Abstract
Hypertrophic cardiomyopathy (HCM) is the most common form of inherited cardiac disease and the leading cause of sudden cardiac death in young people. HCM is caused by mutations in genes encoding contractile proteins. Cardiac myosin binding protein-C (cMyBP-C) is a thick filament contractile protein that regulates sarcomere organization and cardiac contractility. About 200 different mutations in the cMyBP-C gene (MYBPC3) have thus far been reported as causing HCM. Among them, a 25 base pair deletion in the branch point of intron 32 of MYBPC3 is widespread, particularly in South Asia, where it affects ≈4% of South Asian descendants worldwide. This polymorphic mutation results in skipping of exon 33 and a reading frame shift, which, in turn, replaces the last 65 amino acids of the C-terminal C10 domain of cMyBP-C (cMyBP-CC10mut) with a novel sequence of 58 residues. Carriers of the 25 base pair deletion mutation are at increased risk of developing cardiomyopathy and heart failure. Because of the high prevalence of this mutation in certain populations, genetic screening of at-risk groups might be beneficial. Scientifically, the functional consequences of C-terminal mutations and the precise mechanisms leading to HCM should be defined using induced pluripotent stem cells and engineered heart tissue in vitro, or mouse models in vivo. Most importantly, therapeutic strategies that include pharmacology, gene repair and gene therapy should be developed to prevent the adverse clinical effects of cMyBP-CC10mut. This review article aims to examine the effects of cMyBP-CC10mut on cardiac function, emphasizing the need for the development of genetic testing and expanded therapeutic strategies.
Keywords: Cardiac myosin binding protein-C, Gene Repair, Gene Therapy, Genetic Testing, Hypertrophic cardiomyopathy, iPS Cells, MYBPC3
Introduction
Hypertrophic cardiomyopathy (HCM) is the most common inherited cardiac disease. HCM has a prevalence of 1:500 in the general population [24] and is defined by the presence of left ventricular hypertrophy (LVH) that cannot be explained by cardiac or systemic diseases [9]. The clinical phenotype of HCM is heterogeneous and ranges from diastolic dysfunction and atrial fibrillation to heart failure and sudden cardiac death [22,13]. Interestingly, sudden death among athletes is predominantly associated with HCM [23,20]. Characteristic morphological and histological changes observed in HCM include LVH, which is defined as LV wall thickness greater than, or equal to, 15 mm (although 13–14 mm is considered borderline) [9]. Typically, LVH is not uniform, with most HCM patients only showing hypertrophy of the interventricular septum [18]. Apart from hypertrophy, cardiomyocyte disarray and interstitial fibrosis are observed, which may potentially contribute to the impairment of diastolic function and arrhythmias [25].
HCM is caused by mutations in genes encoding for sarcomeric proteins. These mutations are autosomal dominant, and most patients are heterozygous carriers of the disease-causing mutation. In particular, mutations in MYBPC3 (encoding cardiac myosin binding protein-C) and MYH7 (encoding β-myosin heavy chain) each contribute 30–40% of HCM-associated mutations. Discovering that mutations in MYBPC3 lead to HCM [2,47] has hastened the pace of research aimed at understanding the role of cMyBP-C. Studies recently reported a polymorphic HCM-causing mutation [46,5], a 25 base pair (bp) deletion, in MYBPC3 that is highly prevalent in South Asian countries [41] and is estimated to affect 55 million people. However, the pathogenic mechanism of the mutation is completely unknown. Therefore, this review article will examine this mutation, describe the current literature, and emphasize the need for systematic studies to determine the cause of HCM, develop new diagnostic methods and advance therapeutic care.
