Abstract
Background
Despite the high heritability of alcohol dependence (AD), the genes found to be associated with it account for only a small proportion of its total variability. The goal of this study was to identify and analyze phenotypes based on homogeneous classes of individuals to increase the power to detect genetic risk factors contributing to the risk of AD.
Methods
The 7 individual DSM-IV criteria for AD were analyzed using latent class analysis (LCA) to identify classes defined by the pattern of endorsement of the criteria. A genome-wide association study was performed in 118 extended European American families (n = 2,322 individuals) densely affected with AD to identify genes associated with AD, with each of the seven DSM-IV criteria, and with the probability of belonging to two of three latent classes.
Results
Heritability for DSM-IV AD was 61%, and ranged from 17-60% for the other phenotypes. A SNP in the olfactory receptor OR51L1 was significantly associated (7.3 × 10−8) with the DSM-IV criterion of persistent desire to, or inability to, cut down on drinking. LCA revealed a three-class model: the “low risk” class (50%) rarely endorsed any criteria, and none met criteria for AD; the “moderate risk” class (33) endorsed primarily 4 DSM-IV criteria, and 48% met criteria for AD; the “high risk” class (17%) manifested high endorsement probabilities for most criteria and nearly all (99%) met criteria for AD One single nucleotide polymorphism (SNP) in a sodium leak channel NALCN demonstrated genome-wide significance with the high risk class (p=4.1 × 10−8). Analyses in an independent sample did not replicate these associations.
Conclusion
We explored the genetic contribution to several phenotypes derived from the DSM-IV alcohol dependence criteria. The strongest evidence of association was with SNPs in NALCN and OR51L1.
Keywords: alcohol dependence criteria, latent class analysis, family-based association, GWAS
Introduction
Alcohol dependence (AD) is one of the most common and costly public health problems in the United States and throughout the world, affecting 4-5% of the United States population in a 12-month period (Kessler, 2005; Li et al., 2007), and 12.5% across the lifetime (Hasin et al., 2007). Family, twin, and adoption studies have provided convergent evidence for the role of genetic factors in AD (Goodwin et al., 1974; Heath et al., 1997). Approximately 40% to 60% of the total variance in risk for AD is due to heritable influences (Prescott et al., 2006; Schuckit et al., 2001).
To date, a few genes have been shown to be associated with AD, including ADH1B,ALDH2, ADH4, CHRNA5, CHRM2, GABRA2, and GABRG1 (Agrawal et al., 2012; Covault et al., 2008; Edenberg et al., 2004; Edenberg and Foroud, 2006; Edenberg et al., 2006; Enoch, 2008; Enoch et al., 2009; Luo et al., 2005; Wang et al., 2009; Wang et al., 2004); however, the variation in each of these genes accounts for only a small portion of the vulnerability to AD. This could be due to the study design, the gene coverage, or most likely the complex etiology and heterogeneity of AD.
The DSM-IV diagnosis of AD requires the manifestation of three or more of the following criteria at any time during the same 12-month period: tolerance, withdrawal, using alcohol more than intended, a persistent desire to cut down or inability to do so, spending a great deal of time obtaining alcohol or recovering from its effects, giving up activities to drink, and continuing alcohol use despite physical or psychological problems (DSM-IV) (AmericanPsychiatricAssociation, 1994). The requirement of any three inherently creates a group of AD individuals that is clinically and likely genetically heterogeneous. Therefore, approaches that focus on potentially more homogeneous subgroups may increase the power to identify novel genetic variants by reducing the clinical (and therefore possibly the genetic) heterogeneity.
We used several complementary approaches to examine the genetic contributions to the risk of AD. We first analyzed the dichotomous phenotype of AD, because it is the most widely studied phenotype and most directly reflects the clinically relevant trait. We then examined each of the DSM-IV criteria for dependence individually, as one approach to reduce heterogeneity. We also employed latent class analysis to identify subgroups of individuals defined on the basis of their pattern of endorsement of DSM-IV criteria and used the probability of class assignment as a quantitative phenotype for analysis. This approach may also reduce heterogeneity and increase the power to identify genetically similar subgroups.
We utilized a genome-wide approach to detect common genetic variants associated with the phenotypes. Unlike most studies which employ a case-control design, we performed a family-based genome-wide association study (GWAS) using extended families densely affected by AD. The sample was limited to European American (EA) families to reduce one source of potential heterogeneity.
Methods
Sample
Alcoholic probands were recruited from inpatient and outpatient treatment facilities through six sites participating in the Collaborative Study on the Genetics of Alcoholism (COGA) (Begleiter et al., 1995; Foroud et al., 2000). The probands and their family members were administered a validated poly-diagnostic interview, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994; Hesselbrock et al., 1999). Individuals below the age of 18 were administered an adolescent version of the SSAGA. Institutional review boards at all sites approved the study.
A sub-sample of COGA families was selected to maximize the contribution of genetic variants to the risk for AD. All families recruited by COGA were reviewed and prioritized based on, in preferential order: 1) the largest number of AD family members with DNA; 2) the largest number of family members with electrophysiology data and DNA; and 3) the largest number of family members with DNA regardless of other phenotypes. The final sample consisted of 118 large, multiplex European American families, with DNA available from 2,322 individuals.
Phenotypes and Statistical Analysis
Phenotypic information for the members of these 118 families was collected using the SSAGA, with some individuals having multiple assessments across time. If an individual was interviewed more than once, data from the SSAGA interview with the maximum total number of endorsed DSM-IV AD criteria was utilized.
Only individuals who reported drinking at least once per month for 6 months or more at any evaluation (regular drinking) were included in the analysis. Subjects who were 15 years or older and met DSM-IV criteria at any evaluation were classified as AD (n=687), and individuals who were 23 years or older and did not meet criteria for DSM-IV AD were classified as unaffected (n=773). All other individuals were classified as unknown (n=326), including self-reported AD children at a very early age (< 15 years) and unaffected individuals who were younger than 23 years old and thus had not fully passed through the primary age of risk for AD and therefore might still develop it. These 326 individuals were included in analyses of all other phenotypes.
Latent class analysis (LCA) was conducted to identify subgroups of individuals based on the pattern of endorsement of the 7 DSM-IV AD criteria (Muthen and Muthen, 1998-2011). LCA may be viewed as a version of non-parametric cluster analysis, and makes the assumption of conditional independence (i.e. endorsement of one criterion within a class is uncorrelated with other criteria). There are two important estimates recovered from LCA: (a) the prevalence of individual classes of individuals with similar item endorsement profiles and (b) the likelihood of endorsing an item conditional on class membership. The Bayesian Information Criteria (BIC) and entropy (reflecting level of misclassification) were utilized to determine which model (i.e., number of latent classes) provided the best fit to the data.
A critical assumption of LCA is conditional independence and violations can result in extraction of more classes than are required. To address this concern, we examined a factor mixture model where a single factor was nested within each latent class, relaxing the conditional independence assumption. The fit of this model was compared to the LCA. As the objective of this study was to classify individuals (i.e. person-centered) rather than symptoms (i.e. item-centered), we did not conduct factor analyses in the absence of LCA.
