Abstract
Mitochondrial dysfunction is implicated in the etiology and pathogenesis of numerous human disorders involving tissues with high energy demand. Murine models are widely used to elucidate genetic determinants of phenotypes relevant to human disease, with recent studies of C57BL/6J (B6), DBA/2J (D2) and B6xD2 populations implicating naturally occurring genetic variation in mitochondrial function/dysfunction. Using blue native polyacrylamide gel electrophoresis, immunoblots, and in-gel activity analyses of complexes I, II, IV and V, our studies are the first to assess abundance, organization, and catalytic activity of mitochondrial respiratory complexes and supercomplexes in mouse brain. Remarkable strain differences in supercomplex assembly and associated activity are evident, without differences in individual complexes I, II, III, or IV. Supercomplexes I1III2IV2-3 exhibit robust complex III immunoreactivity and complex I and IV activities in D2, but with little detected in B6 for I1III2IV2, and I1III2IV3 is not detected in B6. I1III2IV1 and I1III2 are abundant and catalytically active in both strains, but significantly more so in B6. Furthermore, while supercomplex III2IV1 is abundant in D2, none is detected in B6. In aggregate, these results indicate a shift toward more highly assembled supercomplexes in D2. Respiratory supercomplexes are thought to increase electron flow efficiency and individual complex stability, and to reduce electron leak and generation of reactive oxygen species. Our results provide a framework to begin assessing the role of respiratory complex suprastructure in genetic vulnerability and treatment for a wide variety of mitochondrial-related disorders.
Keywords: respiratory chain, mitochondrial supercomplexes, complex I (NADH dehydrogenase), complex IV (cytochrome c oxidase), complex III (cytochrome c reductase/cytochrome bc1 complex), brain, genetic, mouse, C57BL/6J, DBA/2J
INTRODUCTION
Mitochondria play a vital role in energy production, Ca+2 buffering, apoptotic events, and production of reactive oxygen species (ROS) (Sayer, 2002; Schapira, 1996; Smith et al., 2000; Wallace, 1999). Given these essential functions, it is not surprising that mitochondrial dysfunction is implicated in the etiology and pathogenesis of a variety of disorders (Coskun et al., 2012; Finsterer, 2006; Orth & Schapira, 2002; Zeviani & Carelli, 2003), with brain pathologies as common clinical phenotypes (Lessing & Bonini, 2009). Considering the relevance to human disease, understanding the mechanisms that impact mitochondrial function/dysfunction is of critical importance.
The bulk of cellular energy is derived through oxidative phosphorylation mediated by multi-protein oxidoreductase complexes I (NADH dehydrogenase), II (succinate dehydrogenase), III (cytochrome c reductase/cytochrome bc1 complex), and IV (cytochrome c oxidase). Electron transfer down the chain creates an electrochemical proton gradient across the mitochondrial inner membrane, which is subsequently used by complex V (ATPase) to generate ATP. These complexes can be isolated individually (monomers), or as assembled dimers, oligomers, and high molecular weight suprastructures containing multiple complexes e.g., supercomplexes (Cruciat et al., 2000; Lenaz & Genova, 2009). Supercomplex architecture is readily apparent after separation by blue native polyacrylamide gel electrophoresis (BN-PAGE) and in-gel catalytic activity assays or immunoblotting (Schagger & Pfeiffer, 2000), and single-particle image analysis (Althoff et al., 2011; Dudkina et al., 2011). In yeast, where complex I is lacking, supercomplexes contain, among other proteins, both complexes III and IV, and perhaps complex II under some conditions (Stuart, 2008). In mammals, where NADH is the most important electron donor, supercomplex formation is imperative to complex I function (Acin-Perez et al., 2008; Moreno-Lastres et al., 2012; Schagger et al., 2004; Wittig & Schagger, 2009). Although the precise function has yet to be demonstrated, supercomplex formation is postulated to regulate respiration, enabling more efficient electron flow between complexes and promoting complex stability (Suthammarak et al., 2010), thereby discouraging electron loss and ROS generation (Genova et al., 2008; reviewed in Lenaz & Genova, 2012).
Despite the importance of mitochondrial dysfunction to human disease, there is a dearth of information on the phenotypic impact of genetic variation on respiratory complex organization and function. Murine models, including the C57BL/6J (B6) and DBA/2J (D2) strains and B6D2-derived populations, have been instrumental in elucidating genetic determinants of a variety of phenotypes relevant to human physiology and pathology. Interestingly, a growing body of evidence implicates naturally occurring variation in mitochondrial oxidative phosphorylation and/or ROS generation in genetically influenced differences among B6, D2 and B6D2 populations, disproportionately affecting tissues with high energy demand, particularly brain (Bhave et al., 2006; Denmark & Buck, 2008; Huang et al., 2006; Lynn et al., 2005; Misra et al., 2007; Rebrin et al., 2007; Rebrin et al., 2011). Here, using BN-PAGE analyses of isolated B6 and D2 brain mitochondria, we simultaneously assess respiratory complexes and supercomplexes with parallel enzymatic activity and immunoblot analyses. These are the first analyses of respiratory complex abundance, organization, and catalytic activity in mouse brain.
MATERIALS AND METHODS
Animals
Seven week-old C57BL/6J and DBA/2J (JAX order numbers 000664 and 000671, respectively) male mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and group housed (3-4 per cage) by strain in our colony at the Portland VA Medical Center for two weeks. Mouse chow (Purina #5001) and water were provided ad libitum in a room maintained at 22±1°C, with lights on from 6:00 to 18:00. All animals were sacrificed at 13:00 by cervical dislocation on the same day (at age 9 weeks), the brain rapidly removed, cut in half, flash frozen in liquid nitrogen, and stored at −80°C. All procedures were approved by the VAMC and OHSU Care and Use Committees in accordance with USDA and USPHS guidelines.
