novel mesoporous, silica-coated, hollow MnO nanoparticles have also been developed as positive T1 contrast agents for the labeling and MRI tracking of MSCs (Fig. 1). Mesoporous silica material is known for its excellent stability in aqueous solution and high labeling efficiency. It also allows easy access for water molecules to the magnetic center, which significantly improves the longitudinal water proton relaxation. These MnO nanoparticles proved to be a better positive T1 contrast for MR cell tracking as compared to conventional MnO nanoparticle-based contrast agents [40].
Fig. 1.
Manganese (Mn)-based contract agents for stem cell tracking. a Labeling of MSCs with silica-coated, hollow MnO nanoparticles as MR contrast agent. b No hyperintense signal (red arrow) was detected in a mouse transplanted with unlabeled MSCs. c Hyperintense signals (green arrows) were detected in a mouse transplanted with HMnO@mSiO2-labeled MSCs and were still visible 14 days after injection. (Reproduced from Ref. 40, with permission)
Labeling with Superparamagnetic Iron Oxide Nanoparticles
One of the major limitations of paramagnetic agents for cell tracking is that the MRI signal generated by these agents is not strong enough to make small numbers of cells visible. SPIO particles, however, have a stronger effect on MR reflexivity than paramagnetic agents [41]. SPIOs are based on magnetite (Fe3O4) or maghemite (γ-Fe2O3) cores embedded and stabilized with a hydrophilic shell of dextran [42], siloxan [43], citrate [44], or other polystyrene [45]. The SPIO core contains thousands to millions of iron atoms, which enables the detection of small numbers of cells by increasing the iron concentration in the cells. These particles create a large dipolar magnetic field [46] and signal spin-spin dephasing due to the local field inhomogeneity induced in water molecule near particles [47]. This results in negative contrast on T2-weighted MRI. The unique properties of SPIO were initially utilized to label and track transplanted cells in the rat brain [48]. Since then, several advancements have been made in this field, including the development of dextran-coated monocrystalline iron-oxide nanoparticles (MION) [42, 49] and the detection of the migration of oligodendrocyte progenitors using MION [50]. In 2001, it was shown that magnetodendrimer-encapsulated superparamagnetic iron oxide could be used to label and track stem cells in vivo [51]. In a phase 1/2 open-safety clinical trial SPIOs were used to track the CNS homing of injected MSCs by MRI in multiple sclerosis (MS) and amyotrophic lateral sclerosis (ALS) patients [52] (Fig. 2).
Fig. 2.
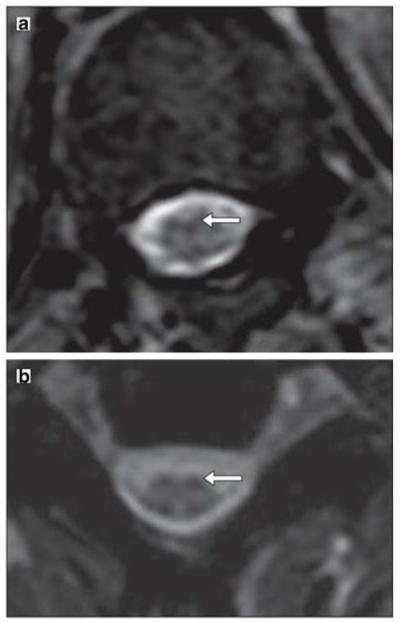
MRI after injection of Feridex®-labeled mesenchymal stem cells. a An axial T2-weighted gradient echo scan through the inferior thoracic cord shows a hypointense pial signal coating the cord similar to that of superficial siderosis, characteristic of Feridex®-labeled cells. b Axial T2-weighted gradient echo scan through the cervical cord shows hypointensity of the dorsal roots and their entry zone and a similar hypointensity of the ventral root entry zones, suggesting the presence of Feridex®-labeled cells. (Reproduced from Ref. 52, with permission)
SPIOs were found to have no effect on the pluripotency or differentiation capacity of cells [53, 54]. Clinical trials about the safety and feasibility of in vivo mononuclear cell tracking using SPIOs (http://www.clinicaltrials.gov; unique identifier: NCT00972946, NCT01169935) demonstrated that SPIO labeling of human peripheral inflammatory cells does not affect their viability or function [55]. However, in mice, although the labeling of bone marrow-derived dendritic cells with SPIO did not affect phenotype or cytokine profiles, overall cell migration was reduced. The reduction in cell migration might be due to nanoparticle size [56]. SPIOs also do not affect the therapeutic efficacy of labeled stem cells after transplantation [57] To date, two iron oxide-based agents have been developed clinically and approved for MR imaging of the liver: ferumoxide (Endorem® in Europe, Feridex®in USA), developed by AMAG Pharma, with a particle size of 50 to 180 nm; and ferucarbotran (SHU555 A, Resovist®), developed by Schering AG, with a particle size of about 60 nm. Endorem®/Feridex® consists of SPIO nanoparticles coated with dextran, while Resovist® consists of SPIO nanoparticles coated with low molecular weight carboxydextran. With regard to the safety of these agents when injected I.V. for MRI of the liver, the USA package insert for Feridex® reports an overall incidence of adverse events of 11/2240 (0.5 %) of the patients who received Feridex I.V. These events include dyspnea, other respiratory symptoms, angioedema, generalized urticaria, and hypotension, which required treatment. For ferucarbotran, the overall incidence of adverse events was 7.1 % (75/1053 subjects), with vasodilatation and paraesthesia the most common event reported (< 2 %). Ferucarbotran was also found to have a unique ability to promote the cell growth of bone marrow-derived hMSCs by diminishing intracellular H2O2 [58]. Unfortunately, while these SPIO formulations have been used off-label for clinical studies, they are no longer manufactured [59].
To facilitate the internalization of SPIO contrast agents in cells, several strategies have been used, such as monoclonal antibody conjugation [50, 60], magnetodendrimers [51], transfection agents [61], and electroporation [62]. Limited in vitro stability and species specificity are major limitations with the use of monoclonal antibodies. An anaphylactic shock may also occur as a result of human immune response to foreign immunoglobulin. Magnetodendrimers, however, can increase the possibility of cell internalization without triggering an immune response. Embedding iron-oxide nanoparticles in carboxyl-terminated dendrimers increases the efficiency of internalization and accumulation of magnetodendrimers in cellular endosomes. These dendrimers did not affect the viability, differentiation, or proliferation capabilities of cells [51].
Another popular and currently most widely used method is the use of transfection agents. Here, poly-L-lysine or protamine sulfate are first multiplexed with SPIOs via electrostatic interactions to provide the formation of SPIO oligomers with a highly positive surface charge, which induces endocytosis of the transfection agent-conjugated SPIO [24, 63]. Because of particle clustering of SPIOs within endosomes, T2* effects are more pronounced, and, thus, the signal is hypointense [64]. This method does not affect the viability and proliferation of cells, but can inhibit the chondrogenic differentiation of human MSCs when poly-L-lysine is used [65].
To avoid the use of any additional agents with SPIOs, and to achieve an instant intracytoplasmatic magnetic labeling of cells, another method called magnetoelectroporation (MEP) can be used. In MEP, small-pulsed voltage is used to internalize SPIO in the cell [62]. This method does not require any transfection agent, which makes this method more attractive for clinical use. This method also provides the ability to label several million cells in a few seconds. This is especially helpful in case of tagging of cells that are prone to morphological changes due to adhesion to the plastic surface of tissue culture dishes. MEP does not affect the viability or proliferation of cells in vivo. Feridex®-labeled mouse neuronal stem cells maintained 98±2 % viability right after a single 130 V, 17-ms pulse MEP. These magnetoelectroporated cells were also able to maintain their proliferative capacity in vivo [66].
One of limitation of SPIO-labeling method is the extracellular deposition of the iron label following death of engrafted cells [67] or by active exocytosis [68]. When SPIO-labeled neural stem cells and human pluripotent stem cells were transplanted into mouse brain, rapid exocytosis of SPIO by live cells was observed as early as 48 h post-transplantation [68]. Exocytosed and deposited iron particles are scavenged by macrophages and may generate a false-positive signal on MRI, particular when these now indirectly labeled macrophages migrate away from the injection site. Nevertheless, direct labeling of cells SPIO is well suited for real-time image-guided cell delivery [59, 69] and for short-term monitoring of immediate cell engraftment.
Labeling with Micron-Sized Iron Oxide Particles
The size and number of iron particles greatly affect the MRI contrast enhancement. Micron-sized iron oxide (MPIO) particles have an approximately three-fold higher iron content per particle (≈100 pg) compared to SPIO. The use of MPIO can significantly increase the possibility of detecting smaller numbers of labeled cells or cells with fewer particles. Labeling of only one or a few MPIO particles per cell is sufficient to produce detectable signal attenuation within one imaging voxel. MPIOs have been used for MRI detection of single cells in vivo [70]. Despite the bigger size, internalization of these particles in cells is easy. Shapiro and colleagues reported that murine hepatocytes can readily endocytose MPIOs from 0.96 μm in diameter to as large as 5.80 μm without the need for lipofection [71]. Several types of cells have been labeled with MPIOs, including hematopoietic progenitor cells, MSCs [72], T cells [73], glioma cells [74], macrophages [75], adult neural progenitors [76], and dendritic cells [77]. The viability of all cells remained unchanged after MPIO labeling. The main limitation with the use of MPIOs, however, is that inert, non-biodegradable polymer polystyrene is used to construct MPIOs. Polystyrene is not FDA-approved for use in humans. To overcome this limitation Nkansah et al. developed and characterized biodegradable, superparamagnetic microparticles of poly(lactide-co-glycolide) (PLGA) and cellulose for cell labeling. Both polymers, commonly used for drug delivery and oral tablet formulations, are FDA-approved for use in humans. PLGA- and cellulose-based particles exhibited very high R2 molar relaxivity. Labeling of MSCs and human breast adenocarcinoma cells with these particles did not result in any detectable cytotoxicity [78]. As in the case of SPIO, the use of poly-L-lysine or coating of MPIOs with −NH2 improves the labeling efficiency of particles [79].
Labeling with 19F-Rich Perfluorocarbon Particles
In addition to 1H nuclei, several other nuclei, such as 31P, 23Na, 13C, and 19F, also exhibit MR phenomena. However, 31P, 23Na, 13C do not qualify for cell tracking studies because of their lower ratio of magnetic dipole moment to angular moment (gyromagnetic ratio) and abundance in the body. A natural absence of 19F atoms in the body and the mere 6 % difference in gyromagnetic ratio with 1H fluorine makes 19F MRI a promising tool for selective detection of labeled cells with MRI [80]. Perfluorocarbon (PFC) nanoparticles have been widely employed within the field of MRI and have been used for 19F MRI cell tracking of labeled progenitor, neuronal stem, immune, and dendritic cells [81–86]. These nanoparticles have little effect on viability or proliferation and differentiation of labeled cells. However, large particles (560 nm) have been found to alter the immunological properties of dendritic cells [87].