Based on the best-fitting model, class membership was utilized in a two-step process. First, an individual was assigned to the latent class with the highest probability, and discrete class memberships were compared using post hoc validators such as gender, age at onset of regular drinking (from the first interview in which the individual began drinking regularly), and DSM-IV AD. Interpretation of the classes was based on the validators. Gender and DSM-IV AD differences between classes were assessed with a chi-squared test; number of DSM-IV criteria, age and age at onset of regular drinking were tested using an analysis of variance (ANOVA), with gender and birth cohort as covariates. Tukey-adjusted t-tests were employed to test for pairwise differences when there was a significant (p<0.05) main effect of latent class assignment.
To optimize the power for genetic analyses, we constructed secondary continuous phenotypes based on the probability of assignment to each of the three latent classes. As a result, information describing each latent class provided an interpretation of the three classes while the probability of being in each class was employed as the phenotype in analysis. This approach avoids multiple pairwise comparisons of the three classes in analyses, avoids misclassification inherent in assigning individuals to a particular latent class, maximizes the number of subjects, and was shown previously to improve power to detect genetic association (Bureau et al., 2011).
Heritability was estimated using the polygenic option in SOLAR (Almasy and Blangero, 1998) for the following phenotypes: DSM-IV AD, each individual DSM-IV criterion, the three continuous phenotypes based on the LCA probabilities, and membership in each latent class. Sex and birth cohort, defined by year of birth (<1930<1930-1949-1950-1969, and ≥1970), were used as covariates for all heritability calculations to account for secular trends (Grucza et al., 2008) in alcohol use and dependence.
Genotyping and Association Analysis
Genotyping for 2,105 subjects in these 118 families was performed at the Genome Technology Access Center at Washington University School of Medicine in St. Louis using the Illumina Human OmniExpress array 12.VI (Illumina, San Diego, CA, USA). In addition, genotypes previously generated on the Illumina Human 1M-Duo BeadChip (Illumina, San Diego, CA, USA)by the Center for Inherited Disease Research were also included for 224 subjects from these families (Edenberg et al., 2010). For quality control purposes, 51 of the 2,105 subjects were genotyped on both arrays and available for analysis, prior to sample exclusions (n=7) due to Mendelian inconsistencies.
Quality control (QC) was first performed on genotype data generated from the OmniExpress. Genotypes for individuals with a genotype rate <98% were excluded from analysis, and SNPs with a genotyping rate <98% were excluded from analysis. Further details can be found in Kang et al (Kang et al., 2012). Genotypes of the 51 overlapping individuals were then compared. There were 571 of 544,276 SNPs genotyped on both arrays with two or more non-missing and different genotypes, which were excluded from all further analyses. There were 442 genotyped founders used to remove SNPs which violated Hardy-Weinberg equilibrium (HWE; p<10−6). SNPs with minor allele frequency less than 5% were also removed from further analysis. After combining the data generated from the two arrays in the members of these 118 families, pedigree structure was reviewed and modified, as described in Kang et al (Kang et al., 2012).
The family-based Generalized Disequilibrium Test (GDT), which employs information from all discordant relative pairs (Chen et al., 2009), was used to test for association with the qualitative traits: AD and each DSM-IV criteria. A linear mixed effects model was employed using the kinship library (lmekin) implemented in R (http://www.inside-r.org/packages/cran/kinship/docs/print.lmekin) to test for association with the quantitative traits reflecting probability of membership in the latent classes. The linear mixed effects model in the kinship function allows for the covariance matrix to be completely specified for the random effects. The result is that each family has a different covariance pattern based on the kinship coefficients, to model the familial genetic random effects. Sex, age at last interview and birth cohort as well as the first principal component (PC1) from the EIGENSTRAT analysis were evaluated as potential covariates. Any significantly associated covariate (p<0.05) was included in all final association models. As described in Wang et al (Wang et al., 2012) imputed data were generated using BEAGLE 3.3.1(Browning and Browning, 2009) in the chromosomal regions providing the greatest evidence of association with the genotyped SNPs.
Replication Study
The Study of Addiction: Genetics and Environment (SAGE) was used as an independent replication sample to evaluate the SNPs showing the greatest evidence of association in the COGA sample. This is a case-control sample derived from three large, complementary studies: COGA, the Family Study of Cocaine Dependence and the Collaborative Genetics Study of Nicotine Dependence (Bierut et al., 2010). The 129 individuals who overlapped between the 118 pedigrees and SAGE were removed from the replication dataset. The remaining SAGE samples included 2,647 individuals of European American descent. AD and individual criteria were obtained from SSAGA data. Imputed dosage data were obtained using the same method as described above. Plink was used in all analyses, employing age at interview and gender as covariates.
Results from LCA were derived separately in this sample using Mplus, as described above. LCA was also conducted in the SAGE+COGA combined sample and in the SAGE sample including the 129 individuals to examine if latent class assignment was sample-dependent. This was tested with McNemar's change test.
Results
Sample
Following quality control and the exclusion of some individuals after correction of pedigree inconsistencies, the final analytic sample included 2,322 genotyped individuals from 118 extended families, of which 1,786 individuals met phenotypic criteria for inclusion (Table 1). The average number of genotyped members per family was 19.6, with an average of 5.9 genotyped individuals per family meeting criteria for DSM-IVAD. A total of 707,557 autosomal SNPs passed quality review. 115,772 SNPs were excluded due to a low minor allele frequency (<5%). The final dataset consisted of 591,785 SNPs, yielding a genome-wide threshold of p=8.45 × 10−8. Since the phenotypes examined represent some facet of AD and additional Bonferroni correction of related phenotypes would be unnecessarily stringent, this p-value based on the number of SNPs was considered as the significance threshold.
Table 1. Demographic and phenotypic characteristics of the sample. Percentages ≥50 are italicized. Percentages ≥90 are italicized bold.