Mutations in MYBPC3 cause HCM
Mutations in MYBPC3 are the most common cause of HCM and account for around 40% of all reported mutations [34]. Thus far, at least 197 HCM-associated mutations in MYBPC3 have been identified [15]. Most MYBPC3 mutations are predicted to result in protein truncation of the C-terminus, either by interfering with normal mRNA splicing, leading to exon skipping, or by causing a reading frame shift resulting in a nonsense coding sequence followed by a premature stop in translation [2,47]. Mutations in other sarcomeric genes typically result in the expression and incorporation of dominant negative mutant proteins, or poison polypeptides, into the sarcomere where they affect function. In human patients harboring a truncation mutation, mutant MYBPC3 is transcribed to mRNA; however, mutant cMyBP-C proteins are not detectable, and, overall, cMyBP-C in the sarcomere is significantly reduced [35,26,44]. These findings strongly support the idea that most MYBPC3 mutations cause HCM through haploinsufficiency. Certain mouse models of MYBPC3 truncation mutations provide additional evidence that haploinsufficiency is sufficient to cause HCM, as these animals express no, or very little, mutant protein, causing reduced overall levels of cMyBP-C. Moreover, these mouse models exhibit hypertrophy along with mild septal thickening [3,45]. In contrast, other animal models demonstrate either robust expression of truncated protein along with sarcomere disorganization [49], very low expression of truncated protein with no reduction of wild-type (WT) protein [50], or reduced expression of total cMyBP-C with incorporation of mutant protein into the sarcomere [17,29]. Furthermore, in mice hemizygous for cMyBP-C (+/−) or heterozygous for a functionally null allele (+/t), sarcomere organization and contractile function are normal [28,27,14,32]. Thus, further investigation is necessary to clearly distinguish the HCM causing mechanisms of individual mutations in order to tailor therapeutic approaches.
Role of C-terminal Region of cMyBP-C in thick filament organization
cMyBP-C is a thick filament-associated protein in the cardiac sarcomere that plays both regulatory and structural roles in cardiomyocyte contraction [39,40,37,38,11,12,36,21]. The cardiac isoform of MyBP-C consists of 12 domains, including one phosphorylation (M) domain, eight immunoglobulin (IgC2)-like domains, and three fibronectin type-III (FN3) domains. In mouse models where cMyBP-C is knocked out or not expressed, animals are viable, but they exhibit abnormal misaligned sarcomeric structure, contractile dysfunction, and cardiomyopathy [14,28]. While these data suggest that cMyBP-C is not essential for cardiac development, it is necessary for normal sarcomere structure and cardiac function. The precise positioning of cMyBP-C in the sarcomere is unclear; however, two models, including the trimeric collar model and the axial model, have been proposed to explain the specific arrangement of cMyBP-C and the interaction of its C-terminus with myosin and titin. The collar model proposes that three molecules of cMyBP-C form collar-like rings every 43 nm around the thick filament which are stabilized by domain interactions in C5–C10 [31,6,7]. The axial model proposes that the C-terminal domains of cMyBP-C run parallel along the thick filament with the N-terminus extended towards the thin filament [42]. Supporting data exist for both models, but more high-resolution structural data are needed to confirm which model is correct.
While the N-terminal region of cMyBP-C functions as a critical regulator of contractile function, the C-terminal region is thought to play a structural anchoring role. Domains C7–C10 of the C-terminal region of cMyBP-C bind to the thick filament and are required for incorporation into the sarcomere [10]. The C10 domain of cMyBP-C interacts with the light meromyosin (LMM) portion of myosin rods, forming the backbone of the thick filament [7]. Domains C8–C10 of cMyBP-C have been shown to bind to immunoglobulin domains of titin that are repeated approximately every 42 nm within the C zone of the thick filament, and it is likely that the position of cMyBP-C in the thick filament is dictated by the localization of these titin domains [19]. Any mutation producing a truncation or modification of the myosin and titin binding sites within the C-terminus of cMyBP-C would therefore be expected to result in a reduced or aberrant incorporation of the mutant protein into the sarcomere.
A highly prevalent C-terminal mutation in cMyBP-C
Previously, we described a polymorphic deletion of 25 base pairs in intron 32 of the cMyBP-C gene (MYBPC3Δ25bp) that is associated with hypertrophic and dilated cardiomyopathies [46]. Interestingly, the distribution of this mutation is almost exclusively restricted to South Asian countries [5,41]. It is estimated that 55 million people in South Asian populations are affected by this deletion and are at increased risk of developing contractile dysfunction and heart failure. To date, this mutation has been found to occur primarily in India, Pakistan, Sri Lanka, Malaysia, and Indonesia [5,33,41]. While most young and middle-aged individuals exhibit a mild phenotype or are asymptomatic, individuals over the age of 40 years present with more moderate to severe symptoms. Out of 28 unrelated carrier families, 90% of the oldest members were found to be symptomatic. The fact that disease symptoms are typically dormant until the third decade of life, generally a time beyond child-bearing years, helps to explain the high prevalence of this mutation, which originated at least 10,000 years ago [48]. Carrying the mutation leads to increased susceptibility to worse outcome following cardiac disease. For example, patients with coronary artery disease also carrying the deletion had significantly worse systolic function [43].