PFCs are the most biologically inert and stable C-F polymers. There are no known enzymes that metabolize PFCs in vivo, and PFCs do not degrade at typical lysosomal pH values. PFCs are water insoluble and do not attach to the cells very easily because of their “non-stick” property. For effective labeling of cells, PFCs must be formulated into a biocompatible nanoemulsion. PFC nanoemulsions can label a wide range of cell types, causing no toxicity or changes in phenotype, morphology, or function. Cells can be labeled by two methods using PFCs: (i) ex vivo labeling with PFC nanoemulsions. The labeled cells are tracked using 19F MRI post-transplantation as shown in Fig. 3. This method is called ‘in vivo cytometry’ [88] as the fluorine signal is linear to its concentration, allowing quantification of cell numbers. With this method, a wide variety of cells can be tracked in a cell-specific manner. (ii) Direct injection of PFC-nanoemulsions. The PFCs are taken up by cells of the reticuloendothelial system, including monocytes and macrophages, post-injection. This approach is mainly helpful in noninvasive visualization of inflammation [89] in different disease conditions and to monitor inflammatory responses associated with graft rejection.
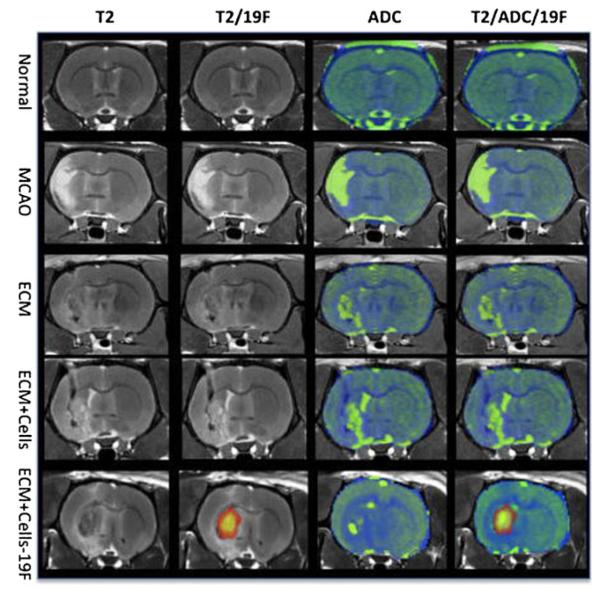
Fig. 3.
Ex vivo 19F and diffusion MR imaging. A middle cerebral artery occlusion induces a hyperintense signal on a T2-weighted MR image compared to a normal control. In the absence of 19F-labeled cells, no signal is detected on the 19F scan. The apparent diffusion coefficient (ADC) image shows an increase in diffusion in the area of stroke infarction. Injection of extracellular matrix (ECM) derived from decellularized tissue affects both the hyperintense T2-weighted signal and the diffusion scan. On the T2-weighted image, the lesion cavity almost completely disappeared, although the ADC image still exhibits areas of high diffusivity. Only 19F-labeled cells can be detected using the 19F channel. It is evident here that injection of the ECM and 19F F-labeled cells extensively cover the lesion cavity. Transplantation of unlabeled cells with the ECM did not significantly affect the ADC compared to injection of just ECM, indicating that the ECM was primarily responsible for significant changes in the lesion cavity’s characteristics. (Reproduced from Ref. 86, with permission)
A major disadvantage of 19F MRI is its low sensitivity. Cell labeling with PFCs generally results in a cell loading of 1011–1013 19F atoms per cell, with the levels higher in phagocytic than in non-phagocytic cells [80, 88, 90]. This uptake of 19F atoms per cell results in a minimum detection sensitivity of around 2,000 cells/voxel at 7 T; however, the detection limit of SPIO-labeled cells was shown to be 1,000 cells/mm3 at 3 T and 500 cells/mm3 at 7 T [91]. Another disadvantage of this technique is that imaging with PFCs can be about 10-fold slower than with other contrast agents [92]. A combination of pulse sequences with high SNR per image acquisition time may reduce the total imaging acquisition time. Further studies are required to improve image acquisition time in 19F MRI.
Labeling with MRI-Detectable pH Nanosensors
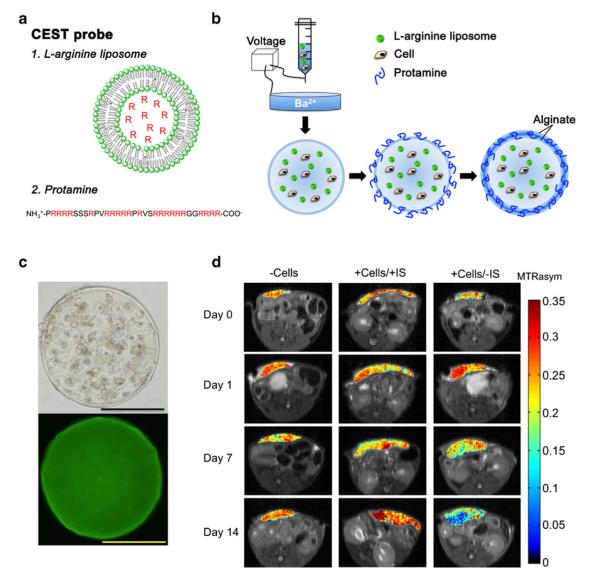
The assessment of cell death following in vivo transplantation of stem cells is challenging. A new technique for non-invasive monitoring of in vivo cell death in capsule hydrogels has been developed using MRI. In this technique, L-arginine, which is a molecule with multiple exchangeable NH protons, is used as a pH-sensitive chemical exchange saturation transfer (CEST) contrast agent. The proton exchange rate with water strongly depends on the pH, and when pH decreases due to cell death, the exchange rate decreases for base-catalyzed guanidyl NH exchangeable protons in L-arginine, leading to decreases in CEST contrast [93]. Arginine-rich LipoCEST microcapsules were synthesized by incorporating an L-arginine-filled liposome inside the capsule and using protamine sulfate as an arginine-rich cross-linker in the capsule coating. It was demonstrated that apoptotic encapsulated hepatocytes can be detected with CEST MRI in vivo (Fig. 4). As these nanosensors are clinical-grade, this technique should be translatable to the clinic.
Fig. 4.
Use of LipoCEST microcapsules for the detection of apoptotic encapsulated hepatocytes by MRI. a The L-arginine (R) moieties (red) entrapped in liposomes (1) and in protamine (2) have exchangeable protons that provide CEST contrast. b A mixture of cells, alginate and L-arginine liposomes is passed through a needle using a nanoinjector pump. The single-layered charged alginate droplets are collected, washed and resuspended in a crosslinker solution (protamine sulfate), followed by a coating with a second layer of alginate. c Phase-contrast image (top) and fluorescent image (bottom; with NBD-PE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl))-labelled liposomes) showing a uniform distribution of cells and liposomes within the microcapsule. Scale bar=200 μm. d BALB/c mice were subcutaneously transplanted with 2,500 empty LipoCEST capsules (−Cells), with LipoCEST capsules containing Luc-transfected hepatocytes while receiving immunosuppression (+Cells/+IS), and with capsules containing cells but no immunosuppression (+Cells/−IS). CEST/MTw (magnetization transfer-weighted) overlays using a frequency offset of 2 ppm at days 0, 1, 7, and 14 after transplantation. (Reproduced from Ref. 93, with permission)
Direct Labeling for PET and SPECT
The nuclear imaging modalities PET and SPECT provide images of the in vivo distribution of administered radionuclides. These modalities have better clinical potential in that they have superior tissue penetration capability and are more highly quantitative than optical imaging [94]. PET and SPECT mainly differ in the radionuclide used and the mechanism for signal generation and detection. For cell labeling and tracking, different radionuclides, such as 64Cu-PTSM and 18F-FDG (for PET), and 111In-oxine, 99mTc- HMPAO (for SPECT), are widely used [94–96]. Due to their nanomolar sensitivity (PET; 10−11–10−12 M and SPECT; 10−10 M), these radionuclides are able to measure biological processes at non-pharmacological doses. For cell labeling, these radionuclides are introduced into the cells by a lipophilic chelator and subsequently bind to intracellular proteins, whereas the chelator leaves the cells. Cells are simply incubated with radio-isotopes in vitro, washed, and transplanted. Labeling efficiency varies from 60 to 90 %, depending upon the labeling condition and cell type [97, 98]. 111In and 99mTc are used as the “gold-standard” clinical procedure for the detection of sites of occult infections and inflammation by imaging the leukocyte distribution [99, 100]. 111In-oxine was approved by the FDA in 1985 for white blood cell (WBC) scintigraphy, and among the several options, has been used most extensively for stem cell labeling due to its relatively long-decay half-life of 2.8 days [101]. In a study in patients with advanced cirrhosis, intravenously administered MSCs labeled with 111In-oxine could be detected up to day 10-post injection [102]. This radionuclide usually does not have any adverse effect on cell viability or function; however, human MSCs have shown moderate functional impairment after labeling [103]. But, 111In-oxine labeling does not affect the stemness of those cells [104]. 99mTc-HMPAO is another clinically used tracer that has a different biodistribution than 111In-oxine 99. 99mTc-HMPAO-labeled, bone marrow-derived MSCs were visualized up to 4 h after cell transplantation in a rat model of myocardial infarction [105]. However, the trade-off between half-life and long-term exposure to ionizing radiation and a possible transfer of radiometal to other non-stem cells are the major limitations of using 99mTc-HMPAO for cell tracking. Similarly 18F-FDG may also be used for cell labeling, but the disadvantage of the short half-life of 18F (only 2 h) makes this tracer less than ideal for follow-up studies [106].
The detachment and efflux of radioisotopes into the extracellular space are obvious disadvantages with this method. For example, 111In-oxine showed a significant efflux within 24 h after labeling [107]. This may cause an inaccurate estimation of the number of labeled cells. However, detached radiotracers are not retained in body for a long period, so pose no apparent health risk [108].
Labeling by Microencapsulation
Prevention of transplanted cells from host immune attack and avoidance of immunosuppression by encapsulating cells with biomaterials, such as an alginate hydrogel, are the primary goals of microencapsulation. Later, it was realized that different contrast agents could also be incorporated in the microcapsule [109, 110]. MRI-visible ‘magnetocapsules’ were made by modifying the alginate–poly-L-lysine–alginate microencapsulation method to include SPIO. Infusion of these ‘magnetocapsules’ through the portal vein of the liver enabled MR–guided, real-time, targeted delivery and imaging of pancreatic islet cells [69], as well as MRI visualization of encapsulated hepatocytes engrafted within the abdomen [111]. Simultaneous immunoprotection and trimodal, non-invasive monitoring of cell engraftment could be achieved by co-encapsulating gold nanoparticles functionalized with Gd-DTDTPA with pancreatic islet cells using protamine sulfate as a clinical-grade alginate cross-linker. Encapsulated cells were readily visualized with high-field MRI, micro-computed tomography (μ-CT), and ultrasound imaging, both in vitro and in vivo [112]. Other examples include incorporation of barium or bismuth sulfate for X-ray imaging [113], perfluorocarbons for 19F MRI [114], and trimodal capsule-in capsules [109].