| DSM-IV criterion | N = # individuals who endorsed this criterion | % individuals who endorsed this criterion | % who endorsed this criterion were male | % who endorsed this criterion were DSM-IV alcohol dependent | % who endorsed this criterion were in low-risk latent class | % who endorsed this criterion were in moderate-risk latent class | % who endorsed this criterion were in high-risk latent class |
|---|---|---|---|---|---|---|---|
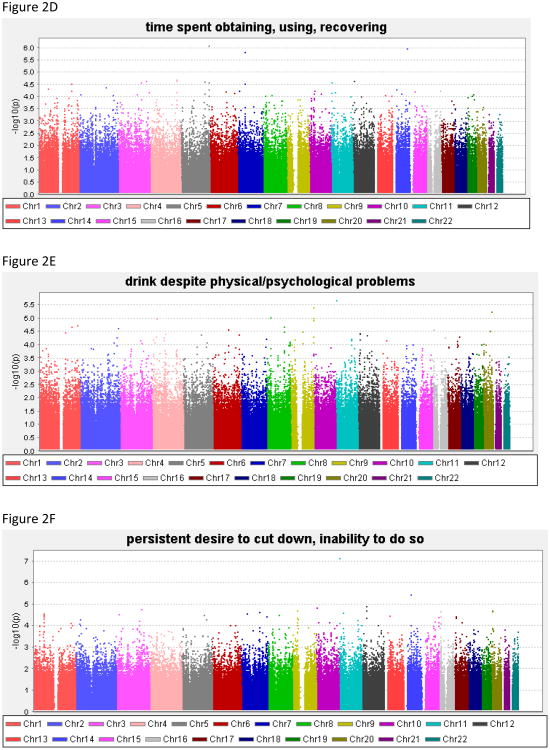
| Withdrawal | 327 | 18.3 | 66.1 | 94.8 | 0.0 | 22.3 | 77.7 |
| Gave up activities to drink | 357 | 20.0 | 64.7 | 95.0 | 0.3 | 26.3 | 73.4 |
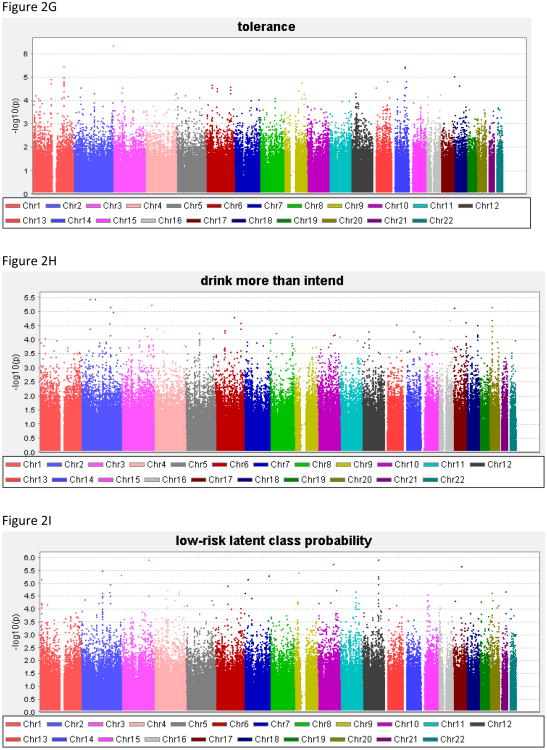
| Time spent obtaining, using or recovering from alcohol | 427 | 23.9 | 63.2 | 91.6 | 4.0 | 30.9 | 65.1 |
| Drink despite physical/psychological problems | 635 | 35.6 | 59.7 | 84.7 | 1.7 | 52.6 | 45.7 |
| Persistent desire to cut down, or inability to do so | 829 | 46.4 | 57.4 | 71.5 | 13.0 | 52.2 | 34.7 |
| Tolerance | 947 | 53.0 | 58.9 | 63.2 | 28.3 | 42.6 | 28.6 |
| Drink more than intend | 1,085 | 60.8 | 54.9 | 60.8 | 23.0 | 50.3 | 26.6 |
Latent Class Analysis
The optimum latent class model (by BIC model fitting criteria) was a 3-class model. The sample-size-adjusted (ssadj) BIC decreased from 13,849 for a 3-class model to 13,827 for a 4-class model, with a corresponding drop in entropy of 0.77 to 0.68. The ssadj BIC from the factor mixed model was 13,830, with an entropy of 0.768. Due to the similar results between the two models, a more parsimonious 3-class model which assumed conditional independence was used in subsequent analyses.
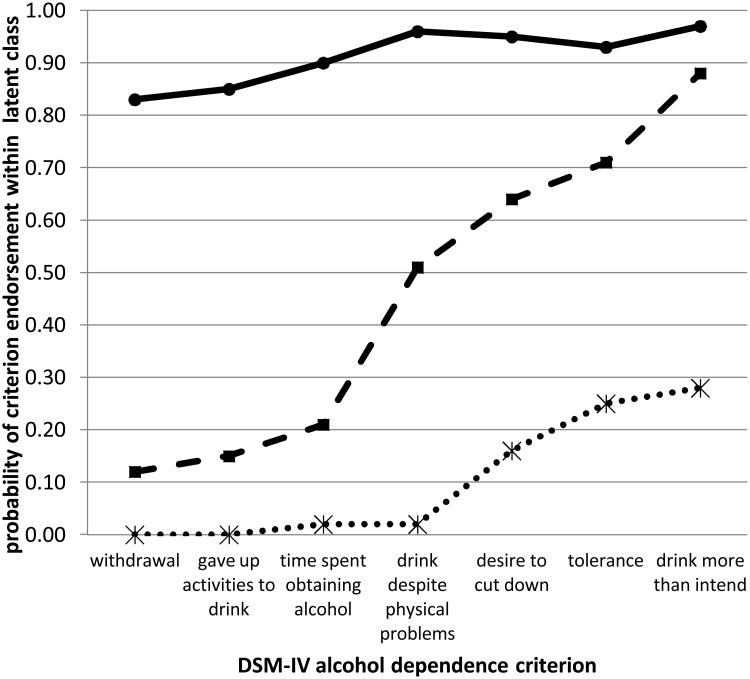
The criterion endorsement probabilities, conditional on class membership, are shown in Figure 1 and reveal a range of severity. A low-risk class (50% of individuals) included individuals with negligible to low endorsement of the dependence criteria. A moderate-risk class (33% of individuals) was distinguished by individuals with low endorsement probability of withdrawal, gave up activities to drink and time spent obtaining or recovering from the effects of alcohol, but higher (>0.51) probabilities of endorsing each of the remaining criteria. A high-risk class (17% of individuals), included individuals with high (>0.86) probability of endorsing the 7 dependence criteria.
Figure 1. Average probability of DSM-IV criterion endorsement within each latent class.
The X-axis indicates the DSM-IV criteria. The Y-axis denotes the probability of endorsing each criterion within each latent class. The low-risk class is depicted with a dotted line. The moderate-risk class is depicted with a dashed line. The high-risk class is depicted with a solid line.
Table 2a summarizes the characteristics of each of the latent classes and Table 2b provides summary statistics for the DSM-IV criteria for each latent class. As expected, there were significantly more DSM-IV AD individuals (X2=1222.3, df=2, p<0.0001) and significantly more males (X2=66.2, df=2, p<0.0001) in the moderate and high-risk classes. The mean number of DSM-IV criteria was significantly different among the three classes (overall F(6, 1779)=2043.7, p<0.0001; p<0.0001 for each pairwise comparison), with higher scores in the high-risk class, indicating that classes reflected a continuum of severity as indexed by number of DSM-IV criteria. There was a main effect of latent class for age at regular drinking onset (overall F(6,1779)=83.0, p<0.0001; latent class p<0.0001), and a marginal effect for age (overall F(6,1779)=2394.0, p<0.0001; latent class p=0.045). The low-risk class contained no individuals meeting criteria for AD, although some individuals endorsed tolerance (30%) or drink more than intended (28%). Individuals in this class began drinking regularly at a significantly older age (19.5 years) than those in the moderate (p<0.0001) or high-risk (p<0.0001) classes. In the moderate risk class, 66% were AD, with 92% of this class endorsing drink more than intended, and more than 50% endorsing use despite physical/psychological problems (56%), persistent desire/inability to cut down (73%), or tolerance (68%). On average they began regular drinking earlier (17.6 years) than their counterparts assigned to the low risk class (p<0.0001). The third class was composed primarily of AD (99%) individuals, with the majority endorsing use despite physical/psychological problems (98%), drink more than intended (98%), or persistent desire/inability to cut down (97%). These individuals began drinking at the youngest age (16.8 years) compared to their counterparts in the moderate (p=0.01) and low-risk (p<0.0001) classes. Individuals in this class were significantly older (55.0 years) than individuals in the low-risk (46.3 years, p=0.03) but not the moderate-risk (49.3 years, p=0.19) classes. There were no significant differences in age between the low- and moderate-risk classes (p=0.61) or between the moderate- and high-risk classes (p=0.19).
