Abstract
Cardiac myosin binding protein C (cMyBP-C) is an integral sarcomeric protein that associates with the thick, thin and titin filament systems in the contractile apparatus. Three different isoforms of MyBP-C exist in mammalian muscle: slow skeletal (MYBPC1), fast skeletal (MyBP-C2, with several variants), and cardiac (cMyBP-C). Genetic screening studies show that mutations in MYBPC3 occur frequently and are responsible for as many as 30–35% of identified cases of familial hypertrophic cardiomyopathy. The function of cMyBP-C is stringently regulated by its post-translational modification. In particular, the addition of phosphate groups occurs with high frequency on certain serine residues that are located in the cardiac-specific regulatory M domain. Phosphorylation of this domain has been extensively studied in vitro and in vivo. Phosphorylation of the M domain can regulate the manner in which actin and myosin interact, affecting the cross bridge cycle and ultimately, cardiac hemodynamics.
Keywords: phosphorylation, kinase, sarcomere, heart, myosin
Introduction
The eukaryotic cell has many ways of regulating protein activity, ranging from control of synthesis to controlled degradation and turnover. However, these are relatively slow processes, on the order of minutes or even hours and the cell has devised ways to modulate protein activity more quickly, either by effectively sequestering the protein, or by post-translational modification and thereby affecting its activity. Post-translational modification of proteins is a functionally powerful tuning process in a wide variety of cellular systems and serves to modulate the downstream effects of many important signaling networks. For example, phosphorylation, which results in modification of an amino acid such as serine to a highly charged residue, has been studied for over half a century and can modulate enzyme activity and protein function [15]. There are many post-translational modifications that a protein can undergo, with major implications for its location within subcellular compartments or its intrinsic activity. For example, acetylation can control protein function, protein-protein interactions, gene expression, and protein localization [3]. Lysine ubiquitination serves as the primary signal for targeting a protein for degradation through the proteasome, regulating the protein’s turnover. Many proteins are substrates for a number of these different post translational processes, which can have a major impact on a protein’s overall metabolism [21] and these potential combinatorial processes provide the cell to generate a single protein of different activities as a result of a single translational process.
This review focuses on the phosphorylation of the sarcomeric protein, cardiac myosin binding protein C (cMyBP-C). The contractile apparatus in general, and the heart in particular, must respond on a beat to beat basis to a changing environment and rapidly changing demands. The sarcomere represents a central target for adrenergic signaling and it is well established that adrenergic stimulation and the normal and abnormal modulation of catecholamine levels result in dramatic changes of the chronotropic (overall change in heart rate), lusitropic (relaxant) and inotropic (contractile) cardiac parameters [36]. Two important pathways activated by adrenergic signaling have, as their major players, protein kinase C (PKC) and protein kinase A (PKA). Although there are other critical targets for both PKC and PKA in modulating contractility [31], it is well established that multiple contractile proteins are important substrates [32], and cMyBP-C contains residues that are phosphorylatable and are phosphorylated in vivo by PKC, PKA, PKD, CaMKII, CK2, GSK3β, and RSK [20, 1, 5, 10, 23]. Thus, cMyBP-C phosphorylation represents a target at the contractile apparatus level for adrenergic activation and potentially, activation by other signaling pathways (eg SUMOylation) as well.
