Abstract
The glaucoma-associated olfactomedin domain of myocilin (myoc-OLF) is a recent addition to the growing list of disease-associated amyloidogenic proteins. Inherited, diseasecausing myocilin variants aggregate intracellularly instead of being secreted to the trabecular meshwork (TM), which is a scenario toxic to TM cells and leads to early onset of ocular hypertension, the major risk factor for glaucoma. Here we systematically structurally and biophysically dissected myoc-OLF to better understand its amyloidogenesis. Under mildly destabilizing conditions, wild-type myoc-OLF adopts non-native structures that readily fibrillize when incubated at a temperature just below the transition for tertiary unfolding. With buffers at physiological pH, two main end-point fibril morphologies are observed: (a) straight fibrils common to many amyloids and (b) unique micron-length, ~300 nm or larger diameter species that lasso oligomers, which also exhibit classical spectroscopic amyloid signatures. Three disease-causing variants investigated herein exhibit non-native tertiary structures under physiological conditions, leading to accelerated growth rates and a variety of fibril morphologies. In particular, the well-documented D380A variant, which lacks calcium, forms large circular fibrils. Two amyloid forming peptide stretches have been identified, one for each of the main fibril morphologies observed. Our study places myoc-OLF within the larger landscape of the amylome and provides insight into the diversity of myoc-OLF aggregation that plays a role in glaucoma pathogenesis.
Keywords: Amyloid, protein misfolding, protein structure, circular dichroism, atomic force microscopy
Myocilin glaucoma is a recent addition to the family of protein misfolding disorders. When it harbors one of numerous non-synonymous mutations, myocilin, the gene product most closely linked to inherited forms of primary open glaucoma1, forms cytotoxic aggregates2; 3; 4; 5 within human trabecular meshwork (HTM) cells located in the anterior segment of the eye. The toxic gain-of-function pathophysiology leading to the early onset of ocular hypertension, the major risk factor for glaucoma that leads to retina degeneration, is not well understood. On a cellular level, mutant myocilin interacts with endoplasmic reticulum (ER)-resident chaperones in a way that sabotages its ability to be retrotranslocated and then cleared by the proteasome6. Upon HTM cell death, debris is presumed to block outflow of fluid through the trabecular meshwork tissue, which in turn affects intraocular pressure. Elevated levels of wild type myocilin are also associated with glaucoma induced by steroid treatment7, but the pathogenic mechanism for this glaucoma subtype, is likewise unclear. Biophysical characterization of the olfactomedin domain of myocilin (myoc-OLF)8, the location of nearly all known glaucoma-causing mutations9,10, has revealed that glaucoma-causing mutations lead to decreased thermal stability that correlate with age of disease diagnosis11,12. Moreover, aggregates of this calcium-binding domain13 were recently found to exhibit hallmarks of amyloid fibrils in vitro, and sequestered mutant myocilin forms thioflavin T (ThT) positive aggregates in cell culture 14.
Viewed through the new lens of disease-relevant amyloid proteins, which share a non-native cross-p core structure15, myocilin has intriguing features. First, mutant myocilin cytotoxicity originates in aggregation of mutant myocilin within the ER3; 16 whereas amyloid proteins are generally found to accumulate in the cytosol or extracellularly17. Second, although glaucoma is a neurodegenerative disease of the retina leading to irreversible vision loss18, thus far myocilin has not been directly implicated in neurotoxicity like numerous other well-studied amyloid proteins. Relatedly, myocilin is expressed in tissues throughout the body, and is conserved among higher eukaryotes where homologs are expressed in neural tissues19; aggregation in such tissues may be relevant but is currently unexplored. Third, beyond intracellular aggregation of mutant myocilin, extracellular, wild-type myocilin is subjected to the full complement of age-onset misfolding triggers: repeated mechanical insults, pH imbalance, UV radiation, and reactive oxygen species20; 21. Even though such conditions induce myoc-OLF to fibrillize in vitro14, and glaucoma is highly prevalent worldwide22, the vast majority of the aging population does not have glaucoma, suggesting that myocilin can generally withstand these insults in the extracellular milieu. Fourth, recombinant ~30 kDa myoc-OLF is well-folded, thermally stable protein8, and does not aggregate under physiological pH and temperature except with agitation14. The core structural domain, ~ 25 kDa, contains high levels of beta-sheet secondary structure8, in a yet-unknown three dimensional architecture. The remaining ~5 kDa is sensitive to proteolysis, and circular dichroism (CD) deconvolution corroborates that the myoc-OLF contains a significant contribution from unstructured polypeptide8. Amyloid formation is well-known for both folded and natively unfolded proteins, and the combination of the two in a single domain presents an interesting platform to study nuances of amyloidogenesis.
To gain further insight into myocilin amyloidogenesis we undertook a detailed molecular biophysical study of myoc-OLF. Here we describe a spectrum of populated states of wild-type myoc-OLF from native to the acid-induced unfolded, and identify key features and the conditions prone to fibrillization or non-specific aggregation. Of the states that readily form fibrils, different experimental conditions such as pH, salt, temperature, and details of experimental materials and equipment, lead to different fibril morphologies. Two main structures have been identified by atomic force microscopy (AFM), typical long straight fibrils and a highly unusual, micron-length circular species that encase oligomers. Both fibril forms are ThT positive, Congo Red sensitive, and exhibit vibrational spectroscopic signatures of amyloid. The three disease-causing myoc-OLF variants investigated here form analogous fibrils under physiological buffers and temperatures; the calcium-depleted D380A variant forms the circular morphology. Two peptide stretches identified by bioinformatics fibrillize into these disparate structures, suggesting they contain the core sequences responsible for these fibrils in myoc-OLF. Besides expanding our knowledge of the amylome superfamily, our findings raise some new issues regarding myocilin glaucoma pathogenesis.
RESULTS
Overview
We systematically perturbed the myoc-OLF structure to identify features that lead to facile amyloid formation. Structural and stability changes of wild-type myoc-OLF at five pH values above and below the calculated pI (~5) were monitored by far- and near-UV CD spectra and thermal melts. Effects due to charge screening were accounted for by comparing results with and without 200 mM NaCl. The corresponding extent of exposed hydrophobicity was tested using anilinonaphthalene-1-sulfonate (ANS) fluorescence at room temperature. To relate the identified partial unfolded states with amyloid propensity, kinetics of fiber formation were monitored using the fluorescent amyloid indicator dye ThT23. Experiments were carried out over the course of 18–90 h in a cuvette or plate reader held at 36 °C to approximate the physiological temperature of the eye24 or at a slightly elevated temperature of 42 °C (see below). End point morphologies were then visualized by AFM. Neutral pH wild-type fibers were further characterized by Fourier Transform Infrared Spectroscopy (FTIR) and Congo Red absorbance. In addition to wild-type myoc-OLF, experiments were conducted on four representative variants and selected peptides at physiological pH and temperature.
pH screen of wild-type myoc-OLF fibrillization in buffers lacking NaCl
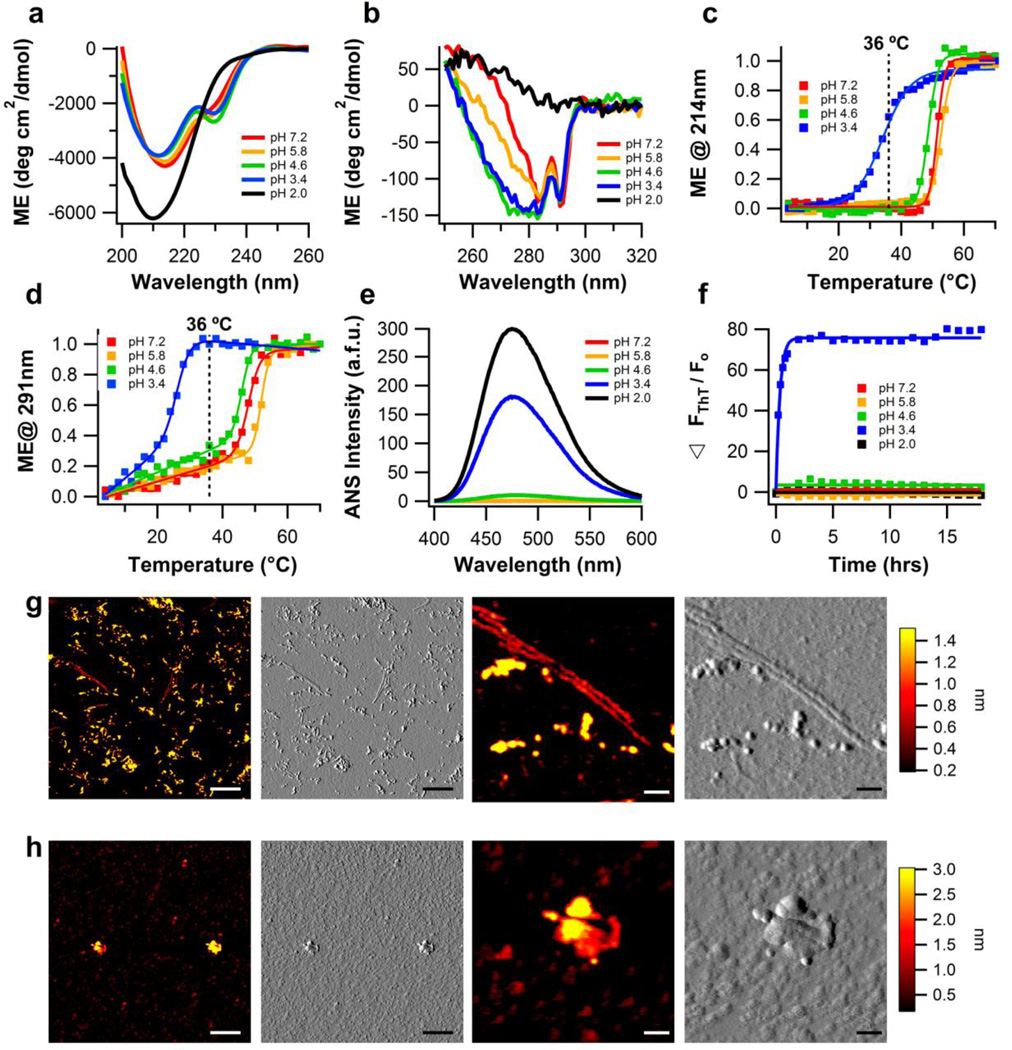
The secondary structure of myoc-OLF is relatively unchanged over the pH range of 7.2 to 3.4 but notable changes in tertiary structure are seen. Across this pH range, the far-UV CD spectrum exhibits the same, previously reported8 minimum around 215–217 nm typical of β-sheet structure and a shoulder at 230 nm (Fig. 1a). The more pronounced 230 nm trough in the pH 4.6 and pH 3.4 spectra is consistent with changes to the environment of aromatic residues25, as detailed in the near UV spectrum where the shoulder between 280 - 250 nm26; 27 appears to broaden (Fig. 1b). At pH 2, myoc-OLF is reminiscent of an acid-induced unfolded state as evidenced by the altered secondary structure in the far-UV CD spectrum (Fig. 1a) and loss of tertiary structure indicated by the flat near-UV CD signal28; 29 (Fig. 1b). CD thermal melts reveal that whereas secondary structure unfolding appears to follow a sigmoidal transition (Fig. 1c), tertiary thermal transitions exhibit a shallow pre-transition phase followed by a sharp main unfolding event (Fig. 1d). Thermal melts following changes in intrinsic tryptophan fluorescence confirm this pre-transition slope, with a 0.4 nm shift in maximum fluorescence to lower energies every 10 °C (Supplemental Fig. S1). Myoc-OLF is least stable in acidic buffers where the protein is highly positively charged, and thermal stability decreases as the protein is shifted to buffers with pH values below its predicted pI (Table 1). ANS fluorescence is highest for myoc-OLF at pH 2 and pH 3.4, consistent with high levels of exposed hydrophobic residues (Fig. 1e). ANS measurements reveal a 17-fold higher fluorescence (Table 1) for myoc-OLF destabilized at pH 3.4 compared to the more compact state at pH 4.6 (Fig. 1e).