Table 2a. Phenotypic characteristics of the three latent classes. Percentages ≥50 are in italicized. Percentages ≥90 are italicized bold.
| Latent Class | N | % male | % DSM-IV AD | age of onset regular drinking (years) | current age (years) | % class who endorsed withdrawal | % class who endorsed gave up activities to drink | % class who endorsed time spent obtaining/recovering from alcohol | % class who endorsed use despite physical/psychological problems | % class who endorsed persistent desire/inability to cut down | % class who endorsed tolerance | % class who endorsed drink more than intend |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low-risk | 896 | 40.5 | 0 | 19.5 | 46.3 | 0.0 | 0.1 | 1.9 | 1.2 | 12.1 | 30.5 | 27.9 |
| Moderate-risk | 594 | 53.0 | 66.3 | 17.6 | 49.3 | 12.3 | 15.8 | 22.2 | 56.2 | 72.9 | 67.9 | 91.9 |
| High-risk | 296 | 66.6 | 99.0 | 16.8 | 55.0 | 85.8 | 88.5 | 93.9 | 98.0 | 97.3 | 91.6 | 97.6 |
| N = sample size, AD = alcohol dependent | ||||||||||||
| Table 2b: Summary statistics describing DSM-IV criteria endorsement for the three latent classes. | ||||||
|---|---|---|---|---|---|---|
| Latent Class | N | mean # criteria | standard error | median # criteria | minimum # criteria | maximum # criteria |
| Low-risk | 896 | 0.74 | 0.025 | 1 | 0 | 2 |
| Moderate-risk | 594 | 3.39 | 0.014 | 3 | 1 | 5 |
| High-risk | 296 | 6.53 | 0.037 | 7 | 4 | 7 |
Genetic Analysis
Heritability was estimated for DSM-IV AD, each of the DSM-IV criteria, and the three traits defined by the probability of belonging to each of latent class (Table 3), after covarying for effects of sex and birth cohort on the phenotype. Heritability for AD was estimated to be 61%. Estimates of heritability for the individual criteria ranged from 59% (drinking more than intended) to 29% (tolerance). Heritability of membership in the low and high-risk classes was 0.52 and 0.53. Due to the redundancy of including all three probabilities and the low heritability of the moderate risk class (0.17), this phenotype was not included in the GWAS.
Table 3. Heritability estimates and significance of covariates.
| Phenotype | h2 | se(h2) | h2 p-value | sex p-value | 1930-1949 cohort p-value | 1950-1969 cohort p-value | ≥1970 cohort p-value | r2 |
|---|---|---|---|---|---|---|---|---|
| DSM-IV alcohol dependence | 0.61 | 0.08 | 2.8 × 10−17 | 1.6 × 10−24 | 2.7 × 10−3 | 1.4 × 10−9 | 2.4 × 10−10 | 0.07 |
| Withdrawal | 0.49 | 0.08 | 1.5 × 10−11 | 5.2 × 10−11 | 0.36 | 0.03 | 0.28 | 0.05 |
| Gave up activities to drink | 0.50 | 0.08 | 6.1 × 10−14 | 1.3 × 10−10 | 0.11 | 4.2 × 10−4 | 0.77 | 0.06 |
| Time spent obtaining/using or recovering from alcohol | 0.41 | 0.07 | 2.4 × 10−12 | 3.7 × 10−12 | 0.50 | 1.1 × 10−4 | 0.39 | 0.05 |
| Use despite physical/psychological problems | 0.51 | 0.07 | 7.8 × 10−17 | 2.3 × 10−14 | 0.03 | 4.1 × 10−7 | 0.05 | 0.08 |
| Persistent desire to cut down, or inability to do so | 0.47 | 0.06 | 3.8 × 10−17 | 5.8 × 10−16 | 0.04 | 0.04 | 0.07 | 0.04 |
| Tolerance | 0.29 | 0.06 | 6.7 × 10−9 | 1.0 × 10−24 | 9.1 × 10−3 | 8.2 × 10−9 | 2.7 × 10−8 | 0.05 |
| Drink more than intended | 0.59 | 0.09 | 1.8 × 10−18 | 1.2 × 10−14 | 8.7 × 10−6 | 5.6 × 10−16 | 5.3 × 10−5 | 0.06 |
| Probability of assignment to low risk latent class | 0.34 | 0.04 | 2.1 × 10−31 | 0.09 | 6.7 × 10−5 | 1.4 × 10−16 | 3.0 × 10−3 | 0.08 |
| Membership in low risk latent class | 0.52 | 0.06 | 4.4 × 10−25 | 1.9 × 10−17 | 6.7 × 10−4 | 7.9 × 10−13 | 4.2 × 10−3 | 0.06 |
| Probability of assignment to moderate risk latent class | 0.12 | 0.03 | 2.0 × 10−7 | 1.1 × 10−5 | 2.6 × 10−3 | 1.7 × 10−6 | 6.2 × 10−3 | 0.02 |
| Membership in moderate risk latent class | 0.17 | 0.03 | 1.8 × 10−5 | 3.3 × 10−5 | 0.03 | 1.9 × 10−4 | 0.025 | 0.01 |
| Probability of assignment to high risk class latent class | 0.19 | 0.03 | 2.9 × 10−18 | 1.2 × 10−14 | 0.24 | 8.8 × 10−6 | 0.58 | 0.07 |
| Membership in high risk latent class | 0.53 | 0.08 | 3.5 × 10−15 | 3.5 × 10−12 | 0.11 | 9.9 × 10−5 | 0.72 | 0.07 |
Sex and birth cohort were included as covariates in all analyses, with <1930 as the reference cohort. r2=proportion of variance accounted for by the covariates. Significant heritabilities (p<0.05) are bolded.