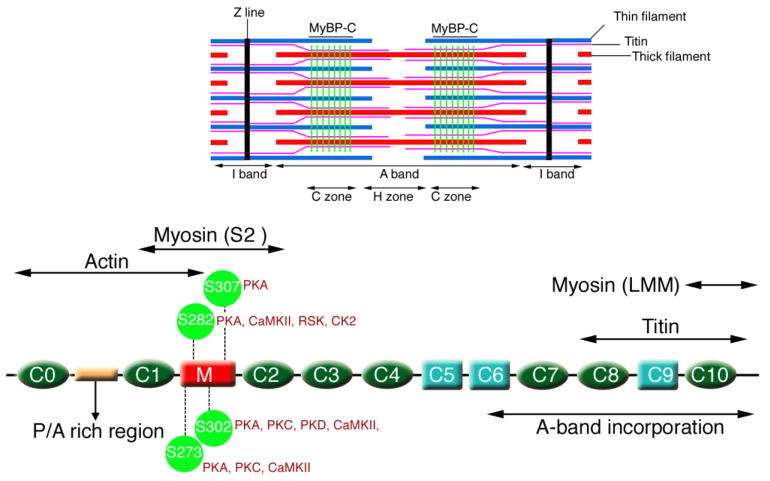
While the slow skeletal muscle isoform has a single site that can be phosphorylated via the cAMP dependent protein kinase (PKA) and/or fiber-associated Ca2+/calmodulin dependent (CaMKII) kinase, recent data was obtained showing that up to 17 sites could be phosphorylated to varying degrees in the cardiac isoform [19]. The sites are clustered to some extent in the N terminal part of the molecule, with up to 6 sites present in a cardiac-specific domain, termed “M” (Fig. 1): H:T274/M:T272, H:S275/M:S273, H:S284/M:S282, H:S286/M:S284, H:T290/Not in M, and H:S311/M:S307, where H is the residue in the human sequence and M the analogous site in the mouse. While cMyBP-C is subject to several other post-translational modifications, this review focuses almost exclusively on cMyBP-C’s phosphorylation and the role it plays on controlling cardiac mechanics and hemodynamics. Data from previous studies showed that cMyBP-C phosphorylation has dramatic effects on both filament orientation and contractile mechanics [6, 7, 10], and so we and others have focused on understanding the structure-function characteristics of the protein and the role these post-translational modifications might play in altering or maintaining normal cardiac hemodynamics.
Figure 1.
Structure of cMyBP-C. Schematic diagram showing the protein’s restricted sarcomeric location (green lines, top), its domain structure with 8 IgG domains (green), 3 fibronectin domains (light blue) and the cardiac specific M domain (red), with the three phosphorylatable serines discussed in this review indicated. The approximate locations of the binding sites for myosin heavy chain (head and rod regions; S2 and LMM, respectively) are also shown as well as the binding sites for titin and actin.
This review focuses on three, phosphorylatable sites present in the M domain of the protein: Ser-273, Ser-282 and Ser-302 and the effects of phosphorylation on thick-thin filament interactions and ultimately, cardiac hemodynamics.
cMyBP-C phosphorylation is functionally important
The function of cMyBP-C can be affected by the coordinated phosphorylation of three serines in the M domain (Fig. 1). Initial studies reported three phosphorylation sites (Ser-273, Ser-282, and Ser-302) in the M domain of cMyBP-C [23] although, as noted above, other sites (eg, Ser-307; Fig. 1) in the M domain can also be substrates for kinase activity [19]. These sites are highly conserved across diverse species, implying significant selective pressures and functional significance. In vitro biochemical studies suggest that PKA can phosphorylate the three serines (Ser-273, Ser-282, Ser-302) as well as Ser-307 [17]. PKC can phosphorylate Ser-273 and Ser-302, CaMKII can phosphorylate Ser-273, Ser-282, Ser-302, Ser 307 [28] and PKD can phosphorylate Ser-302 [1]. RSK can phosphorylate Ser-282, GSK3 can phosphorylate Ser-302, and CK2 can phosphorylate Ser-282 [5, 18, 20]. However, the actual kinases that actively phosphorylate these sites in vivo remain obscure and it is not clear which kinases act upon the different residues or even if multiple kinases can phosphorylate a single residue in vivo.
Because of this limitation and the potential functional disconnects between in vitro and in vivo kinase activity directed at this protein, it became critical to study the functionality of these post-translational events in vivo. Fortunately, it is possible to replace endogenous cMyBP-C with a transgenically encoded species [40], allowing investigators to carry out experiments in which a particular residue or residues could be modified and subsequently determining the functional and long term physiological effects. Coupled with the genetic modifications, a set of immunological tools were developed that could specifically detect ser-273, -282 and 302) [27]. These two developments enabled a series of investigations in which the critical residues could be mutated, either individually or in combinations, to determine the functional consequences of either their chronic phosphorylation or inability to be post-translationally phosphorylated.
