Summary
Dengue virus infection elicits a spectrum of clinical presentations ranging from asymptomatic to severe disease. The mechanisms leading to severe dengue are not known, however it has been reported that the complement system is hyper-activated in severe dengue. Screening of complement proteins demonstrated that C1q, a pattern recognition molecule, can bind directly to Dengue Virus Envelope protein and to whole Dengue Virus serotype 2. Incubation of Dengue Virus serotype 2 with C1q prior to infection of THP-1 cells led to decreased virus infectivity and modulation of mRNA expression of immunoregulatory molecules suggesting reduced inflammatory responses.
Keywords: Dengue, Dengue Hemorrhagic Fever, Dengue Fever, C1q, Classical Complement Pathway, Viral Persistence
Dengue virus (DENV) is a significant human pathogen, causing 250–500 million new infections per year and 2.5 billion of people live in endemic areas, making it an important public health problem (Bhatt et al., 2013). Dengue disease presents in a range of manifestations from a relatively benign, self-limiting febrile illness (dengue fever, DF) to life-threatening vascular leakage (dengue hemorrhagic fever/dengue shock syndrome, DHF/DSS) (World Health Organization, 2012). Mechanisms which might lead to severe clinical outcomes in DENV infections have been described, such as Antibody Dependent Enhancement (ADE) (Halstead and O’Rourke, 1977), Original Antigenic Sin in T cells (Mongkolsapaya et al., 2003) and Cytokine Storm (Dong et al., 2007).
The complement system, an arm of the innate immune, consists of three main pathways: the classical complement pathway, the alternative complement pathway, and the mannose-binding lectin pathway. It comprehends several proteins found in the blood, which normally circulate as inactive precursors that, when stimulated, initiate an amplifying cascade of further cleavages, resulting in release of chemokines, opsonization and activation of the cell-killing membrane attack complex (Ricklin et al., 2011). The expression of complement-related genes has also been found altered in DHF as compared to DF (Nascimento et al., 2009a; Ubol et al., 2008). In addition, it has been previously observed that DHF patients have hyper-active complement activation (Bokish et al., 1973a, 1973b; Nascimento et al., 2009b). These data support that the complement system is activated to a greater extent in DHF patients than in subjects developing DF. However the mechanisms leading to hyper-activation are not completely understood. In addition, direct interactions of DENV with complement factors have been poorly investigated. It has been observed that at least one complement factor, Mannose-Binding Lectin, binds directly to DENV (Avirutnan et al., 2011; Fuchs et al., 2011). Such could be a mechanism of defense similar to other pathogens, i.e., to be covered by molecules of the immune system to avoid detection and elimination from the host (Hilleman, 2004). C1q activates the Classical Complement Pathway, either by directly binding specific structures on the surface of pathogens and apoptotic cells or by binding immunoglobulins, immunocomplexes and pentraxins (Ricklin et al., 2011). It was recently reported that C1q binds to DENV non-structural protein 1 (NS1) (Silva et al., 2013). Also, C1q is able to inhibit ADE in vitro and in the mouse model in an IgG subclass-specific manner (Mehlhop et al., 2008).
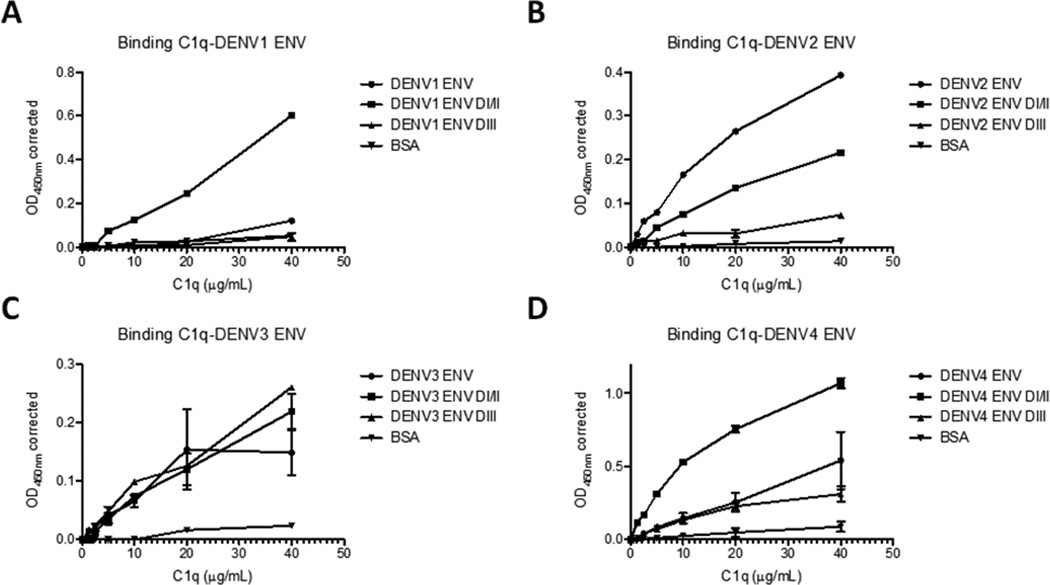