Figure 1. Biophysical characterization of wild-type myoc-OLF as a function of pH, without NaCl.
(a) Secondary structure of myoc-OLF as a function of pH measured by far-UV CD. (b) Tertiary structure of myoc-OLF as a function of pH measured by near-UV CD. (c) Thermal unfolding of secondary structure monitored by CD at 214 nm. Fit is sigmoidal. (d) Thermal unfolding of tertiary structure monitored by CD at 291 nm. Fit is linear plus sigmoidal. (e) ANS fluorescence as a function of pH. (f) ThT fluorescence as a function of pH at 36 °C monitored for 18 hours. (g) Fibrillar end point morphology for samples incubated at 36 °C in pH 3.4 buffer (h) Disordered aggregates from samples incubated at pH 2.0 buffer at 36 °C, upon deposition (see text). For (g) and (h), two left panels scale bar = 300nm and two right panels scale bar = 50nm.
Table 1.
Summary of biophysical properties measured for myoc-OLF samples in this study.
| Secondary Structure Tm ( °C) |
Tertiary Structure Tm ( °C) |
ANS Fl. | ΔFThT/Foa | Fibril Growth t1/2 (hrs) |
Fibril Morphology |
|
|---|---|---|---|---|---|---|
|
Wild-type myoc-OLF |
||||||
| pH 7.2 | 51.5 ± 0.1 | 48.9 ± 0.6 | 0.9 ± 0.5 | 2.4 | ---- | |
| 70.6b | 2.0 | Straight | ||||
| pH 5.8 | 52.7 ± 0.1 | 52.0 ± 0.4 | 1.1 ± 0.7 | −0.4 | ---- | |
| pH 4.6 | 48.7 ± 0.1 | 46.0 ± 0.3 | 10.6 ± 0.4 | 2.3 | ---- | |
| pH 3.4 | 34.0 ± 0.6 | 29.0 ± 1.3 | 181.5 ± 2.3 | 79.8 | 0.4 ± 0.2 |
Long, straight |
| pH 2 | N/A | N/A | 298.3 ± 6.9 | −1.7 | ---- | |
| pH 7.2/NaCl | 52.2 ± 0.6 | 50.9 ± 0.2 | 1.1 ± 1.1 | −0.7 | ||
| 89.4b | 27.7 ± 2.6 | Circular | ||||
| pH 5.8/NaCl | 54.0 ± 0.1 | 52.8 ± 0.3 | 1.4 ± 1.4 | −0.5 | ---- | |
| pH 4.6/NaCl | 46.4 ± 0.1 | 43.1 ± 0.2 | 4.5 ± 2.3 | −0.5 | 9.5 ± 3.9 |
Long, straight |
| pH 3.4/NaCl | N/A | N/A | 234.7 ± 3.9 | 21.5 | ---- | |
| pH 2/NaCl | N/A | N/A | 346.8 ± 24.0 | 16.0 | ---- | |
|
Myoc-OLF variants |
||||||
| I499F | 45.0 ± 1.1 | 43.2 ± 0.5 | 3.4 ± 1.1 | 88.0 | 3.5 ± 0.2 |
Straight fibrils and oligomers |
| D380A | 46.8 ± 0.1 | 44.4 ± 0.6 | 2.1 ± 0.4 | 102.2 | 11.9 ± 4.7 | Circular |
| A427T | 48.9 ± 0.1 | 45.9 ± 0.2 | 1.3 ± 0.1 | 28.5 | >25c |
Straight fibrils and oligomers |
| K398R | 51.1 ± 0.4 | 49.5 ± 2.4 | 0.6 ± 0.2 | −0.7 | ---- |
Bold: Fibrillar structures confirmed by AFM.
N/A: Not available because secondary structure is already disordered at 4 °C.
ThT fluorescence recorded after incubation for 18 h (no salt) or 50 h (200mM NaCl) at 36 °C.
ThT fluorescence recorded after incubation for 18 h (no salt) or 90 h (200mM NaCl) at 42 °C.
Steady-state maximum not reached by termination of experiment at 50 h.
Of the conditions tested, only the distinct partially unfolded state accessible at pH 3.4 is primed for fibril formation at 36 °C (Fig. 1f). None of the other conditions led to fibril growth at this temperature, including the acid-induced unfolded state at pH 2 and the natively folded state at pH 7.2 (Fig. 1f). Corresponding AFM images of fibrils grown in pH 3.4 buffer at 36 °C reveal characteristic straight amyloid morphology (average contour length: 271 ± 164 nm, height: 0.9 ± 0.4 nm, and actual width: 8 ± 2 nm), as well as less ordered laterally assembling oligomers (average contour length: 126 ± 71 nm, height: 2.0 ± 0.5 nm, and actual width: 10 ± 2 nm) (Fig. 1g). AFM images confirm that no fibrils grew from experiments conducted at pH 7.2, pH 5.8, or pH 4.6 at 36 °C (Supplemental Fig. S2 a, b, c). Finally, although samples deposited from conditions at pH 2 appear as disordered aggregate (Fig. 1h), we believe that these are an artifact of the deposition of unstructured monomers on the mica surface because the average particle height (4.2 ± 0.3 nm and 1.3 ± 0.4 nm) is comparable to monomeric myoc-OLF prior to aggregation (Supplemental Fig. S3) and no increase in scattering at 600 nm during the incubation period was seen (Supplemental Fig. S4). Shorter fibril heights of ~ 1 nm, such as those seen in AFM images at pH 3.4 (Fig. 1g), are consistent with dehydrated myoc-OLF monomer deposited on mica for AFM (Supplemental Fig. S3 b, c).
