Abstract
The recent emergence of multimodality imaging, particularly the combination of PET and MRI, has led to excitement over the prospect of improving detection of disease. Iron oxide nanoparticles (IONPs) have become a popular platform for the fabrication of PET/MRI probes due to their advantages of high MRI detection sensitivity, biocompatibility, and biodegradability. In this paper, we report the synthesis of dextran coated iron oxide nanoparticles labeled with the positron emitter 64Cu to generate a PET/MRI probe, and modified with small molecular maleic anhydride to increase negative surface charge. The modified nanoparticulate PET/MRI probe (MDIO-64Cu-DOTA) bears repetitive anionic charges on the surface that facilitate recognition by scavenger receptor type A (SR-A), a ligand-receptor found on activated macrophages but not on normal vessel walls. MDIO-64Cu-DOTA has an average iron oxide core size of 7-8 nm, an average hydrodynamic diameter of 62.7 nm, an r1 relaxivity of 16.8 mM−1·s−1, and an r2 relaxivity of 83.9 mM−1·s−1 (37 °C, 1.4 T). Cell studies confirmed that the probe was nontoxic and was specifically taken up by macrophages via SR-A. In comparison with the non-modified analog, the accumulation of maleylated DIO in macrophages was substantially improved. These characteristics demonstrate the promise of MDIO-64Cu-DOTA for identification of vulnerable atherosclerotic plaques (VAP) via the targeting of macrophages.
Keywords: Iron, Nanoparticles, MRI contrast agents, Macrophages, Inflammation
Introduction
Accurate and early diagnosis of disease is an ongoing challenge for current medical imaging technologies. Ideally, detection of disease requires the imaging modality to be capable of detecting pathological events at early stages of progression. Moreover, the imaging method should also be able to provide detailed anatomical context (in high spatial resolution) on the location of these events. The combination of both high detection capability and resolution is often beyond the scope of any available single imaging modality [1]. For example, amongst the noninvasive imaging methodologies, positron emission tomography (PET) has extremely high detection capability (nM-pM) which allows it to locate molecular signatures (biomarkers) of certain diseases usually expressed at low levels (nM-pM/g tissue) [2], but its spatial resolution is low (1-2 mm). In contrast, magnetic resonance imaging (MRI) can provide high spatial resolution (μm) and excellent soft tissue contrast but has inherently low detection capability (mM-μM). Although PET and MRI are quite complementary and the combination of PET and MRI has been considered among the most promising pairings to satisfy the ideal traits required for diagnostic imaging, the first prototype MRI/PET was developed and the first in vivo images reported only a handful of years ago [3]. Since then there has been an explosive interest in developing hybrid PET/MRI systems with the aim of maximizing the synergy of combining the two modalities for early disease diagnosis in vivo [4-8].
Parallel to the rapid growth of PET/MRI instrumentation has been a surge in the development of multifunctional probes for the dual-modality imaging [9-15]. The majority of PET/MRI probes reported to date have been in the nano-scale. Nanoparticulate probes hold a number of desirable traits for integrating diverse functionalities into a single entity, including the ability to carry a high payload of signal-enhancing/generating materials, easy surface functionalization chemistry for biocompatibility and biomarker targeting, and long blood circulation half-lives. These capabilities could substantially improve the sensitivity, specificity and quality of medical/clinical diagnostic imaging. The biocompatibility and biodegradability profiles of iron oxide nanoparticles (IONPs) have led them to become an important class of materials for fabricating PET/MRI probes [11-15]. PET radionuclide labeled IONPs have been investigated with PET/MRI to detect molecular biomarkers of specific diseases.
For example, IONPs have been developed to image plaque-associated macrophages, which are one of the hallmarks of vulnerable atherosclerotic plaques (VAP) [16]. VAP rupture is considered as the primary cause of acute clinical events such as myocardial infarction and stroke etc [17-20]. And macrophage accumulation in plaques has been associated with degree of instability. Radiolabeled, nonspecific IONPs have been found to accumulate in macrophages found in atherosclerotic plaques [16]. In most cases, macrophage uptake of IONPs relies on macrophage phagocytosis, which is a non-specific process. Nonspecific nanoparticulate probes show a differential distribution in the body based primarily on relative tissue permeability but they lack targeting specificity, and this results in low labeling efficiency [21].
Efficient labeling of VAP macrophages can be achieved by decorating nanoparticulate probes with ligands that have both affinity and specificity for macrophage surface ligand scavenger receptors (SR). Macrophage SR biology was highlighted three decades ago when Brown and Goldstein found that the rate of 125I-labeled-acetyl-LDL (AcLDL; LDL: low-density lipoprotein) uptake and degradation by resident mouse peritoneal cells was 20 times higher than that of 125I-labeled native LDL [22, 23]. SRs are not down-regulated with ligand concentration. They mediate very efficient and rapid internalization of bound ligands, and repeatedly recycle via endocytic compartments allowing accumulation of SR-targeted probes inside the cell; thus enabling the probes to be enriched in activated macrophages [24]. Scavenger receptor type A (SR-A) is a receptor that is over-expressed on the surface of activated atheroma macrophages but not found on normal vessel walls. SR-A localizes in the cap and interior in lesions at all stages of atheromas and therefore is an attractive target for diagnostic imaging probes [25].
The common characteristic of all known SR-A ligands, such as oxidized LDL, maleylated LDL, maleylated BSA, maleylated polysaccharides, sulfated polysaccharides, and polyinosinic acid, etc., is the repetitive anionic surface charge distribution. SR-A contains a number of overlapping binding sites for ligands, with different binding affinities for each [25-28]. We have previously developed positron emitter 64Cu labeled, dextran sulfate coated, IONPs for PET/MRI of macrophages in inflamed plaques [29, 30]. Cell studies confirmed that these probes were specifically taken up by activated macrophages in comparison with non-targeted probes [30]. In vivo PET/MRI of a rat injury model showed that the probes enhanced contrast was observed at sites of vascular inflammation, but not in normal vessel walls, in both PET and MR images [31]. Encouraged by these results, we describe herein the development of a SR-A targeted, dual-modal PET/MRI probe that utilizes a different targeting ligand, maleylated dextran coated IONPs (MDIO). The ability of MDIO to target activated macrophages was tested in vitro in cell studies, and the efficiency of 64Cu labeling of IONPs was evaluated in solutions.
Materials and methods
Materials were obtained from commercial suppliers and used directly, unless otherwise noted. Dextran (from leuconostoc, average MW 9,000-11,000) was purchased from Sigma-Aldrich. p-Isothiocyanatobenzyl-1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (p-SCN-Bn-DOTA, C24H33N5O8S·2.5HCl·2.5H2O, purity ≥ 94%) was purchased from Macrocyclics. 5-(and -6)-Carboxytetramethylrhodamine, succinimidyl ester (5(6)-TAMRA, SE; Abs/Em = 546/575 nm) was purchased from AnaSpec Inc. Cupric-64 chloride solution (half life = 12.7 h) (Washington University) was used as received. Spectra/por® dialysis membrane (MW cut-off 50,000) was purchased from Spectrum Laboratories, Inc. P388D1 macrophages and RPMI-1640 medium were obtained from American Type Culture Collection (ATCC). Fetal bovine serum (FBS), L-glutamine, PBS (1×) and C12 – resazurin were purchased from GIBCO. Lipoprotein deficient bovine serum (LPDS) was obtained from Biomedical Technologies, Inc. (Stoughton, MA). Water was purified using a Millipore Milli-Q Synthesis purifier (18.0 MΩcm, Barnstead).