Reagents
Colloidal Blue, XCell SureLock® Mini-Cell, 4-16% native gels (Life Technologies, Grand Island, NY, USA); Digitonin (EMD Millipore, Billerica, MA, USA); 1-Step TMB-Blotting Solution and bioinchoninic acid assay (BCA; Pierce Protein Research, Rockford, IL, USA); anti-ubiquinol-cytochrome c reductase core protein 1 (Santa Cruz Biotechnology, Dallas, TX, USA); anti-fluorescein alkaline phosphatase (Vector Laboratories, Burlingame, CA, USA); ECF™ substrate (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA); Coomassie Brilliant Blue G-250 (SERVA, Heidelberg, Germany), Immun-Blot PVDF Membrane (BioRad, Hercules, CA, USA). All other reagents are from Sigma-Aldrich (St. Louis, MO, USA).
Sample preparation
Naïve half brain samples from individual B6 (n=10) and D2 (n=10) were flash frozen in liquid N2 (Grazina, 2012; Spinazzi et al., 2011) and then prepared in parallel by the method of Wittig et al. (2005). Additionally, in a separate experiment, brains from naïve B6 (n=2) and D2 (n=2) were cut in half, with one half flash frozen, and then each half prepared in parallel. Individual half-brains were homogenized for five pulses (OMNI International TH homogenizer, Kennesaw, GA, USA) in 3.5 ml ice-cold sucrose buffer (440 mM sucrose, 1 mM ethylenediaminetetraacetic acid [EDTA], 20 mM 3-(N-morpholino)-propanesulfonic acid, 2 nM butylated hydroxytoluene, pH 7.3) and frozen (−80°C) overnight. An aliquot (200 μl) was centrifuged (22,000 × g, 10 min, 4°C), and the pellet resuspended in 140 μl solubilization buffer (50 mM NaCl, 50 mM imidazole hydrogen chloride, 2 mM 6-aminohexanoic acid, 1 mM EDTA, pH 7.0). 10 μl was used for protein quantitation by BCA, and the remainder frozen (−80°C) overnight. 440 μg protein (diluted in solubilization buffer to 160 μl total volume) was solubilized with digitonin at 8g/g detergent to protein as in previous work (Schagger & Pfeiffer, 2001) on ice for 10 min, centrifuged (100,000 × g, 15 min, 4°C), and the supernatant retained for BN-PAGE. 10 μl was removed and used for final protein quantitation by BCA.
Blue Native Polyacrylamide Gel Electrophoresis
BN-PAGE was performed as described by Wittig et al. (2006) with some modifications (Leary, 2012). Each sample was prepared as 5:1 v/v sample:dye (1:1, 90% glycerol, 5% Coomassie blue), loaded on 4-16% native gels and electrophoresed (125 V, 4°C) in cathode buffer B/10 buffer (50 mM Tricine, 7.5 mM imidazole, 0.002% Coomassie blue, pH 7.0) and anode buffer (25 mM imidazole, pH 7.0). In order to mitigate the potential confound of residual Coomassie staining on in-gel activity assays (Leary, 2012; Wittig & Schagger, 2005), cathode buffer was replaced after 20 min with buffer B/0 (50 mM Tricine, 7.5 mM imidazole, pH 7.0) and run for another 4 h or 18 h (for improved resolution). Gels were soaked immediately in deionized water (10 min), incubated with Colloidal Blue (18 h, 25°C), and destained in water (24 h, 25°C) before imaging and densitometry.
Catalytic activity
Established in-gel assays of complex I, II, IV (Jung et al., 2000), V (Diaz et al., 2009; Wittig et al., 2007), and III (Smet et al., 2011) specific activities were used. For quantitative activity determinations of complexes I, II, and IV, parameters were based initially on previous results (Jung et al., 2000), and validated empirically for mouse brain (data not shown). The conditions used for linear (quantitative) in-gel activity analyses were as follows: complex I (40 min, 25°C), II (40 min, 25°C), and IV (120 min, 25°C). We also assessed activity of complexes I, II, IV and V under conditions thought to saturate the enzymatic reactions as follows: complex I (24 h, 25°C), II (24 h, 25°C), IV (24 h, 25°C), and V (24 h, 37°C). Complex I, II, and IV reactions were quenched using 45% methanol/10% acetic acid (10 min, 25°C), complex V reactions were stopped with 50% methanol, and all were transferred to deionized water until imaging.
Immunoblot analyses
BN-PAGE gels were transferred to polyvinylidene difluoride membrane (20V, 4°C, 20 h). TBST buffer (20 mM Tris, 1.37 M NaCl, 0.05% Tween20, pH 7.6) was used for all rinses, antibody dilutions and incubations, with 1% nonfat dry milk and 1% bovine plasma albumin included for blocking steps. After rinsing and blocking, membranes were incubated overnight (4°C) with primary antibody to ubiquinol-cytochrome c reductase core protein 1 (UQCRC1) (1:500). After rinsing, the membranes were incubated (1 h, 25°C) with fluorescein anti-mouse IgG (1:770), rinsed, and incubated (1 h, 25°C) with anti-fluorescein alkaline phosphatase (1:2500). After a final rinse, 1 ml ECF substrate was applied for visualization and densitometry. Data for III2IV1 and III2 were empirically validated as quantitative (data not shown), while signal for larger supercomplexes is qualitative only.
Densitometry and quantitation
Densitometric quantitation of bands visualized by BN-PAGE and Coomassie staining were performed. Parallel gels were used for quantitation of catalytic activity and western blot. For reliable quantitation, each set of gels was scanned under the same settings for Coomassie and in-gel activity stains (CanonScan 9000F) and western blots (Ultra Lum Imaging, 600 nm), and densitometric analyses performed using UltraQuant 6.0 (Ultra Lum, Inc., Claremont, CA, USA) and Quantity One 4.6.5 (BioRad, Hercules, CA, USA) to determine the absolute Integrated Optical Density (IOD) and Trace Quantity (TQ) values, respectively, for each band. These values were then corrected to μg protein loaded for each sample. For further validation, band IOD values were also normalized within samples using three additional approaches as in previous work, i.e., to total within-lane Coomassie signal (Gadicherla et al., 2012), complex II1 catalytic activity (Smet et al., 2011; Yang et al., 2011), and complex V1 Coomassie signal (Suthammarak et al., 2010; Wittig & Schagger, 2007). As all four normalization methods yielded the same conclusions, only IOD or TQ values normalized to μg protein loaded are given.
