Abstract
Stimuli-responsive nanomaterials are increasingly important in a variety of applications such as biosensing, molecular imaging, drug delivery and tissue engineering. For cancer detection, a paramount challenge still exists in search of methods that can illuminate tumors universally regardless of their genotypes and phenotypes. Here we capitalized on the acidic, angiogenic tumor microenvironment to achieve broad detection of tumor tissues in a wide variety of mouse cancer models. This was accomplished using ultra-pH sensitive fluorescent nanoprobes that have tunable, exponential fluorescence activation upon encountering subtle, physiologically relevant pH transitions. These nanoprobes were silent in the circulation, then dramatically activated (>300 fold) in response to neovasculature or to the low extracellular pH in tumors. Thus, we have established non-toxic, fluorescent nanoreporters that can non-linearly amplify tumor microenvironmental signals, permitting identification of tumor tissue independently of histological type or driver mutation, and detection of acute treatment responses much more rapidly than conventional imaging approaches.
Responsive polymer materials are of great interest and importance in a variety of optical, electrical, thermal and mechanical systems in a wide range of applications such as sensing, adaptable surface adhesion, self-healing and drug delivery1,2. In biology and medicine, high performance and bioresponsive materials that can respond and furthermore, amplify patho-physiological signals, have shown great promise to differentiate diseased and healthy tissues, a major challenge in any diagnostic or therapeutic applications3,4. In tumor visualization, a variety of nanomaterials have been reported with functionalities in fluorescence, Raman, magnetic resonance imaging, and photoacoustics5–9. Compared to small molecular tracers, one of the major advantages of nanoprobes is the ultra-sensitive detection at nM-pM particle concentrations7,8. While this increased sensitivity improves the physical detection limit, achieving high biological specificity to differentiate tumors from normal tissues remains a significant challenge. Many current cancer imaging agents target cancer cell-specific biomarkers such as Her2/neu, EGFR, and folate receptors to achieve specificity10–12. While successful imaging outcomes are reported to stratify patients toward personalized therapy, broad tumor applicability in a wide range of cancers is often not possible as cancer cell-specific biomarkers are frequently expressed in only a subset of patient (for example, <25% of breast cancer patients have Her2/neu expression)13,14. In addition, antibody-dye conjugates require long time clearance (e.g. >24 h) due to the persisted blood circulation of humanized antibody and high blood background from the always-ON mode of probe design.
In this study, we report a non-linear signal amplification strategy to greatly increase the detection accuracy of patho-physiological signals of tumor microenvironment to achieve a broad specificity of tumor visualization (Fig. 1). We chose two established tumor microenvironment signals, namely angiogenic tumor vasculature15,16 and low extracellular pH (pHe)17, to demonstrate the proof of principle. Tumor angiogenesis and aerobic glycolysis (aka Warburg effect) are recognized hallmarks of cancer, which are ubiquitous in solid tumors, regardless of cancer types.