DENV Envelope protein (ENV) is a structural protein involved in attachment and fusion of the virus to host cells. ENV proteins have 3 domains, DI, DII and DIII. DI connects DII and DIII through flexible hinges that participate in the conformational changes that drive DENV fusion process, DII interacts with the membranes of the target cell during fusion and DIII is an immunoglobulin-like domain that is thought to mediate interactions between the virus and structures on the host cell involved in virus attachment (Modis et al., 2004). To determine which complement proteins bind DENV, we performed binding assays using purified complement proteins and ENV proteins from all 4 serotypes of DENV. Complement factors that have been found in altered levels in patient sera during severe DENV infections (C1q, C3, C4, C5, factor B, factor D and factor H; Quidel, San Diego CA, USA) (Bokish et al., 1973a; Nascimento et al., 2009b) or in cellular infection in vitro (CD46; Sino Biological, Beijing, China) (Nascimento, unpublished data) were selected for this study. The complement factors were tested at several concentrations, including their physiological concentrations (as determined in Nascimento et al., 2009b). Recombinant ENV proteins were obtained commercially (Prospec, Ness-Ziona, Israel; MyBioSource, San Diego CA, USA). We observed that full ENV proteins from all DENV serotypes bound to C1q (Fig. 1A–D). No significant interactions were detected with the remaining complement factors tested (data not shown). The binding to C1q occurred preferentially in the DI/II of ENV proteins within each serotype except DENV3 (Figure 1A–D). For the latter, C1q appears to bind similarly to both DI/II and DIII (Fig. 1C). The functionality of all recombinant proteins used was previously validated by assessing their binding to antibodies derived from plasma samples of a Dengue cohort study using both Luminex® and flow citometry (Soares de Melo, 2012; Soares de Melo, manuscript in preparation).
Fig. 1.
C1q binds to full recombinant Envelope proteins of all Dengue Virus (DENV1-4) serotypes (A–D, respectively) but not to Bovine Serum Albumin (BSA; negative control). Within each serotype, binding of C1q occurs preferentially to the portion of ENV containing DI/II (A, B and D), except for serotype 3 (C). ELISA plates were coated with DENV ENV full length, ENV DI/II and ENV DIII (aa 1–413, aa 1–296 and aa 297–413, respectively; Table S1) fragments and BSA, and incubated with C1q concentrations ranging from 1 up to 40 µg/ml. Results obtained are representative of 5 independent experiments. Statistical analysis was done using GraphPad program and Student’s paired, 2 tails t-test. Values were considered significant when p<0.05.
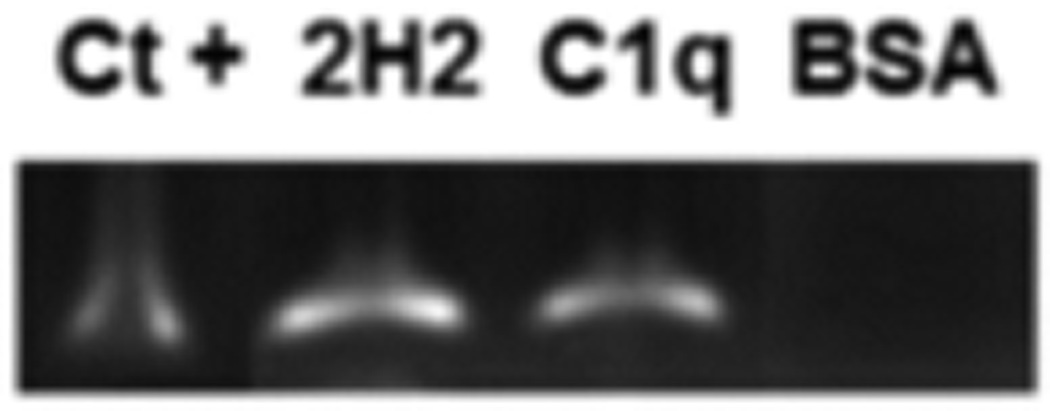
To determine if C1q could bind whole, intact virus we employed a modified ELISA-capture protocol pioneered to detect HCV association with CD59 which has been shown to be much more sensitive than the conventional colorimetric ELISA system (Amet et al., 2012). ELISA plates were coated with a commercial monoclonal mouse antibody (IgG2a) against DENV (2H2, Millipore, Billerica MA, USA) with C1q or with BSA (negative binding control) and then incubated with DENV2 strain 16681 (BEI Resources, Manassas VA, USA). Virus were previously expanded and tittered as described elsewhere (de Melo et al., 2011). Plates were washed to remove any unbound DENV and RNA was extracted from each well. The specific virus cDNA synthesis was performed and followed by PCR with specific primers to DENV2 (Table S2). As expected, DENV bound to the wells containing 2H2 antibody and C1q, but not BSA (Fig. 2).