Labeling with Gold and Multimodal Gold and Silica Nanoparticles
An alternative approach for non-invasive stem cell tracking, with high spatial and temporal resolution at sufficient depths, is the use of gold and silica nanoparticles. These nanoparticles also provide an advantage over radionuclides, as they can be used for long-term imaging and allow for repeated imaging over time. Gold nanoparticles or nanotracers possess optically tunable properties and inert characteristics. Nam and colleagues used gold nanotracers to trace MSCs in vivo with CT and ultrasound-guided photoacoustic (US/PA) imaging [115]. Gold nanoparticles provide sufficient radiopacity to be detected, and MSCs have been successfully labeled and detected by CT follow their ex vivo labeling. Loading MSCs with gold nanotracers was found to have no adverse effects on cell function [116]. However, due to the lack of sensitivity and low contrast-to-noise, CT imaging of stem cells has been pursued in a limited fashion. This has also provided an alternative way for their detection by US/PA imaging [115].
Similarly, silica nanoparticles (SiNPs) can also be used as an alternative to MRI-based stem cell imaging. The use of SiNPs for cell tracking is particularly attractive because these particles can be detected by fluorescent, MR, and ultrasound imaging. Jokerst et al. reported that silica-based nanoparticles can be used for cell sorting (fluorescence), real-time guided cell implantation with ultrasound, and high-resolution, long-term monitoring by MRI. They found that SiNPs labeling could increase the ultrasound and MRI contrast of labeled hMSCs 700% and 200% versus unlabeled cells, respectively, and allowed cell imaging in animal models for 13 days after implantation. The SiNPs were found to have no effect on the proliferation or pluripotency of hMSCs [117].
Labeling with Microbubbles
Another approach for non-invasive cell tracking is the use of gas-filled microbubbles (MBs)-based contrast agents. These contrast agents are clinically approved and have been successfully used to label and track stem cells by ultrasound (US) imaging [118]. Because of the strong signal induced by MBs, a large acoustic impedance mismatch with tissues, a single MB and thus a single cell can be detected with this imaging modality [118]. However, the use of US imaging is still in its infancy, and further studies will have to demonstrate its overall applicability in cell tracking.
Indirect Labeling Methods
In this method, target cells are genetically manipulated to introduce a reporter gene. The reporter gene is translated into an enzyme, a receptor, or a fluorescent or bioluminescent protein. The interaction between the substrate and the encoded reporter protein will result in a signal that can be detected noninvasively. Indirect labeling methods have a clear advantage over direct labeling methods in terms of assessment of cell viability and division. The biggest limitation of any direct labeling method is that it does not provide accurate information about cell viability. In addition, cell numbers increase after every cell division, but the numbers of labeled probes remain the same. This results in “dilution” of the signal over time. Since reporter genes can pass on to the progenies after cell division, imaging of dividing cells is possible. In addition, in contrast to direct methods, where a probe is present and detectable even after cell death, a reporter gene does not continue to be expressed after cell death, and thus, produces no signal. However, epigenetic gene silencing may affect the expression of a reporter gene and suppress the signal. This limitation can be overcome by treating target cells with DNA methyltransferase inhibitor.
Reporter genes are currently mainly used for ex vivo studies (beta-galactosidase and alkaline phosphatase) to assess the trans-gene expression in cells, tissues, and organs [119, 120] by histology. The use of firefly luciferase (for bioluminescent imaging) and green fluorescent protein (for fluorescent imaging) genes are initial evidence that reporter genes can be used to monitor trans-gene expression in living organisms [121, 122]. Just like direct labeling methods, indirect labeling methods can also be used with several different imaging modalities.
Reporter Genes for Optical Imaging
Optical imaging techniques using reporter genes can be separated into two types: (i) fluorescence and (ii) bioluminescence. Several reporter gene strategies are based on fluorochromes, e.g., green fluorescent protein (GFP- the first fluorescent protein gene isolated from Aequorea victoria jelly fish), mutants of the GFP gene emitting blue, cyan, or yellow light, and fluorochromes with red and far-red spectra isolated from different species [123, 124], have been in use for many years. However, a number of issues have been identified with in vivo optical fluorescent imaging. As mentioned earlier in this review, excitation and emission wavelengths of fluorochromes have limited penetration in tissues and a poor signal- to-noise ratio limits the use of fluorochromes in vivo, particularly in deep tissues. Novel technologies, such as diffuse optical tomography and optical coherence tomography, may overcome these problems; however, their current use is limited to small animal studies, and further development is needed to transfer these technologies to clinical settings.
In contrast to fluorescence imaging, where an external light source excites the fluorochrome, bioluminescence imaging (BLI) is based on the emission of photons in reactions catalyzed by luciferase enzymes. Luciferases emit photons during the oxidation of a substrate, such as D-luciferin, in the presence of oxygen and ATP. The most commonly utilized luciferases for in vivo imaging are Firefly (isolated from Photinus pyralis, the North American firefly) and Renilla (isolated from Renilla reniformis, the click beetle) luciferases. The Firefly and Renilla luciferase reporter systems, in combination with their corresponding luminescent substrates, luciferin and coelenterazine, have several advantages for small animal imaging. There is no cross-reactivity between luciferin and coelenterazine substrates, so both luciferases can be imaged simultaneously. BLI has been extensively used for the noninvasive in vivo tracking of transplanted stem cells, including embryonic [125, 126] and neural stem cells [127], in small animals. Target cells can be stably transfected with luciferases gene and be tracked for very long time. This gives an edge to this technique for long-term sequential studies. For example, a regenerative cell population derived from human subcutaneous adipose tissue (hASCs), implanted into the peri-infarct region of a mouse model of myocardial infarction, could be tracked over a period of 10 weeks [128]. This is because BL signal generation is ATP-dependent and BLI can be used to monitor cell survival post-transplantation. Transplantation site-dependent survival of neuronal progenitor allografts in the mouse brain [129] and neuronal progenitor cell survival within hydrogels can be determined in vivo by BLI (Fig. 5) [130]. Recently, BLI has also been used for the non-invasive imaging of disease progression in muscular dystrophy. Maguire and colleagues monitored luciferase activity in mice expressing an inducible luciferase reporter gene in muscle stem cells. These stem cells proliferated in response to muscle degeneration, thus increasing the level of luciferase expression in dystrophic muscle [131].
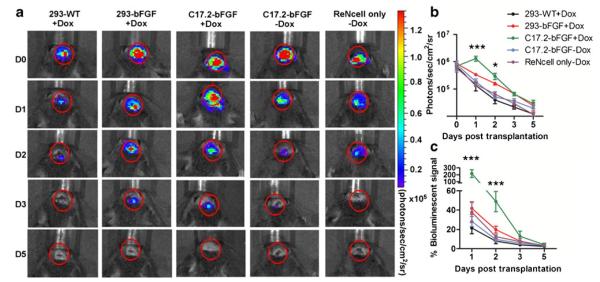
Fig. 5.
Detection of in vivo survival of neuronal progenitor cell by BLI (a). BLI of representative animals transplanted with 1×105 neuronal progenitor cell (ReNcells) with or without helper cells into Rag2−/− immunodeficient mice show strong BLI signal on the same day after transplantation, which decreases progressively over the following days. b Quantification of BLI signal for each group (n =5 each), with the intensity expressed as photon/s/cm2/sr. c Estimation of percent donor cell survival plotted as % signal activity (normalized to day 0) over the 5-day period following transplantation. (Reproduced from Ref. 130, with permission)
One of the major limitations of in vivo BLI is light absorbance from hemoglobin, particularly with a high blood-to-tissue ratio where maximal emission overlaps with the maximal absorption of hemoglobin, and selectively transports the substrate back to the blood system. Multi-layer anatomical barriers also limit the emission. The photon emission per cell can be maximized by using a strong promoter or by using luciferase with a red-shifted spectrum to overcome some of the limitations [132]. Physiological parameters, such as anesthesia [133] and the route of substrate delivery (i.p. vs. subcutaneous) [134], also affect signal intensity. Volatile anesthetics, such as isoflurane, sevoflurane and desflurane, have an inhibitory effect on the luciferase activity in rodents. Pentobarbital was found to have a less inhibitory effect. The possible reasons for this inhibitory effect might be attributable to the hemodynamic effects of anesthetics [135] and the binding of anesthetics to the domain that regulates the opening and closing of the enzymatic pocket, and to the inhibition of the binding of D-luciferin to luciferase [136]. Concentrations of serum proteins may also have negative effects on BLI signal intensity. For example, a hypoalbuminemic condition, where levels of albumin in blood serum are abnormally low, is associated with a higher BLI signal intensity [137].
Reporter Genes for MRI
High spatial resolution and the ability to gather accurate anatomical and physiological information simultaneously are two of the biggest advantages of MRI reporter gene imaging. In addition, unlike the optical reporter gene imaging approach where there is a limit on light tissue penetration, there is no limit on the size of the subject to be imaged as long as it fits into the magnet. In the past decade, several MRI reporter genes have been developed and used in neurological, cardiac, and cancer research [138–141]. For MRI reporter gene imaging, cells are genetically modified to either increase their affinity for a contrast agent, or produce iron-containing proteins, or provide an endogenous contrast agent. Based on the mode of action, MRI reporter gene imaging can be mainly divided into the following types: enzyme-based; iron-based; and chemical exchange saturation transfer (CEST)-based.
Enzyme-Based MRI Reporter Genes
Louie and colleagues pioneered the enzyme-based MRI approach by developing a gadolinium-based substrate (EgadMe) that contains a galactose group. In the presence of lacZ-transfected cells expressing β-galactosidase, the galactopyranose moiety is cleaved, which allows increased water molecule diffusion to the gadolinium, thus increasing the T1 signal [142]. More recently, lacZ-transfected tumor cells, combined with 3,4-cyclohexenoesculetinb-D-galactopyranoside and iron, resulted in T2* relaxation on MRI [143]. Another example of the enzyme-based approach is genetically manipulated cells overexpressing tyrosinase. Tyrosinase is a rate-limiting enzyme that controls the production of melanin. Melanin binds paramagnetic iron ions to produce metallomelanin, and thus, cells overexpressing tyrosinase exhibit high signal intensity on T1-weighted MRI [144]. Some of the potential pitfalls of this approach are tissue delivery barriers, false MR signals due to the presence of “leftover” galactose, or persistence of metallomelanin in the cells, even when the reporter gene is not activated [145].