Data from mouse, dog and human studies all point to the importance of phosphorylation in maintaining normal function and modulating cMyBP-C’s activity. As early as 1984, Hartzell recognized that MyBP-C phosphorylation could be correlated with changes in the rate of relaxation in twitch tension in amphibian muscle [14]. cMyBP-C phosphorylation overall is decreased in cardiac tissue obtained from patients suffering a variety of ailments, including atrial fibrillation, heart failure and cardiomyopathy [8, 16, 35]. For example, using the Ser-282 specific antibody, phosphorylation levels were decreased relative to controls in tissue obtained from patients in terminal heart failure [8, 19]. These data were buttressed with data obtained from a canine heart failure model induced by rapid pacing [8] and are consistent with the phosphorylation levels observed in different murine models of heart failure that were induced either by genetic or surgical manipulation [29, 30]. During ischemia-reperfusion injury, overall phosphorylation of cMyBP-C was dramatically decreased while chronically phosphorylated cMyBP-C was cardio-protective when the hearts were subjected to ischemic injury [30]. Finally and most conclusively, mice were created in which the three serines were changed to non-phosphorylatable alanines. When these mice were crossed into a mouse line that could not produce cMyBP-C (a knockout or “null”) [22], the phosphorylation-deficient cMyBP-C was unable to rescue the null phenotype, despite being present at normal levels and being inserted correctly into the sarcomere [29]. This is in contrast to the effects seen when normal or “wild-type” (WT) cMyBP-C was transgenically expressed [29].
Phosphorylation patterns of serine residues in the m domain of cMyBP-C are hierarchal
The M domain (Fig. 1) is particularly intriguing in terms of its potential for phosphorylation because these residues are found only in the cardiac isoform. Gautel et al first showed that the three serines might not be equivalent [10]. By performing phosphorylation assays on purified rabbit cMyBP-C, and subsequently making a series of modified constructs in which the serines were replaced with alanines, they showed that phosphorylation of Ser-282 increased the phosphorylation levels of the two neighboring serines. In a series of studies carried out in the mouse using residue-specific phospho-mimetics or phosphorylation site ablation, it was confirmed that phosphorylation of Ser-282 acts as a trigger to phosphorylate the neighboring phosphorylatable Ser-273 and -302 sites [28]. Interestingly, in human heart failure patients, while cMyBP-C was significantly hypo-phosphorylated as expected, Ser-282 was particularly under-phosphorylated, relative to Ser-273 and Ser-302 [4].
Ser-282 is phosphorylated by Ca2+ calmodulin-activated kinase (CaMKIIδ) at high calcium levels [28]. Phosphorylation at this site appears to be critical for the subsequent phosphorylation of Ser-302, and abolishing the ability of Ser-282 to be phosphorylated completely inhibited Ser-302’s ability to act as substrate for CaMKIIδ [28]. While these data are intriguing and support the hypothesis that each of these phosphorylation sites is unique, they have little bearing on their functionality in determining thick-thin filament interactions and cardiac hemodynamics: as noted above, different kinases can phosphorylate some or all of the residues and abolishing one kinase’s ability to phosphorylate a residue does not, in of itself, mean that the residue will not be a substrate for an alternate kinase such as PKC, PKA or PKD.
cMyBP-C phosphorylation modulates thick-thin filament interactions
cMyBP-C can slow cross-bridge kinetics, acting as an “anchor” and recently in a series of elegant studies using a novel native filament system, Warshaw et al showed that this ability is restricted to the central portion of the sarcomere, where cMyBP-C is differentially located [25]. This implies that cMyBP-C can effectively modulate critical thick-thin filament interactions and it appears likely that the protein’s phosphorylation can modulate the extent and strength of these interactions, potentially shifting the equilibria between thick and thin filament conformational states and by extension, their binding to one another.
Biochemical assays utilizing cosedimentation and isothermal titration calorimetry first demonstrated the affinity constant of cMyBP-C for the head region of myosin could be modulated by its phosphorylation state, with cMyBP-C phosphorylation essentially abolishing the interaction [12]. These early studies were confirmed by genetic analyses in which a series of constructs consisting of either full length or N-terminal cMyBP-C fragments were generated, in which the three phosphorylatable serines were replaced either by a charged amino acid (phospho-mimetic) or by alanine, rendering the site non-phosphorylatable. Using a series of yeast two-hybrid assays to demonstrate functional interactions, Sadayappan et al showed that phosphorylation of the M domain effectively abolished all interaction with the myosin heavy chain’s head region (S2) [30].