pH screen of wild-type myoc-OLF fibrillization in buffers supplemented with 200 mM NaCl
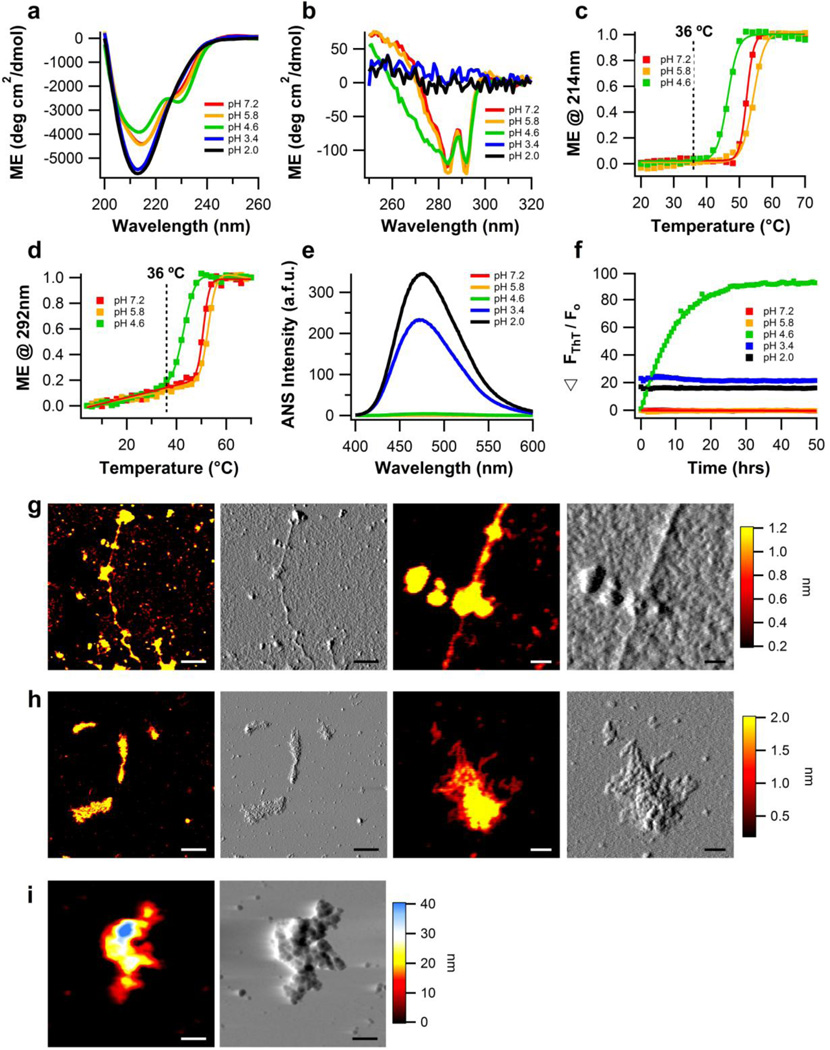
Similar to results obtained with buffers lacking NaCl, the secondary structure of wild-type myoc-OLF is unchanged for myoc-OLF in buffers at pH 7.2, pH 5.8 and pH 4.6 with 200mM NaCl (Fig. 2a). For pH 4.6, there is a slight increase in the feature at 230 nm, again suggesting a change in the aromatic environment (Fig. 2a). This interpretation is supported by a significantly broadened shoulder in the near UV-CD spectrum between 280-250 nm26,27 (Fig. 2 a, b). Unlike the parallel case without salt, however, the addition of 200mM NaCl at pH 3.4 leads to extensive disorder in tertiary structure and altered secondary structure (Fig. 2 a, b). For pH 2, the presence of 200mM NaCl does not change the extent of order compared to samples incubated without salt (Fig. 2 a, b). Secondary structure melts are best modeled as sigmoidal with a melting temperature several degrees Celsius higher than tertiary unfolding, which, as without salt (Fig. 1 c, d), follows a shallow pre-transition step before the main unfolding event (Fig. 2 c, d). In comparison to that in the absence of NaCl, the pre-transition in the presence of 200mM NaCl is less pronounced, most likely due to increased electrostatic screening of charged residues30 (Fig. 1d, 2d). CD results are corroborated by ANS fluorescence, which is highest for pH 2 and pH 3.4, and low for all other pH values, including pH 4.6 (Fig. 2e and Table 1).
Figure 2. Biophysical characterization of wild-type myoc-OLF as a function of pH, in the presence of 200mM NaCl.
(a) Secondary structure of myoc-OLF as a function of pH measured by far-UV CD. (b) Tertiary structure of myoc-OLF as a function of pH measured by near-UV CD. (c) Thermal unfolding of secondary structure monitored by CD at 214 nm. Fit is sigmoidal. (d) Thermal unfolding of tertiary structure monitored by CD at 292 nm. Fit is linear plus sigmoidal. (e) ANS fluorescence as a function of pH. (f) ThT fluorescence as a function of pH at 36 °C monitored for 50 hours. (g) Fibrillar end point morphology for samples incubated at 36 °C in pH 4.6 buffer. (h) Deposits of curvilinear fibrils from samples incubated at 36 °C in pH 3.4 buffer. (i) Disordered aggregates for samples incubated at 36 °C in pH 2.0 buffer. For (g-i), two left panels scale bar = 300nm and two right panels scale bar = 50nm.
Fibril assays (Fig. 2f) combined with AFM imaging (Fig. 2g) reveal that when salt is present, only the partially unfolded state accessed at pH 4.6 is primed for typical fibril growth. Over the 2-day incubation, there is an exponential increase in ThT fluorescence seen, along with primarily long, straight fibrils by AFM (average contour length: 2,643 ± 600 nm, height: 1.1 ± 0.5 nm, actual width: 12 ± 2 nm) (Fig. 2g). Notably, myoc-OLF samples at pH 3.4 and pH 2 showed enhanced ThT fluorescence at the start of the incubation, but no significant change is observed over time. Analysis by size exclusion chromatography confirms that myoc-OLF incubated at pH 3.4/200mM NaCl is aggregated at the start of the incubation while myoc-OLF at pH 4.6/200 mM NaCl remains monomeric (Supplemental Fig. S5). Images from incubation at pH 3.4 reveal some fibril-like aggregates (average contour length: 127 ± 61 nm, height: 0.8 ± 0.3 nm, and actual width: 9 ± 2 nm), but many are clumped together into large disordered structures (Fig. 2h). The unfolded state at pH 2 exhibits a wide distribution of heights ranging from 10.8 to 42.2 nm suggestive of extensive aggregation in the sample prior to deposition (Fig. 2i). The two remaining myoc-OLF samples, at pH 7.2 and pH 5.8, do not form fibrils, even after the 2 day incubation at 36 °C (Supplemental Fig. S2 d, e).
Fibrillization of wild-type myoc-OLF at neutral pH and elevated temperatures
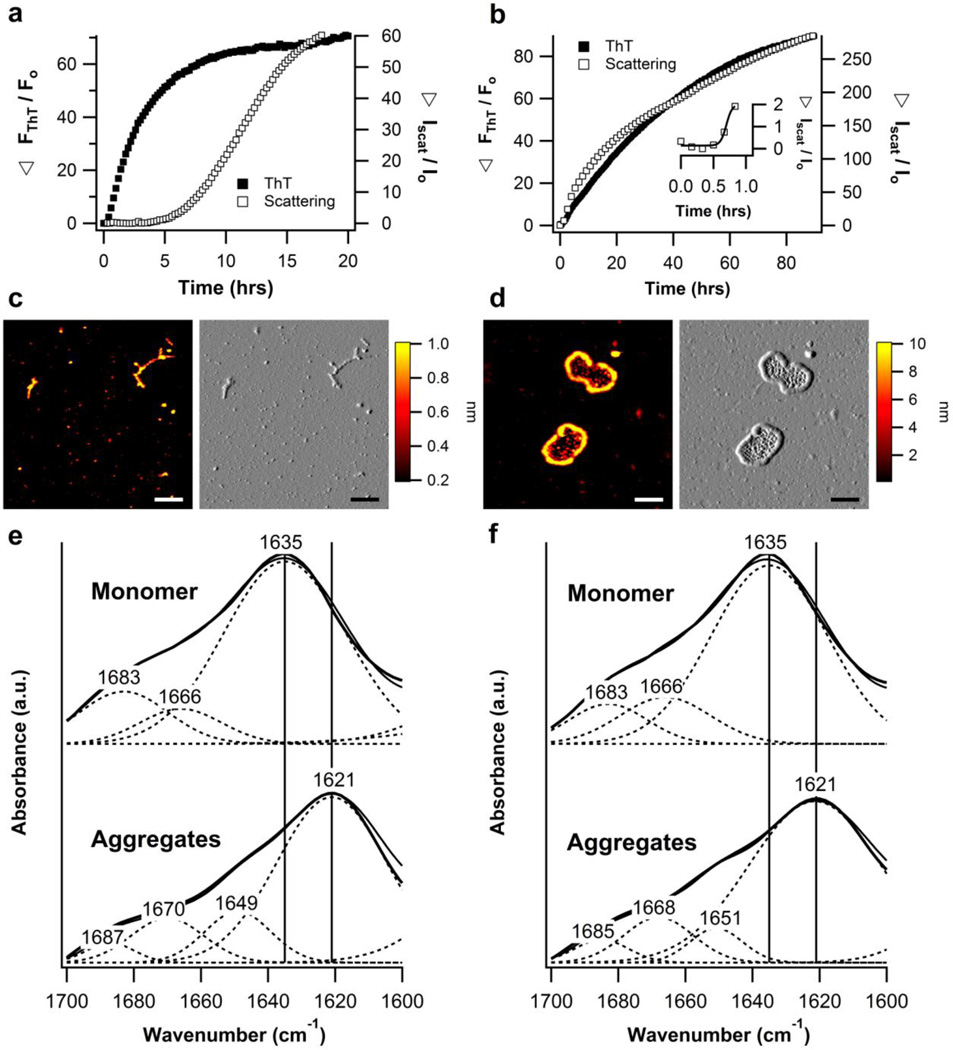
Although myoc-OLF does not readily form fibrils at neutral pH when incubated at 36 °C (Fig. 1f, 2f) or at 37 °C without agitation14, myoc-OLF can form fibrils when incubated at 42 °C, the temperature at the onset of the main tertiary unfolding transition (Fig. 1d, 2d, Supplemental Fig. 6b). In the absence of NaCl, the time to half-maximal ThT fluorescence (t1/2) is 2.0 hours, whereas fibrillization is considerably slower with the addition of 200mM NaCl, with a t1/2 of 27.7 hours (Fig. 3a, 3b Table 1). The increased ThT aggregation rate for wild-type myoc-OLF in the absence of NaCl is likely because of its lower stability (Table 1) due to charge repulsion, not because of an intrinsic difference in the structure at 42 °C prior to aggregation (Supplemental Fig. 6a). In addition, light scattering kinetics in the absence of NaCl show a typical sigmoidal growth curve for amyloid fibrils, with a lag phase of ~5 hours (Fig. 3a). In the presence of salt, the lag phase is reduced to 30 minutes or less (Fig. 3b) with an increase in the ratio of ThT fluorescence to scattering intensity suggesting an increase in non-specific aggregation (Fig. 1b). For both samples, ThT-positive structures are seen by AFM upon the termination of the fibrillization experiment after 90 hours, but unexpectedly, the presence or absence of salt in the buffer leads to different fibril morphologies. In the absence of NaCl, a mixture of fibrils and oligomers are present (fibril average contour length: 334 ± 289 nm and height: 1.0 ± 0.3 nm) (Fig. 3c) while in the presence of 200mM NaCl, the predominant species appears as oligomers (3.9 ± 1.9 nm in height) enclosed by closed, or nearly-closed, loop fibril structure of 10.9 ± 2.5 nm in height and microns in length (average contour length of 2,440 ± 1240 nm and an average end to end distance of 48 ± 35 nm) (Fig. 3d, Supplemental Fig. S7). Both morphologies exhibit the expected amyloid-associated shift in Congo Red absorbance (Supplemental Fig. S8). FTIR spectra also display key shifts in the amide I region from the prototypical native β-sheet signature in the monomeric samples at 1635 cm−1 compared to the corresponding amyloid feature at 1621 cm−1 observed in the aggregated samples (Fig. 3e,f). Deconvolution of the smoothed spectra in this region reveals that for both the straight and curved morphologies, ~70% of the secondary structure in the aggregates is amyloid (Table 2).