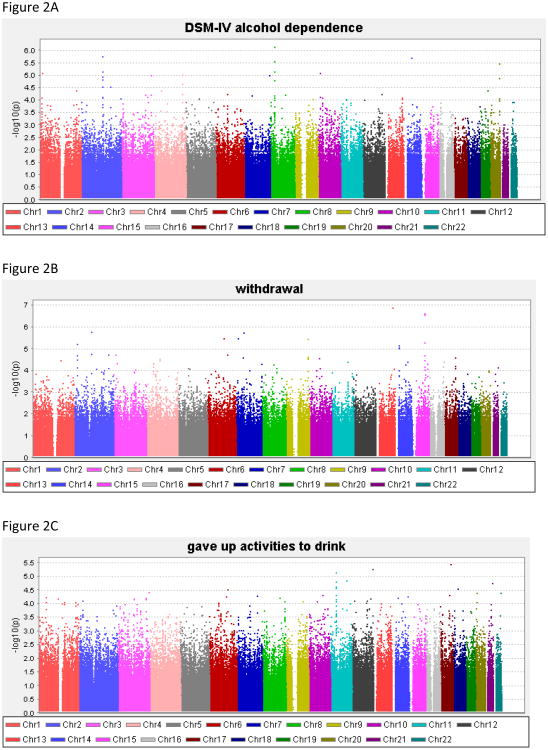
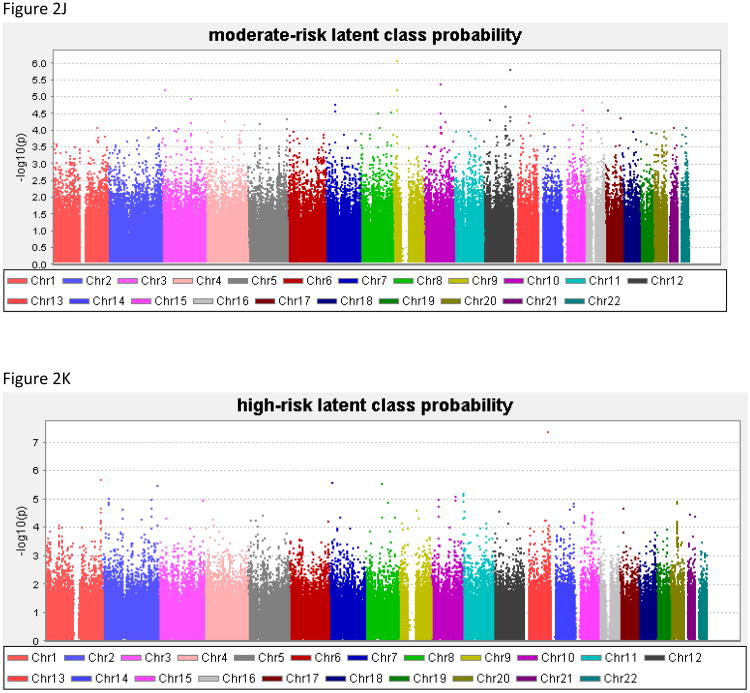
Genome-wide results of the association analyses of each phenotype are summarized in Figure 2, and all associations with p<1.0 × 10−6 are shown in Table 4; Supplemental Table 1 contains results for all SNPs with p<1.0 × 10−5. There was no evidence to suggest inflation of the association p-values (0.984 < λ < 1.01), validating the choice of the significance threshold. QQ plots are provided for each phenotype in the Supplemental Material. For DSM-IV AD the best SNP reached 7.2 × 10−7.
Figure 2. Genome-wide association study results for the phenotypes.
(A) alcohol dependence, (B) withdrawal, (C) gave up activities to drink, (D) time spent obtaining/using/recovering from alcohol, (E) drink despite physical/psychological problems, (F) persistent desire/inability to cut down, (G) tolerance, (H) drink more than intend to, (I) low-risk latent class probability, (J) moderate-risk latent class probability, (K) high-risk latent class probability.
Table 4. Summary of association results with p<10−6 for any trait in the COGA sample, and corresponding results from the SAGE replication sample. Supporting phenotypes are provided in additional rows. Genome-wide significant results are in bold.
| Chra | positionb | SNPc | MAFd | Genee | Phenotype | COGA p-value | SAGE p-value |
|---|---|---|---|---|---|---|---|
| 3 | 3,692,598 | rs1873023 | 0.12 | LRRN1 (148kb) | Tolerance | 4.2 × 10−7 | 0.77 |
| 5 | 175,991,998 | rs6892681 | 0.44 | CDHR2 | Time spent obtaining/recovering from alcohol | 8.3 × 10−7 | 0.16 |
| 8 | 26,894,971 | rs13251780 | 0.14 | LOC100132229 (25kb) | DSM-IV | 7.2 × 10−7 | 0.89 |
| 9 | 18,166,899 | rs12006002 | 0.29 | LOC100421790 (12kb) | Probability of moderate-risk class | 8.3 × 10−7 | 0.35 |
| 11 | 5,029,457 | rs11035102 | 0.13 | OR51L1(6kb) | Desire/inability to cut down | 7.3 × 10−8 | 0.66 |
| 13 | 101,766,174 | rs17484734 | 0.06 | NALCN | Probability of high-risk class | 4.1 × 10−8 | 0.32 |
| 13 | 101,766,174 | rs17484734 | 0.06 | NALCN | Withdrawal | 1.3 × 10−7 | 0.88 |
| 15 | 75,415,962 | rs2029519 | 0.25 | LOC100128721 (8kb) | Withdrawal | 2.6 × 10−7 | 0.22 |
| 15 | 75,422,131 | rs4479194 | 0.24 | LOC100128721 (14kb) | Withdrawal | 2.4 × 10−7 | 0.19 |
| 15 | 75,424,593 | rs7172677 | 0.24 | LOC100128721 (17kb) | Withdrawal | 2.8 × 10−7 | 0.24 |
DSM-IV = alcohol dependence; COGA = Collaborative Study on the Genetics of Alcoholism; SAGE = Study of Addiction: Genes and Environment
Chromosome;
Chromosomal position (base pairs) based on human genome build 19, dbSNP 137
Single Nucleotide Polymorphism;
Minor allele frequency estimated on founders
Gene name and (distance to nearest gene)
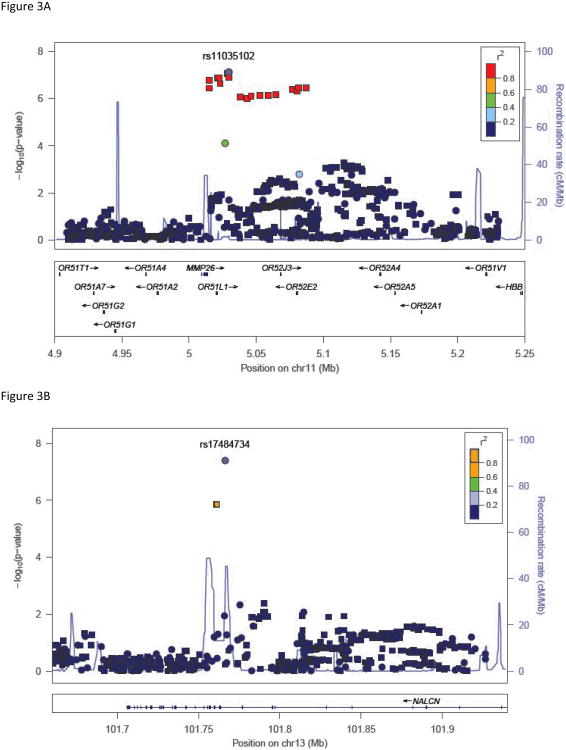
Two genotyped SNPs reached genome-wide significance (p<8.4 × 10−8). The most significant result was for a SNP on chromosome 13 in NALCN, (sodium leak channel, non-selective), rs17484734 (p=4.1 × 10−8, Figure 3A) for the probability of high-risk membership. This SNP also showed evidence of association with withdrawal (p=1.3 × 10−7). This SNP is in a recombination hot spot, and thus is not in linkage disequilibrium (LD) with the surrounding genotyped SNPs. There were no other genotyped SNPs in this region demonstrating association with these phenotypes. One imputed SNP, rs1151384, in moderate LD with rs17484734 (r2= 0.80) provided supporting evidence of association with both phenotypes (p≤1.4 × 10−6 for probability of high risk membership and withdrawal). No other imputed SNPs were in LD with these two SNPs (r2<0.20).