MDIO synthesis
Dextran coated iron oxide nanoparticles (DIO) were synthesized as previously reported with slight modification [32]. Briefly, a solution of reduced dextran (1 eq, 400 mg, 0.04 mmol) and FeCl3·6H2O (27 eq, 292 mg, 1.08 mmol) in degassed de-ionized water (18 mL) was prepared in a 50-mL, 2-necked round-bottom flask. The solution was stirred in an ice-water bath for 30 minutes while bubbling with argon. A solution of FeCl2·4H2O (18 eq, 143 mg, 0.72 mmol) in degassed de-ionized water (4 mL) was prepared and chilled in an ice-water bath for 10 minutes, then the solution was added to the flask using a syringe. Five minutes after adding the Fe2+ solution, chilled (in an ice-water bath) NH4OH (432 eq, 673 uL (606 mg), 17.28 mmol) was added to the flask dropwise with a syringe within a period of 5 minutes under rapid stirring. The ice-water bath was removed. The solution was heated to 85 oC. The reaction was kept at 85 ± 5 °C for 2 h (Argon flow may stop 2-3 minutes after the temperature reaches 85 °C). After cooling to room temperature, the solution was loaded into a Spectra/por® dialysis membrane bag (MW cut-off 50,000) and was dialyzed for 72 h against deionized water (8-10 changes of water). Finally, the solution in the bag was collected, concentrated with centrifugation, and lyophilized to give a brown solid.
Cross-linking and amination of the dextran coating of DIO followed the literature method with slight modifications [33]. In brief, DIO (104 mg), NaOH pellets (209 mg), and deionized water (2 mL) were added to a 10-mL round bottom flask. The mixture was stirred for 30 minutes during which the solution became homogeneous and cooled down to room temperature. Epichlorohydrin (0.4 mL) was added to the flask. After the mixture was stirred for 24 h, the solution was dialyzed for 72 h against deionized water (Spectra/por® dialysis membrane, 8-10 changes of water). The solution was lyophilized to give a brown solid. The solid was transferred into a 50-mL round bottom flask. Ammonium hydroxide (25 mL) was added to the flask and the mixture was stirred for 24 h. The solution was concentrated by rotary evaporation. The residue was diluted and dialyzed for 72 h against deionized water (Spectra/por® dialysis membrane, 8-10 changes of water). The solution was lyophilized to yield a brown solid (103 mg).
Aminated DIO (135 mg), p-SCN-Bn-DOTA (6.71 mg), and 0.1 M sodium borate buffer solution (2 mL) were mixed in a 10-mL round bottom flask. Approximate five drops of NaOH aqueous solution (1N) was added into the flask to bring the pH to 8.5. The mixture was stirred for 24 h. Solid maleic anhydride (2.211 g in total) was added in aliquots to the buffered well-stirred solution in the flask while maintaining pH. The maleic anhydride dissolved slowly, and the rate of addition of the aliquots was governed by the disappearance of the solid material in the reaction system. The pH value of the solution in the flask was monitored continuously and was adjusted to 8.5-9.0 by the addition of solid sodium carbonate. The reaction was complete when no further acid was liberated, i.e., pH value of the solution was stable [34]. The solution was dialyzed against deionized water for 72 h (8-10 changes of water). The solution was lyophilized to give a brown solid (MDIO-DOTA, 143 mg).
Cupric-64 labeling of MDIO
MDIO (26.15 mg) was dissolved in 150 μL of 0.1 M pH 7.0 triethanolamine acetate buffer solution in a 1.5 mL Eppendorf vial. Then cupric-64 chloride solution (79.67 MBq) was added to the vial and the mixture was vortexed for 5 seconds to obtain a homogeneous solution. The solution was incubated in a dry bath incubator (Fisher Scientific) for 45 minutes at 55-60 °C. EDTA aqueous solution (22 μL, 100 mM) was added to the vial. After vortexing for 5 seconds to get a homogeneous solution, the solution was incubated in the same incubator for 15 minutes at 55-60 °C. The crude product was purified by centrifuge filtration with 10 kDa Nanosep filtration tube (Millipore Inc., Billerica, MA) (15 min @14,000 rpm) and washed 3 times with saline (0.9%). Each time the washing was removed by centrifuge filtration with 10 kDa Nanosep filtration tube (10 min @14,000 rpm). After three washings the filtration tube was turned over and inserted into a new vial, and the nanoparticles, MDIO-64Cu-DOTA (~ 15 μL), were collected by centrifuge (2 min @ 1,000 rpm). The radioactive nanoparticles were diluted to 650 μL with saline (0.9%) and measured to contain 46.59 MBq of radioactivity. Therefore, the labeling efficiency was 68% after correction for the natural decay of 64Cu.
MDIO sizes, zeta potential, spectroscopy, and relaxivity
The iron oxide core size of MDIO and DIO were measured by transmission electron microscopy (TEM) on a Philips CM-12, operating at 80 kV. The average core diameter of MDIO or DIO is calculated based on an average of 500 particles from different regions of the grid. The average hydrodynamic particle size and size distribution of MDIO and DIO were measured using dynamic light scattering (DLS) with a Nanotrac 150 particle size analyzer (Microtrac, Inc., Montgomeryville, PA). The zeta potential (ζ) values of MDIO and DIO were measured by determining the electrophoretic mobility using a NICOMP™ 380 ZLS device (Particle Sizing Systems, Inc., Santa Barbara, CA, USA) in deionized water at room temperature. The amount of iron in MDIO or DIO was measured on an atomic absorption (AA) spectrophotometer Varian AA 220FS using an air/acetylene flame. FT-IR spectra were collected on a Shimadzu IR Prestige 21 spectrophotometer. Absorption spectra were measured on a Varian Cary 100-Bio UV-vis spectrophotometer at room temperature. Fluorescent spectra were measured on a JobinYvon Horiba FluoroMax-P spectrophotometer at room temperature.
Longitudinal (T1) and transverse (T2) relaxation times of MDIO and DIO solutions and cell lysates containing MDIO or DIO were measured at 60 MHz (1.4 T) and 37 °C on a Bruker Minispec mq60 (Bruker, Billerica, MA) by the methods we previously used [30]. Briefly, six stock solutions, three for MDIO and the other three for DIO, were prepared by dissolving 5.00 mg of MDIO or DIO in 1 mL of pH 7.0 deionized water. The concentration of Fe was determined by atomic absorption (AA). All solutions were prepared by weight. The MDIO stock solution was diluted 20 times to give the most concentrated MDIO working solution. The dilution factor of DIO stock solution to the most concentrated DIO working solution was calculated according to the most concentrated MDIO solution, which was 33.5 times dilution. T1 values were measured using an inversion recovery sequence with 10-15 data points and T2 values were measured using a Carr–Purcell–Meiboom–Gill (CPMG) sequence with τ = 1 ms, and 200 data points. Each solution was incubated at 37 °C for 10 min before measurement to reach thermal equilibrium. The longitudinal (r1) and transverse (r2) relaxivity were determined as the slope of the line for plots of 1/T1 or 1/ T2, respectively, against increasing iron concentration with a correlation coefficient greater than 0.99.