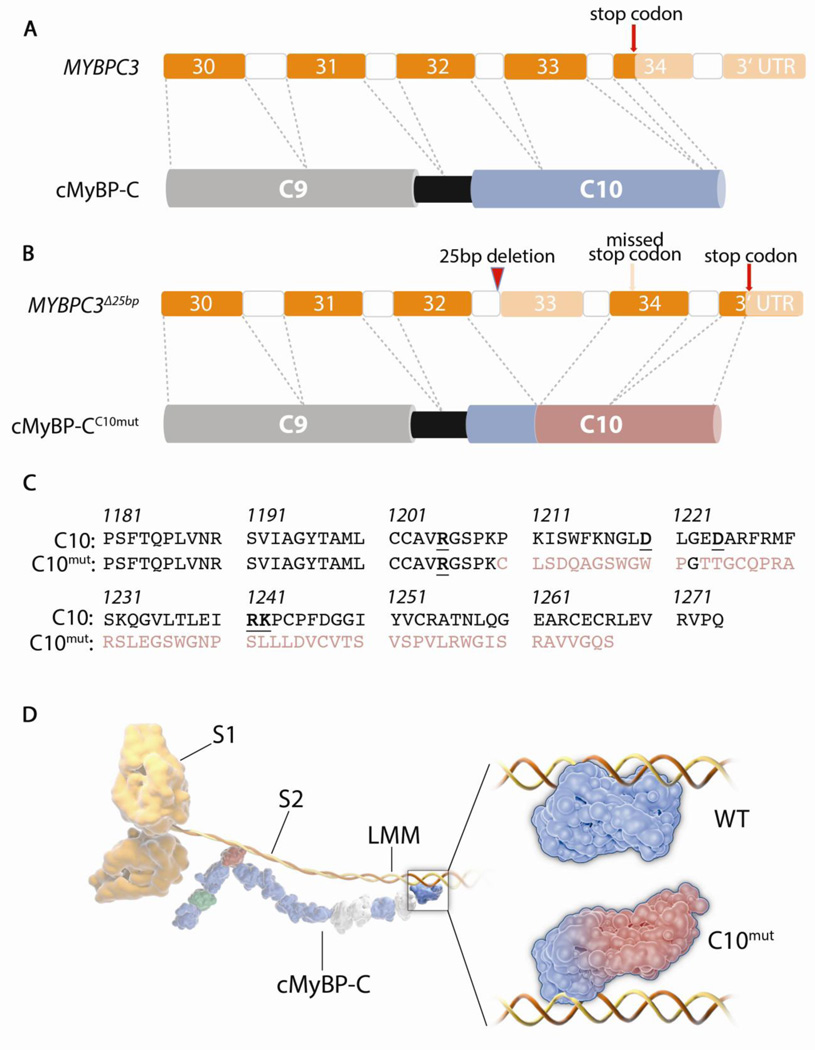
Transcriptional studies demonstrated that MYBPC3Δ25bp causes the skipping of exon 33 and a reading frame shift, resulting in the replacement of 65 wild-type amino acid residues with a novel sequence of 58 residues in the C10 domain (cMyBP-CC10mut) [46] (Figure 1A–C). Expression of cMyBP-CC10mut protein in neonatal cardiomyocytes resulted in the aberrant incorporation of mutant protein in the sarcomere and the disruption of normal sarcomeric structure by a still unknown mechanism [5]. The most likely consequence of the cMyBP-CC10mut mutation is an alteration of the interaction with LMM (Figure 1D). Miyamoto et al. showed that 5 charged amino acids spread throughout the C10 domain are needed for cMyBP-C/LMM interaction (bold in Figure 1C), all of which are conserved between MyBP-C isoforms [30]. In cMyBP-CC10mut, only 1 out of 5 of these key residues is conserved, suggesting a decreased affinity for LMM binding. Given that this MYBPC3Δ25bp mutation affects nearly 1% of the world’s population, it is clinically urgent to fully elucidate the function of this cMyBP-C mutant protein. Further in vitro and in vivo investigation into the mechanisms by which the MYBPC3Δ25bp mutation leads to disorganization of sarcomere structure and cardiomyopathy will greatly contribute to an understanding of the pathology of other cMyBP-C mutations and sarcomeric gene mutations in general.