Iron-Based MRI Reporter Genes
Genetically engineered iron binding proteins, other than metallomelanin, have also extensively been studied for reporter gene MRI. The engineered transferrin receptor (ETR) generates contrast by the receptor-mediated internalization of iron-bound transferrin [146]. However, overexpression of etR could lead to iron-catalyzed free radical formation via the Fenton reaction, and could be toxic to cells [147]. Overexpression of the ubiquitous protein ferritin, which stores and releases the iron to maintain iron homeostasis, traps excess intracellular iron and causes significant shortening of the T2 relaxation time [148]. Recent studies showed that a ferritin reporter gene could be used to track stem/progenitor cells in vivo [149, 150]. Differences in tissue type, molecular aggregation, strength of the MR field, and quantification methods may affect the ferritin-related relaxation rate [151].
Intracellular particles of biogenic magnetite (Fe3O4), synthesized by the MagA gene (a putative iron transporter), found in some fresh-water magnetotactic bacteria of the genus Magnetospirillum sp., also have properties similar to that of SPIO nanoparticles and can also be used as MRI reporter genes. It was observed that MagA-positive cells show a significant signal drop on T2*-weighted MRI [152, 153].
Chemical-Exchange Saturation Transfer (CEST)-Based MRI Reporter Genes
This class of MR reporter genes utilize a process called chemical-exchange saturation transfer (CEST). In CEST, applying a saturation radiofrequency (RF) pulse at the exchangeable proton resonance frequency for a long duration saturates the proton’s magnetization, creating a chemical exchange. Since these protons constantly exchange with bulk water protons, they can be detected as a reduction in the water proton MR signal. Our group has designed a non-metallic, biodegradable, lysine-rich protein (LRP) reporter (containing high-density amide protons), which can be successfully used as a MR CEST reporter [138]. The major advantages of CEST contrast agents are: (i) the CEST contrast is switchable. This contrast is only detectable when a saturation pulse is applied at a specific frequency characteristic of an agent’s exchangeable protons. This unique feature allows the CEST contrast to be undeletable when a saturation pulse is switched off. Thus, the CEST contrast does not interfere with other MRI contrasts. (ii) The ability to create multiple colors by using a saturation pulse at different frequencies. This may allow simultaneous MR imaging of more than two target cells [139, 154].
Reporter Genes for Radionuclide Imaging
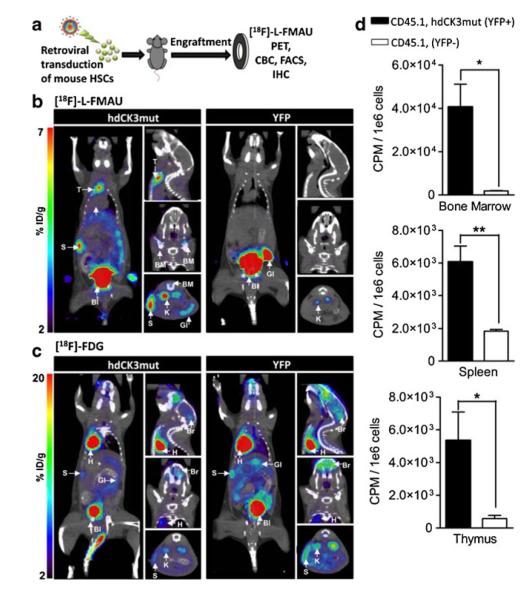
Radionuclide reporter genes encode for receptors or transporters that promote the uptake or accumulation of radiolabeled tracers in target cells. Reporter genes are transferred to the target cells via viral or non-viral methods. Herpes simplex virus thymidine kinase type 1 (HSV1-tk) is the most commonly used radionuclide reporter gene. Thymidine kinase (TK) adds a negative charge to the cell surface by phosphorylating radiolabeled nucleoside substrates, and thereby prevents the radiolabeled tracer from exiting the cell. Thus, the tracer accumulates in the cell [155]. HSV1-tk has been used to track tumor-specific lymphocytes [156], T-cell activation [157], bone marrow MSCs [158], and hESCs [159]. However, as HSV1-tk is a non-human gene, it poses the threat of generating an immune response against the cells. This immunogenicity has prevented the routine use of PET reporter genes clinically [160]. To avoid immunogenicity, the human nucleoside kinases deoxycytidine kinase (dCK) and thymidine kinase 2 (TK2) have been used. Both human kinases have a substrate specificity similar to HSV1-tk [161]. These reporter genes have been successfully used in mouse models [162, 163] and a tumor patient [164]. A mutant of dCK (hdCK3mut) with three amino acid substitutions within the active site has been used with the PET probe 18F-L-FMAU to track mouse and human hematopoietic stem cells after transplantation for long-term observation (Fig. 6) [165]. Another disadvantage of radionuclide imaging is the inability of certain tracers to cross the blood–brain barrier. This limits the ability to track cells transplanted intracerebrally. The use of a somatostatin receptor, with its ligand, 111In-octreotide [47], a sodium-iodine symporter [166], or dopamine receptors [167], can be used improve the uptake of tracers. These reporter genes have been used to track neural stem cells [168] and cardiac stem cells [169], and also to monitor cellular differentiation [170]. An interesting novel approach has transformed HSV1-tk into an MRI reporter gene. To this end, nucleoside analogues were synthesized that are rich in imino protons. Upon accumulation of this substrate, that acts as a CEST contrast agent, implanted glioma cells could be tracked in vivo by MRI [171].
Fig. 6.
Radionuclide imaging using reporter genes. a Lethally irradiated C57BL/6 (CD45.2) mice were transplanted with retrovirally transduced 5-FU–enriched HSCs (CD45.1). Animals were monitored for hematopoietic reconstitution over their total lifespan. MicroPET scans shown from Left, coronal; Right Upper, sagittal; Center, coronal; and Lower, transverse. b [18F]-L-FMAU at 4 week post-transplant. Reporter signal observed in hdCK3mut animals in spleen (S), thymus (T), and bone marrow (BM). Probe metabolism in both cohorts seen in gastrointestinal (GI), bladder (Bl), and kidney (K). c [18F]-FDG MicroPET scan at 4 week post-transplant. Signal unrelated to the reporter was sen in both cohorts in heart (H), spleen (S), gastrointestinal (GI), brain (Br), with metabolism in kidneys (K) and bladder (Bl). d In vivo accumulation of [18F]-L-FMAU in sorted hematopoietic cells from hdCK3mut animals. Reporter positive: CD45.1+, YFP+ and reporter negative: CD45.1+, YFP−. (P <0.05). (Reproduced from Ref. 165, with permission)
Conclusions
Stem cell-based therapies promises a new mode of care that has the potential to greatly reduce the burden of disease. Imaging technologies have become a driving force in stem cell research. Extraordinary efforts are currently being made to improve cellular imaging techniques. The biocompatibility of molecular probes, temporal resolution, detection thresholds, need for real-time image-guided injection, and cost effectiveness are a few important criteria when selecting the appropriate imaging modality for cellular imaging. While optical imaging techniques have demonstrated a clear advantage in imaging small animals and are firmly established in preclinical studies, the use of these techniques is not practically feasible in bigger animals and humans, given the poor tissue penetration and low spatial resolution of signals. The use of MRI and PET/SPECT in cell tracking has an advantage over optical imaging in terms of higher resolution and sensitivity. However, these techniques have their own advantages and disadvantages. The first clinical MRI cell-tracking study on humans demonstrated the key advantages of this technique, being that anatomical information is provided [172], dictating the need for monitoring the accuracy of cell injections, preferable in real-time [59]. PET, however, may possess more sensitivity, but suffers from low spatial resolution and lack of anatomical information. Therefore, employing multimodal imaging techniques with double- or triple-reporter systems for optical, MR, and PET imaging will greatly facilitate the transition of cellular imaging techniques from the bench to the bed.
Footnotes
Conflict of interest The authors declare no conflict of interest.