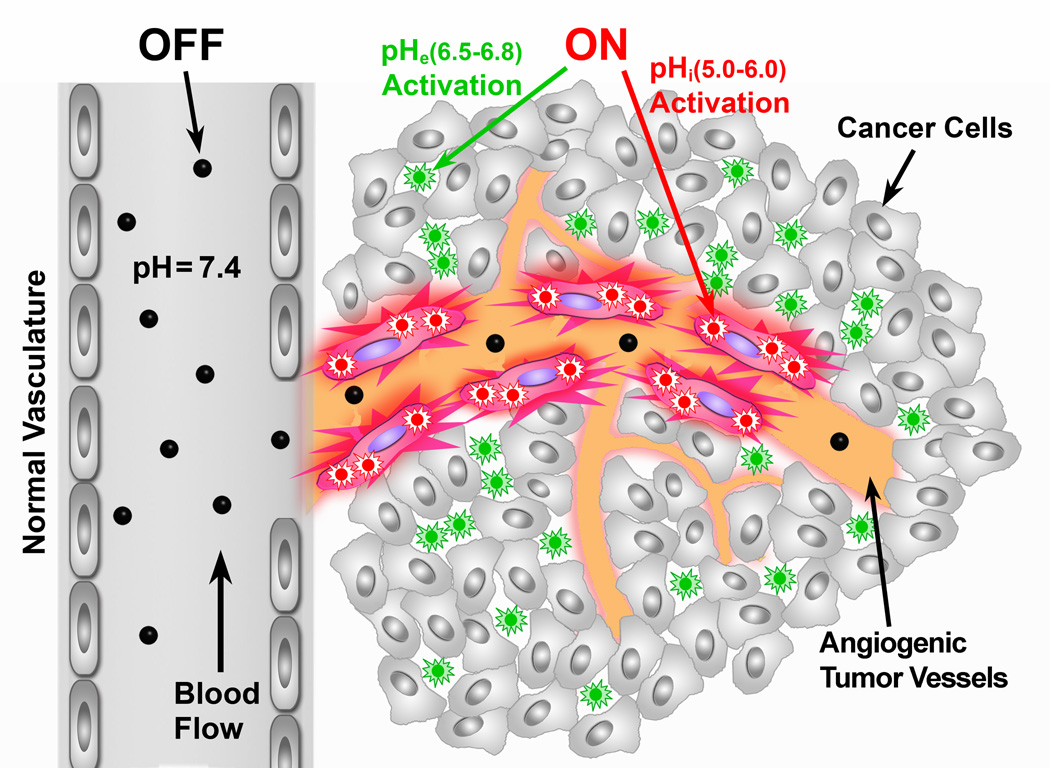
Figure 1. Schematic of imaging tumor microenvironment by ultra-pH sensitive (UPS) nanoprobes.
The UPS nanoprobes stay ‘OFF’ at pH 7.4 during blood circulation. After reaching tumors, the UPS nanoprobes are turned ON by acidic extracellular pHe (6.5–6.8) in the tumor milieu, or endocytic organelles (pHi, 5.0–6.0) in the tumor endothelial cells after receptor-mediated endocytosis.
To accomplish this goal, we established a series of ultra-pH sensitive (UPS) nanoprobes to specifically image the tumor extracellular milieu and angiogenic tumor vessels. The UPS platform is comprised of three independently controlled functional components: (1) an ultra pH-sensitive core that renders a tunable sharp pH response (ΔpHON/OFF<0.2518, as compared to 2 pH for small molecular pH sensors). This unique hydrophobic micellization-induced nanoscale phenomenon is essential for imaging acidic tumor pHe (6.5–6.8)17, which is not drastically different from blood pH (7.4). Many previously reported pH-sensitive nanosystems don’t have sharp response in this pH span and in many cases, take long time (e.g. 24 h) to respond19–23. (2) A series of fluorophores (e.g. TMR and Cy family dyes) with a large emission range from green to near IR (500–820 nm). HomoFRET-induced fluorescence quenching results in large fluorescence activation, crucial for suppressing blood signals and for achieving non-linear amplification of signals in the tumor. The multicolored design also allows simultaneous imaging of multiple tumor targets in space and time. (3) A targeting unit (e.g. cRGD), which binds to cell surface receptors and internalizes nanoprobes to allow signal amplification in acidic endocytic organelles. Our current UPS nanoprobes carry ~1,600 dye molecules per micelle24. Assuming it takes 10 αvβ3 integrins to internalize one micelle, this represents >100-fold dye payload amplification on a per αvβ3 basis. Consequently, we demonstrate broad tumor specificity with large tumor-to-blood ratio (>300 fold) in a diverse set of animal tumor models with different cancer types and organ sites. Tumor-specific imaging was accomplished in the first hour after intravenous nanoprobe injection and in tumors as small as 1 mm3. These capabilities, together with the broad cancer specificity, make the current strategy particularly powerful in image-guided resection of tumors and post-therapy monitoring of drug efficacy.
We first performed a series of experiments to systematically investigate the influence of pH-sensitive segment and dye content on the performance of the nanoprobes (detailed results are summarized in Supplementary Tables S1–3 and Fig. S1). Based on these data, we selected UPS compositions that meet the following criteria: sharp pH transition (ΔpH10–90% < 0.25), large fluorescence ON/OFF ratio, high reproducibility, low critical micelle concentration, relatively small particle size (<30 nm) to allow tumor tissue penetration, and optimal pH transitions.