Figure 1. The MYBPC3Δ25bp mutation and cMyBP-CC10mut protein.
(A) 3’ end of normal MYBPC3 that codes for the C-terminal C9 and C10 domains of cMyBP-C. Coding exons in bright orange. Normal stop codon exists at the beginning of exon 34. (B) Location of MYBPC3Δ25bp mutation is shown in intron 32, at the branch point, which causes skipping of exon 33 and subsequent reading frame shift. This leads to translation of the entire exon 34 and part of the 3’ UTR. This mutation leads to the expression of cMyBP-CC10mut, which has a partly altered C10 domain (indicated in red). (C) Amino acid sequences of wild-type (WT)-C10 and cMyBP-CC10mut domains. First 29 amino acids are encoded by exon 32 and are therefore identical between WT-C10 and cMyBP-CC10mut (black). The mutation leads to the replacement of the last 65 amino acids with a novel sequence of 58 amino acids. In the WT-C10 residues, the 5 charged amino acids that were indispensable for LMM binding [30] are indicated bold and underlined. Only 1 out of 5 of these was preserved in the cMyBP-CC10mut residues. (D) Schematic illustration of cMyBP-C/myosin heavy chain (MHC) interaction. The MHC S2 domain interacts with the M-domain (red) of cMyBP-C on the N-terminal end of the protein. The C10 domain interacts with the LMM domain of MHC, and the WT-C10 domain and putative cMyBP-CC10mut interactions with LMM are enlarged on the insert. We propose that the cMyBP-CC10mut has weaker interaction with the LMM domain. On cMyBP-C, IgC2 domains are displayed in blue, FN3 domains in off-white, M-domain in red and proline-alanine-rich linker in green.
Necessity for genetic testing
The role of genotyping in inherited cardiomyopathies has been a hot button issue. Arguments can be made that knowing the mutation has no implications for treatment in these patients and therefore does not justify the costs; because 1) HCM can be caused by mutations in >11 genes and 2) the frequency of individual causal mutations is generally low [34]. In current clinical practice, genetic testing is used for screening of family members, rather than screening of the general population [4]. Although most mutations are infrequent, a number of mutations occur with much more frequency than others in some populations. Examples of these are the MYBPC3Δ25bp mutation and the 2373InsG founder mutation in MYBPC3 that accounts for nearly 25% of all HCM patients in the Netherlands [1]. The high prevalence of the MYBPC3Δ25bp mutation in South Asia [5,41] may make general population screening for this particular mutation both beneficial and feasible. With a relatively simple PCR-based screening method, this mutation can be easily detected (Figure 2). Identification of MYBPC3Δ25bp carriers at a presymptomatic stage would enable unprecedented monitoring opportunities. Although therapeutic interventions to prevent the onset of symptoms are still not available in patients, recent studies in animal models show that prevention of hypertrophy and cardiac dysfunction [8,16] might be achievable in the not too distant future.