A series of in vitro studies provided a more nuanced view of how phosphorylation influences force development and the rate at which this occurs. Using bacterially generated fragments containing different combinations of residues 273, 282 and 302 that were either mutated to alanine or the charged amino acid, aspartate, the polypeptides were subjected to single molecule studies in a laser trap. The effect of these short, N-terminal fragments on actomyosin motility was characterized using an in vitro motility assay, which measures force and velocity under unloaded conditions and in the load-clamped laser trap assay, in order to measure the force:velocity relationships. The in vitro motility assay confirmed that the phosphomimetic fragments reduced the slowing of actin velocity in proportion to the total number of phosphomimetic replacements. Under load in the laser trap, the N terminal fragment depressed the force:velocity relationship but maximal force was unaffected. When the phosphomimetic fragments were used, they were able to reverse this depression with the degree of reversal dependent upon the total number of replacements [39].
The study of cMyBP-C’s phosphorylation-dependent binding was extended to the thin filament as well. Using both single molecule studies [38] and yeast-based genetic studies, the actin binding regions of cMyBP-C were defined [2]. Although there is some controversy as to their location [26], the preponderance of data point to a series of unique, closely linked and overlapping sites located in the N terminal region as containing multiple actin-binding sites. Strikingly, in the absence of phosphorylation, yeast 3-hybrid analyses indicated that binding to the myosin’s head region was mutually exclusive with actin binding, implying that cMyBP-C phosphorylation, which abolishes binding to the myosin head, might increase cMyBP-C-actin interactions [2]. However, this remains unproven at this time.
Alterations in serine phosphorylation status affects cardiac structure and hemodynamics
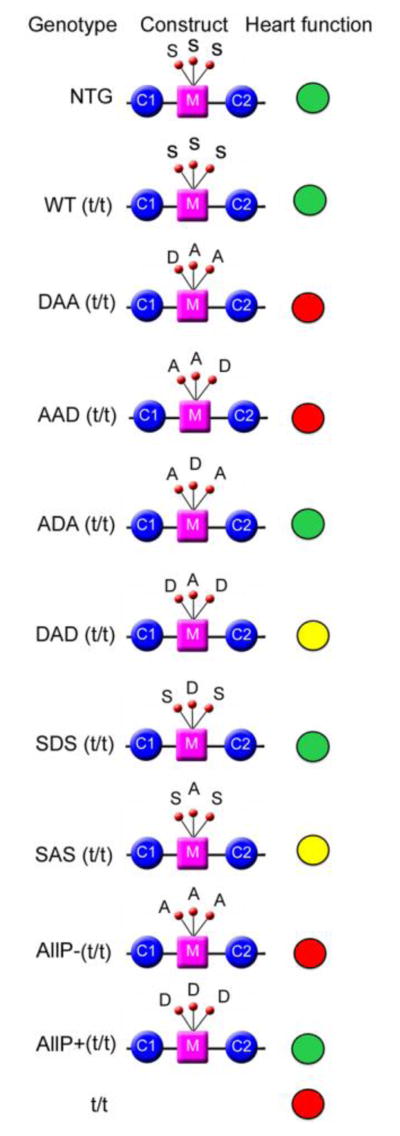
The genetic, biochemical and yeast-hybrid based functional studies all point to the phosphorylation status of cMyBP-C as playing a critical role in determining the protein’s binding characteristics to the myosin’s force-generating region: as cMyBP-C phosphorylation increases, it becomes unable to interact productively with myosin S2 [30]. This implies that cMyBP-C could play critical roles in determining the basic thick and thin filament interactions and, indeed, data using isolated fibers obtained from mouse myocardium lacking cMyBP-C showed that the protein’s absence dramatically affected the stretch activation response, with both force decay and the delayed force transient being substantially accelerated [33]. When present, but fully phosphorylated (and therefore lacking the ability to interact with myosin S2), the development of crossbridge force is both enhanced and accelerated, with the observed kinetics reminiscent of the cMyBP-C null muscle [34]. In a comprehensive series of studies using transgenic replacement strategies, a series of mice were made in which Ser-273, -282 and 302 were replaced either with phosphomimetics or alanines in order to render them non-phosphorylatable. Functional and structural alterations at the molecular and organ levels were also measured [1, 2, 24, 25, 28–30, 37–39, 13]. These experiments allowed the in vitro kinetic data to be extended to the intact heart’s hemodynamics and contractile mechanics. The mutated constructs are depicted in Fig. 2, along with a pictorial representation of the effects on cardiac hemodynamics. The comprehensive datasets, which are beyond the limited