To distinguish the small pH differences between acidic tumor pHe (6.5–6.8)17 and blood (7.4), we synthesized a UPSe nanoprobe from the poly(ethylene glycol)-b-poly(2-(hexamethyleneimino)ethyl methacrylate) copolymer (Fig. 2a, Table S4). A near-infrared dye, Cy5.5 (λex/λem=675/710 nm), was conjugated to the ionizable block of the copolymer (Scheme S1). The UPSe nanoprobe had a pH transition at 6.9 and sharp pH response (ΔpH10–90%, the pH difference between 10% to 90% fluorescence activation was 0.23). The fluorescence activation ratio (RF) was 102-fold between pH 6.7 and 7.4. In contrast, theoretical calculation based on the Henderson-Hasselbach equation for a small molecular pH sensor (pKa = 6.9) yielded only 2.6-fold fluorescence increase in this pH range (Fig. 2b). At blood pH, UPSe nanoprobes were present as self-assembled micelles with a diameter of 25.3±1.5 nm and a spherical morphology (left panel, Fig. 2d). HomoFRET-induced fluorescence quenching was responsible for complete silencing of the fluorophores in the micelle state (Fig. 2c)24. Micelle dissociation at acidic pHe (right panel, Fig. 2d) resulted in dramatic increase in fluorescence signals (Fig. 2c, e).
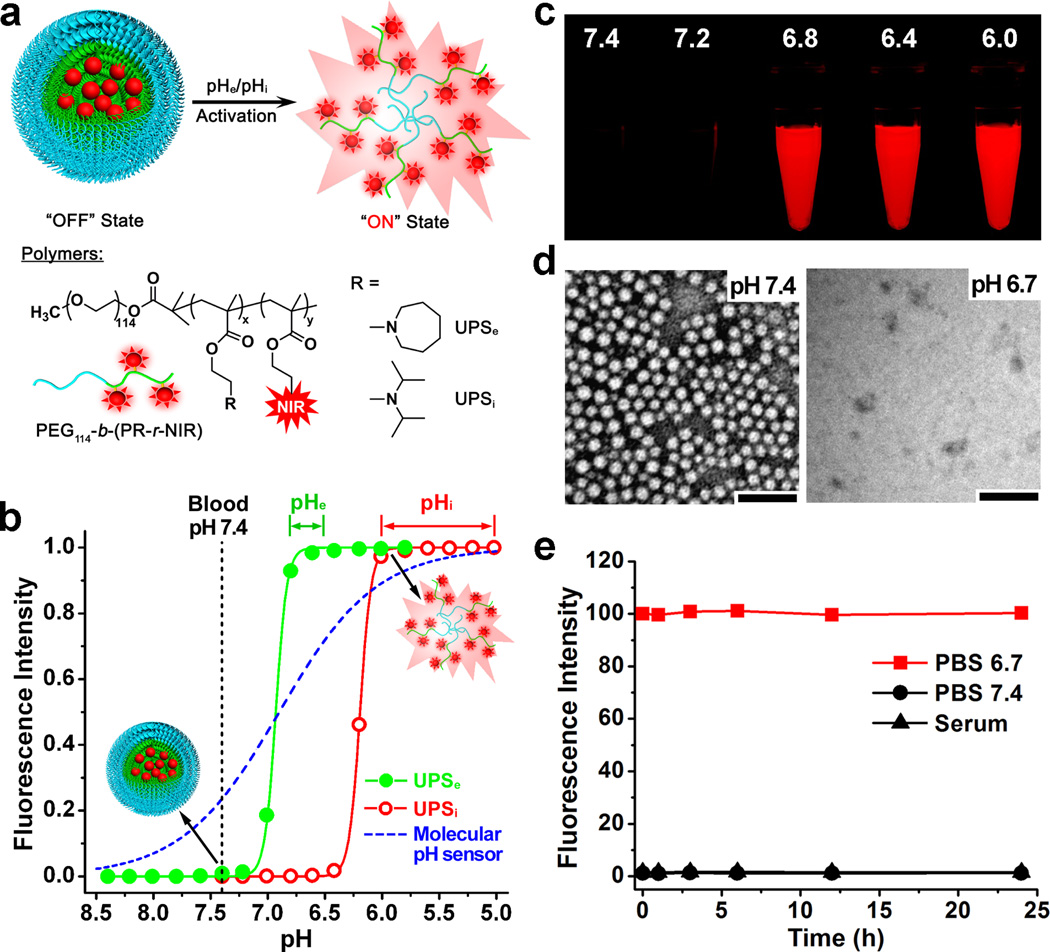
Figure 2. Syntheses and characterization of UPS nanoprobes.