Fig. 2.
Dengue viruses (DENV2) bind to C1q. DENV2 bind to 2H2 antibody and to C1q, but not to BSA. Ct+, positive control used was an extract of Vero Cells infected with DENV2. Amplicon size, 189 bp. To capture the viruses, wells were coated with 2H2 antibody, C1q and BSA and incubated with DENV2. Viral RNA was extracted by Trizol and DENV2 presence was confirmed by PCR using specific primer sequences for this virus. Results obtained are representative of 3 independent experiments.
A structural protein of human astrovirus type 1, the coat protein (CP), has been demonstrated to bind C1q (Bonaparte et al., 2008), inhibiting activation of classical complement pathway in vitro (Hair et al., 2010). However, in similar conditions, ENV protein from all DENV serotypes did not lead to any alteration in the activation of the classical complement pathway (data not shown).
To determine if incubation of DENV with C1q would affect virus infection, DENV2 (strain 16681) was incubated with C1q prior to exposure to permissive THP-1 cells, a human acute monocytic leukemia cell line (Tassaneetrithep et al., 2003). We observed that C1q pre-incubation with DENV2 significantly decreased viral infectivity by 30%, when compared to either viruses incubated with buffer or with buffer containing other complement protein, factor D, which does not bind to ENV, by RT-PCR (Fig. 3A). Analysis of infectivity using flow cytometer based approach led to similar results (data not shown).
Fig. 3.
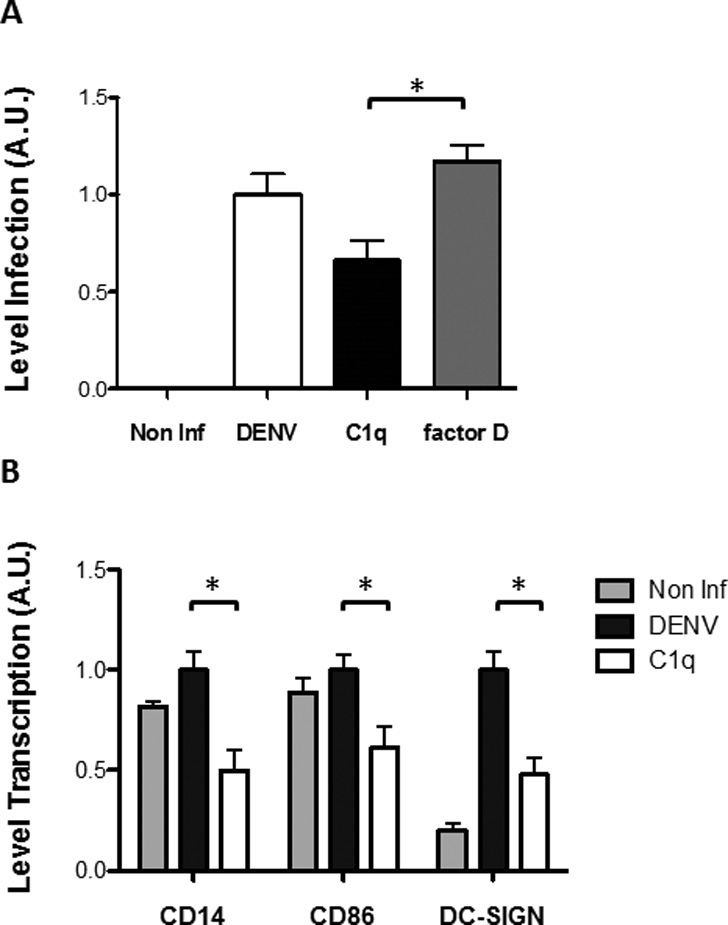
Incubation of DENV2 with C1q leads to a decrease in infection and in the levels of transcription of cellular factors. (A) DENV2 strain 16681 viruses were incubated either with vehicle (DENV), C1q and factor D at 37°C for 1 hour prior to infection of THP-1 cells and the level of infection was measured by RT-PCR 24 hours later. Cells infected with DENV2 incubated with C1q, but not with factor D, had a significant decrease in infection of around 30%. DENV2 viruses were incubated with the complement molecules in their physiological concentration, i.e., the concentrations they are normally found in human blood, 10 µg/ml and 750 ng/ml for C1q and factor D, respectively (Nascimento et al., 2009b). (B) Levels of transcription of several cellular factors are altered by infection of THP-1 with DENV2 previously incubated with C1q. DENV2 strain 16681 viruses were incubated either with vehicle (DENV) or C1q at 37°C for 1 hour prior to infection of THP-1 cells and the level of transcription of several proteins involved in viral clearance was measured by RT-PCR 24 hours later. Levels of CD14 (involved in monocyte maturation), CD86 (involved in the induction of a T cell response) and DC-SIGN (a cellular receptor involved both in viral infection and modulation of innate immune response) show, are significantly decreased in cells infected with virus pre-incubated with C1q. Results obtained are representative of 4 independent experiments. Statistical analysis was done using GraphPad program and Student’s paired, 2 tails t-test. Values were considered significant when p<0.05.
DENV2 bound to C1q was less infectious to THP-1 cells. The reduction of infectivity may be mediated by cellular factors. Therefore, we investigated which host cell molecules could be involved in this process, by assessing the expression of several factors during DENV infection in the presence or absence of C1q. Our results show a significant decrease on CD14, CD86 and DC-SIGN levels of transcription when the THP-1 cells are incubated with DENV2-C1q complex (Fig. 3B).