scope of this mini review, are given in a recent publication [13]. However, a single example is illustrative of the level of detail that can be gleaned from the general approach. Four groups of mice consisting of nontransgenic animals, cMyBP-C functional nulls [22], transgenic mice expressing a normal cMyBP-C (as a second control group) and mice that expressed cMyBP-C that could not be phosphorylated at Ser-273, -282 and -302 were used. Left ventricular function was assessed using a miniature pressure-volume catheter such that the end systolic pressure-volume relationships could be determined as well as the end-systolic elastance, chamber activation and relaxation kinetics [24]. These measurements as well as others allowed the investigators to determine that cMyBP-C could control the rate-dependent shortening of the diastolic time period. Surprisingly, the cMyBP-C phosphorylation-deficient animals exhibited normal chamber activation kinetics but comparison between the normal cohorts and this group showed that phosphorylation modulated the early pressure rise and adrenergic reserve. That is, phosphorylation mediated the dependence of early systolic pressure rise on chamber preload and the inability to be phosphorylated at the three serines markedly suppressed adrenergic and rate-dependent contractile reserve [24].
Figure 2.
Schematic description of the effects on cardiac hemodynamics when the three serines are mutated either to non-phosphorylatable alanine or the phosphomimetic aspartate. Heart function was analyzed by invasive catheterization. Detailed data are presented in [13]. Green, yellow and red indicates normal, moderately compromised and poor cardiac function, respectively.
Conclusions and future directions
The ability of cMyBP-C to be post-translationally modified offers the heart an opportunity to tune contractility precisely on a beat to beat basis. In terms of the protein’s ability to be phosphorylated, the field is now quite mature, with >20 peer reviewed studies being published in the last 10 years dealing directly with the issue. The data have been illuminating and, in some cases, surprising, as they point to the essential nature of this post-translational process on maintaining the protein’s functionality in the whole organ. Additionally, as detailed elsewhere in this volume and in earlier studies, phosphorylation of the serines in the M domain can be cardioprotective [30] and help maintain the protein’s structural integrity [11].
Future studies will necessarily deal with the additional sites subject to phosphorylation and with sites that can be post-translationally modified in other ways. For example, cMyBP-C can also be post-translationally modified by the small ubiquitin-related modifier (SUMO). SUMOylation is highly dynamic, reversible and has a long list of targets, including cMyBP-C. SUMOylation can have diverse effects on different proteins, including regulating spatial location and stability [9]. Our laboratory has recently confirmed that the N-terminal domain of cMyBP-C is sumoylated at specific lysines and this has a major effect on protein half-life (Gupta and Robbins, unpublished observations). It will be critical to understand the interplay of these post-translational modifications with one another and their modulation during normal function and during the development of cardiac disease.
Acknowledgments
This work was supported by National Heart, Lung and Blood Institute grants P01HL077101, P01HL059408, R01HL105924 and The Transatlantic Network of Excellence Program grant from Le Fondation Leducq (to J.R.). MKG is a postdoctoral fellow of American Heart Association (Great Rivers Affiliate).
Footnotes
Disclosures: none declared.