Figure 3. Association results with key phenotypes.
(A) High-risk latent class probability with SNPs in the region around rs17484734 in NALCN. (B) Persistent desire/inability to cut down with SNPs in the region around rs11035201 in OR51L1. Y-axis denotes the –log10(p-value) for association. X-axis is the physical position on the chromosome (Mb). The most significantly associated SNP is shown in purple. The extent of linkage disequilibrium (as measured by r2) between each SNP and the most significantly associated SNP is indicated by the color scale at top right. Larger values of r2 indicate greater linkage disequilibrium. Genotyped SNPs are indicated as circles, and imputed SNPs by squares.
The DSM-IV criterion persistent desire/inability to cut down on drinking also exceeded genome-wide significance thresholds for rs11035102, in the olfactory receptor OR51L1 (olfactory receptor, family 51, subfamily L, member 1; p=7.3 × 10−8). Evaluation of imputed SNPs in this region revealed supporting evidence of genome-wide association for rs11035097 (p=8.3 × 10−8) in addition to moderate association with six other SNPs all in high LD with the genotyped SNP (r2≥0.80, all p<9.7 × 10−7). OR51L1 is located in a region of chromosome 11 with many other olfactory receptors (Figure 3B). The genotyped SNP also demonstrated suggestive association with drinking despite physical problems (p=2.1 × 10−6) and with the high-risk latent class probability trait (p=4.9 × 10−5).
Replication of Association Results
LCA was performed in the SAGE sample with comparable results to the COGA sample, including probabilities of criteria endorsement. When examining class assignments of (a) SAGE subjects in the SAGE+COGA versus the primary SAGE sample, and (b) the 106 overlapping COGA subjects in SAGE (23 individuals were excluded in the COGA analysis due to the restriction on regular drinking), a McNemar's change test demonstrated no significant change in class assignment in the SAGE sample compared to SAGE+COGA nor in the 106 overlapping individuals (all p>0.72). Nine of the 10 SNPs with top association results in Table 4 were genotyped in the SAGE data, imputed dosage data was used for rs12006002, which had not been genotyped in SAGE. The SNPs did not demonstrate association with any of the phenotypes (all p>0.16).
Discussion
This is the first GWAS to our knowledge that examines each individual criterion of the DSM-IV AD diagnosis as well as phenotypes defined by the probability of latent class assignment. As such, it is the first association study demonstrating genome-wide significance with one of the DSM-IV AD criteria as well as with the phenotype derived from the probability of membership in the high-risk class. This suggests that characterizing more homogeneous subgroups of individuals, defined either by a specific AD criterion or by the probability of belonging to a latent class, increased the power to detect genetic association relative to the diagnosis of AD, a particularly heterogeneous phenotype. For example, estimated power was equivalent for the two phenotypes DSM-IV and persistent desire/inability to cut down, yet one of the strongest association results was with the persistent desire/inability to cut down, p=7.3 × 10−8, while there was modest evidence of association with DSM-IV (p=1.5 × 10−5, Supplementaltal Table 1).
We estimated the heritability of AD, the individual DSM-IV criteria, as well as two continuous phenotypes based on the LCA probabilities. The estimated heritability for AD (61%) was similar to previous reports (Prescott et al., 2006). However, the heritability estimates for the individual DSM-IV criteria were slightly larger than previously reported (Slutske et al., 1999). This may reflect the effects of shared environment, which is known to inflate the estimate of the genetic contribution to the trait (Tenesa and Haley, 2013). Although the phenotypes based on the probabilities of class assignment were less heritable (≤ 34%), actual membership in each class was more heritable (≥52%). This may be related to the entropy such that misclassification may have favored the assignment of related individuals into the same class. The less heritable moderate risk class (17%) could be a result of environmental influences, and therefore was not examined further.
As seen in Table 4, the individual DSM-IV criteria provided stronger evidence of association than DSM-IV AD. This suggests that individuals who endorse a specific criterion may have a more common genetic etiology compared with the heterogeneous group of AD individuals, who are diagnosed by endorsing different combinations of DSM-IV criteria. Furthermore, association was observed with a quantitative trait derived from the probability of belonging to the high-risk latent class, despite its lower heritability estimate compared to the heritability of AD. This is consistent with Kuo et al. (Kuo et al., 2008), who found that the first factor derived in a factor analysis was a better fit to a general population of social drinkers when combined with a latent class framework of three classes of drinkers (severe, moderate, and non-problem drinkers).
Our goal was to classify individuals, not symptoms and hence, we employed a latent class approach rather than a factor analysis model defining one continuous latent trait. It is noteworthy however that the pattern of LCA results obtained here (high, moderate, low, versus subtypes) are typical of data that are well-suited to factor analysis. The use of latent class analysis has successfully been employed to examine patterns of alcohol abuse and dependence symptoms (Beseler et al., 2012; Bucholz et al., 1996; Ko et al., 2010; Moss et al., 2008) as well as other addictions such as nicotine dependence (Agrawal et al., 2011), and opioid dependence (Shand et al., 2011). The majority of these studies also found a 3-class model was the best fit, with the classes representing a severity range from low-risk to high-risk. There are several advantages to examining individuals according to their risk profile, including identifying individuals in a trajectory towards future dependence, distinguishing the best clinical or pharmacological treatment for moderate or at-risk individuals, and -- importantly for this study – reducing genetic and phenotypic heterogeneity.
One SNP, rs11035102, which is located in a region of several olfactory genes, showed genome-wide significant association with having a persistent desire to cut down/inability to do so criteria for AD, with strong support from several imputed SNPs in high LD (Figure 3B). This SNP was also moderately associated with the criterion of drinking alcohol despite physical/psychological consequences in the present study and with the total number of DSM-IV criteria (symptom count) in the same COGA family sample (p=3.8 × 10−5, Wang et al., 2012; Supplemental Table 2). This gene is near two olfactory receptors (OR52E2, OR52J3, seen in Figure 3B) shown previously to be significantly associated with number of cigarettes smoked per day in a pathway analysis across three independent cohorts ascertained on nicotine or AD, one of which was the COGA sample (Harari et al., 2012). In a study of opioid-dependent individuals, the desire/inability to cut down was found to be one of the most prevalent substance use disorder criteria across five substances of abuse, including alcohol (Wu et al., 2012). Although this was not the most prevalent criterion in our sample, it was the criterion with the most diverse group of people, with 7% of the individuals endorsing this criterion being in the low-risk class, 53% from the moderate-risk class and 40% from the high-risk class. There is strong evidence supporting a relationship between learned olfactory sensory cues and alcohol consumption and dependence. It was shown that after alcohol odor stimuli, alcoholics found it more difficult to resist a drink than before the stimuli (Stormark et al., 1995). Priming subjects with an alcohol odor also increased activation of the limbic system (Bragulat et al., 2008) and led to enhanced craving for alcohol (Kareken et al., 2010; Kareken et al., 2004).