Stability of MDIO-64Cu-DOTA
MDIO (7.43 mg) was dissolved in 50 μL of 0.1 M pH 7.0 triethanolamine acetate buffer solution in a 1.5 mL Eppendorf vial. As for labeling described above, cupric-64 (27.1 MBq) was added to the vial and the mixture was incubated for 45 minutes at 55-60 °C. The solution was centrifuged in 10 kDa Nanosep filtration tube and washed 3 times with the pH 7.0 buffer solution. The labeling efficiency was 67%. The purified MDIO-64Cu-DOTA was diluted with 200 μL of 0.1 M pH 7.0 triethanolamine acetate buffer solution in an Eppendorf vial. EDTA aqueous solution (25 μL, 100 mM) was added to the vial. The solution was incubated in a dry bath incubator at 55-60 °C for up to 48 h. An aliquot (50 μL) was taken at each time point (1, 4, 24, and 48 h) and the activity (before purification) was measured using a dose calibrator. Each aliquot was centrifuged and washed 3 times through a 10 kDa Nanosep filtration tube in order to remove EDTA and EDTA-64Cu. Final radioactivity (post purification) was measured for the purified MDIO-64Cu-DOTA on a dose calibrator. Comparisons between values were made using least squares means t test. Three parallel solutions of MDIO-64Cu-DOTA were prepared for the stability test to provide statistical significance.
In vitro cellular uptake of MDIO and DIO
The P388D1 macrophages were maintained in medium (RPMI-1640 with 1% L-glutamine and 10% fetal bovine serum (FBS)) at 37 °C in a humidified environment of 5% CO2 atmosphere. When the cells reached 80-90% confluence, the medium was removed. The cells were scraped down and used for either sub-culture or cell studies. The P388D1 cells were plated to 60 mm diameter tissue culture dishes at 1 × 106 cells/mL in RPMI-1640 with 1% L-glutamine and 10% lipoprotein deficient bovine serum (LPDS) (2 mL per dish), and were maintained at 37 °C in a humidified environment of 5% CO2 atmosphere overnight which allowed cells to adhere to the bottom of dishes. Three stock solutions of MDIO or DIO were prepared by dissolving an appropriate amount of nanoparticulate probes in RPMI-1640 with 1% L-glutamine and 10% LPDS. The stock solutions were diluted with the same media to give a series of three solutions with decreasing iron concentration. All solutions were prepared by weight and iron concentrations were calculated based on the concentration of the stock solution and appropriate dilution factors. After removal of maintenance media, the MDIO or DIO medium solutions (37 °C) were introduced to the cells and cells were incubated at 37 °C in 5% CO2 atmosphere for 2 h. After removal of the media, cells were washed with PBS (37 °C) (3 × 2 mL for each dish, ~ 2 min for each washing) and were in PBS for confocal imaging or for the MRI samples, deionized water was added in dishes (1.2 mL for each dish), and the freeze-thaw cycle was repeated twice to lyse cells. The cell lysates were put into 1.5 mL conical tubes and lyophilized. Deionized water was added to the residue (0.3 mL each) to generate solutions for relaxation time measurement or MR imaging [30].
Specificity of Uptake
The P388D1 cells were prepared following the procedure described in the section of “In vitro cellular uptake of MDIO and DIO”. The P388D1 cells were incubated with MDIO ([Fe] = 100 μM) in the presence of maleylated dextran, dextran sulfate, or dextran in 0, 0.02, 0.1, 0.4, 1, and 10-fold excess concentrations, respectively. Cells were incubated at 37 °C in a 5% CO2 atmosphere for 2 h and then prepared for relaxation time measurements.
Cytotoxicity
P388D1 cells in RPMI-1640 were plated in 96-well dishes at a concentration of 1.1 × 104 cells per well. After overnight incubation in 5% CO2 atmosphere at 37°C, the existing RPMI-1640 was replaced with fresh media containing varying amounts of MDIO. Cells were incubated with MDIO for either 4 or 24 h. The media were removed and cells were washed with PBS three times, then media containing C12-resazurin (5 μM) was added. After incubation at 37 °C in a 5% CO2 atmosphere for 15 minutes, fluorescence was measured using a Safire2 monochromator microplate reader (Tecan Austria G.M.B.H., Austria) with excitation of 563 nm and an emission of 587 nm. Linearity and LOD of the plate reader were checked using fluorescein. Samples were performed in triplicate to provide statistical significance.
Confocal Microscopy
P388D1 macrophages containing MDIO or DIO were imaged with a Zeiss LSM 5 Pascal confocal microscope equipped with a LD-neofluar 40×0.6 corr objective. An excitation wavelength of 543 nm (50% power) was used with a 560 nm low pass filter. A 1024 × 1024 matrix (230.3 × 230.3 μm2 in-plane resolution) and a scan speed of 2 (56.2 μs pixel time) were used.
In vitro MRI
MRI was performed on a Bruker Biospec 7T system (300 MHz, Bruker, Billerica, MA). The magnet was equipped with the standard gradient set (95 mTm−1 maximum gradient) and 72 mm internal diameter (ID) volume coil. Cell lysates of MDIO or DIO were prepared following literature methods [30]. Parameters were TR = 1,000 ms and TE = 40 ms for T2 weighted (T2W) images. For all images a spin echo sequence was used with a field view (FOV) of 3.44 × 3.06 cm2, slice thickness 2.0 mm, and a 128 × 128 matrix.