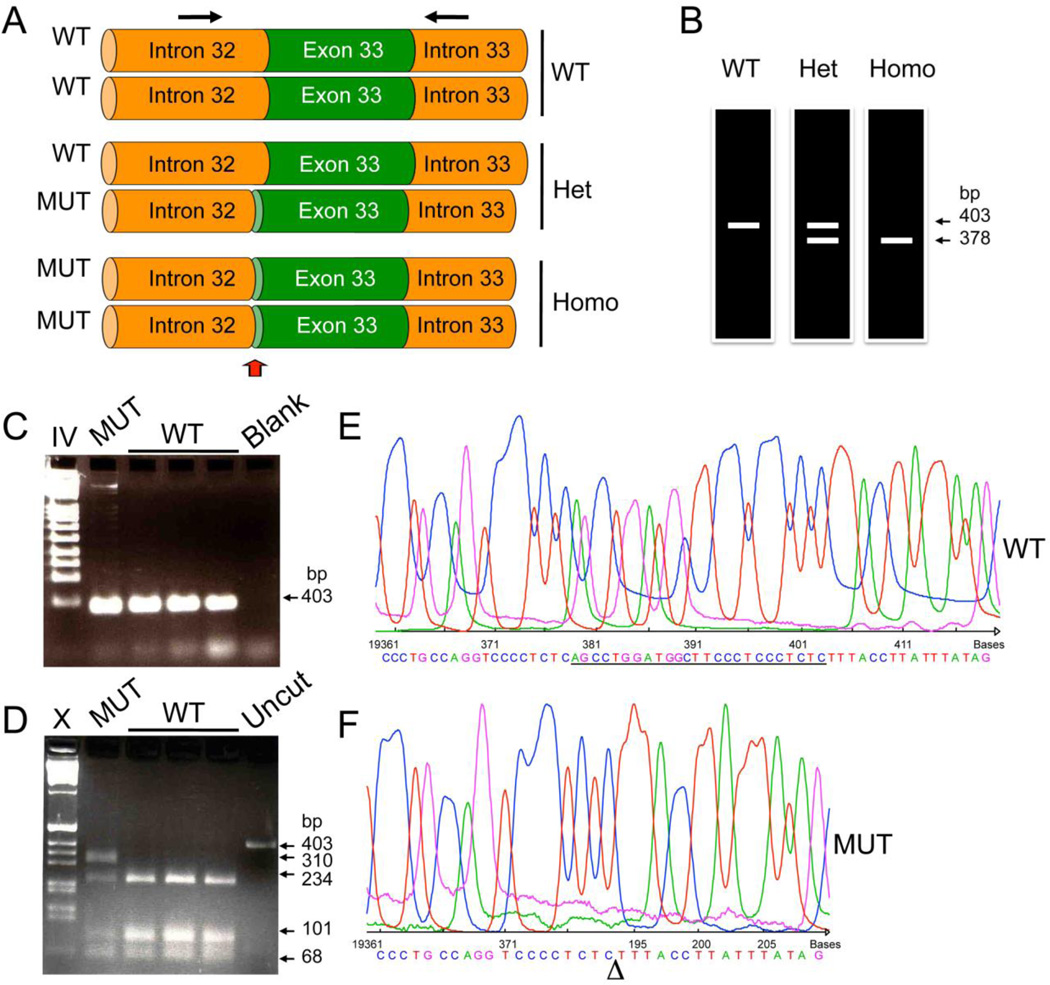
Figure 2. Genetic testing to determine the presence of the 25 bp deletion mutation.
(A) Schematic representation of part of MYBPC3 gene representing wild-type (WT), heterozygous and homozygous mutation carries. The arrows indicate both forward and reverse primers used in the PCR reaction (Forward primer: 5'-GTT TCC AGC CTT GGG CAT AGT C-3' and Reverse primer: 5'-GAG GAC AAC GGA GCA AAG CCC-3') to obtain a 403 bp and 378 bp from WT and mutant alleles, respectively. (B) Schematic representation of expected PCR products run on an acrylamide gel (7%) with silver staining as previously described [46]. Non-mutation carriers have a 403 bp PCR product, while homozygous MYBPC3Δ25bp carriers have only the shorter 378 bp PCR product. Heterozygous MYBPC3Δ25bp carriers have both products. In contrast, when PCR products were resolved in 1% agarose gel, the difference of 25 bp is not resolved (C). However, by digesting them with BglI enzyme (D), MYBPC3Δ25bp carriers can be detected. Within the 403 bp sequence, two BglI sites are present, resulting in three fragments (234, 101 and 68 bp). However, MYBPC3Δ25bp loses one of the two BglI sites, resulting in two fragments (310 and 68 bp). Blank reaction resulted in the absence of PCR products (Blank). Uncut PCR products without digestion with BglI are shown (Uncut). Marker lanes IV and X indicate Roche IV and Roche X markers, respectively. DNA sequences of wild-type (E) and mutant (F) alleles are shown. PCR products from heterozygous carriers were cloned into pCR-Blunt II TOPO vector using Zero Blunt TOPO PCR cloning kit (Invitrogen) and then sequenced using T7 forward and M13 reverse primers.