Statistical analyses
B6 and D2 comparison data were not normally distributed based upon a significant Shapiro-Wilks test; thus, a non-parametric t-test was performed to generate Mann-Whitney U statistics and corresponding p values. All data were analyzed using Prism5 statistical software (GraphPad Software, Inc., San Diego, CA, USA) and are given as the mean ± S.E.M. For all statistical comparisons, the significance level was set at p < 0.05 (two-tailed).
RESULTS
Detection and quantitation of mitochondrial respiratory complexes and supercomplexes and enzymatic activities
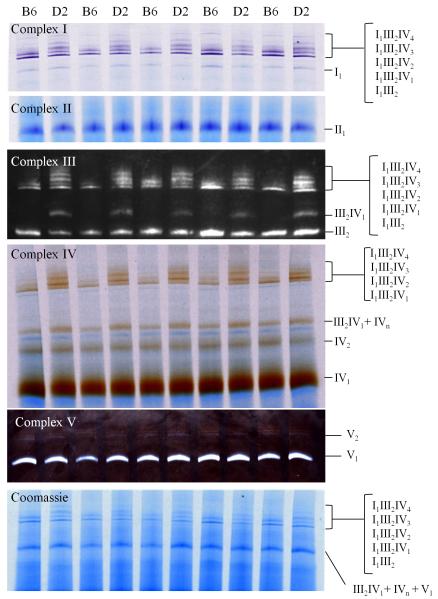
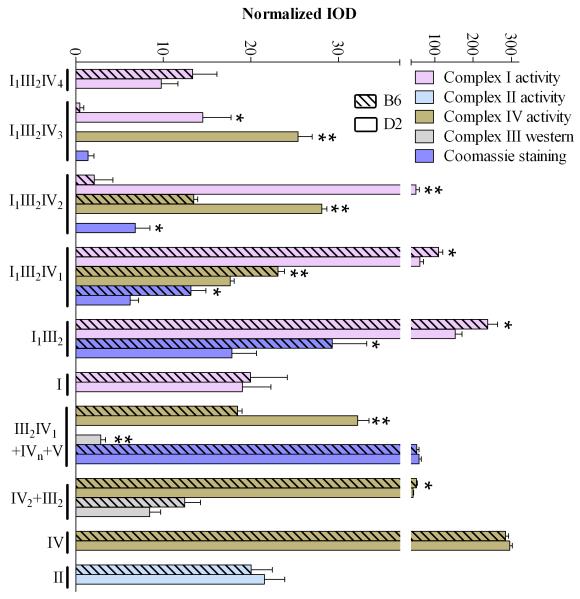
Solubilization with the mild detergent digitonin followed by BN-PAGE allows simultaneous evaluation of respiratory complex monomers and higher order organization, including supercomplexes (Schagger & Pfeiffer, 2000; Wittig et al., 2006). Identification was based on our empirical data (below and Supplemental Figure 1) and previous results, including one- and two-dimensional BN-PAGE analyses (Acin-Perez et al., 2008; Reifschneider et al., 2006; Schagger et al., 2004; Schagger & Pfeiffer, 2001). Results for our complex I, II, IV, and V specific activity assays, complex III UQCRC1 immunoblot analyses, and Coomassie staining for 10 representative samples from individual animals (5 B6 and 5 D2) are given in Figure 1. Ten additional, independent samples (from different animals) were analyzed on separate gels (Supplemental Figure 2). Quality control analyses indicate that results using flash frozen or fresh brain are comparable for studies assessing supercomplex organization and in-gel catalytic activities (Supplemental Figure 3). Importantly, protein yields before and after solubilization did not differ between the D2 and B6 samples (p>0.70, NS), nor did the amount of sample protein loaded (mean ± SEM [range] = 15.1 ± 0.98 [8.8-19.7] and 15.6 ± 0.9 [12.0-22.5] μg protein/lane, respectively). Furthermore, the amount of sample protein loaded was within the linear (quantitative) range for Coomassie staining and in-gel activity assays for complexes I, II and IV based on previous work (Jung et al., 2000; Sabar et al., 2005) and our empirical results for mouse brain (not shown). Our empirical data also validate that immunoblot results for complex III core protein 1 (UQCRC1) are quantitative for III2IV1 and III2 and qualitative for other complex III-containing supercomplexes. Results for complex V activity and abundance are qualitative only. The quantitative densitometric analyses, based on 20 individual mice tested (n=10 per strain), are summarized in Figure 2.
Figure 1.
Respiratory complex organization and band identification in naïve B6 and D2 mouse brain after BN-PAGE and quantitative assessment of activities for complex I (purple), II (indigo), IV (brown), and V (white), immunoblots for complex III core protein UQCRC1 (III2IV1 and III2; others are qualitative), and Coomassie staining. Half of the samples assessed are shown here; the other half are shown in Supplemental Figure 2.
Figure 2.
Quantitative assessment of activities for complex I, II, and IV, immunoblots for complex III core protein UQCRC1 (III2IV1 and III2), and Coomassie staining. Densitometry and quantitation based on results for all 20 individual mice (n=10 B6, n=10 D2). Integrated optical density (IOD) values were normalized to μg protein loaded for each sample. *p < 0.05, **p < 0.0001.
Additionally, because detection of some respiratory complexes and supercomplexes was modest or absent in one or both strains, we also assessed activity of complexes I, II, IV and V under saturating conditions. With the exception of detecting activity for individual (V1) and dimeric (V2) complex V, and complex IV activity in I1III2IV4, these results did not change any of the conclusions. These qualitative data are reported, but only the quantitative data wereincluded in our statistical analyses.
Complex I
The vast majority of complex I activity was associated with high molecular weight supercomplexes, five of which were readily apparent in D2 (I1III2IV1-4 and I1III2). Two of these showed little or no complex I activity in B6 (I1III2IV2 and I1III2IV3, respectively). Conversely, I1III2 and I1III2IV1 had 55% (U18=18, p=0.01) and 81% (U18=13, p=0.004), respectively, higher complex I activity in B6 than D2. No strain difference was detected for the monomer, which accounted for only 6±0.8% of total complex I activity in both strains. Nor was a strain difference detected for supercomplex I1III2IV4 (U18=39, p=0.44, NS). Where strain differences in supercomplex-associated complex I activity existed, differential Coomassie staining was also apparent, suggesting that divergent catalytic activities were due, in large part at least, to differences in abundance. However, allelic variation (Keane et al., 2011) affecting the structure and function of complex I core protein constituents (e.g., Ndufs2, Denmark & Buck, 2008) may also contribute to B6/D2 differences.