Results and discussion
Synthesis of 64Cu labeled MDIO
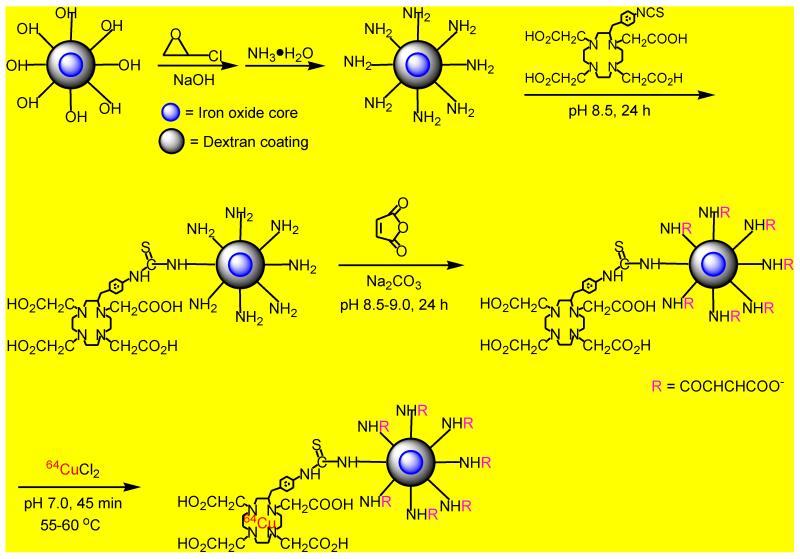
The synthetic route for 64Cu labeled MDIO is illustrated in Scheme 1. Dextran coated iron oxide nanoparticles (DIO) were synthesized as previously reported with slight modification [32]. The dextran coating of the nanoparticles was cross-linked and aminated to generate a stronger nucleophile, amino group functionalized DIO, for conjugation with the 64Cu chelator, p-SCN-Bn-DOTA [33]. The acylation reaction of DIO-DOTA by maleic anhydride was performed in pH 8.5 borate buffer solution. Because the electron-rich double bond in maleic anhydride hinders the nucleophilic attack of the amino group, and the reaction of maleic anhydride with amines is competitive with hydrolysis of maleic anhydride, particularly at high pH values, high concentration of maleic anhydride was used by addition of excess solid maleic anhydride in aliquots to push the acylation reaction forward, while the pH value of the system was kept at ~ 8.5 by addition of small amounts of solid sodium carbonate to neutralize large amount of acids generated during maleylation [35]. The isotope 64Cu has been used as a tracer in studies of multimodality PET/MR imaging of atherosclerosis because of its relatively long half-life (half life t½ = 12.7 h) [20, 29, 34], which allows repeated imaging over a few days. Copper is easily and stably coordinated to multi-dentate macrocyclic compounds such as DOTA (log KML = 22.3) [36]. Usually the chelation of 64Cu by DOTA derivatives is performed in pH 5.5 acetic acid-acetate buffer solution [10, 12, 20]. However, maleylation is a reversible reaction and maleyl groups could be removed by hydrolysis under acidic circumstances [35]. Therefore, to minimize the loss of macrophage targeting ligands caused by demaleylation, the chelation of 64Cu ions by MDIO-DOTA was run in a neutral (pH 7.0) buffer solution. Uncoordinated free 64Cu ions were extracted by applying excess ethylenediamine tetraacetic acid (EDTA). EDTA solution was added into the reaction vial after MDIO and cupric-64 chloride were mixed and incubated for 45 minutes at 55-60 °C. Then, the solution was incubated for 15 minutes at 55-60 °C. Because the coordination stability constant of DOTA-Cu2+ (log K = 22.3) is much higher than that of EDTA-Cu2+ ion (log K = 18.7) [36, 37], uncoordinated free 64Cu ions in the vial would easily coordinate with EDTA but the 64Cu ions of DOTA-64Cu chelates would rarely be pulled out by EDTA under the applied experimental conditions. Therefore, the desired dual-modality PET/MRI probe MDIO-64Cu-DOTA was obtained. After purification the radiolabeling yield was found to be 68%, and the specific activity of MDIO-64Cu-DOTA was calculated as 48 mCi/ug. The corresponding probe MDIO-Cu-DOTA, which contains non-radioactive copper, was also synthesized and used for probe characterization in solution and cell studies to reduce personnel exposure to radiation.
Scheme 1.
Synthesis of macrophage targeted, dual-modality PET/MRI probe MDIO-64Cu-DOTA.
MDIO formation verification
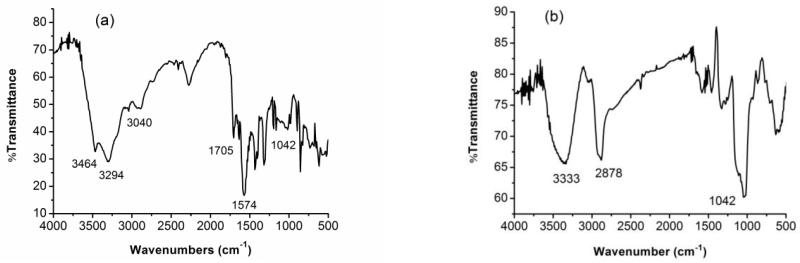
The formation of MDIO was confirmed by FT-IR spectroscopy. Compared to the IR spectrum of DIO (Fig. 1b), new absorptions at 3464, 3294 cm−1 (asymmetrical N–H stretching of secondary NH groups), 3040 cm−1 (asymmetrical =C–H stretching), 1705 cm−1 (asymmetrical C=O stretching), and 1574 cm−1 (asymmetrical –C=C– stretching) were observed, while the absorptions at 2878 cm−1 (dextran backbone methylene asymmetrical C–H stretch) and 1042 cm−1 (C–O stretch) were substantially decreased in the FT-IR spectrum of MDIO (Fig. 1a) [38, 39]. The decrease of iron content (from 17.53% in DIO to 10.48% in MDIO) in MDIO further verified successful formation of MDIO.
Fig. 1.
FT-IR spectra of (a) MDIO and (b) DIO.
MDIO size, surface charge, relaxivity, and emission
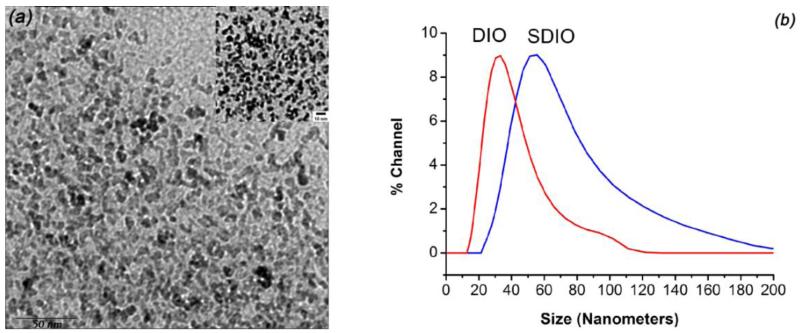
Fig. 2a shows a representative TEM image, which reveals the uniform cores in MDIO. The average core diameter (7-8 nm) and morphology of MDIO are similar to those of non-maleylated DIO [32], indicating that surface modification has little influence on the iron oxide core of the particles. However, the average hydrodynamic diameter in water increased from 38.1 nm for DIO to 62.7 nm for MDIO (Fig. 2b). MDIO has an r1 relaxivity of 16.8 mM−1·s−1 and an r2 relaxivity of 83.9 mM−1·s−1 (37 °C, 1.4 T) in pH 7.0 deionized water. The r2 to r1 ratio of MDIO was 5.0, indicating that MDIO could be used, preferably, as a T2-weighted MRI contrast agent [40]. The relaxivities of MDIO are similar to those of dextran sulfate coated IONPs (SDIO: r1 relaxivity 18.1 and r2 relaxivity 95.8 mM−1·s−1 (37 °C, 1.4 T)) we reported before [30]. This is not unexpected as the size of the iron oxide cores was similar for MDIO and SDIO. The properties of the MDIO, SDIO [30], and DIO are summarized in Table 1 to differentiate the three. The zeta potential (ζ) value of MDIO was – 37.65 mV in water, a 22.95 mV decrease in comparison with the ζ value of DIO measured under the same conditions (− 14.7 mV). The highly charged surface indicates that MDIO should have a stable dispersion and emulsification in water because the double layers with high ζ values would have powerful electrostatic repulsion and consequently prevent aggregation resulting from collisions caused by Brownian motion [41]. Repeat measurements on MDIO after one-year storage on the shelf at room temperature did not exhibit measurable changes in its properties.
Fig. 2.
(a) TEM (top-right corner inset: amplified TEM image (10 nm bar)) of MDIO, and (b) DLS of MDIO and DIO.
Table 1.
Properties of MDIO, SDIO, and DIO.
| Nanop- articles |
Iron Oxide Core size (nm) |
Coating Materials |
Average hydrodynamic diameter (nm) |
Relaxivity (mM−1s−1) (1.4 T, 37 °C) |
% iron | ζ (mV) | |
|---|---|---|---|---|---|---|---|
| r 1 | r 2 | ||||||
| MDIO | 7-8 | Maleylated | 62.7 | 16.8 | 83.9 | 10.48 | − 37.65 |
| Dextran | |||||||
| SDIO | 7-8 | Sufated | 62.4 | 18.1 | 95.8 | 11.34 | − 40.05 |
| Dextran | |||||||
| DIO | 7-8 | Dextran | 38.1 | 15.7 | 89.2 | 17.53 | − 14.70 |
SDIO data is from reference [30].