Contributor Information
Amit K. Srivastava, Russell H. Morgan Department of Radiology and Radiological Science, Division of MR Research, The Johns Hopkins University School of Medicine, 217 Traylor Building, 720 Rutland Avenue, Baltimore, MD 21205-1832, USA; Cellular Imaging Section, Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD 21205-1832, USA
Jeff W. M. Bulte, Russell H. Morgan Department of Radiology and Radiological Science, Division of MR Research, The Johns Hopkins University School of Medicine, 217 Traylor Building, 720 Rutland Avenue, Baltimore, MD 21205-1832, USA; Cellular Imaging Section, Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD 21205-1832, USA
References
- 1.Jacobson LO, Simmons EL, Bethard WF. Studies on hematopoietic recovery from radiation injury. The Journal of Clinical Investigation. 1950;29(6):825. [PubMed] [Google Scholar]
- 2.Simmons EL, Jacobson LO, Marks EK, Gaston EO. Long-term survival of irradiated mice treated with homologous tissue suspensions. Nature. 1959;183(4660):556. doi: 10.1038/183556a0. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson LO, Simmons EL. Comparison of the effects of isologous, homologous, and heterologous hematopoietic tissues on post-irradiation survival. Radiology. 1960;75:6–10. doi: 10.1148/75.1.6. [DOI] [PubMed] [Google Scholar]
- 4.Barnes DW, Loutit JF. Protective effects of implants of splenic tissue. Proceedings of the Royal Society of Medicine. 1953;46(4):251–252. [PubMed] [Google Scholar]
- 5.Main JM, Prehn RT. Fate of skin homografts in xirradiated mice treated with homologous marrow. Journal of the National Cancer Institute. 1957;19(6):1053–1064. [PubMed] [Google Scholar]
- 6.Thomas ED, Lochte HL, Jr., Cannon JH, Sahler OD, Ferrebee JW. Supralethal whole body irradiation and isologous marrow transplantation in man. The Journal of Clinical Investigation. 1959;38:1709–1716. doi: 10.1172/JCI103949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatzistergos KE, Blum A, Ince T, Grichnik JM, Hare JM. What is the oncologic risk of stem cell treatment for heart disease? Circulation Research. 2011;108(11):1300–1303. doi: 10.1161/CIRCRESAHA.111.246611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocha V, Wagner JE, Jr., Sobocinski KA, et al. Graft-versus-host disease in children who have received a cord-blood or bone marrow transplant from an HLA-identical sibling. Eurocord and International Bone Marrow Transplant Registry Working Committee on Alternative Donor and Stem Cell Sources. The New England Journal of Medicine. 2000;342(25):1846–1854. doi: 10.1056/NEJM200006223422501. [DOI] [PubMed] [Google Scholar]
- 9.Nair G, Tanahashi Y, Low HP, Billings-Gagliardi S, Schwartz WJ, Duong TQ. Myelination and long diffusion times alter diffusion-tensor-imaging contrast in myelin-deficient shiverer mice. NeuroImage. 2005;28(1):165–174. doi: 10.1016/j.neuroimage.2005.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickers C, Gallegos R, Seethamraju RT, et al. Applications of magnetic resonance imaging for cardiac stem cell therapy. Journal of Interventional Cardiology. 2004;17(1):37–46. doi: 10.1111/j.1540-8183.2004.01712.x. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Zhu J, Baeslack J, et al. Longitudinal PET imaging for monitoring myelin repair in the spinal cord. Annals of Neurology. 2013 doi: 10.1002/ana.23965. doi:10.1002/ana.23965. [DOI] [PubMed] [Google Scholar]
- 12.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nature Medicine. 2004;10(9):993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 13.Lei Y, Tang H, Yao L, Yu R, Feng M, Zou B. Applications of mesenchymal stem cells labeled with Tat peptide conjugated quantum dots to cell tracking in mouse body. Bioconjugate Chemistry. 2008;19(2):421–427. doi: 10.1021/bc0700685. [DOI] [PubMed] [Google Scholar]
- 14.Ohyabu Y, Kaul Z, Yoshioka T, et al. Stable and nondisruptive in vitro/in vivo labeling of mesenchymal stem cells by internalizing quantum dots. Human Gene Therapy. 2009;20(3):217–224. doi: 10.1089/hum.2008.100. [DOI] [PubMed] [Google Scholar]
- 15.Sugiyama T, Kuroda S, Osanai T, et al. Near-infrared fluorescence labeling allows noninvasive tracking of bone marrow stromal cells transplanted into rat infarct brain. Neurosurgery. 2011;68(4):1036–1047. doi: 10.1227/NEU.0b013e318208f891. [DOI] [PubMed] [Google Scholar]
- 16.Rak-Raszewska A, Marcello M, Kenny S, Edgar D, See V, Murray P. Quantum dots do not affect the behaviour of mouse embryonic stem cells and kidney stem cells and are suitable for short-term tracking. PLoS One. 2012;7(3):e32650. doi: 10.1371/journal.pone.0032650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li K, Qin W, Ding D, et al. Photostable fluorescent organic dots with aggregation-induced emission (AIE dots) for noninvasive long-term cell tracing. Scientific Reports. 2013;3:1150. doi: 10.1038/srep01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenblatter M, Ehrchen J, Varga G, et al. In vivo optical imaging of cellular inflammatory response in granuloma formation using fluorescence-labeled macrophages. Journal of Nuclear Medicine. 2009;50(10):1676–1682. doi: 10.2967/jnumed.108.060707. [DOI] [PubMed] [Google Scholar]
- 19.Ruan J, Song H, Li C, et al. DiR-labeled embryonic stem cells for targeted imaging of in vivo gastric cancer cells. Theranostics. 2012;2(6):618–628. doi: 10.7150/thno.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan L. Molecular Imaging and Contrast Agent Database (MICAD) (Internet) National Centre For Biotechnology Information (US); Bethsda (MD): 2004. Near-infrared fluorescence 1,1-dioctadecyl-3,3,3,3-tetramethylindotricarbocyanine iodide (DiR)-labeled macrophages for cell imaging; pp. 2004–2013. [PubMed] [Google Scholar]
- 21.Frangioni JV. In vivo near-infrared fluorescence imaging. Current Opinion in Chemical Biology. 2003;7(1):626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Sykova E, Jendelova P. Migration, fate and in vivo imaging of adult stem cells in the CNS. Cell Death and Differentiation. 2007;14(7):1336–1342. doi: 10.1038/sj.cdd.4402140. [DOI] [PubMed] [Google Scholar]
- 23.Bulte JW, Hoekstra Y, Kamman RL, et al. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228(2):480–487. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 24.Frank JA, Miller BR, Arbab AS, et al. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228(2):480–487. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 25.Hedlund A, Ahren M, Gustafsson H, et al. Gd(2)O(3) nanoparticles in hematopoietic cells for MRI contrast enhancement. International Journal of Nanomedicine. 2011;6:3233–3240. doi: 10.2147/IJN.S23940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agudelo CA, Tachibana Y, Hurtado AF, Ose T, Iida H, Yamaoka T. The use of magnetic resonance cell tracking to monitor endothelial progenitor cells in a rat hindlimb ischemic model. Biomaterials. 2012;33(8):2439–2448. doi: 10.1016/j.biomaterials.2011.11.075. [DOI] [PubMed] [Google Scholar]
- 27.Guenoun J, Koning GA, Doeswijk G, et al. Cationic Gd-DTPA liposomes for highly efficient labeling of mesenchymal stem cells and cell tracking with MRI. Cell Transplantation. 2012;21(1):191–205. doi: 10.3727/096368911X593118. [DOI] [PubMed] [Google Scholar]
- 28.Modo M, Beech JS, Meade TJ, Williams SC, Price J. A chronic 1 year assessment of MRI contrast agent-labelled neural stem cell transplants in stroke. NeuroImage. 2009;47(Suppl 2):T133–T142. doi: 10.1016/j.neuroimage.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudelius M, Daldrup-Link HE, Heinzmann U, et al. Highly efficient paramagnetic labelling of embryonic and neuronal stem cells. European Journal of Nuclear Medicine and Molecular Imaging. 2003;30(7):1038–1044. doi: 10.1007/s00259-002-1110-0. [DOI] [PubMed] [Google Scholar]
- 30.Bhorade R, Weissleder R, Nakakoshi T, Moore A, Tung CH. Macrocyclic chelators with paramagnetic cations are internalized into mammalian cells via a HIV-tat derived membrane translocation peptide. Bioconjugate Chemistry. 2000;11(3):301–305. doi: 10.1021/bc990168d. [DOI] [PubMed] [Google Scholar]
- 31.Tseng CL, Shih IL, Stobinski L, Lin FH. Gadolinium hexanedione nanoparticles for stem cell labeling and tracking via magnetic resonance imaging. Biomaterials. 2010;31(20):5427–5435. doi: 10.1016/j.biomaterials.2010.03.049. [DOI] [PubMed] [Google Scholar]
- 32.Ghaghada KB, Ravoori M, Sabapathy D, Bankson J, Kundra V, Annapragada A. New dual mode gadolinium nano-particle contrast agent for magnetic resonance imaging. PLoS One. 2009;4(10):e7628. doi: 10.1371/journal.pone.0007628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klasson A, Ahren M, Hellqvist E, et al. Positive MRI contrast enhancement in THP-1 cells with Gd2O3 nanoparticles. Contrast Media & Molecular Imaging. 2008;3(3):106–111. doi: 10.1002/cmmi.236. [DOI] [PubMed] [Google Scholar]
- 34.Faucher L, Tremblay M, Lagueux J, Gossuin Y, Fortin MA. Rapid synthesis of PEGylated ultrasmall gadolinium oxide nanoparticles for cell labeling and tracking with MRI. ACS Applied Materials & Interfaces. 2012;4(9):4506–4515. doi: 10.1021/am3006466. [DOI] [PubMed] [Google Scholar]
- 35.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) Journal of Magnetic Resonance. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 36.Aime S, Carrera C, Delli Castelli D, Geninatti Crich S, Terreno E. Tunable imaging of cells labeled with MRI-PARACEST agents. Angewandte Chemie International Edition England. 2005;44(12):1813–1815. doi: 10.1002/anie.200462566. [DOI] [PubMed] [Google Scholar]
- 37.Ferrauto G, Castelli DD, Terreno E, Aime S. In vivo MRI visualization of different cell populations labeled with PARACEST agents. Magnetic Resonance in Medicine. 2013;69(6):1703–1711. doi: 10.1002/mrm.24411. [DOI] [PubMed] [Google Scholar]
- 38.Silva AC, Bock NA. Manganese-enhanced MRI: an exceptional tool in translational neuroimaging. Schizophrenia Bulletin. 2008;34(4):595–604. doi: 10.1093/schbul/sbn056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilad AA, Walczak P, McMahon MT, et al. MR tracking of transplanted cells with “positive contrast” using manganese oxide nanoparticles. Magnetic Resonance in Medicine. 2008;60(1):1–7. doi: 10.1002/mrm.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim T, Momin E, Choi J, et al. Mesoporous silica-coated hollow manganese oxide nanoparticles as positive T1 contrast agents for labeling and MRI tracking of adipose-derived mesenchymal stem cells. Journal of the American Chemical Society. 2011;133(9):2955–2961. doi: 10.1021/ja1084095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Josephson L, Lewis J, Jacobs P, Hahn PF, Stark DD. The effects of iron oxides on proton relaxivity. Magnetic Resonance Imaging. 1988;6(6):647–6453. doi: 10.1016/0730-725x(88)90088-4. [DOI] [PubMed] [Google Scholar]