References
- 1.Bardswell SC, Cuello F, Rowland AJ, Sadayappan S, Robbins J, Gautel M, Walker JW, Kentish JC, Avkiran M. Distinct sarcomeric substrates are responsible for protein kinase D-mediated regulation of cardiac myofilament Ca2+ sensitivity and cross-bridge cycling. J Biol Chem. 2010;285:5674–5682. doi: 10.1074/jbc.M109.066456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhuiyan MS, Gulick J, Osinska H, Gupta M, Robbins J. Determination of the critical residues responsible for cardiac myosin binding protein C’s interactions. J Mol Cell Cardiol. 2012;53:838–847. doi: 10.1016/j.yjmcc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/Science.1175371. [DOI] [PubMed] [Google Scholar]
- 4.Copeland O, Sadayappan S, Messer AE, Steinen GJ, van der Velden J, Marston SB. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J Mol Cell Cardiol. 2010;49:1003–1011. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Cuello F, Bardswell SC, Haworth RS, Ehler E, Sadayappan S, Kentish JC, Avkiran M. Novel role for p90 ribosomal S6 kinase in the regulation of cardiac myofilament phosphorylation. J Biol Chem. 2011;286:5300–5310. doi: 10.1074/jbc.M110.202713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker RS, Decker ML, Kulikovskaya I, Nakamura S, Lee DC, Harris K, Klocke FJ, Winegrad S. Myosin-binding protein C phosphorylation, myofibril structure, and contractile function during low-flow ischemia. Circulation. 2005;111:906–912. doi: 10.1161/01.CIR.0000155609.95618.75. [DOI] [PubMed] [Google Scholar]
- 7.Decker RS, Nakamura S, Decker ML, Sausamuta M, Sinno S, Harris K, Klocke FJ, Kulikovskaya I, Winegrad S. The dynamic role of cardiac myosin binding protein-C during ischemia. J Mol Cell Cardiol. 2012 doi: 10.1016/j.yjmcc.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 8.El-Armouche A, Pohlmann L, Schlossarek S, Starbatty J, Yeh YH, Nattel S, Dobrev D, Eschenhagen T, Carrier L. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J Mol Cell Cardiol. 2007;43:223–229. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 10.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govindan S, Sarkey J, Ji X, Sundaresan NR, Gupta MP, de Tombe PP, Sadayappan S. Pathogenic properties of the N-terminal region of cardiac myosin binding protein-C in vitro. J Muscle Res Cell Motil. 2012;33:17–30. doi: 10.1007/s10974-012-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 13.Gupta MK, Gulick J, James J, Osinska H, Lorenz JN, Robbins J. Functional dissection of myosin binding protein C phosphorylation. J Mol Cell Cardiol. doi: 10.1016/j.yjmcc.2013.08.006. http://dx.doi.org/10.1016/j.yjmcc.2013.08.006. [DOI] [PMC free article] [PubMed]
- 14.Hartzell HC. Phosphorylation of C-protein in intact amphibian cardiac muscle. Correlation between 32P incorporation and twitch relaxation. J Gen Physiol. 1984;83:563–588. doi: 10.1085/jgp.83.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes NE, Ingham PW, Lim WA, Marshall CJ, Massague J, Pawson T. Signalling change: signal transduction through the decades. Nat Rev Mol Cell Biol. 2013;14:393–398. doi: 10.1038/nrm3581. [DOI] [PubMed] [Google Scholar]
- 16.Jacques AM, Copeland O, Messer AE, Gallon CE, King K, McKenna WJ, Tsang VT, Marston SB. Myosin binding protein C phosphorylation in normal, hypertrophic and failing human heart muscle. J Mol Cell Cardiol. 2008;45:209–216. doi: 10.1016/j.yjmcc.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Jia W, Shaffer JF, Harris SP, Leary JA. Identification of novel protein kinase A phosphorylation sites in the M-domain of human and murine cardiac myosin binding protein-C using mass spectrometry analysis. J Proteome Res. 2010;9:1843–1853. doi: 10.1021/pr901006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooij V, Boontje N, Zaremba R, Jaquet K, dos Remedios C, Stienen GJ, van der Velden J. Protein kinase C alpha and epsilon phosphorylation of troponin and myosin binding protein C reduce Ca2+ sensitivity in human myocardium. Basic Res Cardiol. 2010;105:289–300. doi: 10.1007/s00395-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kooij V, Holewinski RJ, Murphy AM, Van Eyk JE. Characterization of the cardiac myosin binding protein-C phosphoproteome in healthy and failing human hearts. J Mol Cell Cardiol. 2013;60:116–120. doi: 10.1016/j.yjmcc.