Stability of MDIO-64Cu-DOTA
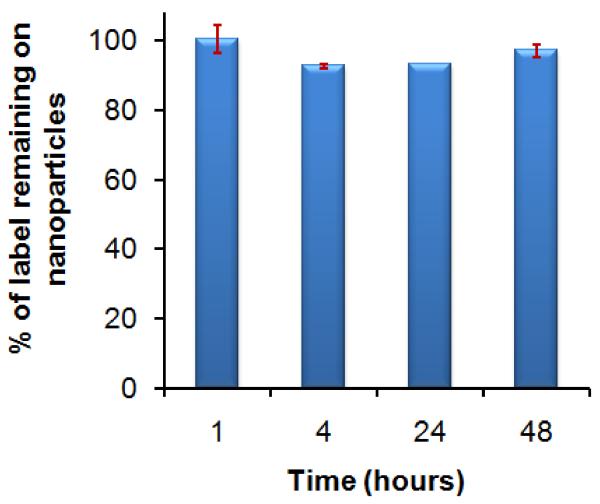
While it is necessary to employ a 64Cu chelate with high thermodynamic stability, kinetic inertness of the chelate is probably more important in determining long-term in vivo stability of the chelate. To confirm that 64Cu ions remain attached to the MDIO during imaging over the 48-h time period, the stability of MDIO-64Cu-DOTA was tested by subjecting MDIO-64Cu-DOTA to a competitive ligand exchange with EDTA, which is known to have a high affinity for Cu (log KCu-EDTA = 18.7) [37]. Results showed that over time, the MDIO-64Cu-DOTA at neutral pH did not lose radiolabel, confirming the exceptional stability of the chelate (Fig. 3).
Fig. 3.
Stability of MDIO-64Cu-DOTA in pH 7.0 trienthanol amine acetate buffer solution over the 48-h time period.
MDIO targeting to macrophages P388D1
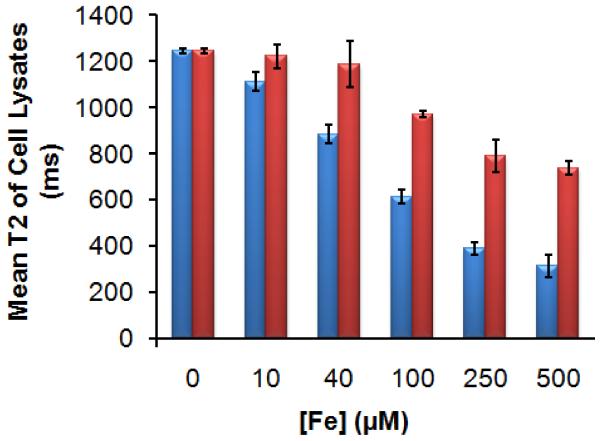
In vitro uptake of MDIO-Cu-DOTA by P388D1 murine macrophages was performed to test the targeting ability of MDIO to macrophages. As a control, we also evaluated uptake of untargeted DIO, whose properties are similar to the commercially available, ultrasmall superparamagnetic iron oxide nanoparticulate MRI contrast agent AMI-227 (Sinerem) [42], under the same conditions. The P388D1 cells were activated by maintaining the cells with RPMI-1640 media with 10% lipoprotein deficient bovine serum (LPDS) overnight. Fig. 4 presents the T2 values of the lysates of macrophages incubated with increasing concentrations of MDIO or DIO for 2 hours, and T2 values for these lysates in comparison with the lysates of macrophages that were not incubated with particles ([Fe] = 0). The mean T2 values for cell lysates incubated with MDIO were significantly (p < 0.05) shorter than the T2 decrease for DIO which was not as efficient and required higher concentrations to produce same effect as the MDIO, indicating that there was limited uptake of DIO (not a ligand of SR-A) compared to MDIO (a ligand of SR-A).
Fig. 4.
Mean T2 values of cell lysates incubated for 2 h with MDIO (blue, left column) or DIO (red, right column) of different iron concentrations.
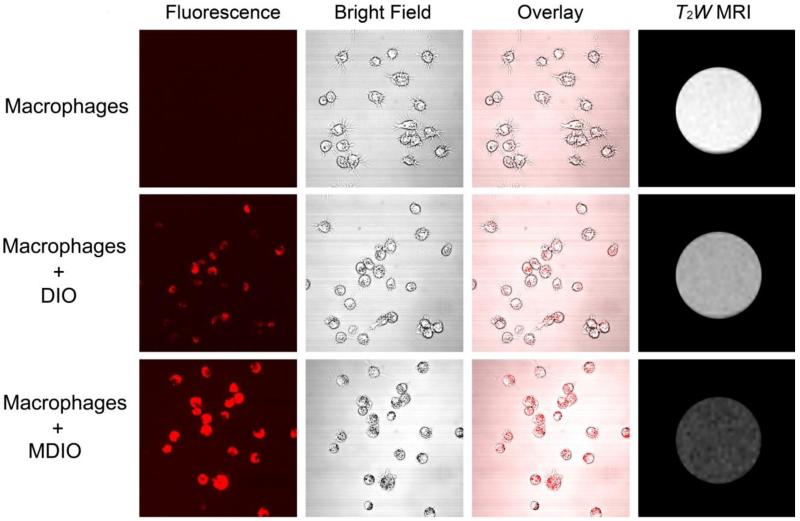
The different uptake of MDIO and DIO by macrophages was also demonstrated by results obtained from fluorescence and T2-weighted (T2W) MR imaging. A modified dye, 5(6)-TAMRA-SE, was added to the reaction system when conjugating DOTA to MDIO. We choose TAMRA as the fluorophore because it is nontoxic and offers a combination of desirable properties such as good photostability, high extinction coefficient, and high fluorescence quantum yield [43, 44]. The resultant fluorophore-labeled MDIO had a peak emission at 588 nm when excited at 541 nm, a 13 nm red-shift in comparison with that of 5(6)-TAMRA [45]. After 2-h incubation with MDIO, strong fluorescence is visible in the P388D1 cells, as shown in Fig. 5c. No fluorescence is observed from blank P388D1 cells (Fig. 5a), verifying that the fluorescence observed in Fig. 5c arises from the probes and not autofluorescence from the cells. Cells from a parallel experiment were lysed for T2W-MRI. The contrast between cell lysate containing MDIO and blank cell lysate is obvious, as shown in Fig. 5a and 5c. On the other hand, both fluorescence imaging and T2W MRI images (Fig. 5b) of P388D1 incubated with DIO show that there was limited uptake of DIO compared to the images of MDIO.
Fig. 5.
Confocal imaging and T2W MRI of blank P388D1 macrophage cells, DIO in P388D1 macrophage cells, and MDIO in P388D1 macrophage cells. ([Fe] = 2 × 10−4 M).