a, Structural composition of two types of nanoprobes, UPSe and UPSi, with pH transitions at 6.9 and 6.2, respectively. The UPSe is specifically designed to activate in acidic tumor extracellular fluid (pHe = 6.5–6.8). The UPSi can be activated inside acidic endocytic organelles (e.g. pHi = 5.0–6.0). Cy5.5 is used as the NIR fluorophore in most of the animal studies. b, Normalized fluorescence intensity as a function of pH for UPSe and UPSi nanoprobes. At high pH (e.g. 7.4), both probes stay silent. At pH below their transitions (i.e. 6.9 and 6.2), the nanoprobes can be activated as a result of micelle dissociation. The blue dash-line simulates the pH response of a small molecular pH sensor with a pKa of 6.9 based on Henderson-Hasselbach equation. For UPS, the pH response (ΔpH10–90%) is extremely sharp (<0.25 pH unit between ON/OFF states) with >100-fold signal amplification. In contrast, small molecular pH sensors require 3 pH units for comparable signal change. c, Fluorescent images of UPSe-Cy5.5 nanoprobe solution in different pH buffers (λex/λem=675/710 nm). d, Transmission electron micrographs of UPSe nanoprobes at pH 7.4 and 6.7 (polymer concentration = 1 mg/mL, scale bar = 100 nm). e, UPSe nanoprobes remain stable in fresh mouse serum over 24 h at 37 °C.
To achieve selective activation in the acidic endocytic organelles (e.g. endosomes and lysosomes, pH = 5.0–6.0), we established a UPSi nanoprobe from the poly(ethylene glycol)-b-poly(2-(diisopropyl amino)ethyl methacrylate) copolymer (Fig. 2a). For imaging of αvβ3-expressing angiogenic tumor endothelial cells, we functionalized the UPSi surface with 10% cRGDfK (cRGD) peptide through thiol-maleimide linkage (Scheme S2 and Fig. S2). The cRGD-UPSi nanoprobe had a pH transition at 6.2 with ΔpH10–90% value of 0.21. The fluorescence ON/OFF activation ratio was 128-fold (Table S4). The cRGD-encoded UPSi nanoprobes (24.5±1.1 nm) were stable at blood pH and acidic tumor pHe, but can be selectively activated inside the lysosomes of tumor endothelial cells upon receptor-mediated endocytosis (see data below). Both UPSe and UPSi nanoprobes were stable in freshly prepared mouse serum as indicated by the negligible change in fluorescence intensity over 24 h incubation (Fig. 2e and Fig. S2e).
To investigate whether UPS dilution (e.g. in blood after injection) affects the pH response, we examined the concentration-dependence of UPSe/UPSi properties (Fig. S3). For UPSe and UPSi, the plasma concentrations 24 h after i.v. injection are approximately 15 and 100 µg/mL, respectively (calculated from the 10 mg/kg injection dose and micelle pharmacokinetics). Data show that fluorescence intensity in the ON state (lower pH) decreased at lower probe concentrations as expected; however, the RF values remained high (>60-fold) even at 10 µg/mL. Normalization of fluorescence signals showed superimposable, sharp pH transitions, indicating high fidelity of UPSi/UPSe in the physiologically relevant concentration range.
A 3-D plot of fluorescence intensity vs. probe concentration and pH illustrates the pH-modulated signal amplification/suppression strategy orthogonal to probe concentration (Fig. S4). Along the concentration axis, higher probe concentration results in larger fluorescence signals (tumor accumulation is still a prerequisite of achieving UPS signals). Along the pH axis, homoFRET-induced quenching abolished fluorescence signals at the normal tissue or blood pH (7.2–7.4). Upon reaching the targeted pH environments, UPS probes are activated, leading to exponentially increased signals.
To investigate the specificity of UPSe nanoprobes for pHe imaging, we first evaluated two inhibitors of tumor glycolysis and examined their effects on extracellular pH in cell culture. The first agent, 2-deoxy-D-glucose (2-DG), competitively inhibits glucose uptake through cell surface glucose transporters (GLUT) and subsequent phosphorylation by hexokinases; the second agent, α-cyano-4-hydroxycinnamate (CHC), is a suicide inhibitor of monocarboxylate transporter (MCT) that prevents the secretion of lactic acid from cancer cells (Fig. 3a)25. In vitro cell culture experiments show that both agents significantly decreased lactate secretion in A549 lung cancer cells (Fig. 3b). Moreover, we also observed that 2-DG and CHC treatment retarded the acidification of cell culture medium (ΔpH was 0.41, 0.17, 0.16 for vehicle, 2-DG, CHC, respectively. Fig. 3c).