Complex II
Detection of complex II and quantitation of associated catalytic activity was limited to the monomer, consistent with most results (reviewed in Winge, 2012 but see Acin-Perez et al., 2008 for exception). Activity did not differ between strains (U18=47, p=0.85, NS), suggesting that genetic variation in mitochondrial respiratory complexes may exclude complex II among B6, D2 and B6D2-derived mice, at least in experimentally naïve animals.
Complex III
No complex III catalytic activity was detected, potentially due to incompatibility with digitonin solubilization. However, complex III was readily evident by immunoblot for core protein 1 UQCRC1. In D2, robust UQCRC1 immunoreactivity was evident in five high molecular weight supercomplexes that also exhibited complex I activity (I1III2, I1III2IV1-4). In B6, however, UQCRC1 signal was detected in only the two smallest of these (I1III2 and I1III2IV1), consistent with diminished or absent Coomassie staining for the three larger supercomplexes (I1III2IV2-4) compared to D2. UQCRC1 immunoreactivity was also readily apparent in two lower molecular weight species (III2IV1 and III2) in D2, but in only one of these bands (III2) in B6, which we conclude lacks III2IV1. A trend (U18=26, p=0.07) was detected for more III2 (47%) in B6 than D2. Not surprisingly, Coomassie staining reflected the immunoblot data, except where comigration (i.e., III2IV1 and IVn in D2) precluded accurate quantitation of individual entities.
Complex IV
As in previous studies (Schagger, 2001), catalytic activity was predominantly associated with complex IV monomer, accounting for 70% of total complex IV activity, with no strain difference (U18=36, p=0.32, NS). In D2, activity was apparent in three high molecular weight supercomplexes (I1III2IV1-3), all of which also demonstrated complex I activity and complex III immunoreactivity. In B6, complex IV activity was not detected for I1III2IV3 and was detected for I1III2IV2, but is significantly lower than in D2 (48%, U18=0, p<0.0001). However, complex IV activity for I1III2IV1 was greater in B6 than D2 (131%, U18=1, p<0.0001). Trace complex IV activity was detected in I1III2IV4 (as identified in previous work, Reifschneider et al., 2006) under saturating conditions in D2, but was not detected in B6. Complex IV activity was also apparent in two lower molecular weight bands. Not surprisingly, the larger exhibited more (75%) activity in D2 (U18=0, p<0.0001), due at least in part to comigration of III2IV1 and IVn; while III2IV1 is lacking in B6, so signal was from IVn only. The smaller band (IV2) had somewhat more (21%) activity in B6 than D2 (U18=12, p=0.006), while. Where strain differences in supercomplex-associated complex IV activity were found, differential Coomassie staining was also apparent, suggesting that differential catalytic activities were due, in large part at least, to differences in supercomplex abundance between B6 and D2. However, allelic variation affecting the structure and function of complex IV constituents (e.g., Cox 11) might also play a role (Chen et al., 2012).
Complex V
Detection of catalytic activity was limited to the monomer and dimer, and only under saturating conditions, so activity was assessed qualitatively only. Consequently, its potential contribution to genetic variation among B6, D2 and B6D2-derived animals remains to be elucidated.
DISCUSSION
The present studies are the first analyses of mouse brain mitochondrial respiratory complex suprastructure and associated catalytic activity. Additionally, our studies compare two of the most extensively studied genetic animal models, the B6 and D2 mouse strains. Considering that supercomplex associations are apparently tightly regulated (Chen et al., 2012; Ramirez-Aguilar et al., 2011), involve multiple non-respiratory accessory proteins (Hatle et al., 2013; Lapuente-Brun et al., 2013; reviewed in Winge, 2012 and Lenaz & Genova 2012), and are altered in some pathological situations (Mckenzie et al., 2006; Rosca et al., 2008) including aging (Frenzel et al., 2010; Gomez et al., 2009), the remarkable strain differences (qualitative and quantitative) in organization and activity we observed may have profound implications for brain mitochondrial function in a number of contexts. Additionally, our results indicate that B6, D2, and B6D2-derived populations will be valuable genetic models with which to assess the potential role of naturally occurring genetic variation in supercomplex formation and/or stability in a wide variety of phenotypes (e.g., GeneNetwork [genenetwork.org]) and Mouse Phenome Database at The Jackson Laboratory [phenome.jax.org]).
Large supercomplex I1III2IV2-3 assemblies are readily apparent in D2 brain, but little or none is detected in B6, indicating a shift in D2 toward more highly ordered suprastructure, i.e., with greater incorporation of complex IV. These deficits may be compensated in B6, at least in part, by higher abundance and catalytic activity for different supercomplexes (i.e., I1III2IV1) and IV dimer. Neither the physiological importance of specific supercomplexes nor increased incorporation of complex IV has been precisely defined, though several lines of evidence point to kinetic advantages in facilitating integrated electron transfer and improving flow efficiency (Rosca et al., 2008; Van Raam et al., 2008). Seminal work by Acin-Perez and colleagues (2008) demonstrated that some supercomplexes with a I+III+IV core also contain the mobile electron carriers ubiquinone (CoQ) and cytochrome c, and transfer electrons from NADH to oxygen, i.e., are true functional “respirasomes.” Importantly, this group recently showed through genetic modulation in mouse fibroblasts that formation of supercomplexes determines electron flux by segmenting complex IV into pools receiving electrons from different sources (NADH, FAD, or both), allowing optimization of respiration to substrate availability (Lapuente-Brun et al., 2013). Furthermore, segmentation was genotype-dependent and associated with an assembly factor isoform, whereby C57BL/6 (Harlan and Charles River) alleles were associated with less supercomplex formation in fibroblasts. Our strain comparison data parallel these results, and are thus likely relevant to further elucidation of specific supercomplex function, particularly for the critical substrate requirements of brain tissue.