Our results show that C1q binds to the recombinant ENV proteins of all DENV serotypes and to whole, infectious DENV2. The binding of C1q occurs predominantly in the DI/II regions of ENV proteins within each serotype, except for DENV3, where similar binding levels were observed for both DI/II and DIII. Differences in the sequences of ENV proteins for all serotypes may account for the different levels of binding to C1q observed (Fig. 1). C1q might have more affinity for a region which varies among the 4 serotypes or, even if that region is conserved, it may not be as available due to protein different spatial conformations resultant from such variations in sequence.
We also observed a significant decrease in the levels of transcription of CD14, CD86 and DC-SIGN. CD14 is a molecule involved in monocyte activation and is thought to be involved in DENV2 cell entry (Alhoot et al., 2011; Chen et al., 1999). It has been also observed that interaction of free C1q with C1q-receptors expressed in the surface of monocytes could modulate the differentiation of these cells in immature dendritic cells (DC), increasing the expression of CD14 (Hosszu et al., 2010). Since C1q was bound to the virus, it could have caused similar monocyte activation or induced differentiation into DC, in a process similar to when it is bound to immunocomplexes, therefore also resulting in the decrease observed for the levels of CD14. These hypotheses are consistent with the decrease of infection of the THP-1 cells when incubated with DENV2-C1q complex. CD86 plays roles both in DC maturation and T cell activation (Banchereau et al., 2000). Similar to CD14, infection of THP-1 cells with DENV2-C1q complex resulted in a significant decrease in CD86 expression. These results suggest an immune modulatory mechanism that can potentially inhibit DC - T cells interactions. DC-SIGN (CD209) is a cellular receptor expressed on DC that binds to ICAM-3 in T cells, facilitating the interaction between these two types of cells and inducing proliferation of the latter (Geijtenbeek et al., 2000). However several pathogens, including DENV, use DC-SIGN as a receptor to invade host cells (Anderluh et al., 2012; Tassaneetrithep et al., 2003). As observed for CD14 and CD86, DC-SIGN transcription is also significantly decreased in cells infected with DENV incubated with C1q (Fig. 3B) and, similar to CD14, reduced infectivity by DENV is, most probably, due to the loss of another of its receptors in the host cell. A recent report shows that C1q, gC1qR and DC-SIGN form a heterotrimeric receptor complex on the surface of DC thought to be involved in the differentiation of these cells (Hosszu et al., 2012), suggesting these three molecules could act together as the DENV receptor and triggering of the trimeric complex could have a role in modulating antiviral responses. Also, DC-SIGN binds DENV through DII (Pokidysheva et al., 2006). That would explain the observed preferential binding of C1q to DI/II, immediately in the vicinity of DC-SIGN, as suggested by the authors for the interaction of an antigen or, in our case, DENV, with the referred trimeric complex (Hosszu et al., 2012). We postulate that this mechanism is also involved in the impairment on the maturation of DC as mentioned above. In agreement, with this hypothesis, our preliminary data showed a decrease in the levels of transcription of several pro-inflammatory molecules, such as IL-6, IL-8, NF-κB and TNF-α in the cells infected with DENV pre-incubated with C1q (data not shown). Further investigation would determine if the reduction observed in the levels of transcription translate in the levels of expression of the referred immunomodulatory molecules.