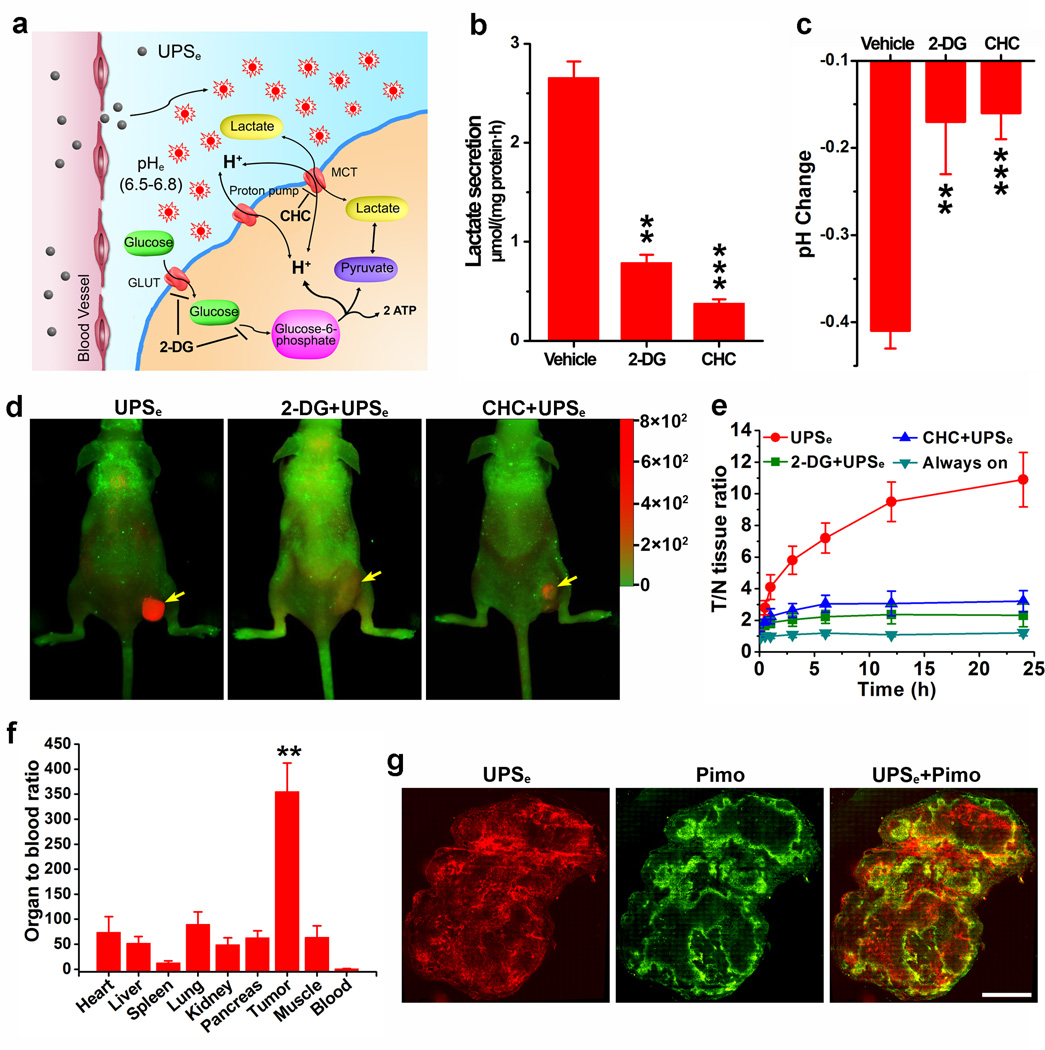
Figure 3. UPSe nanoprobes can specifically image acidic tumor pHe.