As the terminal respiratory chain enzyme, complex IV transfers electrons from cytochrome c to rapidly convert molecular oxygen to water. In the process, four protons are translocated across the inner mitochondrial membrane, thereby contributing to the electrochemical proton gradient used by complex V to synthesize ATP. Thus, genetic differences in supercomplex-associated complex IV abundance and catalytic activity could markedly impact respiratory chain efficiency and ATP synthesis. In the aforementioned studies, assembly factor isoform and associated formation of large (I+III+IV) and small (III+IV) supercomplexes affected respiration rates and maximal ATP production in mouse liver mitochondria, and varied by substrate, demonstrating that complex IV incorporation is advantageous in minimizing inhibitory competition and promoting optimal oxidation of multiple substrates (Lapuente-Brun et al., 2013). Similar to data for larger complex IV-containing supercomplexes, we also observed abundant III2IV1 in D2 and absence in B6, which may thus reflect comparable genotype-dependent partitioning in brain mitochondria that could significantly impact respiration and energy production. A direct conformational advantage of complex IV incorporation has also been suggested (Schafer et al., 2006), as well as alternative roles for mammalian small supercomplexes as adjoining modules linking chains of multiple respirasomes (Wittig & Schagger, 2009) or pre-assembled intermediates (Acin-Perez et al., 2004).
Supercomplex formation strongly depends on membrane phospholipid composition, particularly cardiolipin, which is also an integral component of individual complexes (Bazan et al., 2013 and references therein). Cardiolipin is the signature mitochondrial phospholipid, constituting ~20% of the inner mitochondrial membrane total lipid composition (Mileykovskaya & Dowhan, 2009). In most animal tissues, the four cardiolipin side chains are predominantly 18-carbon fatty acids with two unsaturated bonds on each, suggesting that the 18:2 acyl chain configuration is an important structural requirement of its high affinity for inner membrane proteins in mammalian mitochondria (Schlame et al., 1990). Interestingly, D2 and B6 brain may differ in fatty acid composition, with higher linoleic (18:2) content detected and representing more of total synaptosomal phospholipids in D2 over B6 (0.94% and 0.66%, respectively) (La Droitte et al., 1984), suggesting that genetic variation may be an important influence on fatty acid composition in these strains. Furthermore, some differences appear region-specific and independent of diet (Mcnamara et al., 2009). It is therefore tempting to speculate that more 18:2 linoleic acid in D2 brain reflects higher cardiolipin content in the inner mitochondria membrane that would facilitate supercomplex formation and/or stabilization, thus contributing to the observed shift toward more highly assembled supercomplexes compared to B6. Considering the vast molecular diversity exclusive to brain cardiolipin (Cheng et al., 2008; Ji et al., 2012), future strain comparisons assessing content characteristics (e.g., using gas chromatography), and the spatial limitations of supercomplex organization (e.g., synaptic and/or nonsynaptic, discrete brain regions), will provide important insights to understanding the role of cardiolipin in genetic variation of mitochondrial respiratory complexes.
A clear functional consequence of supercomplex I1III2 disruption is a 2-4 fold increase in ROS generation by complex I, with comparable observations in cellular and animal models linking dissociation and increased ROS production (see Maranzana et al., 2013 and references therein). In addition to acting as direct damaging agents, ROS also modulate some intracellular signaling pathways, depending on the amounts produced (Anilkumar et al., 2009; Ide et al., 2000; Ray et al., 2012). Several observations support the idea that supercomplex association affords enhanced complex I stability and activity, including the detrimental effects of complex III and IV mutations on complex I structural and functional integrity (Acin-Perez et al., 2004; D’aurelio et al., 2006; Diaz et al., 2006; Schagger et al., 2004; Stroh et al., 2004), and the necessity of respirasome assembly for full complex I activity (Moreno-Lastres et al., 2012). Furthermore, ROS levels appear to directly affect complex I and supercomplex stability (Diaz et al., 2012). The markedly reduced NADH-ubiquinone oxidoreductase activity observed in cell models lacking supercomplexes (Acin-Perez et al., 2004; D’aurelio et al., 2006; Diaz et al., 2006; Schagger et al., 2004; Stroh et al., 2004) is supportive of a critical role in mammalian complex I-driven respiration and emphasizes the value of future studies exploring the impact of naturally occurring complex I variation.
MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) is a neurotoxin precursor to MPP+, which interferes with complex I leading to the buildup of free radicals and cell death (Fonck & Baudry, 2003; Obata, 2002). The degree to which MPTP impacts neuron survival in mice is strain-dependent; B6 is particularly sensitive to MPTP-induced cell loss in the substantia nigra pars compacta, while D2 shows intermediate cell loss compared to more resistant strains (Hamre et al., 1999), suggesting the presence of both protective and risk alleles. B6 are also highly susceptible to MPTP-induced reduction in striatal dopamine concentrations and midbrain dopaminergic cells (German et al., 1996). This high MPTP susceptibility is autosomal dominant for an anonymous B6 allele (Hamre et al., 1999), recapitulates the regional neuronal degeneration underlying Parkinson’s disease in humans, and is most likely not explained by greater MPTP uptake, since striatal dopamine transporter levels are actually lower than in D2 (Janowsky et al., 2001). Thus, it is plausible that our observations may be related to genetic differences in MPTP susceptibility through variation in complex I-related supercomplex organization, abundance, and/or activity. Future strain comparisons of complex I-associated ROS and consequent lipid and protein modifications (e.g., isoprostanes, 4-hydroxynonenal [HNE]) will be important in elucidating the extent to which genetic respiratory chain differences contribute to vulnerability to MPTP, and potentially other complex I toxins.