- 42.Shen T, Weissleder R, Papisov M, Bogdanov A, Jr., Brady TJ. Monocrystalline iron oxide nanocompounds (MION): physicochemical properties. Magnetic Resonance in Medicine. 1993;29(5):599–604. doi: 10.1002/mrm.1910290504. [DOI] [PubMed] [Google Scholar]
- 43.Jung CW. Surface properties of superparamagnetic iron oxide MR contrast agents: ferumoxides, ferumoxtran, ferumoxsil. Magnetic Resonance Imaging. 1995;13(5):675–691. doi: 10.1016/0730-725x(95)00023-a. [DOI] [PubMed] [Google Scholar]
- 44.Wagner S, Schnorr J, Pilgrimm H, Hamm B, Taupitz M. Monomer-coated very small superparamagnetic iron oxide particles as contrast medium for magnetic resonance imaging: pre-clinical in vivo characterization. Investigative Radiology. 2002;37(4):167–177. doi: 10.1097/00004424-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(30):10901–10906. doi: 10.1073/pnas.0403918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao R, Xing R, Xu Z, Hou Y, Gao S, Sun S. Synthesis, functionalization, and biomedical applications of multifunctional magnetic nanoparticles. Advanced Materials. 2010;22(25):2729–2742. doi: 10.1002/adma.201000260. [DOI] [PubMed] [Google Scholar]
- 47.Rogers WJ, Meyer CH, Kramer CM. Technology insight: in vivo cell tracking by use of MRI. Nature clinical practice. Cardiovascular Medicine. 2006;3(10):554–562. doi: 10.1038/ncpcardio0659. [DOI] [PubMed] [Google Scholar]
- 48.Norman AB, Thomas SR, Pratt RG, Lu SY, Norgren RB. Magnetic resonance imaging of neural transplants in rat brain using a superparamagnetic contrast agent. Brain Research. 1992;594(2):279–283. doi: 10.1016/0006-8993(92)91135-2. [DOI] [PubMed] [Google Scholar]
- 49.Bulte JW, Brooks RA, Moskowitz BM, Bryant LH, Jr., Frank JA. T1 and T2 relaxometry of monocrystalline iron oxide nanoparticles (MION-46L): theory and experiment. Academic Radiology. 1998;5(Suppl 1):S137–S140. doi: 10.1016/s1076-6332(98)80084-6. [DOI] [PubMed] [Google Scholar]
- 50.Bulte JW, Zhang S, van Gelderen P, et al. Neurotransplantation of magnetically labeled oligodendrocyte progenitors: magnetic resonance tracking of cell migration and myelination. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):15256–15261. doi: 10.1073/pnas.96.26.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bulte JW, Douglas T, Witwer B, et al. Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nature Biotechnology. 2001;19(12):1141–1147. doi: 10.1038/nbt1201-1141. [DOI] [PubMed] [Google Scholar]
- 52.Karussis D, Karageorgiou C, Vaknin-Dembinsky A, et al. Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Archives of Neurology. 2010;67(10):1187–1194. doi: 10.1001/archneurol.2010.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Wang L, Che Y, Li Z, Kong D. Preparation and evaluation of magnetic nanoparticles for cell labeling. Journal of Nanoscience and Nanotechnology. 2011;11(5):3749–3756. doi: 10.1166/jnn.2011.3822. [DOI] [PubMed] [Google Scholar]
- 54.Nejadnik H, Henning TD, Castaneda RT, et al. Somatic differentiation and MR imaging of magnetically labeled human embryonic stem cells. Cell Transplantation. 2012;21(12):2555–2567. doi: 10.3727/096368912X653156. [DOI] [PubMed] [Google Scholar]
- 55.Richards JM, Shaw CA, Lang NN, et al. In vivo mononuclear cell tracking using superparamagnetic particles of iron oxide: feasibility and safety in humans. Circulation. Cardiovascular Imaging. 2012;5(4):509–517. doi: 10.1161/CIRCIMAGING.112.972596. [DOI] [PubMed] [Google Scholar]
- 56.de Chickera SN, Snir J, Willert C, et al. Labelling dendritic cells with SPIO has implications for their subsequent in vivo migration as assessed with cellular MRI. Contrast Media & Molecular Imaging. 2011;6(4):314–327. doi: 10.1002/cmmi.433. [DOI] [PubMed] [Google Scholar]
- 57.Dunning MD, Lakatos A, Loizou L, et al. Superparamagnetic iron oxide-labeled Schwann cells and olfactory ensheathing cells can be traced in vivo by magnetic resonance imaging and retain functional properties after transplantation into the CNS. Journal of Neuroscience. 2004;24(44):9799–9810. doi: 10.1523/JNEUROSCI.3126-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang DM, Hsiao JK, Chen YC, et al. The promotion of human mesenchymal stem cell proliferation by superparamagnetic iron oxide nanoparticles. Biomaterials. 2009;30(22):3645–3651. doi: 10.1016/j.biomaterials.2009.03.032. [DOI] [PubMed] [Google Scholar]
- 59.Bulte JW. In vivo MRI cell tracking: clinical studies. American Journal of Roentgenology. 2009;193(2):314–325. doi: 10.2214/AJR.09.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahrens ET, Feili-Hariri M, Xu H, Genove G, Morel PA. Receptor-mediated endocytosis of iron-oxide particles provides efficient labeling of dendritic cells for in vivo MR imaging. Magnetic Resonance in Medicine. 2003;49(6):1006–1013. doi: 10.1002/mrm.10465. [DOI] [PubMed] [Google Scholar]
- 61.Modo M, Hoehn M, Bulte JW. Cellular MR imaging. Molecular Imaging. 2005;4(3):143–164. doi: 10.1162/15353500200505145. [DOI] [PubMed] [Google Scholar]
- 62.Walczak P, Kedziorek DA, Gilad AA, Lin S, Bulte JW. Instant MR labeling of stem cells using magnetoelectroporation. Magnetic Resonance in Medicine. 2005;54(4):769–774. doi: 10.1002/mrm.20701. [DOI] [PubMed] [Google Scholar]
- 63.Frank JA, Zywicke H, Jordan EK, et al. Magnetic intracellular labeling of mammalian cells by combining (FDA-approved) superparamagnetic iron oxide MR contrast agents and commonly used transfection agents. Academic Radiology. 2002;9(Suppl 2):S484–S487. doi: 10.1016/s1076-6332(03)80271-4. [DOI] [PubMed] [Google Scholar]
- 64.Kraitchman DL, Bulte JW. Imaging of stem cells using MRI. Basic Research in Cardiology. 2008;103(2):105–113. doi: 10.1007/s00395-008-0704-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bulte JW, Kraitchman DL, Mackay AM, Pittenger MF. Chondrogenic differentiation of mesenchymal stem cells is inhibited after magnetic labeling with ferumoxides. Blood. 2004;104(10):3410–3412. doi: 10.1182/blood-2004-06-2117. [DOI] [PubMed] [Google Scholar]
- 66.Walczak P, Ruiz-Cabello J, Kedziorek DA, et al. Magnetoelectroporation: improved labeling of neural stem cells and leukocytes for cellular magnetic resonance imaging using a single FDA-approved agent. Nanomedicine : Nanotechnology, Biology, and Medicine. 2006;2(2):89–94. doi: 10.1016/j.nano.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Terrovitis J, Stuber M, Youssef A, et al. Magnetic resonance imaging overestimates ferumoxide-labeled stem cell survival after transplantation in the heart. Circulation. 2008;117(12):1555–1562. doi: 10.1161/CIRCULATIONAHA.107.732073. [DOI] [PubMed] [Google Scholar]
- 68.Cromer Berman SM, Kshitiz, Wang CJ, et al. Cell motility of neural stem cells is reduced after SPIO-labeling, which is mitigated after exocytosis. Magnetic Resonance in Medicine. 2013;69(1):255–262. doi: 10.1002/mrm.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barnett BP, Arepally A, Karmarkar PV, et al. Magnetic resonance-guided, real-time targeted delivery and imaging of magnetocapsules immunoprotecting pancreatic islet cells. Nature Medicine. 2007;13(8):986–991. doi: 10.1038/nm1581. [DOI] [PubMed] [Google Scholar]
- 70.Shapiro EM, Sharer K, Skrtic S, Koretsky AP. In vivo detection of single cells by MRI. Magnetic Resonance in Medicine. 2006;55(2):242–249. doi: 10.1002/mrm.20718. [DOI] [PubMed] [Google Scholar]
- 71.Shapiro EM, Skrtic S, Koretsky AP. Sizing it up: cellular MRI using micron-sized iron oxide particles. Magnetic Resonance in Medicine. 2005;53(2):329–338. doi: 10.1002/mrm.20342. [DOI] [PubMed] [Google Scholar]
- 72.Hinds KA, Hill JM, Shapiro EM, et al. Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood. 2003;102(3):867–872. doi: 10.1182/blood-2002-12-3669. [DOI] [PubMed] [Google Scholar]
- 73.Shapiro EM, Medford-Davis LN, Fahmy TM, Dunbar CE, Koretsky AP. Antibody-mediated cell labeling of peripheral T cells with micron-sized iron oxide particles (MPIOs) allows single cell detection by MRI. Contrast Media & Molecular Imaging. 2007;2(3):147–153. doi: 10.1002/cmmi.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernas LM, Foster PJ, Rutt BK. Magnetic resonance imaging of in vitro glioma cell invasion. Journal of Neurosurgery. 2007;106(2):306–313. doi: 10.3171/jns.2007.106.2.306. [DOI] [PubMed] [Google Scholar]
- 75.Foley LM, Hitchens TK, Ho C, et al. Magnetic resonance imaging assessment of macrophage accumulation in mouse brain after experimental traumatic brain injury. Journal of Neurotrauma. 2009;26(9):1509–1519. doi: 10.1089/neu.2008.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sumner JP, Shapiro EM, Maric D, Conroy R, Koretsky AP. In vivo labeling of adult neural progenitors for MRI with micron sized particles of iron oxide: quantification of labeled cell phenotype. NeuroImage. 2009;44(3):671–678. doi: 10.1016/j.neuroimage.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rohani R, de Chickera SN, Willert C, Chen Y, Dekaban GA, Foster PJ. In vivo cellular MRI of dendritic cell migration using micrometer-sized iron oxide (MPIO) particles. Molecular Imaging and Biology. 2011;13(4):679–694. doi: 10.1007/s11307-010-0403-0. [DOI] [PubMed] [Google Scholar]
- 78.Nkansah MK, Thakral D, Shapiro EM. Magnetic poly(lactide-co-glycolide) and cellulose particles for MRI-based cell tracking. Magnetic Resonance in Medicine. 2011;65(6):1776–1785. doi: 10.1002/mrm.22765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang KS, Shapiro EM. Enhanced magnetic cell labeling efficiency using -NH2 coated MPIOs. Magnetic Resonance in Medicine. 2011;65(6):1564–1569. doi: 10.1002/mrm.22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahrens ET, Flores R, Xu H, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nature Biotechnology. 2005;23(8):983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 81.Partlow KC, Chen J, Brant JA, et al. 19F magnetic resonance imaging for stem/progenitor cell tracking with multiple unique perfluorocarbon nanobeacons. FASEB Journal. 2007;21(8):1647–1654. doi: 10.1096/fj.06-6505com. [DOI] [PubMed] [Google Scholar]