Figure 3. Fibrillization of wild-type myoc-OLF incubated at 42 °C in neutral pH buffer.
(a) ThT fibril growth curves including scattering intensity kinetics for incubation at 42 °C without NaCl and (b) with 200mM NaCl. (c) Extended fibril morphology observed for samples incubated in the absence of NaCl. (d) Loop morphology observed for samples incubated in the presence of 200mM NaCl. Scale bar for both conditions (c, d) is 300nm. (e) FTIR spectral analysis for monomers and aggregates without NaCl and (f) with 200mM NaCl.
Table 2.
FTIR analysis of beta sheet content for myoc-OLF wild-type monomer and aggregates.
| Amide I’ maximum (cm−1) |
Native β- sheet (%) |
Amyloid β- sheet (%) |
Turns (%) |
Disorder (%) |
|
|---|---|---|---|---|---|
| Monomer (7.2) | 1635 | 75 | --- | 25 | --- |
| Monomer (7.2/NaCl) | 1635 | 74 | --- | 26 | --- |
| Aggregate (7.2) | 1621 | --- | 68 | 18 | 14 |
| Aggregate (7.2/NaCl) | 1621 | --- | 75 | 18 | 7 |
Native β-sheet bands were assigned from 1630–1641 cm−1 while amyloid β-sheet bands were assigned from 1611–1630 cm−1 as described in Ref. 62.
Non-native structure, fibril growth, and fibril morphologies of myoc-OLF variants under physiological conditions
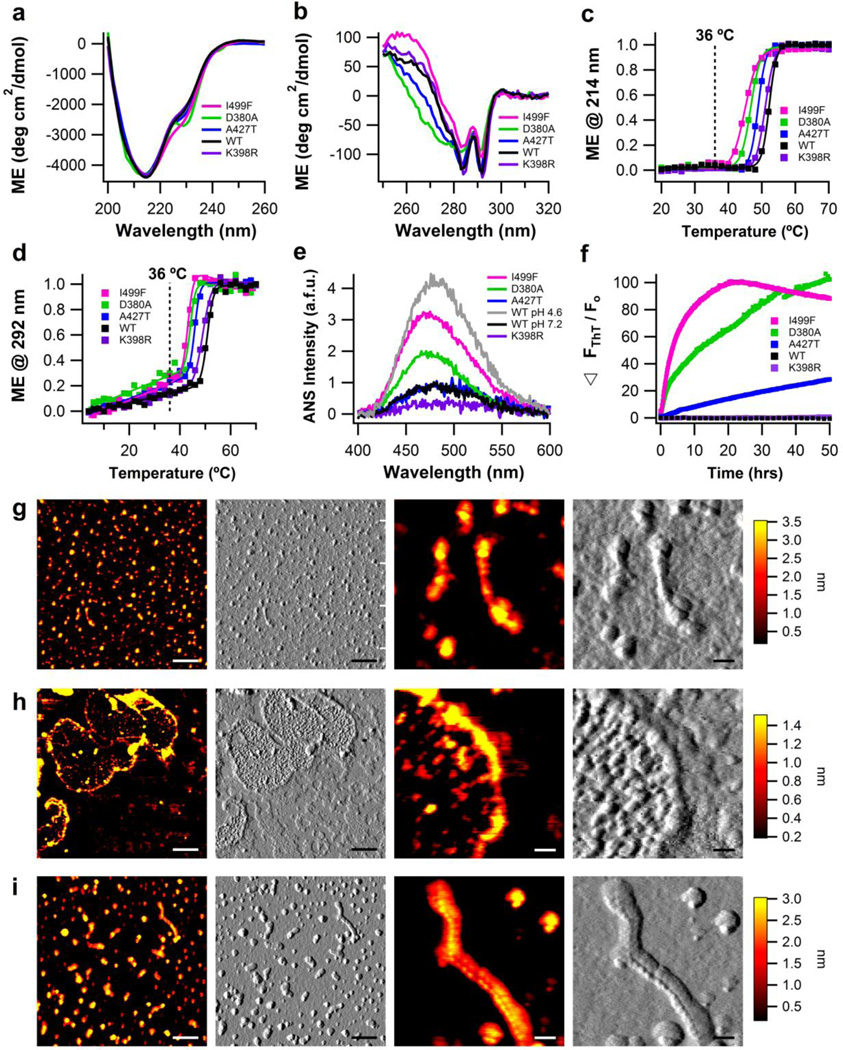
Next, four representative myoc-OLF variants9 were evaluated for their structure, stability, and amyloid fibrillization propensity under physiological conditions (pH 7.2/200 mM NaCl, 36 °C): (a) K398R, a wild-type-like single-nucleotide polymorphism variant,12 (b) A427T, a disease-causing variant with near wild-type stability,12 (c) D380A, a disease-causing variant lacking a calcium ion cofactor,13 and (d) I499F, a disease-causing variant with significantly reduced thermal stability compared to wild-type.12 Purity and monomeric state of each myoc-OLF variant were confirmed by size exclusion chromatography12 and SDS-PAGE (Supplemental Fig. S9). Whereas for all four variants secondary structure CD spectra are nearly superimposable with wild-type (Fig. 4a) the three disease-causing variants, A427T, D380A, and I499F exhibit significant non-native tertiary features (Fig. 4b). The tertiary structure of K398R is indistinguishable from wild-type (Fig. 4b), but both A427T and D380A have broader negative features in the phenylalanine range (270-250 nm)26 (Fig. 4b), correlating with their reduced thermal stability (Table 1). The I499F variant is likewise non-native but positive in this region of the near-UV spectrum (Fig. 4b), which may be due to the additional phenylalanine residue. Like wild-type myoc-OLF, unfolding of tertiary structure for all four variants involves a pre-transition followed by a major unfolding event (Fig. 4 d); secondary structures melt in an apparent sigmoidal transition 2–3 °C higher (Fig. 4c and Table 1). Melting temperatures are consistent with previously reported values determined by other methods.12; 13 Comparison of ANS fluorescence levels reveals that the three disease variants are less compact proteins compared to wild type myoc-OLF or K398R to an extent comparable with wild-type at pH 4.6/200mM NaCl, but less unfolded than wild-type is at pH 2 (Table 1, Fig. 2e, 4e).
Figure 4. Biophysical characterization of myoc-OLF variants K398R (SNP), D380A, A427T, and I499F (disease-causing) in physiological buffers.
(a) Secondary structure measured by far-UV CD. (b) Tertiary structure measured by near-UV CD. (c) Thermal unfolding of secondary structure monitored by CD at 214 nm. Fit is sigmoidal. (d) Thermal unfolding of tertiary structure monitored by CD at 292 nm. Fit is linear plus sigmoidal. For (a)-(d), wild-type myoc-OLF at neutral pH with 200 mM NaCl is overlayed for comparison. (e) ANS fluorescence as a function of pH, with overlay of wild-type myoc-OLF at pH 7.2/200 mM NaCl and 4.6/200mM NaCl. (f) ThT fluorescence at 36 °C monitored for 50 hours. (g) End-point morphologies seen for myoc-OLF(A427T) are fibrils and oligomers. (h) Deposits of myoc-OLF(D380A) appear as curvilinear and circular fibrils enclosing smaller globular aggregates. (i) Morphologies seen for myoc-OLF(I499F) are fibrils and oligomers, similar to myoc-OLF(A427T). For (g)-(i), two left panels scale bar = 300nm and two right panels scale bar = 50nm.
Kinetics of amyloid formation were evaluated next for each variant at 36 °C. For K398R, low constant levels of ThT fluorescence indicate no fibrillization, like wild-type myoc-OLF under these conditions. By contrast, all three disease-causing variants form ThT-positive structures (Fig. 4f). The less stable variants fibrillize faster, as measured by t1/2 (Fig. 4f, Table 1). AFM imaging confirms that A427T, D380A, and I499F aggregate into diverse, amyloid-like morphologies (Fig. 4 g, h, i), while the K398R does not (Supplemental Fig. S2f). For A427T and I499F, a significant population of the aggregates appears as oligomers as well as a few fibrils with heights and contour lengths of 3.3 ± 0.4 nm and 166 ± 101 nm (A427T) and 3.0 ± 0.4 nm and 282 ± 143 nm (I499F), respectively. Fibrils of D380A, resemble wild-type myoc-OLF incubated at pH 7.2 with salt 42 °C (Fig. 3d), appearing as closed or nearly closed fibril loops surrounding oligomers (Fig. 4h). The oligomers have a height of 1.2 ± 0.3 nm and a surrounding fibril border 1.4 ± 0.4 nm tall with an average contour length of 2,119 ± 887 nm.