We postulate that direct binding of DENV to C1q and phagocytosis of DENV-C1q opsonized complex via complement receptors can inhibit the virus infection without inducing exacerbated inflammation and that dysregulation of this mechanism may lead to abnormal inflammatory responses, harmful both for the host and for the virus itself. Our results underscores the importance and complexity of the interactions between DENV and the complement system as already been pointed out in previous publications (Avirutnan et al., 2011; Bokish et al., 1973a; Fuchs et al., 2011; Mehlhop et al., 2008; Nascimento et al., 2009b),. A better understanding of these interactions is important to develop effective prophylactic and therapeutic interventions against Dengue.
Supplementary Material
Highlights.
-
-
C1q binds to Dengue Envelope protein of all 4 serotypes
-
-
C1q binds to Dengue serotype 2 virus
-
-
Dengue serotype 2 virus bound to C1q is less infective to THP-1, a human monocytic leukemia cell line
-
-
The levels of transcription of immunoregulatory molecules in such infected THP-1 cells is altered
Acknowledgements
We thank the reviewers for their helpful comments and suggestions. B.D. is a Fondazione Ri.MED (Palermo, Italy) postdoctoral fellow. This work was partially supported by a National Institutes of Health grant R21 AI095750 to N.K.K. J.D.E. is supported by a grant from the Fine Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alhoot MA, Wang SM, Sekaran SD. Inhibition of dengue virus entry and multiplication into monocytes using RNA interference. PLoS Negl Trop Dis. 2011;5:e1410. doi: 10.1371/journal.pntd.0001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amet T, Ghabril M, Chalasani N, Byrd D, Hu N, Grantham A, Liu Z, Qin X, He JJ, Yu Q. CD59 incorporation protects hepatitis C virus against complement-mediated destruction. Hepatology. 2012;55:354–363. doi: 10.1002/hep.24686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderluh M, Jug G, Svajger U, Obermajer N. DC-SIGN antagonists, a potential new class of anti-infectives. Curr Med Chem. 2012;19:992–1007. doi: 10.2174/092986712799320664. [DOI] [PubMed] [Google Scholar]
- Avirutnan P, Hauhart RE, Marovich MA, Garred P, Atkinson JP, Diamond MS. Complement-Mediated Neutralization of Dengue Virus Requires Mannose-Binding Lectin. MBio. 2011;2:e00276-11. doi: 10.1128/mBio.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y, Pulendran B, Palucka K. Immunobiology of Dendritic Cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokish VA, Jr FHT, Russell PK, Dixon FJ, Muller-Eberhard HJ. The Potential Pathogenic Role of Complement in Dengue Hemorrhagic Shock Syndrome. N Eng J Med. 1973a;289:996–1000. doi: 10.1056/NEJM197311082891902. [DOI] [PubMed] [Google Scholar]
- Bokish VA, Muller-Eberhard HJ, Dixon FJ. The role of complement in hemorrhagic shock syndrome (dengue) . Trans Assoc Am Physicians. 1973b;86:102–110. [PubMed] [Google Scholar]
- Bonaparte RS, Hair PS, Banthia D, Marshall DM, Cunnion KM, Krishna NK. Human astrovirus coat protein inhibits serum complement activation via C1, the first component of the classical pathway. J Virol. 2008;82:817–827. doi: 10.1128/JVI.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang S, King C. Bacterial Lipopolysaccharide Inhibits Dengue Virus Infection of Primary Human Monocytes / Macrophages by Blockade of Virus Entry via a CD14-Dependent Mechanism Bacterial Lipopolysaccharide Inhibits Dengue Virus Infection of Primary Human Monocytes / Macro. J Virol. 1999;73:2650–2657. doi: 10.1128/jvi.73.4.2650-2657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Melo AB, da Silva MDPC, Magalhães MCF, Gonzales Gil LHV, Freese de Carvalho EM, Braga-Neto UM, Bertani GR, Marques ET, Cordeiro MT. Description of a prospective 17DD yellow fever vaccine cohort in Recife, Brazil. Am J Trop Med Hyg. 2011;85:739–747. doi: 10.4269/ajtmh.2011.10-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Moran E, Vinh Chau N, Simmons C, Luhn K, Peng Y, Wills B, Phuong Dung N, Thi Thu Thao L, Hien TT, McMichael A, Farrar J, Rowland-Jones S. High pro-inflammatory cytokine secretion and loss of high avidity cross-reactive cytotoxic T-cells during the course of secondary dengue virus infection. PLoS One. 2007;2:e1192. doi: 10.1371/journal.pone.