Conclusion
cMyBP-C is a key regulator of cardiac contractility. Although mutations in the gene encoding cMyBP-C are a leading cause of HCM, little is known about the molecular mechanisms underlying the disease process. The availability of cardiac tissue from myectomy operations has resulted in a relatively large body of evidence on contractile function in human HCM patients. Nonetheless, additional experimentation, including animal models, induced pluripotent stem cells and engineered heart tissue, is required to clearly establish the detrimental effects of mutated proteins, all of which comprise the focus of current studies in the authors’ laboratory. Gaining more insight into these fundamental questions will lead to the development of novel therapeutic strategies for the treatment of HCM and save millions of patients from heart failure.
ACKNOWLEDGMENT
The authors were supported by National Institutes of Health grants R01HL105826 and K02HL114749 (SS), as well as the American Heart Association Midwest Postdoctoral Fellowship 13POST17220009 (DWDK) entitled “Pathophysiology of Hypertrophic Cardiomyopathy-associated Cardiac Myosin Binding Protein-C Mutation” (July 2013–June 2015).
Abbreviations
- HCM
Hypertrophic cardiomyopathy
- LV
left ventricle
- LVH
left ventricular hypertrophy
- MYH7
myosin heavy chain gene
- MYBPC3
cardiac myosin binding protein-C gene
- cMyBP-C
cardiac myosin binding protein-C
- MYBPC3Δ25bp
25 basepair deletion mutation in MYBPC3
- cMyBP-CC10mut
protein product of MYBPC3Δ25bp gene
Footnotes
DISCLOSURES
A full patent application is pending (Application Serial No. 13/464,466, Pub. No. U.S. 2012/0282618 A1 and Date: 05/04/12) for diagnosing the presence of 25 bp deletion mutation in MYBPC3.
Contributor Information
Diederik W. D. Kuster, Department of Cell and Molecular Physiology, Health Sciences Division, Loyola University Chicago, Maywood, IL 60153-5500, USA, dkuster@lumc.edu.
Sakthivel Sadayappan, Department of Cell and Molecular Physiology, Health Sciences Division, Loyola University Chicago, Maywood, IL 60153-5500, USA, Phone: 708-216-7994, Fax: 708-216-6308, ssadayappan@lumc.edu.
References
- 1.Alders M, Jongbloed R, Deelen W, van den Wijngaard A, Doevendans P, Ten Cate F, Regitz-Zagrosek V, Vosberg HP, van Langen I, Wilde A, Dooijes D, Mannens M. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. Eur Heart J. 2003;24(20):1848–1853. doi: 10.1016/s0195-668x(03)00466-4. [DOI] [PubMed] [Google Scholar]
- 2.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, Weissenbach J, Vosberg HP, Fiszman M, Komajda M, Schwartz K. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet. 1995;11(4):438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 3.Carrier L, Knoll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J, Jr, Schwartz K, Chien KR. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res. 2004;63(2):293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Charitakis K, Basson CT. Can genetic testing improve our aim in hypertrophic cardiomyopathy? Circ Res. 2010;106(9):1446–1448. doi: 10.1161/CIRCRESAHA.110.220343. [DOI] [PubMed] [Google Scholar]
- 5.Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41(2):187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ Res. 2004;94(10):1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 7.Flashman E, Watkins H, Redwood C. Localization of the binding site of the C-terminal domain of cardiac myosin-binding protein-C on the myosin rod. Biochem J. 2007;401(1):97–102. doi: 10.1042/BJ20060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gedicke-Hornung C, Behrens-Gawlik V, Reischmann S, Geertz B, Stimpel D, Weinberger F, Schlossarek S, Precigout G, Braren I, Eschenhagen T, Mearini G, Lorain S, Voit T, Dreyfus PA, Garcia L, Carrier L. Rescue of cardiomyopathy through U7snRNA-mediated exon skipping in Mybpc3-targeted knock-in mice. EMBO Mol Med. 2013;5(7):1060–1077. doi: 10.1002/emmm.201202168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124(24):2761–2796. doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert R, Kelly MG, Mikawa T, Fischman DA. The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J Cell Sci. 1996;109(Pt 1):101–111. doi: 10.1242/jcs.109.1.101. [DOI] [PubMed] [Google Scholar]