a, Aerobic glycolysis converts glucose to lactate in cancer cells. 2-DG and CHC are metabolic inhibitors for glucose uptake and lactic acid secretion, respectively. b, Effect of 2-DG or CHC on the rate of lactic acid secretion in A549 cells. c, Acidification of A549 cell culture medium in the presence of 2-DG or CHC after 6 h incubation. *P < 0.05, **P < 0.01, ***P < 0.001, compared with vehicle group. d, Overlaid fluorescent images of A549 tumor-bearing mice at 24 h postinjection of UPSe nanoprobes (10 mg/kg). In the control groups, 2-DG (250 mg/kg) or CHC (250 mg/kg) was injected 12 h before UPSe nanoprobe administration. Cy5.5 (red) and autofluorescence (green) are separately shown in the composite images. Yellow arrows indicate the tumor location. e, NIR fluorescence intensity ratio between tumor and normal tissues (T/N ratio) as a function of time after UPSe injection. Data are presented as mean ± s.d. (n = 4). f, Organ to blood ratios (see data in Table S5) 24 h post-injection of UPSe (n = 4). A549 tumor has 355-fold of signal amplification over blood by UPSe. **P < 0.01, compared with other organs. g, Hypoxia bands qualitatively correlate with activation pattern of UPSe in A549 tumor xenograft. Whole mount images of tumor slices stained for hypoxia (green). All images were obtained from the adjacent sections at ×200 magnification. Scale bar is 2 mm.
For in vivo tumor imaging studies, UPSe nanoprobes (10 mg/kg) were injected intravenously in mice bearing subcutaneous A549 lung cancer xenografts (n=4 for each group). For glycolysis inhibition control, 2-DG or CHC (250 mg/kg) was injected 12 h before the UPSe administration. An always-ON nanoprobe control with the same particle size and core composition, but no pH-sensitive fluorescence response was also used (see Fig. S5 for detailed description). As early as the first hour after UPSe injection, a significant tumor contrast (tumor/normal tissue ratio ~4, P<0.05 compared to the three other controls) was observed (Fig. 3e). Over 24 h, fluorescence signals in the UPSe group increased considerably while the signals from control groups remained relatively the same. The NIR fluorescence intensity in the tumor increased 10.9 ± 1.7 fold from 5 min to 24 h. The always-ON nanoprobe control showed highly elevated fluorescence background in normal tissues and low tumor/tissue contrast (<1.2-fold over 24 h, Fig. S6), which suggest that EPR effect26 alone is not sufficient to yield high tumor contrast. Pretreatment with metabolic inhibitors resulted in significant signal decrease in tumors compared to UPSe alone (e.g. 2.3 ± 0.7 fold decrease for 2-DG (P = 0.004), and 3.2 ± 0.6 fold for CHC (P = 0.007)). After 24 h, mice were sacrificed and the excised organs were imaged (Fig. S7). The fluorescence signal of each organ/tissue was normalized over nanoprobe dose in the corresponding tissue as measured by 3H-labelled nanoparticles. The value was further normalized to blood to yield the organ to blood ratio (OBR, Table S5). A549 tumor had a large OBR (i.e. 355) (Fig. 3f), demonstrating the effectiveness of signal amplification in tumors and background suppression in blood. These data strongly support the hypothesis that UPSe nanoprobes can specifically detect acidic tumor pHe, and that the probes can sense metabolic alterations within 24 h after initiating therapy.
Immunostaining of the whole-mount tumor sections (Fig. 3g) showed a qualitative correlation between the UPSe signal (red) with hypoxia stain by pimonidazole, which agrees with previous reports that hypoxic regions of the tumors are more acidic27. The nanoprobe activation map extended beyond the region of hypoxia detected by pimonidazole, likely because low pH, particularly < 6.8, significantly reduces pimonidazole binding to cancer cells28. Interestingly, UPSe signal did not correlate as well with tumor vascular density (CD31 stain, mostly at tumor periphery) or cell proliferation marker Ki-67 (Fig. S8).
For specific imaging of angiogenic tumor vasculature, we constructed cRGD-encoded UPSi nanoprobes (cRGD-UPSi, Fig. 4a) to image αvβ3 integrins, an established angiogenic biomarker. cRGD peptides bind to αvβ3 integrins, resulting in receptor-mediated endocytosis and uptake into the acidic endocytic organelles (Fig. 4b). Human umbilical vein endothelial cells (HUVECs) were treated with cRGD-UPSi, cRGD-free UPSi (UPSi), and a 50- fold molar excess of free cRGD peptide followed by cRGD-UPSi nanoprobes to demonstrate the proof of concept. Because the nanoprobes were “silent” in cell culture medium, we can directly measure the kinetics of nanoprobe internalization and activation without removing the medium. Thirty minutes after cRGD-UPSi incubation, punctate fluorescence activation was observed inside the HUVEC cells. At 3 h, an 11-fold fluorescence increase in cRGD-UPSi group was observed over the UPSi and cRGD competition control groups (Fig. S9). Almost all the fluorescent punctates in cells treated with cRGD-UPSi colocalized with LysoTracker (Fig. S10), demonstrating that cRGD-UPSi nanoprobes became activated in the endosomes/lysosomes. Finally, A549 and MCF-7 cells with low αvβ3 expressions showed significantly reduced fluorescence signals after incubation with cRGD-UPSi (Fig. S11).