Identification of amyloidogenic peptides within the myoc-OLF sequence
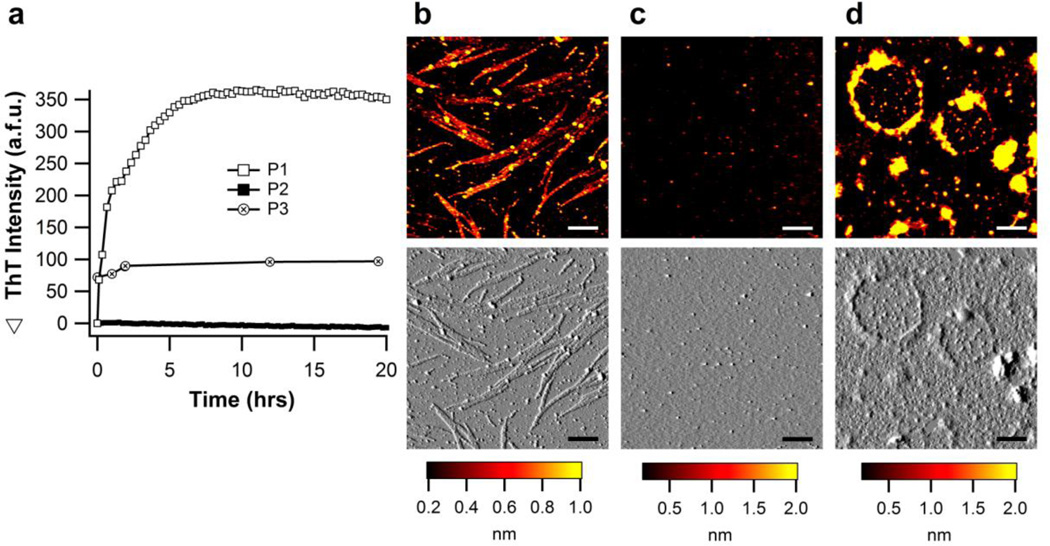
To assess whether different amino acid sequences within myoc-OLF could be responsible for forming the two major mature aggregate morphologies observed, namely, long straight/curvilinear fibrils (Fig. 1g, 2g, 3c, 4g, 4i) or lassoed oligomers (Fig. 3d, 4h), the amino acid sequence myoc-OLF was evaluated using four amyloid-prediction algorithms, Waltz31, PASTA32, AmylPred33, and TANGO34. The three highest scoring sequences, G326AVVYSGSLYFQ (P1), G387LWVIYSTDEAKGAIVLSK (P2), and V426ANAFIICGTLYTVSSY (P3) (Supplemental Fig. S10), were identified with consensus priority for amyloid of P3>P1>P2, and synthesized for further evaluation. Indeed, incubation of P1 and P3, but not P2 for 20 hrs at 36 °C, reveals a high level of ThT fluorescence (Fig. 5a). Additional incubation of P2 for 72 hours still did not lead to a change in ThT fluorescence (data not shown). While the growth of P1 is exponential over the course of the experiment, rates could not be slowed to observe a clear lag phase. P3 fibrillization is nearly instantaneous upon dilution of the DMSO-dissolved peptide into buffer, which could not be slowed by altering experimental conditions like further dilution, temperature, or buffer. In support of our hypothesis, P1 appears as straight amyloid fibrils by AFM, with an average height and contour length of 0.9 ± 0.2 nm and 700 ± 537 nm (Fig. 5b) akin to myoc-OLF (Fig. 1g, 2g, 3c), P2 is not aggregated (Fig. 5c), and P3 aggregates into closed amyloid fibrils with an average height and contour length of 1.4 ± 0.6 nm and 1,566 ± 298 nm (Fig. 5d), similar to wild-type myoc-OLF in pH 7.2/NaCl and 42 °C (Fig. 3c) and D380A (Fig. 4h).
Figure 5. Fibrillization of peptides P1-P3 incubated at 36 °C in physiological buffer.
(a) ThT fibril growth kinetics. (b) Straight fibril morphology observed for P1 (c) No aggregates observed for P2 (d) Closed loop morphology observed for P3. Scale bars for (b, c, d) are 300nm.
DISCUSSION
Amyloid formation is a kinetically driven, self-assembly process that results in common characteristic filaments35 among disparate proteins36. Often associated with diseases, including Alzheimer, Parkinson, and Huntington, where they are neurotoxic, amyloid can also play a functional role17, may represent an ancient protein fold38, and can be formed by model proteins37. Amyloids are believed to share a core interdigitated, so-called cross-beta steric zipper39,40 that forms via an accessible partially folded state and is controlled by colloidal stability and kinetics.36 Bioinformatics approaches can evaluate sequences with high amyloid propensity from chemical aspects of amino acid sequence41. However, observed supramolecular morphologies vary widely and are critically dependent on the precise experimental conditions or amino acid substitutions that perturb protein structures to enable access to one or more amyloidogenic peptide stretches23; 30; 42; 43; 44.
To address the molecular details of amyloid formation by myoc-OLF, a newly identified amyloid protein14, and enable comparison with other highly studied amyloids, we identified and characterized a spectrum of populated conformations. The native state CD signature involves previously identified secondary structural features at ~215 nm and a 230 nm shoulder8; here we add the additional characteristic of native tertiary structure consisting of tight double troughs at 282 and 291 nm. Native state CD spectra are superimposable regardless of the presence of salt. At the other extreme is the acid-induced unfolded state that lacks tertiary structure, maintains some secondary structure, but has a high level of ANS fluorescence (Fig. 1). In the absence of salt at pH 2, the high positive charge (26 Arg, Lys residues) leads to charge repulsion that enables an unfolded state to exist that does not aggregate or form fibrils. The addition of salt at pH 2 (Fig. 2) introduces charge shielding and attractive intermolecular forces commonly described by the second virial coefficient B22 that lead to immediate bulk, disordered aggregation30; 45. 46
At intermediate pH values we have identified a spectrum of partially unfolded myoc-OLF states, and it is these non-native, destabilized states that are prone to fibril formation under distinct experimental conditions. In the presence of NaCl, we characterized one partially folded state with altered tertiary structure at pH 4.6; at other pH values, myoc-OLF was either a largely aggregation-prone unfolded state or indistinguishable from its native conformation (Fig. 2). In the absence of NaCl, we identified several non-native states from pH 3.4 to 5.8 (Fig. 1), likely due to decreased charge screening.46 All of these states are less thermally stable than native, but of variable levels of exposed hydrophobic environments. Fibrillization can proceed from many of these states under our non-agitating assay conditions when incubated at a temperature near the onset of tertiary unfolding. 45
The importance of the non-native and destabilized conformational state requirements for fibril formation under conditions of favorable intermolecular interactions is underscored by our results for wild-type myoc-OLF and variants under physiological conditions. In the case of pH 7.2/NaCl, wild-type myoc-OLF will fibrillize without agitation at 42 °C, the onset of tertiary unfolding of myoc-OLF at this pH value. We previously reported14 that incubation at 37 °C for at least 2 weeks does not lead to fibril formation14. Rather, to grow fibrils at 37 °C, agitation is required14, which likely introduces a combination of mechanical local unfolding, effects due to the liquid/air interface, and concentration fluctuations that create nuclei for amyloid formation.37; 47; 48 We also demonstrated that other destabilizing reagents like acid spike or peroxide, which mimic aging conditions, presumably act similarly to accelerate fibrillization of myoc-OLF under conditions that it would otherwise not form fibrils14. The three disease-causing myoc-OLF variants, A427T, D380A, and I499F investigated in this study (Table 1) are destabilized to variable extents under physiological buffer and temperatures. They exhibit a non-native tertiary structure such that fibrils can form at 36 °C, a temperature consistent with that found in the eye, and within the window where fibrils would be expected to form along the unfolding pathway based on our pH study. We infer that fibrillization at 36 °C is a common feature of the many other disease-causing variants whose measured Tms12 are bracketed by the broad spectrum represented by A427T, D380A, and I499F.
A variety of fibril morphologies were observed in this study under different experimental conditions, including an unprecedented end-point structure of a large fibrillar closed ring that appear to encapsulate oligomers. Myoc-OLF aggregate morphology is highly dependent on pH, salt, and temperature of incubation. In several conditions myoc-OLF forms straight fibrils expected for amyloids, with dimensions common to other amyloid fibrils49. As illustrated for fibrils grown from wild type myoc-OLF at pH 7.2 without salt after incubation at 42 °C, amyloid signatures in FTIR, and the expected shift of Congo Red absorbance, are present. For 30; 42the large fibril loops with enclosed oligomers grown in the presence of salt at neutral pH at 42 °C, the amyloid character was confirmed by a Congo Red absorbance shift and signature resonances in FTIR, but additional studies will be necessary to clarify its three dimensional structure. Annular aggregates have been reported for a few other amyloid proteins including equine lysozyme50 and α-synuclein51, but these are less than 100 nm in circumference, and are short-lived intermediates on the pathway to fibril formation. In the case of myoc-OLF, the closed fibril loops grow with exponential growth, are ThT-positive, and end-point morphologies are on the order of microns in circumference. To the best of our knowledge, there is just one other protein reported to form fibrils with any resemblance to these structures, the non-diseaseassociated N-terminal domain of Escherichia coli HypF (HypF-N)52. HypF-N fibrils are ThT-positive crescents of similar size to myoc-OLF, but there are numerous differences. First, the predominant structure is crescent-shaped, and devoid of material in the center; closed rings, when observed, are transient. Second, HypF-N crescents grow from 30% TFE at pH 5.5 over the course of three days at room temperature, unlike our mild and more biological conditions. Finally, HypF-N crescents or rings disappear and form twisted long ribbons form by five days, whereas closed-ring myoc-OLF fibrils do not morph into such ribbon-like fibrils. Unexpectedly, in addition to wild-type myoc-OLF, the D380A disease variant, which is depleted of its calcium ion13, also forms these ring like structures. Emerging evidence from other amyloids such as equine lysozyme50 and gelsolin53 support the hypothesis that metal-binding, and calcium in particular, affects fibril formation and morphology, by changing local protein structure and/or electrostatics that affect intermolecular interactions; both could be in play in the case of D380A or other variants that affect calcium binding.