Receptor-mediated uptake of MDIO by macrophages P388D1
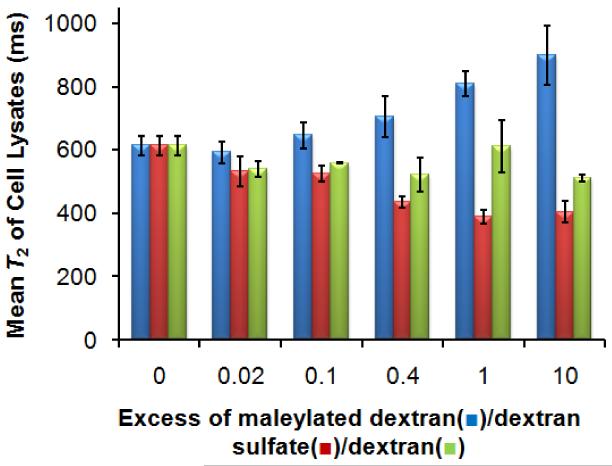
If a substrate’s uptake is receptor-mediated, excess unlabeled SR-A ligands (polyanionic macromolecules or particles) should compete for uptake. Nonspecific uptake is strictly concentration-dependent and will not be affected by additional ligands in the solution [34]. The specificity for the cellular uptake of MDIO was tested by competition studies in which cells were incubated with a fixed (iron) concentration of MDIO and increasing excess of maleylated dextran (a ligand of SR-A), dextran sulfate (a ligand of SR-A) or dextran (not a ligand of SR-A)[27, 28]. Increasing amounts of competitor maleylated dextran, from 0.02 to 10-fold excess, substantially reduced macrophage uptake of MDIO as evidenced by increased T2 values, while increasing amounts of dextran or dextran sulfate had little influence on the macrophage uptake of MDIO, as shown in Fig. 6. This suggests that there may be overlapping but different binding sites for SDIO and MDIO
Fig. 6.
Competitive uptake of MDIO ([Fe] = 100 μM) and maleylated dextran (blue, left column), dextran sulfate (red, middle), or dextran (yellow-green, right) by P388D1 macrophages.
Cytotoxicity of MDIO
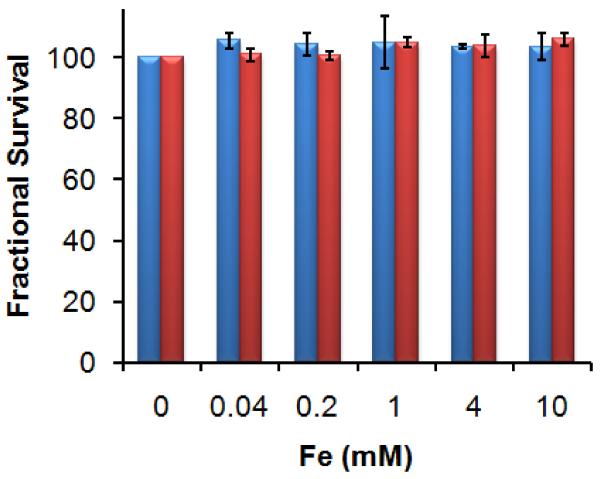
Dextran coated IONPs are classified as biocompatible and a few formulations have been approved for clinical MRI use by the US Food and Drug Administration (FDA) [40]. However, modulation of nanoparticle surfaces may influence particle uptake and biological responses. Therefore the biocompatibility of modified nanoparticles should be assessed even if the parent nanoparticles are nontoxic. C12-resazurin is a commonly used method for the assessment of a material’s toxicity to mammal cells that has shown good reliability. C12-resazurin is added to the system after the material has been incubated with cells for a certain period. Non-fluorescent C12-resazurin is reduced by viable cells to fluorescent C12-resorufin and the resultant signal intensity is proportional to the number of live cells present [46]. MDIO was applied to P388D1 macrophages in culture and the cell viability was evaluated by the C12-resazurin viability assay. Untreated cells served as negative control. The cell viability is above 96% after either 4 h or 24 h incubation with the nanoparticles varying from 0.04 to 10 mM iron, respectively (Fig. 7), indicating that MDIO is nontoxic to macrophages.
Fig. 7.
Cell viability of P388D1 macrophages after 4-h (blue, left column) or 24-h (red, right column) incubation with different iron concentrations of MDIO.
Multimodal probes, visible by different imaging modalities, can aid hybrid approaches for disease diagnosis. For example, a single probe visible by PET and MRI can be used to co-register images. The PET signal may be used to determine, in real time, the location of the probe and aid the selection of the relevant volume to interrogate by MRI. Several examples of 64Cu labeled IONPs for PET/MRI of VAP have been reported [12, 20, 30, 47-49]. In our previous study we have sulfated DIO, thereby targeting the nanoparticulate agent to SR-A of activated macrophages [30]. Co-registration of the PET signal with the MRI signal at 24 h post-injection of SDIO confirms that the PET signal correlates with the change in MR signal, verifying localization of the multimodal probe at the aortic valve [31]. The magnetic/radioactive properties and in vitro targeting behaviors of maleylated DIO are highly analogous to those of sulfated DIO, implying the two agents’ in vivo behaviors should be quite similar. This will be the topic of future study.
We demonstrated that both MDIO and SDIO are taken up by macrophages via SR-A. However, as shown in Table 2, the macrophage targeting ability of MDIO is somewhat weaker than our previously reported probe SDIO. In comparison with the lysates of blank macrophages, lysates of macrophages incubated with MDIO showed a steady decrease in T2 value from a low iron concentration (10 μM Fe, 10.8%) to a high iron concentration (500 uM, 74.8%). By comparison, a T2 value decrease of 80.2% was observed when macrophages were incubated with 100 uM Fe SDIO [30]. These results indicate that MDIO could be an efficient nanoparticulate agent to generate MRI image contrast provided that proper dosage is determined.
Table 2.
Mean T2 values of macrophage P388D1 cell lysates incubated for 2 h with MDIO or SDIO of different iron concentrations.
| [Fe] (μM) | 0 | 10 | 100 | 500 | |
|---|---|---|---|---|---|
| T2 (ms) | 1246 | 1112 | 615 | 315 | |
| MDIO | T2 decrease | 10.8% | 50.7% | 74.8% | |
| (p value) | (0.025) | (0.0003) | (0.0005) | ||
| T2 (ms) | 1246 | 675 | 246 | N/A | |
| SDIO | T2 decrease | 45.8% | 80.2% | ||
| (p value) | (0.002) | (0.0001) |
SDIO data is from reference [30].
Several factors may contribute to the uptake differences observed between the SDIO and MDIO. In competitive inhibition experiments, the SR-A ligand dextran sulfate effectively inhibited the macrophage uptake of SDIO, but barely inhibited the uptake of MDIO by macrophages. SR-A belongs to a growing family of transmembrane glycoproteins. There are three naturally occurring forms of SR-A, type I-III, which are alternative splice variants of the same gene found on chromosome 8p22 [50]. SR-A type I contains a C-terminal scavenger receptor cysteine-rich domain of 110 amino acids, while type II and type III SR-A express a short C-terminus or truncated cysteine-rich domain, respectively. The specific ligand-binding activities usually occur on type I and type II SR-A because SR-A type III is trapped in the endoplasmic reticulum. Although SR-A was originally identified through its specific recognition of modified LDL, it is now known to be able to interact with many diverse ligands containing repetitive anionic charge distribution. SR-A recognizes and binds these polyanionic ligands via a positively charged stretch of lysines in the collagenous binding domain of the receptor [25], therefore, the binding of MDIO and SDIO may occur either at different amino acid segment of the same type of SR-A, or on the different type of SR-A [25]. This is consistent with what is known about ligands for SR-A, which exhibit nonreciprocal cross-competition for some ligands [51]. The detailed binding mechanisms may be complex and the exploration is ongoing.