In summary, these are the first studies of mitochondrial respiratory complex suprastructure and associated activity in mouse brain. Importantly, our comparisons of the widely used B6 and D2 strains suggest that natural variation in supercomplex formation and/or stability may influence a broad array of mitochondrial- and oxidative stress-related phenotypes, including response to global mitochondrial superoxide dismutase deficiency (Huang et al., 2006; Lynn et al., 2005), brain antioxidative capacity (Rebrin et al., 2007), susceptibility to age-dependent pathology (Misra et al., 2007; Rebrin et al., 2011), and susceptibility to severe alcohol withdrawal (Denmark & Buck, 2008). Furthermore, these results indicate that B6D2-derived populations will be highly valuable genetic models with which to further characterize this potential relationship and provide a basis from which to explore the extent to which differences in mitochondrial function/dysfunction are innate, developmentally regulated, a consequence of disease and its progression, and amenable to intervention prior to disease and/or various treatments (e.g., pharmacological and diet adjunct therapies). Our studies contribute significantly to the field, but there are some limitations. First, our analyses are based on whole brain that, while an important first step, will need to be expanded in future studies assessing discrete brain regions and/or cell populations. Regional variation in respiratory supercomplex characteristics likely exist and may contribute to the striking region-specific mitochondrial and functional vulnerability associated with many neuropathological conditions (Dubinsky, 2009). Additionally, our analyses are based on total mitochondria, and will be expanded in future studies assessing specific cellular compartments (e.g., synaptic or nonsynaptic mitochondria). Finally, we used samples from flash frozen tissue. Although not necessarily typical for BN-PAGE analyses, this is sometimes necessary (e.g., to process large numbers of samples for quantitative analyses, as in the present studies; or when using biopsied tissue, as in clinical analyses). While this does not compromise the results reported here, including the respiratory complex enzymatic activity analyses, it did not allow for more direct measurements of mitochondrial function (e.g., membrane potential and pH gradients, and oxygen consumption), all of which will be important to assess in future work. As more information becomes available, the role of respiratory supercomplexes in mitochondrial energetics and the etiology and pathogenesis of a variety of disorders will become apparent.
Supplementary Material
Supplemental Figure 1. Respiratory complex organization and band identification in B6 and D2 mouse brain after BN-PAGE for complex I, II, and IV activities and Coomassie staining from the same gel with NativeMark Unstained Native Protein Marker (Life Technologies, Grand Island, NY, USA).
Supplemental Figure 2. Respiratory complex organization and band identification in B6 and D2 mouse brain after BN-PAGE and quantitative assessment of activities for complex I (purple), II (indigo), IV (brown), and V (white); immunoblots for complex III core protein UQCRC1 (III2IV1 and III2; others are qualitative), and Coomassie staining. The other ten samples are given in Figure 1.
Supplemental Figure 3. Assessment of activities for complex I and IV in fresh vs. flash frozen tissue in B6 and D2 shows that qualitative genetic differences in respiratory complex organization is maintained. 18 μg of protein were loaded in each lane. Complex I in-gel activity (IGA): 40 minutes, and complex IV IGA: 45 minutes. A = fresh, B = flash frozen.
ACKNOWLEDGEMENTS
This work was supported by I01-BX000222, P60AA010760, R01AA011114 (KJB), F30AA017342 and T32AA07468 (DLD). We thank Drs. Isaac Forquer, Michael Riscoe, P. Hemachandra Reddy, and Joseph Quinn for helpful discussions on these studies and a draft of this manuscript; and Dr. Laura Kozell, Maarvi Khawaja, and Melinda Helms for technical support.
REFERENCES
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar N, Sirker A, Shah AM. Redox sensitive signaling pathways in cardiac remodeling, hypertrophy and failure. Front Biosci. 2009;14:3168–3187. doi: 10.2741/3443. [DOI] [PubMed] [Google Scholar]
- Bazan S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem. 2013;288:401–411. doi: 10.1074/jbc.M112.425876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave SV, Hoffman PL, Lassen N, Vasiliou V, Saba L, Deitrich RA, Tabakoff B. Gene array profiles of alcohol and aldehyde metabolizing enzymes in brains of C57BL/6 and DBA/2 mice. Alcohol Clin Exp Res. 2006;30:1659–1669. doi: 10.1111/j.1530-0277.2006.00201.x. [DOI] [PubMed] [Google Scholar]
- Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, Han X. Shotgun lipidomics reveals the temporally dependent, highly diversified cardiolipin profile in the mammalian brain: temporally coordinated postnatal diversification of cardiolipin molecular species with neuronal remodeling. Biochemistry. 2008;47:5869–5880. doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun P, Wyrembak J, Schriner SE, Chen HW, Marciniack C, Laferla F, Wallace DC. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2012;1820:553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- D’Aurelio M, Gajewski CD, Lenaz G, Manfredi G. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum Mol Genet. 2006;15:2157–2169. doi: 10.1093/hmg/ddl141. [DOI] [PubMed] [Google Scholar]
- Denmark DL, Buck KJ. Molecular analyses and identification of promising candidate genes for loci on mouse chromosome 1 affecting alcohol physical dependence and associated withdrawal. Genes Brain Behav. 2008;7:599–608. doi: 10.1111/j.1601-183X.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- Diaz F, Barrientos A, Fontanesi F. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr Protoc Hum Genet. 2009:14. doi: 10.1002/0471142905.hg1904s63. Chapter 19, Unit19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Enriquez JA, Moraes CT. Cells lacking Rieske iron-sulfur protein have a reactive oxygen species-associated decrease in respiratory complexes I and IV. Mol Cell Biol. 2012;32:415–429. doi: 10.1128/MCB.06051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubinsky JM. Heterogeneity of nervous system mitochondria: location, location, location! Exp Neurol. 2009;218:293–307. doi: 10.1016/j.expneurol.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci U S A. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. Central nervous system manifestations of mitochondrial disorders. Acta Neurol Scand. 2006;114:217–238. doi: 10.1111/j.1600-0404.2006.00671.x. [DOI] [PubMed] [Google Scholar]