Individual prediction programs vary widely on how many amyloidogenic sequences myoc-OLF harbors in total, but inspection of the program outputs side-by-side led to the selection of our three top candidate peptides, P1-P3, which have predicted pI values of 5.5 – 6.0 and thus, like myoc-OLF, would be negatively charged at neutral pH. P1 forms long straight fibrils of typical amyloid morphologies similar to myoc-OLF at pH 7.2 without salt, exhibiting exponential growth upon dilution into physiological buffer at 36 °C. P3 aggregated upon dilution, forming large rings similar to myoc-OLF at pH 7.2/NaCl and D380A. These data support the hypothesis that the two main morphologies seen for myoc-OLF derive from these peptide stretches, although we cannot exclude other amyloidogenic peptide stretches within myoc-OLF. While it is surprising that P2 did not form fibrils given the high level of prediction, particularly for the N-terminal sequence GLWVIY, perhaps this is because P2 has the least overall predicted secondary structure. The ring structures seen with P3 suggest that the fibril core is more flexible or kinked than in P1. Perhaps this is a consequence of the presence of an internal Cys; in myoc-OLF, there is a disulfide bond involving this Cys, which may influence the unfolding pathway under certain experimental unfolding scenarios. Computational prediction using PASTA32 also indicates that P3 may form antiparallel β-strands, compared to P1, for which parallel strands are more likely. Further studies to elucidate the closed ring morphology, including the structure(s) of the encased oligomers apparent in both myoc-OLF and D380A, are warranted.
In sum, myoc-OLF is a newly identified disease-related protein that can form bona fide amyloid fibrils with a variety of morphologies initiated by access of a partially folded state via particular experimental parameters. The finding that the three disease variants examined here form different fibril morphologies raises some questions about the molecular recognition of mutant myocilin by ER-resident chaperones like Grp946. Specifically it is of interest whether such recognition is sequence dependent, and if so, if all known disease mutants expose this sequence. Alternatively, chaperone recognition could be dependent on fibril morphology, requiring different chaperones depending on the variant. Regarding wild-type myocilin, it is perplexing that a broad role for aggregation in glaucoma is currently only speculative given that fibrillogenesis is relatively facile in vitro and given the fact that the physiological environment of the TM has abundant known facilitators of fibril formation like glycosaminoglycans54; 55; 56. We would predict that morphologies we reported here could damage TM tissue and in turn influence the balance of fluid flow and intraocular pressure, akin to oligomers of Aβ, which form membrane-spanning channels that release calcium ions57. If indeed protective mechanisms are at play in the TM that suppress extracellular myocilin aggregation, such knowledge could be of high value to other amyloid systems.
METHODS
Protein expression and purification of Myoc-OLF
Myoc-OLF and variants were expressed as MBP-OLF fusion proteins and isolated after cleavage by Factor Xa, as previously described8; 11; 12; 13 or by cleavage with the Tobacco Etch Virus (TEV) protease. For cleavage with the TEV protease, the original pMAL-c4x construct with the Factor XA cleavage site (SSIEGR) between MBP and OLF was mutated to a TEV protease cleavage site (ENLFYQS) using the site directed mutagenesis (QuikChange, Stratagene) kit. Primers are listed in the Supplemental Table S1. All plasmids were verified by DNA sequencing (Operon). Expression and purification of MBP-OLF(TEV) proceeded as previously described for MBP-OLF(Factor Xa). Cleavage of MBP-OLF(TEV) was accomplished by overnight (~18 hours) incubation with TEV protease at room temperature in 10 mM Na2HPO4/KH2PO4, 200mM NaCl, pH 7.2 buffer. TEV protease was produced in house using the pRK793 plasmid (Addgene) as described previously58. After cleavage, the TEV protease (6xHis-tag) was captured by nickel affinity using a 1mL His Trap (GE Healthcare), followed by removal of MBP with amylose affinity chromatography, and isolation of monomeric OLF by gel filtration using the Superdex 75 (GE healthcare).
Buffers for pH study
Myoc-OLF was examined in five buffers, with or without 200 mM NaCl: 10 mM Na2HPO4/KH2PO4 pH 7.2, 10 mM Na2HPO4/KH2PO4 pH 5.8, 10 mM sodium acetate pH 4.6, 10 mM citrate pH 3.4, and 10 mM phosphate (KH2PO4/phosphoric acid) pH 2. Immediately prior to any experiment, purified myoc-OLF was concentrated at 4 °C and re-diluted three times into the appropriate buffer using an Amicon Ultra-15 Centrifugation device. The final protein concentration was determined after buffer exchange using the predicted molar extinction coefficient at 280 nm of 68,425 M−1cm−1. Myoc-OLF variants were purified using 10 mM Na2HPO4/KH2PO4, 200 mM NaCl, pH 7.2 directly and used without further buffer exchange.
Circular dichroism (CD)
CD studies were conducted using a Jasco J-810 or J-815 spectropolarimeter equipped with a Neslab RTE 111 circulating water bath and a Jasco PTC-4245/15 temperature control system. Far-UV CD spectra were acquired in a 0.1 cm cuvette from 300 nm to 200 nm, at 4 °C, with a scan rate of 500 nm min−1 and a data pitch of 1 nm. Protein concentration was 10–12 uM, and for each buffer, the reported spectra are an average of duplicate measurements of 10 scans that were background-subtracted. Far-UV CD melts were performed by increasing the temperature at 2 °C min−1 from 4 °C to 70 °C and a delay time of 60 seconds before each acquisition. The spectrum at each temperature is an average of 10 scans using above parameters. Averaged spectra were converted to mean residue ellipticity Θ= (Mres×Θobs) / (10×d×c), where Mres=112.9 is the mean residue mass calculated from the protein sequence; Θobs is the observed ellipticity (degrees) at wavelength λ; d is the pathlength (cm); and c is the protein concentration (g/ml). The Tm was determined using mean residue ellipticity values recorded at 214 nm via Boltzmann Sigmoid analysis in Igor Pro.
Near-UV CD spectra were acquired using a 0.1 cm cuvette at 4 °C, from 320 nm to 250 nm with a scan rate of 50 nm min−1 and a data pitch of 1 nm. Protein concentration was 40–50 uM and the reported spectra for each buffer are an average of duplicate measurements from 10 scans that were background-subtracted. Near-UV CD melts, performed in duplicate, were conducted by increasing the temperature by 2 °C min−1 from 4 °C to 70 °C, with a delay time of 60 seconds before each acquisition. The spectrum at each temperature is an average of 10 scans using aforementioned scan parameters. The near-UV spectra were baseline-subtracted from an average spectrum value calculated from spectra acquired during the melt at each temperature, to account for aggregation observed due to higher protein concentrations (range of 320 to 310 nm). The Tm was determined using mean residue ellipticity values at 291 nm or 292 nm and fit using a combination of a linear and sigmoidal functions in Igor Pro.59
ANS Fluorescence
ANS fluorescence was measured on a Shimadzu RF-530/PC spectrofluorophotometer at room temperature, with an excitation wavelength of 380 nm (slit width 3 nm) and an emission range of 400 nm to 600 nm (slit width 1.5 nm). Stock solutions of 10 mM ANS in deionized water were diluted to 600 µM working stock solutions in appropriate buffer. Each sample, composed of 10 uM myoc-OLF and 100 µM ANS, was prepared 15 minutes prior to data acquisition. Five accumulations were averaged and background-subtracted for each measurement. Reported spectra are an average of two independent experiments.
Fibrillization assay
Amyloid aggregation was monitored by ThT fluorescence. Myoc-OLF and variants at 30 µM were incubated over time in a variety of buffer conditions. A stock solution of 1 mg/mL ThT in deionized water was diluted to 200 µM working solutions in appropriate buffer, and dispensed for a final concentration of 10 µM for aggregation experiments. For experiments without NaCl, data were acquired on a Shimadzu RF-530/PC spectrofluorophotometer using a 200 µL low head space cell (Starna 26.50LHS-Q-10/Z15). ThT fluorescence was recorded every 10–30 minutes using an excitation wavelength of 440 nm (slit width 3 nm) and emission range of 450 nm to 600 nm (slit width 3 nm). The change in the maximum ThT intensity at 485 nm plotted versus time. Experiments in the presence of NaCl were conducted using a Biotek Synergy microplate reader equipped with a 440 nm excitation filter and 485 nm emission filter. Myoc-OLF samples (150 uL) were prepared in 1.5 mL centrifuge tubes, transferred to a 96-well microplate (Grenier), and then sealed with clear MicroAmp PCR film (Applied Biosystems). Measurements were recorded every 10 minutes and background-subtracted with a corresponding buffer-only solution supplemented with 10 uM ThT. At the termination of the assay (18–90 h), samples were removed from the microplate and stored in microcentrifuge tubes until imaging by AFM (see below). All presented fibrillization assay data are an average of at least two independent experiments rescaled as ΔF/Fo, where Fo is the initial fluorescence. Aggregation was also monitored by light scattering using a Zetasizer Nano S (Malvern Instruments, Worchestershire, UK) with a He-Ne laser (λ = 633 nm) or by monitoring absorbance (λ = 620 nm) using a Biotek Synergy microplate reader. All presented light scattering data are rescaled as ΔI/Io, where Io is the initial scattering intensity or initial absorbance.