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuster DW, Sequeira V, Najafi A, Boontje NM, Wijnker PJ, Witjas-Paalberends ER, Marston SB, Dos Remedios CG, Carrier L, Demmers JA, Redwood C, Sadayappan S, van der Velden J. GSK3beta phosphorylates newly identified site in the proline-alanine-rich region of cardiac myosin-binding protein C and alters cross-bridge cycling kinetics in human: short communication. Circ Res. 2013;112:633–639. doi: 10.1161/circresaha.112.275602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lothrop AP, Torres MP, Fuchs SM. Deciphering post-translational modification codes. FEBS Lett. 2013;587:1247–1257. doi: 10.1016/j.febslet.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1771. doi: 10.1172/JCI7377C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamed AS, Dignam JD, Schlender KK. Cardiac myosin-binding protein C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch Biochem Biophys. 1998;358:313–319. doi: 10.1006/abbi.1998.0857. [DOI] [PubMed] [Google Scholar]
- 24.Nagayama T, Takimoto E, Sadayappan S, Mudd JO, Seidman JG, Robbins J, Kass DA. Control of in vivo left ventricular [correction] contraction/relaxation kinetics by myosin binding protein C: protein kinase A phosphorylation dependent and independent regulation. Circulation. 2007;116:2399–2408. doi: 10.1161/CIRCULATIONAHA.107.706523. [DOI] [PubMed] [Google Scholar]
- 25.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337:1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: characterization and mapping of the binding site. J Biol Chem. 2011;286:2008–2016. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadayappan S, Gulick J, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Robbins J. Cardiac myosin binding protein-C phosphorylation in a {beta}-myosin heavy chain background. Circulation. 2009;119:1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadayappan S, Gulick J, Osinska H, Barefield D, Cuello F, Avkiran M, Lasko VM, Lorenz JN, Maillet M, Martin JL, Brown JH, Bers DM, Molkentin JD, James J, Robbins J. A critical function for Ser-282 in cardiac Myosin binding protein-C phosphorylation and cardiac function. Circ Res. 2011;109:141–150. doi: 10.1161/CIRCRESAHA.111.242560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97:1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16918–16923. doi: 10.1073/Pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solaro RJ, Henze M, Kobayashi T. Integration of troponin I phosphorylation with cardiac regulatory networks. Circ Res. 2013;112:355–366. doi: 10.1161/circresaha.112.268672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solaro RJ, Kobayashi T. Protein phosphorylation and signal transduction in cardiac thin filaments. J Biol Chem. 2011;286:9935–9940. doi: 10.1074/jbc.R110.197731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stelzer JE, Dunning SB, Moss RL. Ablation of cardiac myosin-binding protein-C accelerates stretch activation in murine skinned myocardium. Circ Res. 2006;98:1212–1218. doi: 10.1161/01.RES.0000219863.94390.ce. [DOI] [PubMed] [Google Scholar]
- 34.Stelzer JE, Patel JR, Moss RL. Protein kinase A-mediated acceleration of the stretch activation response in murine skinned myocardium is eliminated by ablation of cMyBP-C. Circ Res. 2006;99:884–890. doi: 10.1161/01.RES.0000245191.34690.66. [DOI] [PubMed] [Google Scholar]
- 35.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/circulationaha.108.838672. [DOI] [PubMed] [Google Scholar]
- 36.Wachter SB, Gilbert EM. Beta-adrenergic receptors, from their discovery and characterization through their manipulation to beneficial clinical application. Cardiology. 2012;122:104–112. doi: 10.1159/000339271. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Grant JE, Doede CM, Sadayappan S, Robbins J, Walker JW. PKC-betaII sensitizes cardiac myofilaments to Ca2+ by phosphorylating troponin I on threonine-144. J Mol Cell Cardiol. 2006;41:823–833. doi: 10.1016/j.yjmcc.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Weith A, Sadayappan S, Gulick J, Previs MJ, Vanburen P, Robbins J, Warshaw DM. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52:219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weith AE, Previs MJ, Hoeprich GJ, Previs SB, Gulick J, Robbins J, Warshaw DM. The extent of cardiac myosin binding protein-C phosphorylation modulates actomyosin function in a graded manner. J Muscle Res Cell Motil. 2012 doi: 10.1007/s10974-012-9312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J Clin Invest. 1998;102:1292–1300. doi: 10.1172/JCI3880. [DOI] [PMC free article] [PubMed] [Google Scholar]