The strongest association was between a SNP in a nonselective sodium leak channel NALCN and the probability of high-risk membership. This SNP, rs17484734, is in a recombination hotspot (Figure 3A). As a result, only one imputed SNP provided supporting evidence for association. This channel can be activated by several peptides including acetylcholine and substance P, both of which have been shown to interact with alcohol. Acetylcholine is released when alcohol binds to the nicotinic acetylcholine receptors as an agonist (Bito-Onon et al., 2011; Feduccia et al., 2012). Substance P, through interaction with the neurokinin 1 receptor, has been implicated in alcohol consumption, preference, and reinstatement, and is a potential therapeutic target for AD (Schank et al., 2012). One of the components of the NALCN complex is UNC79, the homolog of which was shown to increase alcohol consumption in mice and led to increased sensitivity to alcohol in C. elegans (Speca et al., 2010).
Six of the SNPs presented in Table 4 also demonstrated moderate association with symptom count (rs17484734, rs12006002, rs2029519, rs4479194, rs7172677, rs11035102; all p<7.9 × 10−5) (Wang et al., 2012). The most significant association reported with the symptom count phenotype was with rs12903120 in C15orf53. This SNP was not associated with any of the phenotypes examined in this study (0.36≤p≤2.7 × 10−5). Since the symptom count phenotype is the total number of endorsed DSM-IV criteria, we might have expected more overlap between results with the individual criterion. However, there are several possible explanations for the observed differences. First, if our hypothesis is correct that different genetic factors may contribute to each criterion, then analyzing them separately would be a more powerful approach to identify the individual genetic effects. Second, there are some differences in the sample used in these two analyses as well as the analytic methods. For example, in the analyses reported herein, we employed a stricter inclusion criterion that only allowed regular drinkers. Previous analyses of alcohol symptom count utilized a more inclusive criterion that included all individuals reporting to have ever consumed alcohol.
We did not find additional support in the SAGE sample. This may be due to several factors. First, the power to replicate findings of relatively small effect, such as those identified in this sample, is modest in replication samples of the size of SAGE. Second, the COGA family GWAS sample is quite unique in that the families were selected because of their large size and density of AD. Much of the SAGE sample consists of individuals recruited for nicotine and cocaine, then selected for comorbid AD. The risk factors contributing to AD and our AD-related phenotypes may be different from those more commonly found in sporadic AD or AD associated with other substance dependence. Finally, the lack of replication may be that our initial COGA findings are false positive associations. Clearly, further studies, ideally in family samples ascertained for a high level of AD, are necessary.
The design of this study has several strengths. First, because most families have a strong history of AD, the sample is likely enriched for a greater genetic contribution to risk. Second, family-based analysis is robust to population substructures such as slight differences in ethnicity. This family-based design has been previously used to test for association with event-related brain oscillations correlated with deficits in alcoholics and offspring who are at risk for AD, with genome-wide significant association (p = 4.7 × 10−10) of SNPs in KCNJ6 (Kang et al., 2012). We did not correct the significance threshold for the number of phenotypes analyzed; however, there was no indication of inflated association results, and the correlated phenotypes are all related to AD. A weakness of the study is the cross-sectional nature of the phenotypes employed. Thus a longitudinal investigation to follow-up on individuals in the low-risk and especially those in the moderate-risk classes who are not yet AD could provide insight as to clusters of individuals who change latent classes over time, and further evidence of the validity of employing homogenous phenotypes and suggest a genetic mechanism of liability.
In summary, this family-based GWAS demonstrated two genome-wide significant associations, one of a polymorphism in an olfactory receptor with the persistent desire or inability to cut down on alcohol consumption, and the other with a sodium leak channel and the high-risk latent class trait. These results validate the advantage of defining a more homogenous sample in which to analyze genetic associations. These results combined with those of other recent studies (Kang et al., 2012; Wang et al., 2012) also demonstrate the valuable resource of the densely affected European American families of COGA.
Supplementary Material
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. J. Edenberg, L. J. Bierut, includes 10 different centers: University of Connecticut (V. Hesselbrock); Indiana University (H. J. Edenberg, J. Nurnberger Jr, T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. J. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy), Howard University (R. Taylor) and Virginia Commonwealth University (D. Dick). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, S. O'Connor, L. Wetherill, X. Xuei (Indiana University); G. Chan (University of Iowa); N. Manz, M. Rangaswamy (SUNY Downstate); J. Rohrbaugh, J.-C. Wang (Washington University in St. Louis); A. Brooks (Rutgers University); and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions.
This national Collaborative Study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). Funding support for GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
References
- Agrawal A, Scherrer JF, Pergadia ML, Lynskey MT, Madden PA, Sartor CE, Grant JD, Duncan AE, Haber JR, Jacob T, Bucholz KK, Xian H. A latent class analysis of DSM-IV and Fagerstrom (FTND) criteria for nicotine dependence. Nicotine Tob Res. 2011;13(10):972–81. doi: 10.1093/ntr/ntr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Verweij KJ, Gillespie NA, Heath AC, Lessov-Schlaggar CN, Martin NG, Nelson EC, Slutske WS, Whitfield JB, Lynskey MT. The genetics of addiction-a translational perspective. Transl Psychiatry. 2012;2:e140. doi: 10.1038/tp.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders DSM-IV. 4th. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit M, Edenberg H, Rice JP. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health & Research World. 1995;19(3):228–236. [PMC free article] [PubMed] [Google Scholar]
- Beseler CL, Taylor LA, Kraemer DT, Leeman RF. A latent class analysis of DSM-IV alcohol use disorder criteria and binge drinking in undergraduates. Alcohol Clin Exp Res. 2012;36(1):153–61. doi: 10.1111/j.1530-0277.2011.01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bito-Onon JJ, Simms JA, Chatterjee S, Holgate J, Bartlett SE. Varenicline, a partial agonist at neuronal nicotinic acetylcholine receptors, reduces nicotine-induced increases in 20% ethanol operant self-administration in Sprague-Dawley rats. Addict Biol. 2011;16(3):440–9. doi: 10.1111/j.1369-1600.2010.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragulat V, Dzemidzic M, Talavage T, Davidson D, O'Connor SJ, Kareken DA. Alcohol sensitizes cerebral responses to the odors of alcoholic drinks: an fMRI study. Alcohol Clin Exp Res. 2008;32(7):1124–34. doi: 10.1111/j.1530-0277.2008.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. JStudAlcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Heath AC, Reich T, Hesselbrock VM, Kramer JR, Nurnberger JI, Jr, Schuckit MA. Can we subtype alcoholism? A latent class analysis of data from relatives of alcoholics in a multicenter family study of alcoholism. Alcohol Clin Exp Res. 1996;20(8):1462–71. doi: 10.1111/j.1530-0277.1996.tb01150.x. [DOI] [PubMed] [Google Scholar]