Peptide synthesis and fibril formation
Peptides were synthesized by Celtek Peptides (Nashville, TN). Peptides G326AVVYSGSLYFQ (P1) and G387LWVIYSTDEAKGAIVLSK (P2) are >95% pure and peptide V426ANAFIICGTLYTVSSY (P3) was desalted. Peptides were prepared as stock solutions in DMSO (5 mg/ml) and stored at room temperature. For reported fibrillization assays, peptides were diluted to 500 µM in 10 mM Na2HPO4/KH2PO4, 200 mM NaCl pH 7.2 and 10 uM ThT. Aggregation was monitored as above for experiments with myoc-OLF at pH 7.2 without salt.
Atomic force microscopy
A few hours after the termination of the aggregation assay, 40 µL was removed from the bottom of the 1.5 mL microcentrifuge tube and left to be adsorbed onto freshly cleaved mica for 30 minutes, rinsed for 3 seconds with deionized water, and left to dry overnight in a Petri dish. For myoc-OLF at pH 3.4 and pH 2 in the absence of NaCl, pH 7.2 in the absence of NaCl at 42 °C, myoc-OLF(D380A), and P3, samples were diluted 100-fold prior to deposition. Dry samples were imaged in air with a MFP-3D atomic force microscope (Asylum Research) using PPP-FMR (NanoAndMore) silicon tips with nominal tip radii less than 7 nm. The cantilever was driven at 60–70 kHz in alternating current mode and a scan rate of 0.5 Hz with 512 × 512-pixel resolution. Raw image data were corrected for image bow and slope using the software provided by Asylum Research. Dimensional analysis for average height and width was performed using the particle analysis software provided by Asylum Research, ignoring particles with an area less than 25 nm2 and a height less than 0.5 nm. The actual width was deconvoluted from the average apparent width using Wact = Wapp-2[H(2Rt-H)]1/2 where Wact is the actual width of the fibril, Wapp is the apparent width of the fibril, H is the height of fibril, and Rt is the radius of the tip60. Due to the large statistical range in the measured apparent width, the standard deviation reported for the actual width is calculated using the equation for Wact, the average apparent width, and the error in the height. For all calculations, the radius of the tip, Rt, was assumed to be 7 nm as provided by the manufacturer. Contour length and end to end distances were manually measured using the freehand line scan option provided by Asylum research.
Fourier Transform Infrared Spectroscopy (FTIR)
ATR-FTIR spectroscopy was performed on a Bruker Optik Vertex 70 (Ettlingen, Germany) spectrometer equipped with a BioATRcell II accessory (Harrick Scientific Products, Inc) as previously described61. Briefly, 30uL of freshly prepared monomer or pelletized aggregate was placed on a silicon crystal equilibrated to 24 °C. Sample spectra were acquired from 1000 to 3000 cm−1 with 4 cm−1 resolution after background subtraction of the buffer solution, both recorded for 1000 scans. The spectra presented are an average of 5 such sample spectra. Combined peak and deconvolution analysis was performed within the OPUS software analysis package (Bruker Optik). After a horizontal baseline correction and smoothing algorithm (13 to 17 smoothing points), Gaussian curves were fitted from 1500 cm−1 to 1700 cm−1 with fixed peaks identified by second derivative analysis. As previously described62, native β-sheet bands were assigned from 1630–1641 cm−1, amyloid β-sheet bands from 1611–1630 cm−1, turns from 1663–1695 cm−1, and disorder from 1647–1654 cm−1.
Supplementary Material
Highlights.
Aggregates of mutant myocilin are linked to early-onset, inherited glaucoma
The myocilin olfactomedin domain forms amyloid fibrils via partially unfolded states
Fibrils appear as straight/curvilinear and as a circular species of micron length
Two peptide stretches fibrillize into these disparate morphologies
Observed fibril morphologies may contribute to glaucoma pathogenesis
Acknowledgements
We thank the Hud, Sulcheck, and Williams labs for access to their CD spectropolarimeter, AFM, and fluorimeter, respectively. We also thank the USF Department of Physics and the Muschol lab for access to FTIR and DLS instruments. We also acknowledge assistance from Katherine Turnage for generating the MBP-OLF(TEV) construct, as well as Pamela Chi, Dana Freeman, Elaine Nguyen, and Leigh Stafford in cell growth and purification.
Funding Sources
This work was funded by grants from NIH (R01EY021205) and Pew Scholar in Biomedical Sciences program to R. L. L.
Abbreviations
- Myoc-OLF
olfactomedin domain of myocilin
- TM
trabecular meshwork
- HTM
human trabecular meshwork
- ER
endoplasmic reticulum
- ThT
thioflavin T
- CD
circular dichroism
- AFM
atomic force microscopy
- ANS
anilinonapthalene-1-sulfonate
- t1/2
time to half-maximal fluorescence
- P1
amino acid sequence G326AVVYSGSLYFQ
- P2
amino acid sequence G387LWVIYSTDEAKGAIVLSK
- P3
amino acid sequence V426ANAFIICGTLYTVSSY
- Tm
melting temperature
- HypF-N
N-terminal domain of Eschericia coli HypF-N
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 2.Gobeil S, Rodrigue M-A, Moisan S, Nguyen TD, Polansky JR, Morissette J, Raymond V. Intracellular sequestration of hetero-oligomers formed by wild-type and glaucoma-causing myocilin mutants. Invest. Ophthalmol. Vis. Sci. 2004;45:3560–3567. doi: 10.1167/iovs.04-0300. [DOI] [PubMed] [Google Scholar]
- 3.Joe MK, Sohn S, Hur W, Moon Y, Choi YR, Kee C. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem. Biophys. Res. Commun. 2003;312:592–600. doi: 10.1016/j.bbrc.2003.10.162. [DOI] [PubMed] [Google Scholar]
- 4.Yam GH-F, Gaplovska-Kysela K, Zuber C, Roth J. Aggregated myocilin induces russell bodies and causes apoptosis: implications for the pathogenesis of myocilin-caused primary open-angle glaucoma. Am. J. Pathol. 2007;170:100–109. doi: 10.2353/ajpath.2007.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z, Vollrath D. A cellular assay distinguishes normal and mutant TIGR/myocilin protein. Hum. Mol. Genet. 1999;8:2221–2218. doi: 10.1093/hmg/8.12.2221. [DOI] [PubMed] [Google Scholar]
- 6.Suntharalingam A, Abisambra JF, O'Leary JC, 3rd, Koren J, 3rd, Zhang B, Joe MK, Blair LJ, Hill SE, Jinwal UK, Cockman M, Duerfeldt AS, Tomarev S, Blagg BS, Lieberman RL, Dickey CA. Glucose-regulated protein 94 triage of mutant myocilin through endoplasmic reticulum-associated degradation subverts a more efficient autophagic clearance mechanism. J. Biol. Chem. 2012;287:40661–40669. doi: 10.1074/jbc.M112.384800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polansky JR, Fauss DJ, Chen P, Chen H, Lutjen-Drecoll E, Johnson D, Kurtz RM, Ma ZD, Bloom E, Nguyen TD. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997;211:126–139. doi: 10.1159/000310780. [DOI] [PubMed] [Google Scholar]
- 8.Orwig SD, Lieberman RL. Biophysical characterization of the olfactomedin domain of myocilin, an extracellular matrix protein implicated in inherited forms of glaucoma. PLoS ONE. 2011;6:e16347. doi: 10.1371/journal.pone.0016347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong G, Kosoko-Lasaki O, Haynatzki GR, Wilson MR. Genetic dissection of myocilin glaucoma. Hum. Mol. Genet. 2004;13:R91–R102. doi: 10.1093/hmg/ddh074. [DOI] [PubMed] [Google Scholar]
- 10.Hewitt AW, Mackey DA, Craig JE. Myocilin allele-specific glaucoma phenotype database. Hum. Mutat. 2007;0:1–5. doi: 10.1002/humu.20634. [DOI] [PubMed] [Google Scholar]
- 11.Burns JN, Orwig SD, Harris JL, Watkins JD, Vollrath D, Lieberman RL. Rescue of glaucoma-causing mutant myocilin thermal stability by chemical chaperones. ACS Chem. Biol. 2010;5:477–487. doi: 10.1021/cb900282e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns JN, Turnage KC, Walker CA, Lieberman RL. The stability of myocilin olfactomedin domain variants provides new insight into glaucoma as a protein misfolding disorder. Biochemistry. 2011;50:5824–5833. doi: 10.1021/bi200231x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donegan RK, Hill SE, Turnage KC, Orwig SD, Lieberman RL. The glaucoma-associated olfactomedin domain of myocilin is a novel calcium binding protein. J. Biol. Chem. 2012;287:43370–43377. doi: 10.1074/jbc.M112.408906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orwig SD, Perry CW, Kim LY, Turnage KC, Zhang R, Vollrath D, Schmidt-Krey I, Lieberman RL. Amyloid fibril formation by the glaucoma-associated olfactomedin domain of myocilin. J. Mol. Biol. 2012;421:242–255. doi: 10.1016/j.jmb.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zode GS, Kuehn MH, Nishimura DY, Searby CC, Mohan K, Grozdanic SD, Bugge K, Anderson MG, Clark AF, Stone EM, Sheffield VC. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J. Clin. Invest. 2011;121:3542–3553. doi: 10.1172/JCI58183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 18.Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. N. Engl. J. Med. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabelt BAT, Kaufman PL. Changes in aqueous humor dynamics with age and glaucoma. Prog. Retin. Eye Res. 2005;24:612–137. doi: 10.1016/j.preteyeres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Tamm ER, Ethier CR. Current aspects of aqueous humor dynamics and glaucoma. Exp. Eye. Res. 2009;88:618–619. doi: 10.1016/j.exer.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Quigley HA. Number of people with glaucoma worldwide. Br. J. Ophthalmol. 1996;80:389–393. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. Dependence of alpha-synuclein aggregate morphology on solution conditions. J. Mol. Biol. 2002;322:383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 24.Kessel L, Johnson L, Arvidsson H, Larsen M. The relationship between body and ambient temperature and corneal temperature. Invest. Ophthalmol. Vis. Sci. 2010;51:6593–6597. doi: 10.1167/iovs.10-5659. [DOI] [PubMed] [Google Scholar]