- 82.Ruiz-Cabello J, Walczak P, Kedziorek DA, et al. In vivo “hot spot” MR imaging of neural stem cells using fluorinated nanoparticles. Magnetic Resonance in Medicine. 2008;60(6):1506–1511. doi: 10.1002/mrm.21783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waiczies H, Guenther M, Skodowski J, et al. Monitoring dendritic cell migration using 19F/1H Magnetic Resonance Imaging. Journal of Visulaized Experiments. 2013;73:e50251. doi: 10.3791/50251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boehm-Sturm P, Mengler L, Wecker S, Hoehn M, Kallur T. In vivo tracking of human neural stem cells with 19F magnetic resonance imaging. PLoS One. 2011;6(12):e29040. doi: 10.1371/journal.pone.0029040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hitchens TK, Ye Q, Eytan DF, Janjic JM, Ahrens ET, Ho C. 19F MRI detection of acute allograft rejection with in vivo perfluorocarbon labeling of immune cells. Magnetic Resonance in Medicine. 2011;65(4):1144–1153. doi: 10.1002/mrm.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bible E, Dell’Acqua F, Solanky B, et al. Non-invasive imaging of transplanted human neural stem cells and ECM scaffold remodeling in the stroke-damaged rat brain by (19)F- and diffusion-MRI. Biomaterials. 2012;33(10):2858–2871. doi: 10.1016/j.biomaterials.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waiczies H, Lepore S, Janitzek N, et al. Perfluorocarbon particle size influences magnetic resonance signal and immunological properties of dendritic cells. PLoS One. 2011;6(7):e21981. doi: 10.1371/journal.pone.0021981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magnetic Resonance in Medicine. 2007;58(4):725–734. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- 89.Kadayakkara DK, Ranganathan S, Young WB, Ahrens ET. Assaying macrophage activity in a murine model of inflammatory bowel disease using fluorine-19 MRI. Laboratory Investigation. 2012;92(4):636–645. doi: 10.1038/labinvest.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Janjic JM, Srinivas M, Kadayakkara DK, Ahrens ET. Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. Journal of the American Chemical Society. 2008;130(9):2832–2841. doi: 10.1021/ja077388j. [DOI] [PubMed] [Google Scholar]
- 91.Verdijk P, Scheenen TW, Lesterhuis WJ, et al. Sensitivity of magnetic resonance imaging of dendritic cells for in vivo tracking of cellular cancer vaccines. International Journal of Cancer. 2007;120(5):978–984. doi: 10.1002/ijc.22385. [DOI] [PubMed] [Google Scholar]
- 92.Srinivas M, Heerschap A, Ahrens ET, Figdor CG, de Vries IJ. (19)F MRI for quantitative in vivo cell tracking. Trends in Biotechnology. 2010;28(7):363–370. doi: 10.1016/j.tibtech.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan KW, Liu G, Song X, et al. MRI-detectable pH nanosensors incorporated into hydrogels for in vivo sensing of transplanted-cell viability. Nature Materials. 2013;12(3):268–275. doi: 10.1038/nmat3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes & Development. 2003;17(5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 95.Adonai N, Nguyen KN, Walsh J, et al. Ex vivo cell labeling with 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) for imaging cell trafficking in mice with positron-emission tomography. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):3030–3035. doi: 10.1073/pnas.052709599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zanzonico P, Koehne G, Gallardo HF, et al. [131I]FIAU labeling of genetically transduced, tumor-reactive lymphocytes: cell-level dosimetry and dose-dependent toxicity. European Journal of Nuclear Medicine and Molecular Imaging. 2006;33(9):988–997. doi: 10.1007/s00259-005-0057-3. [DOI] [PubMed] [Google Scholar]
- 97.Bhargava KK, Gupta RK, Nichols KJ, Palestro CJ. In vitro human leukocyte labeling with (64)Cu: an intraindividual comparison with (111)In-oxine and (18)F-FDG. Nuclear Medicine and Biology. 2009;36(5):545–549. doi: 10.1016/j.nucmedbio.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brenner W, Aicher A, Eckey T, et al. 111In-labeled CD34+ hematopoietic progenitor cells in a rat myocardial infarction model. Journal of Nuclear Medicine. 2004;45(3):512–518. [PubMed] [Google Scholar]
- 99.Rennen HJ, Boerman OC, Oyen WJ, Corstens FH. Imaging infection/inflammation in the new millennium. European Journal of Nuclear Medicine. 2001;28(2):241–252. doi: 10.1007/s002590000447. [DOI] [PubMed] [Google Scholar]
- 100.Becker W, Meller J. The role of nuclear medicine in infection and inflammation. The Lancet Infectious Diseases. 2001;1(5):326–333. doi: 10.1016/S1473-3099(01)00146-3. [DOI] [PubMed] [Google Scholar]
- 101.Jin Y, Kong H, Stodilka RZ, et al. Determining the minimum number of detectable cardiac-transplanted 111In-tropolone-labelled bone-marrow-derived mesenchymal stem cells by SPECT. Physics in Medicine and Biology. 2005;50(19):4445–4455. doi: 10.1088/0031-9155/50/19/001. [DOI] [PubMed] [Google Scholar]
- 102.Gholamrezanezhad A, Mirpour S, Bagheri M, et al. In vivo tracking of 111In-oxine labeled mesenchymal stem cells following infusion in patients with advanced cirrhosis. Nuclear Medicine and Biology. 2011;38(7):961–967. doi: 10.1016/j.nucmedbio.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 103.Gholamrezanezhad A, Mirpour S, Ardekani JM, et al. Cytotoxicity of 111In-oxine on mesenchymal stem cells: a time-dependent adverse effect. Nuclear Medicine Communications. 2009;30(3):210–216. doi: 10.1097/MNM.0b013e328318b328. [DOI] [PubMed] [Google Scholar]
- 104.Monteiro-Riviere NA, Inman AO, Zhang LW. Limitations and relative utility of screening assays to assess engineered nanoparticle toxicity in a human cell line. Toxicology and Applied Pharmacology. 2009;234(2):222–235. doi: 10.1016/j.taap.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 105.Barbash IM, Chouraqui P, Baron J, et al. Systemic delivery of bone marrow-derived mesenchymal stem cells to the infarcted myocardium: feasibility, cell migration, and body distribution. Circulation. 2003;108(7):863–868. doi: 10.1161/01.CIR.0000084828.50310.6A. [DOI] [PubMed] [Google Scholar]
- 106.Kang WJ, Kang HJ, Kim HS, Chung JK, Lee MC, Lee DS. Tissue distribution of 18F-FDG-labeled peripheral hematopoietic stem cells after intracoronary administration in patients with myocardial infarction. Journal of Nuclear Medicine. 2006;47(8):1295–1301. [PubMed] [Google Scholar]
- 107.Zhou R, Thomas DH, Qiao H, et al. In vivo detection of stem cells grafted in infarcted rat myocardium. Journal of Nuclear Medicine. 2005;46(5):816–822. [PMC free article] [PubMed] [Google Scholar]
- 108.Swirski FK, Pittet MJ, Kircher MF, et al. Monocyte accumulation in mouse atherogenesis is progressive and proportional to extent of disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10340–10345. doi: 10.1073/pnas.0604260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim J, Arifin DR, Muja N, et al. Multifunctional capsule-in-capsules for immunoprotection and trimodal imaging. Angewandte Chemie International Edition England. 2011;50(10):2317–2321. doi: 10.1002/anie.201007494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arifin DR, Kedziorek DA, Fu Y, et al. Microencapsulated cell tracking. NMR in Biomedicine. 2012 doi: 10.1002/nbm.2894. doi:10.1002/nbm.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Link TW, Arifin DR, Long CM, et al. Use of magnetocapsules for in vivo visualization and enhanced survival of xenogeneic HepG2 cell transplants. Cell Medicine. 2012;4(2):77–784. doi: 10.3727/215517912X653337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arifin DR, Long CM, Gilad AA, et al. Trimodal gadolinium-gold microcapsules containing pancreatic islet cells restore normoglycemia in diabetic mice and can be tracked by using US, CT, and positive-contrast MR imaging. Radiology. 2011;260(3):790–798. doi: 10.1148/radiol.11101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barnett BP, Kraitchman DL, Lauzon C, et al. Radiopaque alginate microcapsules for X-ray visualization and immunoprotection of cellular therapeutics. Molecular Pharmaceutics. 2006;3(5):531–538. doi: 10.1021/mp060056l. [DOI] [PubMed] [Google Scholar]
- 114.Barnett BP, Ruiz-Cabello J, Hota P, et al. Fluorocapsules for improved function, immunoprotection, and visualization of cellular therapeutics with MR, US, and CT imaging. Radiology. 2011;258(1):182–191. doi: 10.1148/radiol.10092339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nam SY, Ricles LM, Suggs LJ, Emelianov SY. In vivo ultrasound and photoacoustic monitoring of mesenchymal stem cells labeled with gold nanotracers. PLoS One. 2012;7(5):e37267. doi: 10.1371/journal.pone.0037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ricles LM, Nam SY, Sokolov K, Emelianov SY, Suggs LJ. Function of mesenchymal stem cells following loading of gold nanotracers. International Journal of Nanomedicine. 2011;6:407–416. doi: 10.2147/IJN.S16354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jokerst JV, Khademi C, Gambhir SS. Intracellular aggregation of multimodal silica nanoparticles for ultrasound-guided stem cell implantation. Science Translational Medicine. 2013;5(177):177ra35. doi: 10.1126/scitranslmed.3005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cui W, Tavri S, Benchimol MJ, et al. Neural progenitor cells labeling with microbubble contrast agent for ultrasound imaging in vivo. Biomaterials. 2013;34(21):4926–4935. doi: 10.1016/j.biomaterials.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Forss-Petter S, Danielson PE, Catsicas S, et al. Transgenic mice expressing beta-galactosidase in mature neurons under neuron-specific enolase promoter control. Neuron. 1990;5(2):187–197. doi: 10.1016/0896-6273(90)90308-3. [DOI] [PubMed] [Google Scholar]
- 120.Himes SR, Shannon MF. Assays for transcriptional activity based on the luciferase reporter gene. Methods in Molecular Biology. 2000;130:165–174. doi: 10.1385/1-59259-686-x:165. [DOI] [PubMed] [Google Scholar]
- 121.Contag CH, Jenkins D, Contag PR, Negrin RS. Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2000;2(1–2):41–52. doi: 10.1038/sj.neo.7900079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhuo L, Sun B, Zhang CL, Fine A, Chiu SY, Messing A. Live astrocytes visualized by green fluorescent protein in transgenic mice. Developmental Biology. 1997;187(1):36–42. doi: 10.1006/dbio.1997.8601. [DOI] [PubMed] [Google Scholar]
- 123.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 124.Kremers GJ, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW. Fluorescent proteins at a glance. Journal of Cell Science. 2011;124(2):157–160. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Priddle H, Grabowska A, Morris T, et al. Bioluminescence imaging of human embryonic stem cells transplanted in vivo in murine and chick models. Cloning and Stem Cells. 2009;11(2):259–267. doi: 10.1089/clo.2008.0056. [DOI] [PubMed] [Google Scholar]
- 126.Love Z, Wang F, Dennis J, et al. Imaging of mesenchymal stem cell transplant by bioluminescence and PET. Journal of Nuclear Medicine. 2007;48(12):2011–2020. doi: 10.2967/jnumed.107.043166. [DOI] [PubMed] [Google Scholar]
- 127.Tang Y, Shah K, Messerli SM, Snyder E, Breakefield X, Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Human Gene Therapy. 2003;14(13):1247–1254. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 128.Bai X, Yan Y, Coleman M, et al. Tracking long-term survival of intramyocardially delivered human adipose tissue-derived stem cells using bioluminescence imaging. Molecular Imaging and Biology. 2011;13(4):633–645. doi: 10.1007/s11307-010-0392-z. [DOI] [PubMed] [Google Scholar]