- Bureau A, Croteau J, Tayeb A, Merette C, Labbe A. Latent class model with familial dependence to address heterogeneity in complex diseases: adapting the approach to family-based association studies. Genet Epidemiol. 2011;35(3):182–9. doi: 10.1002/gepi.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Manichaikul A, Rich SS. A generalized family-based association test for dichotomous traits. Am J Hum Genet. 2009;85(3):364–76. doi: 10.1016/j.ajhg.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33(4):837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. AddictBiol. 2006;11(3-4):386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34(5):840–52. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. HumMolGenet. 2006;15(9):1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Enoch MA. The role of GABA(A) receptors in the development of alcoholism. PharmacolBiochemBehav. 2008;90(1):95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34(5):1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feduccia AA, Chatterjee S, Bartlett SE. Neuronal nicotinic acetylcholine receptors: neuroplastic changes underlying alcohol and nicotine addictions. Front Mol Neurosci. 2012;5:83. doi: 10.3389/fnmol.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol ClinExpRes. 2000;24(7):933–945. [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Moller N, Hermansen L, Winokur G, Guze SB. Drinking problems in adopted and nonadopted sons of alcoholics. Arch Gen Psychiatry. 1974;31(2):164–9. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- Grucza RA, Bucholz KK, Rice JP, Bierut LJ. Secular trends in the lifetime prevalence of alcohol dependence in the United States: a re-evaluation. Alcohol Clin Exp Res. 2008;32(5):763–70. doi: 10.1111/j.1530-0277.2008.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. ArchGenPsychiatry. 2007;64(7):830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. PsycholMed. 1997;27(6):1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94(9):1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Jr, Rice J, Schuckit M, Tischfield J, Bierut LJ, Edenberg HJ, Goate A, Foroud T, Porjesz B. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012;11(6):712–9. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareken DA, Bragulat V, Dzemidzic M, Cox C, Talavage T, Davidson D, O'Connor SJ. Family history of alcoholism mediates the frontal response to alcoholic drink odors and alcohol in at-risk drinkers. Neuroimage. 2010;50(1):267–76. doi: 10.1016/j.neuroimage.2009.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareken DA, Claus ED, Sabri M, Dzemidzic M, Kosobud AE, Radnovich AJ, Hector D, Ramchandani VA, O'Connor SJ, Lowe M, Li TK. Alcohol-related olfactory cues activate the nucleus accumbens and ventral tegmental area in high-risk drinkers: preliminary findings. Alcohol Clin Exp Res. 2004;28(4):550–7. doi: 10.1097/01.alc.0000122764.60626.af. [DOI] [PubMed] [Google Scholar]
- Kessler DA. Alcohol marketing and youth: the challenge for public health. J Public Health Policy. 2005;26(3):292–5. doi: 10.1057/palgrave.jphp.3200041. [DOI] [PubMed] [Google Scholar]
- Ko JY, Martins SS, Kuramoto SJ, Chilcoat HD. Patterns of alcohol-dependence symptoms using a latent empirical approach: associations with treatment usage and other correlates. J Stud Alcohol Drugs. 2010;71(6):870–8. doi: 10.15288/jsad.2010.71.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Aggen SH, Prescott CA, Kendler KS, Neale MC. Using a factor mixture modeling approach in alcohol dependence in a general population sample. Drug Alcohol Depend. 2008;98(1-2):105–14. doi: 10.1016/j.drugalcdep.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Hewitt BG, Grant BF. The Alcohol Dependence Syndrome, 30 years later: a commentary. The 2006 H. David Archibald lecture. Addiction. 2007;102(10):1522–1530. doi: 10.1111/j.1360-0443.2007.01911.x. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence and affective disorders: results from an extended case-control structured association study. HumMolGenet. 2005;14(16):2421–2434. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- Moss HB, Chen CM, Yi HY. DSM-IV criteria endorsement patterns in alcohol dependence: relationship to severity. Alcohol Clin Exp Res. 2008;32(2):306–13. doi: 10.1111/j.1530-0277.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- Muthen LK, Muthen BO. Mplus User's Guide. 6th. Muthen & Muthen; Los Angeles CA: 1998-2011. [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. AmJHumGenet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, Robinson VP, Neale MC, van den Oord EJ, Walsh D, Riley BP, Kendler KS. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. MolPsychiatry. 2006;11(6):603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. NatGenet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schank JR, Ryabinin AE, Giardino WJ, Ciccocioppo R, Heilig M. Stress-related neuropeptides and addictive behaviors: beyond the usual suspects. Neuron. 2012;76(1):192–208. doi: 10.1016/j.neuron.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol ClinExp Res. 2001;25(3):323–329. [PubMed] [Google Scholar]
- Shand FL, Slade T, Degenhardt L, Baillie A, Nelson EC. Opioid dependence latent structure: two classes with differing severity? Addiction. 2011;106(3):590–8. doi: 10.1111/j.1360-0443.2010.03217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske WS, True WR, Scherrer JF, Heath AC, Bucholz KK, Eisen SA, Goldberg J, Lyons MJ, Tsuang MT. The heritability of alcoholism symptoms: “indicators of genetic and environmental influence in alcohol-dependent individuals” revisited. Alcohol Clin Exp Res. 1999;23(5):759–69. doi: 10.1111/j.1530-0277.1999.tb04181.x. [DOI] [PubMed] [Google Scholar]
- Speca DJ, Chihara D, Ashique AM, Bowers MS, Pierce-Shimomura JT, Lee J, Rabbee N, Speed TP, Gularte RJ, Chitwood J, Medrano JF, Liao M, Sonner JM, Eger EI, 2nd, Peterson AS, McIntire SL. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormark KM, Laberg JC, Bjerland T, Nordby H, Hugdahl K. Autonomic cued reactivity in alcoholics: the effect of olfactory stimuli. Addict Behav. 1995;20(5):571–84. doi: 10.1016/0306-4603(95)00017-7. [DOI] [PubMed] [Google Scholar]
- Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013;14(2):139–49. doi: 10.1038/nrg3377. [DOI] [PubMed] [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NXH, Bertelsen S, Budde J, Chou YL, Harari O, Koller DL, Wetherill L, Agrawal A, Almasy L, Brooks A, Bucholz K, Dick D, Hesselbrock V, Kang S, Kapoor M, Kramer J, Kuperman S, Manz N, McClintick JN, Nurnberger JJ, Ragaswamy M, Rice J, Schuckit M, Tischfield JA, Xuei X, Porjesz B, Heath AC, Edenberg HJ, Bierut LJ, Goate AM. A genome wide association study of alcohol dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, Fox L, Goldstein E, Reyes O, Saccone N, Saccone S, Xuei X, Bucholz K, Kuperman S, Nurnberger J, Jr, Rice JP, Schuckit M, Tischfield J, Hesselbrock V, Porjesz B, Edenberg HJ, Bierut LJ, Goate AM. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. MolPsychiatry. 2009;14(5):501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, Jones K, Nurnberger JI, Jr, Tischfield J, Porjesz B, Edenberg HJ, Hesselbrock V, Crowe R, Schuckit M, Begleiter H, Reich T, Goate AM, Bierut LJ. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. HumMolGenet. 2004;13(17):1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Wu LT, Blazer DG, Woody GE, Burchett B, Yang C, Pan JJ, Ling W. Alcohol and drug dependence symptom items as brief screeners for substance use disorders: results from the Clinical Trials Network. J Psychiatr Res. 2012;46(3):360–9. doi: 10.1016/j.jpsychires.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.