- Fonck C, Baudry M. Rapid reduction of ATP synthesis and lack of free radical formation by MPP+ in rat brain synaptosomes and mitochondria. Brain Res. 2003;975:214–221. doi: 10.1016/s0006-8993(03)02675-1. [DOI] [PubMed] [Google Scholar]
- Frenzel M, Rommelspacher H, Sugawa MD, Dencher NA. Ageing alters the supramolecular architecture of OxPhos complexes in rat brain cortex. Exp Gerontol. 2010;45:563–572. doi: 10.1016/j.exger.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Gadicherla AK, Stowe DF, Antholine WE, Yang M, Camara AK. Damage to mitochondrial complex I during cardiac ischemia reperfusion injury is reduced indirectly by anti-anginal drug ranolazine. Biochim Biophys Acta. 2012;1817:419–429. doi: 10.1016/j.bbabio.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova ML, Baracca A, Biondi A, Casalena G, Faccioli M, Falasca AI, Formiggini G, Sgarbi G, Solaini G, Lenaz G. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta. 2008;1777:740–746. doi: 10.1016/j.bbabio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- German DC, Nelson EL, Liang CL, Speciale SG, Sinton CM, Sonsalla PK. The neurotoxin MPTP causes degeneration of specific nucleus A8, A9 and A10 dopaminergic neurons in the mouse. Neurodegeneration. 1996;5:299–312. doi: 10.1006/neur.1996.0041. [DOI] [PubMed] [Google Scholar]
- Gomez LA, Monette JS, Chavez JD, Maier CS, Hagen TM. Supercomplexes of the mitochondrial electron transport chain decline in the aging rat heart. Arch Biochem Biophys. 2009;490:30–35. doi: 10.1016/j.abb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazina MM. Mitochondrial respiratory chain: biochemical analysis and criterion for deficiency in diagnosis. Methods Mol Biol. 2012;837:73–91. doi: 10.1007/978-1-61779-504-6_6. [DOI] [PubMed] [Google Scholar]
- Hamre K, Tharp R, Poon K, Xiong X, Smeyne RJ. Differential strain susceptibility following 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration acts in an autosomal dominant fashion: quantitative analysis in seven strains of Mus musculus. Brain Res. 1999;828:91–103. doi: 10.1016/s0006-8993(99)01273-1. [DOI] [PubMed] [Google Scholar]
- Hatle KM, Gummadidala P, Navasa N, Bernardo E, Dodge J, Silverstrim B, Fortner K, Burg E, Suratt BT, Hammer J, Radermacher M, Taatjes DJ, Thornton T, Anguita J, Rincon M. MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations. Mol Cell Biol. 2013;33:2302–2314. doi: 10.1128/MCB.00189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Naeemuddin M, Elchuri S, Yamaguchi M, Kozy HM, Carlson EJ, Epstein CJ. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet. 2006;15:1187–1194. doi: 10.1093/hmg/ddl034. [DOI] [PubMed] [Google Scholar]
- Ide T, Tsutsui H, Kinugawa S, Suematsu N, Hayashidani S, Ichikawa K, Utsumi H, Machida Y, Egashira K, Takeshita A. Direct evidence for increased hydroxyl radicals originating from superoxide in the failing myocardium. Circ Res. 2000;86:152–157. doi: 10.1161/01.res.86.2.152. [DOI] [PubMed] [Google Scholar]
- Janowsky A, Mah C, Johnson RA, Cunningham CL, Phillips TJ, Crabbe JC, Eshleman AJ, Belknap JK. Mapping genes that regulate density of dopamine transporters and correlated behaviors in recombinant inbred mice. J Pharmacol Exp Ther. 2001;298:634–643. [PubMed] [Google Scholar]
- Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RS, Kochanek PM, Wipf P, Kagan VE, Bayir H. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nat Neurosci. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Higgins CM, Xu Z. Measuring the quantity and activity of mitochondrial electron transport chain complexes in tissues of central nervous system using blue native polyacrylamide gel electrophoresis. Anal Biochem. 2000;286:214–223. doi: 10.1006/abio.2000.4813. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Droitte P, Lamboeuf Y, de Saint Blanquat G. Ethanol sensitivity and membrane lipid composition in three strains of mouse. Comp Biochem Physiol C. 1984;77:351–356. doi: 10.1016/0742-8413(84)90027-6. [DOI] [PubMed] [Google Scholar]
- Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- Leary SC. Blue Native Polyacrylamide Gel Electrophoresis: A Powerful Diagnostic Tool for the DEtection of Assembly Defects in the Enzyme Complexes of Oxidative Phosphorylation. Methods in Molecular Biology. 2012;387 doi: 10.1007/978-1-61779-504-6_13. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML. Structural and functional organization of the mitochondrial respiratory chain: a dynamic super-assembly. Int J Biochem Cell Biol. 2009;41:1750–1772. doi: 10.1016/j.biocel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML. Supramolecular organisation of the mitochondrial respiratory chain: a new challenge for the mechanism and control of oxidative phosphorylation. Adv Exp Med Biol. 2012;748:107–144. doi: 10.1007/978-1-4614-3573-0_5. [DOI] [PubMed] [Google Scholar]
- Lessing D, Bonini NM. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nat Rev Genet. 2009;10:359–370. doi: 10.1038/nrg2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn S, Huang EJ, Elchuri S, Naeemuddin M, Nishinaka Y, Yodoi J, Ferriero DM, Epstein CJ, Huang TT. Selective neuronal vulnerability and inadequate stress response in superoxide dismutase mutant mice. Free Radic Biol Med. 2005;38:817–828. doi: 10.1016/j.freeradbiomed.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML. Mitochondrial Respiratory Supercomplex Association Limits Production of Reactive Oxygen Species from Complex I. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- McNamara RK, Able J, Jandacek R, Rider T, Tso P. Inbred C57BL/6J and DBA/2J mouse strains exhibit constitutive differences in regional brain fatty acid composition. Lipids. 2009;44:1–8. doi: 10.1007/s11745-008-3244-8. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V, Lee H, Singh A, Huang K, Thimmulappa RK, Mitzner W, Biswal S, Tankersley CG. Global expression profiles from C57BL/6J and DBA/2J mouse lungs to determine aging-related genes. Physiol Genomics. 2007;31:429–440. doi: 10.1152/physiolgenomics.00060.2007. [DOI] [PubMed] [Google Scholar]