Macrophages are present through all stages of atherosclerosis development, and high densities of macrophages have been associated with plaque instability. This makes them attractive markers for targeting diagnostics and therapeutics [52, 53]. Antibodies and peptides showing high affinity to macrophage scavenger receptors have been attached to imaging probes for targeted imaging of macrophages [54-56]. Antibodies have been the common targeting molecules for decades [57]. Although current development of monoclonal antibodies has been focused on humanized derivatives to decrease their immunogenicity, these large, complex molecules require significant engineering at the molecular level to be effective. The variation from batch to batch further restricts their efficiency as targeting molecules. Peptides can bind targets with high sensitivity and specificity. Compared to antibodies, peptides are smaller, less immunogenic, more stable (without batch-to-batch variations), and easier to manufacture, but they have low serum stability [58].
Due to their small size and low cost of production, small molecules have emerged as a new class of targeting molecules and they have shown comparable targeting ability and efficiency to traditional targeting ligands [59]. For example, folic acid (folate) has a high-affinity for tumors because folate receptors are frequently over-expressed on tumor cells [59]. A variety of folate-decorated nanoparticles have been used for imaging and therapy of cancers. We have modified dextran coated IO NPs with negatively charged small molecules such as maleate and sulfate, to place repetitive anionic charge distribution on the probes’ surface. Results showed that the SR-A mediated uptake of maleate and sulfate modified probes is much more efficient than non-modified analogs, indicating that although maleate and sulfate themselves are not ligands of SR-A, suitable arrangement of them on a nanoparticulate (or a macromolecular) probe could make the probe specifically recognized and effectively taken up by SR-A.
As a research tool, the ability to image macrophages in vivo provides a valuable biomarker to assess plaque stability. The ability to evaluate plaque vulnerability would be a new diagnostic tool for clinicians. How this might be used to inform patient management is a matter still under consideration. Detection of vulnerability could precipitate prescription of systemic treatment with anti-inflammatories. For example the more complex question is whether to intervene with more invasive procedures, should a clinician stent or otherwise treat a plaque that has not yet ruptured but appears to be very vulnerable? Whether to take action on an asymptomatic patient is a question of some debate. Observing inflammatory changes over time has the potential help answer this question by allowing clinicians to better understand the natural history of plaque progression and regression. It has been recognized that plaque progression involves repeated cycles of inflammation and remodeling wherein there may be many ruptures over a period of time before sudden death occurs [60]. However, there is not yet a good mechanism to observe or predict these cycles. Non-invasive imaging provides a powerful tool to observe macrophage burden and plaque size, to help understand plaque progression as well as response to therapies.
Conclusions
Current advances in multimodality imaging allow researchers to gather in vivo anatomic and molecular information from multiple perspectives, providing attractive and practical tools for early and accurate diagnosis of disease. We have synthesized 64Cu labeled IONPs and functionalized the nanoparticulate probe with maleic anhydride, thus targeting the dual-modal PET/MRI probe to SR-A of macrophages. Macrophages are key components in the formation and development of vulnerable atherosclerotic plaques (VAP) whose detection and identification is still a challenge by conventional means. Our results indicate that small molecules could be an effective alternative to target nanoparticulate probes to biomarkers of VAP. This could potentially result in the development of new multi-modal imaging probes with small molecules as ligands targeting to plaque biomarkers, thus may open a new methodology to synthesize targeted and hybrid imaging probes.
Acknowledgements
The authors wish to acknowledge the National Institute of Health (EB008576-01 and EB000993), the Center for Molecular and Genomic Imaging at UC Davis (U24 CA 110804), and the NMR award of the University of California, Davis for support of this work. We thank Dr. Jeongchan Park, Dr. Jai Woong Seo, and Mr. Ray Wong for help in TEM imaging, zeta potential, and IR spectroscopy, respectively.
References
- 1.Fuster V, Lois F, Franco M. Nat. Rev. Cardiol. 2010;7:327–333. doi: 10.1038/nrcardio.2010.54. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Cormode DP, Fayad ZA, Mulder WJM. Wiley Interdiscip. Rev.-Nanomed. Nanobiotechnol. 2011;3:146–161. doi: 10.1002/wnan.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer T, Pichler B. European Journal of Nuclear Medicine and Molecular Imaging. 2009;36:1–2. doi: 10.1007/s00259-008-1041-5. [DOI] [PubMed] [Google Scholar]
- 4.Cherry SR, Louie AY, Jacobs RE. Proceedings of the IEEE. 2008;96:416–438. [Google Scholar]
- 5.Marti-Bonmati L, Sopena R, Bartumeus P, Sopena P. Contrast Media Mol. Imaging. 2010;5:180–189. doi: 10.1002/cmmi.393. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi H, Del Guerra A. Med. Phys. 2011;38:5667–5689. doi: 10.1118/1.3633909. [DOI] [PubMed] [Google Scholar]
- 7.Sauter AW, Wehrl HF, Kolb A, Judenhofer MS, Pichler BJ. Trends Mol. Med. 2010;16:508–515. doi: 10.1016/j.molmed.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Ng TSC, Procissi D, Wu Y, Jacobs RE. Med. Phys. 2010;37:1995–2003. doi: 10.1118/1.3369447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louie A. Chem. Rev. 2010;110:3146–3195. doi: 10.1021/cr9003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu CQ, Ma XC, House A, Kauzlarich SM, Louie AY. ACS Med. Chem. Lett. 2011;2:285–288. doi: 10.1021/ml1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tassa C, Shaw SY, Weissleder R. Accounts Chem. Res. 2011;44:842–852. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie J, Chen K, Huang J, Lee S, Wang JH, Gao J, Li XG, Chen XY. Biomaterials. 2010;31:3016–3022. doi: 10.1016/j.biomaterials.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Rosales RTM, Tavare R, Glaria A, Varma G, Protti A, Blower PJ. Bioconjugate Chemistry. 2011;22:455–465. doi: 10.1021/bc100483k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XQ, Hong H, Grailer JJ, Rowland IJ, Javadi A, Hurley SA, Xiao YL, Yang YA, Zhang Y, Nickles R, Cai WB, Steeber DA, Gong SQ. Biomaterials. 2011;32:4151–4160. doi: 10.1016/j.biomaterials.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Welch MJ. Bioconjugate Chemistry. 2012;23:671–682. doi: 10.1021/bc200264c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang TY, Muller KH, Graves MJ, Li ZY, Walsh SR, Young V, Sadat U, Howarth SPS, Gillard JH. Arterioscler. Thromb. Vasc. Biol. 2009;29:1001–1008. doi: 10.1161/ATVBAHA.108.165514. [DOI] [PubMed] [Google Scholar]