- 11.Govindan S, McElligott A, Muthusamy S, Nair N, Barefield D, Martin JL, Gongora E, Greis KD, Luther PK, Winegrad S, Henderson KK, Sadayappan S. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J Mol Cell Cardiol. 2012;52(1):154–164. doi: 10.1016/j.yjmcc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govindan S, Sarkey J, Ji X, Sundaresan NR, Gupta MP, de Tombe PP, Sadayappan S. Pathogenic properties of the N-terminal region of cardiac myosin binding protein-C in vitro. J Muscle Res Cell Motil. 2012;33(1):17–30. doi: 10.1007/s10974-012-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris KM, Spirito P, Maron MS, Zenovich AG, Formisano F, Lesser JR, Mackey-Bojack S, Manning WJ, Udelson JE, Maron BJ. Prevalence, clinical profile, and significance of left ventricular remodeling in the end-stage phase of hypertrophic cardiomyopathy. Circulation. 2006;114(3):216–225. doi: 10.1161/CIRCULATIONAHA.105.583500. [DOI] [PubMed] [Google Scholar]
- 14.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90(5):594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 15.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108(6):751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342(6154):111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kittleson MD, Meurs KM, Munro MJ, Kittleson JA, Liu SK, Pion PD, Towbin JA. Familial hypertrophic cardiomyopathy in maine coon cats: an animal model of human disease. Circulation. 1999;99(24):3172–3180. doi: 10.1161/01.cir.99.24.3172. [DOI] [PubMed] [Google Scholar]
- 18.Klues HG, Schiffers A, Maron BJ. Phenotypic spectrum and patterns of left ventricular hypertrophy in hypertrophic cardiomyopathy: morphologic observations and significance as assessed by two-dimensional echocardiography in 600 patients. J Am Coll Cardiol. 1995;26(7):1699–1708. doi: 10.1016/0735-1097(95)00390-8. [DOI] [PubMed] [Google Scholar]
- 19.Koretz JF, Irving TC, Wang K. Filamentous aggregates of native titin and binding of C-protein and AMP-deaminase. Arch Biochem Biophys. 1993;304(2):305–309. doi: 10.1006/abbi.1993.1354. [DOI] [PubMed] [Google Scholar]
- 20.Lauschke J, Maisch B. Athlete's heart or hypertrophic cardiomyopathy? Clin Res Cardiol. 2009;98(2):80–88. doi: 10.1007/s00392-008-0721-2. [DOI] [PubMed] [Google Scholar]
- 21.Lin B, Govindan S, Lee K, Zhao P, Han R, Runte KE, Craig R, Palmer BM, Sadayappan S. Cardiac Myosin binding protein-C plays no regulatory role in skeletal muscle structure and function. PLoS One. 2013;8(7):e69671. doi: 10.1371/journal.pone.0069671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. Jama. 2002;287(10):1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 23.Maron BJ. Distinguishing hypertrophic cardiomyopathy from athlete's heart: a clinical problem of increasing magnitude and significance. Heart. 2005;91(11):1380–1382. doi: 10.1136/hrt.2005.060962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 25.Maron BJ, Sato N, Roberts WC, Edwards JE, Chandra RS. Quantitative analysis of cardiac muscle cell disorganization in the ventricular septum. Comparison of fetuses and infants with and without congenital heart disease and patients with hypertrophic cardiomyopathy. Circulation. 1979;60(3):685–696. doi: 10.1161/01.cir.60.3.685. [DOI] [PubMed] [Google Scholar]
- 26.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C, Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105(3):219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 27.McConnell BK, Fatkin D, Semsarian C, Jones KA, Georgakopoulos D, Maguire CT, Healey MJ, Mudd JO, Moskowitz IP, Conner DA, Giewat M, Wakimoto H, Berul CI, Schoen FJ, Kass DA, Seidman CE, Seidman JG. Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ Res. 2001;88(4):383–389. doi: 10.1161/01.res.88.4.383. [DOI] [PubMed] [Google Scholar]
- 28.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104(9):1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, Kittleson JA, Munro MJ, Dryburgh K, Macdonald KA, Kittleson MD. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet. 2005;14(23):3587–3593. doi: 10.1093/hmg/ddi386. [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto CA, Fischman DA, Reinach FC. The interface between MyBP-C and myosin: site-directed mutagenesis of the CX myosin-binding domain of MyBP-C. J Muscle Res Cell Motil. 1999;20(7):703–715. doi: 10.1023/a:1005513312939. [DOI] [PubMed] [Google Scholar]