- 25.Hider RC, Kupryszewski G, Rekowski P, Lammek B. Origin of the positive 225–230 nm circular dichroism band in proteins. Its application to conformational analysis. Biophys. Chem. 1988;31:45–51. doi: 10.1016/0301-4622(88)80007-3. [DOI] [PubMed] [Google Scholar]
- 26.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim. Biophys. Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Ranjbar B, Gill P. Circular dichroism techniques: biomolecular and nanostructural analyses- a review. Chem. Biol. Drug Des. 2009;74:101–120. doi: 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 28.Buchner J, Renner M, Lilie H, Hinz HJ, Jaenicke R, Kiefhabel T, Rudolph R. Alternatively folded states of an immunoglobulin. Biochemistry. 1991;30:6922–6929. doi: 10.1021/bi00242a016. [DOI] [PubMed] [Google Scholar]
- 29.Kelly SM, Price NC. The application of circular dichroism to studies of protein folding and unfolding. Biochim. Biophys. Acta. 1997;1338:161–185. doi: 10.1016/s0167-4838(96)00190-2. [DOI] [PubMed] [Google Scholar]
- 30.Hill SE, Miti T, Richmond T, Muschol M. Spatial extent of charge repulsion regulates assembly pathways for lysozyme amyloid fibrils. PLoS One. 2011;6:e18171. doi: 10.1371/journal.pone.0018171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveberg M. Waltz, an exciting new move in amyloid prediction. Nat. Methods. 2010;7:187–188. doi: 10.1038/nmeth0310-187. [DOI] [PubMed] [Google Scholar]
- 32.Trovato A, Seno F, Tosatto SC. The PASTA server for protein aggregation prediction. Protein Eng. Des. Sel. 2007;20:521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 33.Tsolis AC, Papandreou NC, Iconomidou VA, Hamodrakas SJ. A consensus method for the prediction of 'aggregation-prone' peptides in globular proteins. PLoS One. 2013;8:e54175. doi: 10.1371/journal.pone.0054175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed AB, Kajava AV. Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. FEBS Lett. 2013;587:1089–1095. doi: 10.1016/j.febslet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Wetzel R. Kinetics and thermodynamics of amyloid fibril assembly. Acc. Chem. Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 36.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 38.Greenwald J, Riek R. On the possible amyloid origin of protein folds. J. Mol. Biol. 2012;421:417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U S A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzpatrick AW, Debelouchina GT, Bayro MJ, Clare DK, Caporini MA, Bajaj VS, Jaroniec CP, Wang L, Ladizhansky V, Muller SA, MacPhee CE, Waudby CA, Mott HR, De Simone A, Knowles TP, Saibil HR, Vendruscolo M, Orlova EV, Griffin RG, Dobson CM. Atomic structure and hierarchical assembly of a cross-beta amyloid fibril. Proc. Natl. Acad. Sci. U S A. 2013;110:5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JW, Rousseau F. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 42.Gosal WS, Morten IJ, Hewitt EW, Smith DA, Thomson NH, Radford SE. Competing pathways determine fibril morphology in the self-assembly of beta2-microglobulin into amyloid. J. Mol. Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 43.Kodali R, Williams AD, Chemuru S, Wetzel R. Abeta(1–40) forms five distinct amyloid structures whose beta-sheet contents and fibril stabilities are correlated. J. Mol. Biol. 2010;401:503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersen CB, Hicks MR, Vetri V, Vandahl B, Rahbek-Nielsen H, Thogersen H, Thogersen IB, Enghild JJ, Serpell LC, Rischel C, Otzen DE. Glucagon fibril polymorphism reflects differences in protofilament backbone structure. J. Mol. Biol. 2010;397:932–946. doi: 10.1016/j.jmb.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Raman B, Chatani E, Kihara M, Ban T, Sakai M, Hasegawa K, Naiki H, Rao Ch M, Goto Y. Critical balance of electrostatic and hydrophobic interactions is required for beta 2-microglobulin amyloid fibril growth and stability. Biochemistry. 2005;44:1288–1299. doi: 10.1021/bi048029t. [DOI] [PubMed] [Google Scholar]
- 46.Goto Y, Calciano LJ, Fink AL. Acid-induced folding of proteins. Proc. Natl. Acad. Sci. U S A. 1990;87:573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasahara K, Yagi H, Sakai M, Naiki H, Goto Y. Amyloid nucleation triggered by agitation of beta2-microglobulin under acidic and neutral pH conditions. Biochemistry. 2008;47:2650–2660. doi: 10.1021/bi701968g. [DOI] [PubMed] [Google Scholar]
- 48.Sicorello A, Torrassa S, Soldi G, Gianni S, Travaglini-Allocatelli C, Taddei N, Relini A, Chiti F. Agitation and high ionic strength induce amyloidogenesis of a folded PDZ domain in native conditions. Biophys. J. 2009;96:2289–2298. doi: 10.1016/j.bpj.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirahama T, Cohen AS. High-resolution electron microscopic analysis of the amyloid fibril. J. Cell Biol. 1967;33:679–708. doi: 10.1083/jcb.33.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malisauskas M, Zamotin V, Jass J, Noppe W, Dobson CM, Morozova-Roche LA. Amyloid protofilaments from the calcium-binding protein equine lysozyme: formation of ring and linear structures depends on pH and metal ion concentration. J. Mol. Biol. 2003;330:879–890. doi: 10.1016/s0022-2836(03)00551-5. [DOI] [PubMed] [Google Scholar]
- 51.Ding TT, Lee SJ, Rochet JC, Lansbury PT., Jr Annular alpha-synuclein protofibrils are produced when spherical protofibrils are incubated in solution or bound to brain-derived membranes. Biochemistry. 2002;41:10209–10217. doi: 10.1021/bi020139h. [DOI] [PubMed] [Google Scholar]
- 52.Relini A, Torrassa S, Rolandi R, Gliozzi A, Rosano C, Canale C, Bolognesi M, Plakoutsi G, Bucciantini M, Chiti F, Stefani M. Monitoring the process of HypF fibrillization and liposome permeabilization by protofibrils. J. Mol. Biol. 2004;338:943–957. doi: 10.1016/j.jmb.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 53.Solomon JP, Page LJ, Balch WE, Kelly JW. Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit. Rev. Biochem. Mol. Biol. 2012;47:282–296. doi: 10.3109/10409238.2012.661401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solomon JP, Bourgault S, Powers ET, Kelly JW. Heparin binds 8 kDa gelsolin cross-beta-sheet oligomers and accelerates amyloidogenesis by hastening fibril extension. Biochemistry. 2011;50:2486–2498. doi: 10.1021/bi101905n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Relini A, De Stefano S, Torrassa S, Cavalleri O, Rolandi R, Gliozzi A, Giorgetti S, Raimondi S, Marchese L, Verga L, Rossi A, Stoppini M, Bellotti V. Heparin strongly enhances the formation of beta2-microglobulin amyloid fibrils in the presence of type I collagen. J. Biol. Chem. 2008;283:4912–4920. doi: 10.1074/jbc.M702712200. [DOI] [PubMed] [Google Scholar]
- 56.Perez M, Valpuesta JM, Medina M, Montejo de, Garcini E, Avila J. Polymerization of tau into filaments in the presence of heparin: the minimal sequence required for tau-tau interaction. J. Neurochem. 1996;67:1183–1190. doi: 10.1046/j.1471-4159.1996.67031183.x. [DOI] [PubMed] [Google Scholar]
- 57.Pollard HB, Arispe N, Rojas E. Ion channel hypothesis for Alzheimer amyloid peptide neurotoxicity. Cell. Mol. Neurobiol. 1995;15:513–526. doi: 10.1007/BF02071314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tropea JE, Cherry S, Waugh DS. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. 2009;498:297–307. doi: 10.1007/978-1-59745-196-3_19. [DOI] [PubMed] [Google Scholar]
- 59.Stelea SD, Keiderling TA. Pretransitional structural changes in the thermal denaturation of ribonuclease S and S protein. Biophys. J. 2002;83:2259–2269. doi: 10.1016/S0006-3495(02)73986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fung SY, Keyes C, Duhamel J, Chen P. Concentration effect on the aggregation of a self-assembling oligopeptide. Biophys. J. 2003;85:537–548. doi: 10.1016/S0006-3495(03)74498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foley J, Hill SE, Miti T, Mulaj M, Ciesla M, Robeel R, Persichilli C, Raynes R, Westerheide S, Muschol M. Structural fingerprints and their evolution during oligomeric vs. oligomer-free amyloid fibril growth. J. Chem. Phys. 2013;139:121901. doi: 10.1063/1.4811343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shivu B, Seshadri S, Li J, Oberg KA, Uversky VN, Fink AL. Distinct beta-Sheet Structure in Protein Aggregates Determined by ATR-FTIR Spectroscopy. Biochemistry. 2013;52:5176–5183. doi: 10.1021/bi400625v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.