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Lin T, Beasley DW, Stover CM, Schwaeble WJ, Pierson TC, Diamond MS. Direct Complement Restriction of Flavivirus Infection Requires Glycan Recognition by Mannose Binding Lectin. Cell Host Microbe. 2011;8:186–195. doi: 10.1016/j.chom.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, van Kooyk Y, Figdor CG. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- Hair PS, Gronemus JQ, Crawford KB, Salvi VP, Cunnion KM, Thielens NM, Arlaud GJ, Rawal N, Krishna NK. Human astrovirus coat protein binds C1q and MBL and inhibits the classical and lectin pathways of complement activation. Mol Immunol. 2010;47:792–798. doi: 10.1016/j.molimm.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Halstead SB, O’Rourke EJ. Antibody-enhanced dengue virus infection in primate leukocytes. Nature. 1977;265:739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- Hilleman MR. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc Natl Acad Sci U S A. 2004;101(Suppl):14560–14566. doi: 10.1073/pnas.0404758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu KK, Santiago-Schwarz F, Peerschke EIB, Ghebrehiwet B. Evidence that a C1q/C1qR system regulates monocyte-derived dendritic cell differentiation at the interface of innate and acquired immunity. Innate Immun. 2010;16:115–127. doi: 10.1177/1753425909339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu KK, Valentino A, Vinayagasundaram U, Vinayagasundaram R, Joyce MG, Ji Y, Peerschke EIB, Ghebrehiwet B. DC-SIGN, C1q, and gC1qR form a trimolecular receptor complex on the surface of monocyte-derived immature dendritic cells. Blood. 2012;120:1228–1236. doi: 10.1182/blood-2011-07-369728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhop E, Ansarah-sobrinho C, Johnson S, Engle M, Daved H, Pierson TC, Diamond MS. C1q Inhibits Antibody-Dependent Enhancement of Flavivirus Infection In Vitro and In Vivo in an IgG Subclass Specific Manner. Cell Host Microbe. 2008;2:417–426. doi: 10.1016/j.chom.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J, Dejnirattisai W, Xu X, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus P, McMichael A, Malasit P, Screaton G. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- Nascimento EJM, Braga-Neto U, Calzavara-Silva CE, Gomes ALV, Abath FGC, Brito Caa, Cordeiro MT, Silva AM, Magalhães C, Andrade R, Gil LHVG, Marques ET. Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS One. 2009a;4:e7892. doi: 10.1371/journal.pone.0007892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento EJM, Silva AM, Cordeiro MT, Brito CA, Gil LHVG, Braga-Neto U, Marques ET. Alternative complement pathway deregulation is correlated with dengue severity. PLoS One. 2009b;4:e6782. doi: 10.1371/journal.pone.0006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson Wa, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement - a key system for immune surveillance and homeostasis. Nat Immunol. 2011;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva EM, Conde JN, Allonso D, Nogueira ML, Mohana-Borges R. Mapping the Interactions of Dengue Virus NS1 Protein with Human Liver Proteins Using a Yeast Two-Hybrid System: Identification of C1q as an Interacting Partner. PLoS One. 2013;8:e57514. doi: 10.1371/journal.pone.0057514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares de Melo KM. Production and validation of diagnostic inputs for dengue virus infections. Oswaldo Cruz Foundation. 2012 [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller Ma, Pattanapanyasat K, Sarasombath S, Birx DL, Steinman RM, Schlesinger S, Marovich Ma. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, Kasisith J. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J. Infect. Dis. 2008;197:1459–1467. doi: 10.1086/587699. [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2012 http://www.who.int/mediacentre/factsheets/fs117/en/index.html [WWW Document].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.