- 17.Woollard KJ, Geissmann F. Nat. Rev. Cardiol. 2010;7:77–86. doi: 10.1038/nrcardio.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libby P, DiCarli M, Weissleder R. J. Nucl. Med. 2010;51:33S–37S. doi: 10.2967/jnumed.109.069633. [DOI] [PubMed] [Google Scholar]
- 19.Moore KJ, Tabas I. Cell. 2011;145:341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nahrendorf M, Zhang HW, Hembrador S, Panizzi P, Sosnovik DE, Aikawa E, Libby P, Swirski FK, Weissleder R. Circulation. 2008;117:379–387. doi: 10.1161/CIRCULATIONAHA.107.741181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadat U, Li ZY, Graves MJ, Tang TY, Gillard JH. Nat. Clin. Pract. Cardiovasc. Med. 2009;6:200–209. doi: 10.1038/ncpcardio1455. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein JL, Ho YK, Basu SK, Brown MS. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MS, Goldstein JL, Krieger M, Ho YK, Anderson RGW. Journal of Cell Biology. 1979;82:597–613. doi: 10.1083/jcb.82.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu CQ, Ma XC, Pantazis P, Kauzlarich SM, Louie AY. J. Am. Chem. Soc. 2010;132:2016–2023. doi: 10.1021/ja909303g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neyen C, Pluddemann A, Roversi P, Thomas B, Cai L, van der Westhuyzen DR, Sim RB, Gordon S. Biochemistry. 2009;48:11858–11871. doi: 10.1021/bi9013769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi T, Higashino K, Kurihara Y, Wada Y, Miyazaki T, Nakamura H, Uesugi S, Imanishi T, Kawabe Y, Itakura H, Yazaki Y, Matsumoto A, Kodama T. J. Biol. Chem. 1993;268:2126–2133. [PubMed] [Google Scholar]
- 27.Greaves DR, Gordon S. J. Lipid Res. 2009;50:S282–S286. doi: 10.1194/jlr.R800066-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Hamblin MR. Int. J. Immunopathol. Pharmacol. 2005;18:391–402. doi: 10.1177/039463200501800301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarrett BR, Gustafsson B, Kukis DL, Louie AY. Bioconjugate Chemistry. 2008;19:1496–1504. doi: 10.1021/bc800108v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu C, Ng TSC, Sohi HK, Palko HA, House A, Jacobs RE, Louie AY. Biomaterials. 2011;32:7209–7216. doi: 10.1016/j.biomaterials.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarrett BR, Correa C, Ma KL, Louie AY. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013254. Article Number: e13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarrett BR, Frendo M, Vogan J, Louie AY. Nanotechnology. 2007;18 doi: 10.1088/0957-4484/18/3/035603. Article Number: 035603. [DOI] [PubMed] [Google Scholar]
- 33.Wunderbaldinger P, Josephson L, Weissleder R. Academic Radiology. 2002;9:S304–S306. doi: 10.1016/s1076-6332(03)80210-6. [DOI] [PubMed] [Google Scholar]
- 34.Gustafsson B, Youens S, Louie AY. Bioconjugate Chemistry. 2006;17:538–547. doi: 10.1021/bc060018k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feeney RE, Yamasaki RB, Geoghegan KF. Advances in Chemistry Series. 1982:3–55. [Google Scholar]
- 36.Anderegg G, Arnaud-Neu F, Delgado R, Felcman J, Popov K. Pure Appl. Chem. 2005;77:1445–1495. [Google Scholar]
- 37.Jones-Wilson TM, Deal KA, Anderson CJ, McCarthy DW, Kovacs Z, Motekaitis RJ, Sherry AD, Martell AE, Welch MJ. Nuclear Medicine and Biology. 1998;25:523–530. doi: 10.1016/s0969-8051(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 38.Hargreaves MK, Stevinson EA. Spectrochimica Acta. 1964;20:317–324. [Google Scholar]
- 39.Predoi D. Dig. J. Nanomater. Biostruct. 2007;2:169–173. [Google Scholar]
- 40.Tu CQ, Louie AY. WIREs Nanomed. Nanobiotechnol. 2012 doi: 10.1002/wnan.1170. doi: 10.1002/wnan.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obeidat WM, Schwabe K, Muller RH, Keck CM. Eur. J. Pharm. Biopharm. 2010;76:56–67. doi: 10.1016/j.ejpb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Gossuin Y, Gillis P, Hocq A, Vuong QL, Roch A. Wiley Interdiscip. Rev.-Nanomed. Nanobiotechnol. 2009;1:299–310. doi: 10.1002/wnan.36. [DOI] [PubMed] [Google Scholar]
- 43.Alford R, Simpson HM, Duberman J, Hill GC, Ogawa M, Regino C, Kobayashi H, Choyke PL. Molecular Imaging. 2009;8:341–354. [PubMed] [Google Scholar]
- 44.Longmire MR, Ogawa M, Hama Y, Kosaka N, Regino CAS, Choyke PL, Kobayashi H. Bioconjugate Chem. 2008:19. doi: 10.1021/bc800140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christie RJ, Tadiello CJ, Chamberlain LM, Grainger DW. Bioconjugate Chemistry. 2009;20:476–480. doi: 10.1021/bc800367e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Brien J, Wilson I, Orton T, Pognan F. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 47.de Rosales RTM, Tavare R, Paul RL, Jauregui-Osoro M, Protti A, Glaria A, Varma G, Szanda I, Blower PJ. Angew. Chem.-Int. Edit. 2011;50:5509–5513. doi: 10.1002/anie.201007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barreto JA, Matterna M, Graham B, Stephan H, Spiccia L. New J. Chem. 2011;35:2705–2712. [Google Scholar]
- 49.Glaus C, Rossin R, Welch MJ, Bao G. Bioconjugate Chemistry. 2010;21:715–722. doi: 10.1021/bc900511j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greaves DR, Gordon S. J. Lipid Res. 2005;46:11–20. doi: 10.1194/jlr.R400011-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Platt N, Gordon S. J. Clin. Invest. 2001;108:649–654. doi: 10.1172/JCI13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osborn EA, Jaffer FA. Curr. Opin. Cardiol. 2008;23:620–628. doi: 10.1097/HCO.0b013e328310fc7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw SY. Nat. Rev. Cardiol. 2009;6:569–579. doi: 10.1038/nrcardio.2009.119. [DOI] [PubMed] [Google Scholar]
- 54.Amirbekian V, Lipinski MJ, Briley-Saebo KC, Amirbekian S, Aguinaldo JGS, Weinreb DB, Vucic E, Frias JC, Hyafil F, Mani V, Fisher EA, Fayad ZA. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:961–966. doi: 10.1073/pnas.0606281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamzah J, Kotamraju VR, Seo JW, Agemy L, Fogal V, Mahakian LM, Peters D, Roth L, Gagnon MKJ, Ferrara KW, Ruoslahti E. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7154–7159. doi: 10.1073/pnas.1104540108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchida M, Kosuge H, Terashima M, Willits DA, Liepold LO, Young MJ, McConnell MV, Douglas T. ACS Nano. 2011;5:2493–2502. doi: 10.1021/nn102863y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choudhury RP, Fisher EA. Arterioscler. Thromb. Vasc. Biol. 2009;29:983–991. doi: 10.1161/ATVBAHA.108.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang AZ, Gu F, Zhang LF, Chan JM, Radovic-Moreno A, Shaikh MR, Farokhzad OC. Expert Opin. Biol. Ther. 2008;8:1063–1070. doi: 10.1517/14712598.8.8.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Talekar M, Kendall J, Denny W, Garg S. Anti-Cancer Drugs. 2011;22:949–962. doi: 10.1097/CAD.0b013e32834a4554. [DOI] [PubMed] [Google Scholar]
- 60.Finn AV, Nakano M, Narula J, Kolodgie FD, Virmani R. Arterioscler. Thromb. Vasc. Biol. 2010;30:1282–1292. doi: 10.1161/ATVBAHA.108.179739. [DOI] [PubMed] [Google Scholar]