- 31.Moolman-Smook J, Flashman E, de Lange W, Li Z, Corfield V, Redwood C, Watkins H. Identification of novel interactions between domains of Myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circ Res. 2002;91(8):704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 32.Palmer BM, Georgakopoulos D, Janssen PM, Wang Y, Alpert NR, Belardi DF, Harris SP, Moss RL, Burgon PG, Seidman CE, Seidman JG, Maughan DW, Kass DA. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ Res. 2004;94(9):1249–1255. doi: 10.1161/01.RES.0000126898.95550.31. [DOI] [PubMed] [Google Scholar]
- 33.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 35.Rottbauer W, Gautel M, Zehelein J, Labeit S, Franz WM, Fischer C, Vollrath B, Mall G, Dietz R, Kubler W, Katus HA. Novel splice donor site mutation in the cardiac myosin-binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization Of cardiac transcript and protein. J Clin Invest. 1997;100(2):475–482. doi: 10.1172/JCI119555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadayappan S, de Tombe PP. Cardiac myosin binding protein-C: redefining its structure and function. Biophys Rev. 2012;4(2):93–106. doi: 10.1007/s12551-012-0067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadayappan S, Gulick J, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Robbins J. Cardiac myosin binding protein-C phosphorylation in a {beta}-myosin heavy chain background. Circulation. 2009;119(9):1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadayappan S, Gulick J, Osinska H, Barefield D, Cuello F, Avkiran M, Lasko VM, Lorenz JN, Maillet M, Martin JL, Brown JH, Bers DM, Molkentin JD, James J, Robbins J. A critical function for Ser-282 in cardiac Myosin binding protein-C phosphorylation and cardiac function. Circ Res. 2011;109(2):141–150. doi: 10.1161/CIRCRESAHA.111.242560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97(11):1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein-C phosphorylation is cardioprotective. Proc Natl Acad Sci U S A. 2006;103(45):16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonson TS, Zhang Y, Huff CD, Xing J, Watkins WS, Witherspoon DJ, Woodward SR, Jorde LB. Limited distribution of a cardiomyopathy-associated variant in India. Ann Hum Genet. 2010;74(2):184–188. doi: 10.1111/j.1469-1809.2010.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331(3):713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava A, Garg N, Mittal T, Khanna R, Gupta S, Seth PK, Mittal B. Association of-25 bp deletion in MYBPC3 gene with left ventricle dysfunction in coronary artery disease patients. PLoS One. 2011;6(9):e24123. doi: 10.1371/journal.pone.0024123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119(11):1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 45.Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Pointu H, Mougenot N, Guiard J, Reimer R, Hohenberg H, Schwartz K, Vernet M, Eschenhagen T, Carrier L. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res. 2009;105(3):239–248. doi: 10.1161/CIRCRESAHA.109.201251. [DOI] [PubMed] [Google Scholar]
- 46.Waldmuller S, Sakthivel S, Saadi AV, Selignow C, Rakesh PG, Golubenko M, Joseph PK, Padmakumar R, Richard P, Schwartz K, Tharakan JM, Rajamanickam C, Vosberg HP. Novel deletions in MYH7 and MYBPC3 identified in Indian families with familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2003;35(6):623–636. doi: 10.1016/s0022-2828(03)00050-6. [DOI] [PubMed] [Google Scholar]
- 47.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11(4):434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 48.Xu Q, Dewey S, Nguyen S, Gomes AV. Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. J Mol Cell Cardiol. 48(5):899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J Clin Invest. 1998;102(7):1292–1300. doi: 10.1172/JCI3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. In vivo modeling of myosin binding protein C familial hypertrophic cardiomyopathy. Circ Res. 1999;85(9):841–847. doi: 10.1161/01.res.85.9.841. [DOI] [PubMed] [Google Scholar]