- 129.Janowski M, Engels C, Gorelik M, et al. Survival of neural progenitors allografted into the CNS of immunocompetent recipients is highly dependent on transplantation site. Cell Transplant. 2013 doi: 10.3727/096368912X661328. doi: http://dx.doi.org/10.3727/096368912X661328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liang Y, Agren L, Lyczek A, Walczak P, Bulte JW. Neural progenitor cell survival in mouse brain can be improved by co-transplantation of helper cells expressing bFGF under doxycycline control. Experimental Neurology. 2013;247C:73–79. doi: 10.1016/j.expneurol.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maguire KK, Lim L, Speedy S, Rando TA. Assessment of disease activity in muscular dystrophies by noninvasive imaging. The Journal of Clincal Investigation. 2013;123(5):2298–2305. doi: 10.1172/JCI68458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. Journal of Biomedical Optics. 2013;17(1):016004. doi: 10.1117/1.JBO.17.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ueda I, Kamaya H, Eyring H. Molecular mechanism of inhibition of firefly luminescence by local anesthetics. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(2):481–485. doi: 10.1073/pnas.73.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Inoue Y, Kiryu S, Izawa K, Watanabe M, Tojo A, Ohtomo K. Comparison of subcutaneous and intraperitoneal injection of D-luciferin for in vivo bioluminescence imaging. European Journal of Nuclear Medicine and Molecular Imaging. 2009;36(5):771–779. doi: 10.1007/s00259-008-1022-8. [DOI] [PubMed] [Google Scholar]
- 135.Keyaerts M, Remory I, Caveliers V, et al. Inhibition of firefly luciferase by general anesthetics: effect on in vitro and in vivo bioluminescence imaging. PLoS One. 2012;7(1):e30061. doi: 10.1371/journal.pone.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Szarecka A, Xu Y, Tang P. Dynamics of firefly luciferase inhibition by general anesthetics: Gaussian and anisotropic network analyses. Biophysical Journal. 2007;93(6):1895–1905. doi: 10.1529/biophysj.106.102780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Keyaerts M, Heneweer C, Gainkam LO, et al. Plasma protein binding of luciferase substrates influences sensitivity and accuracy of bioluminescence imaging. Molecular Imaging and Biology. 2011;13(1):59–66. doi: 10.1007/s11307-010-0325-x. [DOI] [PubMed] [Google Scholar]
- 138.Gilad AA, McMahon MT, Walczak P, et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nature Biotechnology. 2007;25(2):217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 139.Gilad AA, Ziv K, McMahon MT, van Zijl PC, Neeman M, Bulte JW. MRI reporter genes. Journal of Nuclear Medicine. 2008;49(12):1905–1908. doi: 10.2967/jnumed.108.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P. Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magnetic Resonance in Medicine. 2011;65(2):564–574. doi: 10.1002/mrm.22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu G, Bulte JW, Gilad AA. CEST MRI reporter genes. Methods in Molecular Biology. 2011;711:271–280. doi: 10.1007/978-1-61737-992-5_13. [DOI] [PubMed] [Google Scholar]
- 142.Louie AY, Huber MM, Ahrens ET, et al. In vivo visualization of gene expression using magnetic resonance imaging. Nature Biotechnology. 2000;18(3):321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 143.Cui W, Liu L, Kodibagkar VD, Mason RP. S-Gal, a novel 1H MRI reporter for beta-galactosidase. Magnetic Resonance in Medicine. 2010;64(1):65–71. doi: 10.1002/mrm.22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Alfke H, Stoppler H, Nocken F, et al. In vitro MR imaging of regulated gene expression. Radiology. 2003;228(2):488–492. doi: 10.1148/radiol.2282012006. [DOI] [PubMed] [Google Scholar]
- 145.Gilad AA, Winnard PT, Jr., van Zijl PC, Bulte JW. Developing MR reporter genes: promises and pitfalls. NMR in Biomedicine. 2007;20(3):275–290. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- 146.Weissleder R, Moore A, Mahmood U, et al. In vivo magnetic resonance imaging of transgene expression. Nature Medicine. 2000;6(3):351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 147.Kotamraju S, Chitambar CR, Kalivendi SV, Joseph J, Kalyanaraman B. Transferrin receptor-dependent iron up-take is responsible for doxorubicin-mediated apoptosis in endothelial cells: role of oxidant-induced iron signaling in apoptosis. The Journal of Biological Chemistry. 2002;277(19):17179–17187. doi: 10.1074/jbc.M111604200. [DOI] [PubMed] [Google Scholar]
- 148.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7(2):109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Naumova AV, Reinecke H, Yarnykh V, Deem J, Yuan C, Murry CE. Ferritin overexpression for noninvasive magnetic resonance imaging-based tracking of stem cells transplanted into the heart. Molecular Imaging. 2010;9(4):201–210. [PMC free article] [PubMed] [Google Scholar]
- 150.Campan M, Lionetti V, Aquaro GD, et al. Ferritin as a reporter gene for in vivo tracking of stem cells by 1.5-T cardiac MRI in a rat model of myocardial infarction. American Journal of Physiology. Heart and Circulatory Physiology. 2011;300(6):H2238–H2250. doi: 10.1152/ajpheart.00935.2010. [DOI] [PubMed] [Google Scholar]
- 151.Gossuin Y, Muller RN, Gillis P. Magnetic resonance imaging of cells overexpressing MagA, an endogenous contrast agent for live cell imaging. Molecular Imaging. 2009;8(3):129–139. [PubMed] [Google Scholar]
- 152.Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magnetic Resonance in Medicine. 2008;59(6):1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldhawk DE, Lemaire C, McCreary CR, et al. Magnetic resonance imaging of cells overexpressing MagA, an endogenous contrast agent for live cell imaging. Molecular Imaging. 2009;8(3):129–139. [PubMed] [Google Scholar]
- 154.McMahon MT, Gilad AA, DeLiso MA, Berman SM, Bulte JW, van Zijl PC. New “multicolor” polypeptide diamagnetic chemical exchange saturation transfer (DIACEST) contrast agents for MRI. Magnetic Resonance in Medicine. 2008;60(4):803–812. doi: 10.1002/mrm.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Alauddin MM, Shahinian A, Gordon EM, Conti PS. Direct comparison of radiolabeled probes FMAU, FHBG, and FHPG as PET imaging agents for HSV1-tk expression in a human breast cancer model. Molecular Imaging. 2004;3(2):76–84. doi: 10.1162/15353500200403160. [DOI] [PubMed] [Google Scholar]
- 156.Koehne G, Doubrovin M, Doubrovina E, et al. Serial in vivo imaging of the targeted migration of human HSV-TK-transduced antigen-specific lymphocytes. Nature Biotechnology. 2003;21(4):405–413. doi: 10.1038/nbt805. [DOI] [PubMed] [Google Scholar]
- 157.Ponomarev V, Doubrovin M, Lyddane C, et al. Imaging TCR-dependent NFAT-mediated T-cell activation with positron emission tomography in vivo. Neoplasia. 2001;3(6):480–488. doi: 10.1038/sj.neo.7900204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pei Z, Lan X, Cheng Z, et al. A multimodality reporter gene for monitoring transplanted stem cells. Nuclear Medicine and Biology. 2012;39(6):813–820. doi: 10.1016/j.nucmedbio.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 159.Pomper MG, Hammond H, Yu X, et al. Serial imaging of human embryonic stem-cell engraftment and teratoma formation in live mouse models. Cell Research. 2009;19(3):370–379. doi: 10.1038/cr.2008.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mercier-Letondal P, Deschamps M, Sauce D, et al. Early immune response against retrovirally transduced herpes simplex virus thymidine kinase-expressing gene-modified T cells coinfused with a T cell-depleted marrow graft: an altered immune response? Human Gene Therapy. 2008;19(9):937–950. doi: 10.1089/hum.2007.156. [DOI] [PubMed] [Google Scholar]
- 161.Serganova I, Ponomarev V, Blasberg R. Human reporter genes: potential use in clinical studies. Nuclear Medicine and Biology. 2007;34(7):791–807. doi: 10.1016/j.nucmedbio.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 162.Campbell DO, Yaghoubi SS, Su Y, et al. Structure-guided engineering of human thymidine kinase 2 as a positron emission tomography reporter gene for enhanced phosphorylation of non-natural thymidine analog reporter probe. The Journal of Biological Chemistry. 2012;287(1):446–454. doi: 10.1074/jbc.M111.314666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ponomarev V, Doubrovin M, Shavrin A, et al. A human-derived reporter gene for noninvasive imaging in humans: mitochondrial thymidine kinase type 2. Journal of Nuclear Medicine. 2007;48(5):819–826. doi: 10.2967/jnumed.106.036962. [DOI] [PubMed] [Google Scholar]
- 164.Yaghoubi SS, Jensen MC, Satyamurthy N, et al. Noninvasive detection of therapeutic cytolytic T cells with 18FFHBG PET in a patient with glioma. Nature Clinical Practice Oncology. 2009;6(1):53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McCracken MN, Gschweng EH, Nair-Gill E, et al. Long-term in vivo monitoring of mouse and human hematopoietic stem cell engraftment with a human positron emission tomography reporter gene. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(5):1857–1862. doi: 10.1073/pnas.1221840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang M, Batra RK, Kogai T, et al. Ectopic expression of the thyroperoxidase gene augments radioiodide uptake and retention mediated by the sodium iodide symporter in non-small cell lung cancer. Cancer Gene Therapy. 2001;8(8):612–618. doi: 10.1038/sj.sgt.7700354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Auricchio A, Acton PD, Hildinger M, et al. In vivo quantitative noninvasive imaging of gene transfer by single-photon emission computerized tomography. Human Gene Therapy. 2003;14(3):255–261. doi: 10.1089/10430340360535805. [DOI] [PubMed] [Google Scholar]
- 168.Kim YH, Lee DS, Kang JH, et al. Reversing the silencing of reporter sodium/iodide symporter transgene for stem cell tracking. Journal of Nuclear Medicine. 2005;46(2):305–311. [PubMed] [Google Scholar]
- 169.Terrovitis J, Kwok KF, Lautamaki R, et al. Ectopic expression of the sodium-iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. Journal of the American College of Cardiology. 2008;52(20):1652–1660. doi: 10.1016/j.jacc.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hwang do W, Kang JH, Jeong JM, et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. European Journal of Nuclear Medicine and Molecular Imaging. 2008;35(1):135–145. doi: 10.1007/s00259-007-0561-8. [DOI] [PubMed] [Google Scholar]
- 171.Bar-Shir A, Liu G, Liang Y, et al. Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. Journal of the American Chemical Society. 2013;135(4):1617–1624. doi: 10.1021/ja312353e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.de Vries IJ, Lesterhuis WJ, Barentsz JO, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nature Biotechnology. 2005;23(11):1407–1413. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]