- Moreno-Lastres D, Fontanesi F, Garcia-Consuegra I, Martin MA, Arenas J, Barrientos A, Ugalde C. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T. Dopamine efflux by MPTP and hydroxyl radical generation. J Neural Transm. 2002;109:1159–1180. doi: 10.1007/s00702-001-0683-2. [DOI] [PubMed] [Google Scholar]
- Orth M, Schapira AH. Mitochondrial involvement in Parkinson’s disease. Neurochem Int. 2002;40:533–541. doi: 10.1016/s0197-0186(01)00124-3. [DOI] [PubMed] [Google Scholar]
- Ramirez-Aguilar SJ, Keuthe M, Rocha M, Fedyaev VV, Kramp K, Gupta KJ, Rasmusson AG, Schulze WX, van Dongen JT. The composition of plant mitochondrial supercomplexes changes with oxygen availability. J Biol Chem. 2011;286:43045–43053. doi: 10.1074/jbc.M111.252544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebrin I, Forster MJ, Sohal RS. Effects of age and caloric intake on glutathione redox state in different brain regions of C57BL/6 and DBA/2 mice. Brain Res. 2007;1127:10–18. doi: 10.1016/j.brainres.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebrin I, Forster MJ, Sohal RS. Association between life-span extension by caloric restriction and thiol redox state in two different strains of mice. Free Radic Biol Med. 2011;51:225–233. doi: 10.1016/j.freeradbiomed.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J Proteome Res. 2006;5:1117–1132. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res. 2008;80:30–39. doi: 10.1093/cvr/cvn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, Balk J, Leaver CJ. Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native polyacrylamide gel electrophoresis. Plant J. 2005;44:893–901. doi: 10.1111/j.1365-313X.2005.02577.x. [DOI] [PubMed] [Google Scholar]
- Sayer RJ. Intracellular Ca2+ handling. Adv Exp Med Biol. 2002;513:183–196. doi: 10.1007/978-1-4615-0123-7_6. [DOI] [PubMed] [Google Scholar]
- Schafer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- Schagger H. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 2001;65:231–244. doi: 10.1016/s0091-679x(01)65014-3. [DOI] [PubMed] [Google Scholar]
- Schagger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem. 2004;279:36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J Biol Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Oxidative stress and mitochondrial dysfunction in neurodegeneration. Curr Opin Neurol. 1996;9:260–264. doi: 10.1097/00019052-199608000-00003. [DOI] [PubMed] [Google Scholar]
- Schlame M, Horvath L, Vigh L. Relationship between lipid saturation and lipid-protein interaction in liver mitochondria modified by catalytic hydrogenation with reference to cardiolipin molecular species. Biochem J. 1990;265:79–85. doi: 10.1042/bj2650079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smet J, De Paepe B, Seneca S, Lissens W, Kotarsky H, De Meirleir L, Fellman V, Van Coster R. Complex III staining in blue native polyacrylamide gels. J Inherit Metab Dis. 2011;34:741–747. doi: 10.1007/s10545-011-9315-7. [DOI] [PubMed] [Google Scholar]
- Smith MA, Nunomura A, Zhu X, Takeda A, Perry G. Metabolic, metallic, and mitotic sources of oxidative stress in Alzheimer disease. Antioxid Redox Signal. 2000;2:413–420. doi: 10.1089/15230860050192198. [DOI] [PubMed] [Google Scholar]
- Spinazzi M, Casarin A, Pertegato V, Ermani M, Salviati L, Angelini C. Optimization of respiratory chain enzymatic assays in muscle for the diagnosis of mitochondrial disorders. Mitochondrion. 2011;11:893–904. doi: 10.1016/j.mito.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, Schagger H. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J Biol Chem. 2004;279:5000–5007. doi: 10.1074/jbc.M309505200. [DOI] [PubMed] [Google Scholar]
- Stuart RA. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Bioenerg Biomembr. 2008;40:411–417. doi: 10.1007/s10863-008-9168-4. [DOI] [PubMed] [Google Scholar]
- Suthammarak W, Morgan PG, Sedensky MM. Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. J Biol Chem. 2010;285:40724–40731. doi: 10.1074/jbc.M110.159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raam BJ, Sluiter W, de Wit E, Roos D, Verhoeven AJ, Kuijpers TW. Mitochondrial membrane potential in human neutrophils is maintained by complex III activity in the absence of supercomplex organisation. PLoS One. 2008;3:e2013. doi: 10.1371/journal.pone.0002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Winge DR. Sealing the mitochondrial respirasome. Mol Cell Biol. 2012;32:2647–2652. doi: 10.1128/MCB.00573-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, Schagger H. Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis. 2007;28:3811–3820. doi: 10.1002/elps.200700367. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Electrophoretic methods to isolate protein complexes from mitochondria. Methods Cell Biol. 2007;80:723–741. doi: 10.1016/S0091-679X(06)80033-6. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim Biophys Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Yang YY, Vasta V, Hahn S, Gangoiti JA, Opheim E, Sedensky MM, Morgan PG. The role of DMQ(9) in the long-lived mutant clk-1. Mech Ageing Dev. 2011;132:331–339. doi: 10.1016/j.mad.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Carelli V. Mitochondrial disorders. Curr Opin Neurol. 2003;16:585–594. doi: 10.1097/01.wco.0000093101.34793.c8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Respiratory complex organization and band identification in B6 and D2 mouse brain after BN-PAGE for complex I, II, and IV activities and Coomassie staining from the same gel with NativeMark Unstained Native Protein Marker (Life Technologies, Grand Island, NY, USA).
Supplemental Figure 2. Respiratory complex organization and band identification in B6 and D2 mouse brain after BN-PAGE and quantitative assessment of activities for complex I (purple), II (indigo), IV (brown), and V (white); immunoblots for complex III core protein UQCRC1 (III2IV1 and III2; others are qualitative), and Coomassie staining. The other ten samples are given in Figure 1.
Supplemental Figure 3. Assessment of activities for complex I and IV in fresh vs. flash frozen tissue in B6 and D2 shows that qualitative genetic differences in respiratory complex organization is maintained. 18 μg of protein were loaded in each lane. Complex I in-gel activity (IGA): 40 minutes, and complex IV IGA: 45 minutes. A = fresh, B = flash frozen.