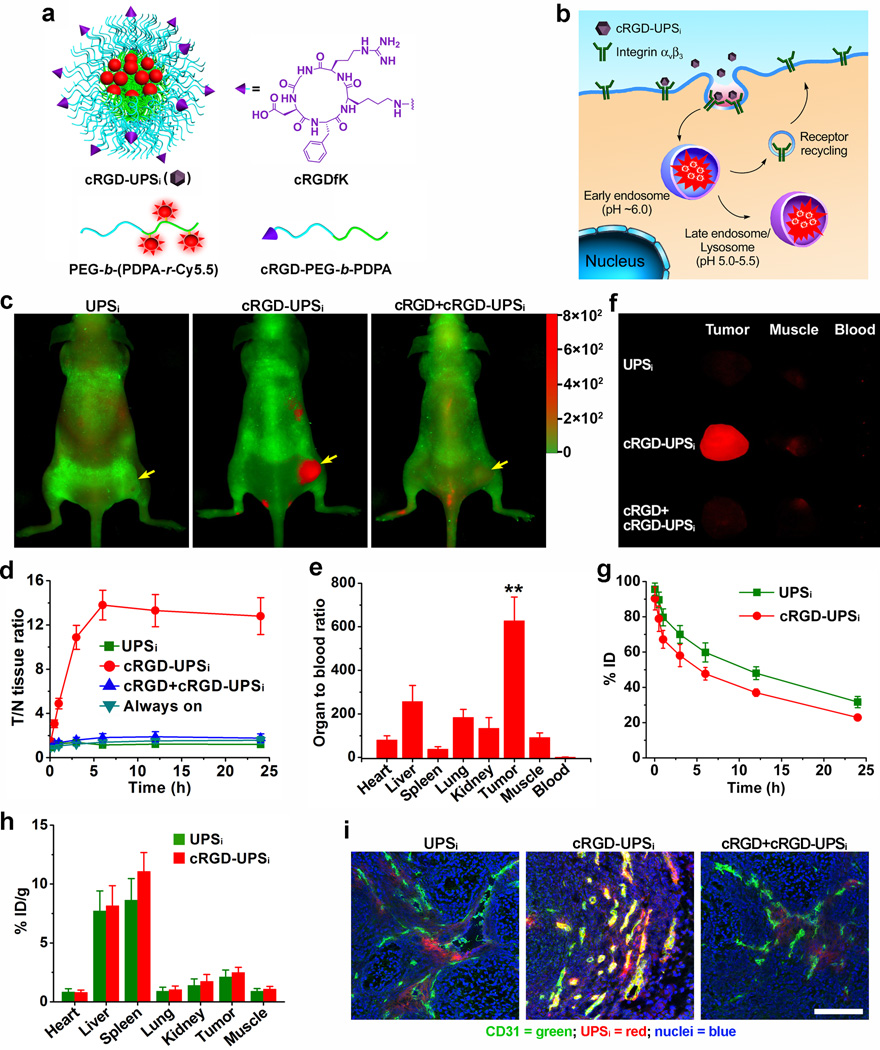
Figure 4. cRGD-UPSi nanoprobes can specifically image angiogenic tumor vasculature.
a, Design of cRGDUPSi nanoprobe. b, Schematic of internalization and activation of cRGD-UPSi nanoprobes after αvβ3-mediated endocytosis in tumor endothelial cells. The nanoprobes are accumulated in the endosomes or lysosomes, where the acidic pH activates the nanoprobes. c, Superimposed fluorescent images of A549 tumor-bearing mice at 6 h post-injection of cRGD-UPSi or UPSi nanoprobe (10 mg/kg). In the competition group, a blocking dose of cRGD peptide (25 mg/kg) was injected 30 min before cRGD-UPSi administration. Cy5.5 (Red) and autofluorescence (Green) are separately shown in the composite images. d, T/N ratio after injection of nanoprobes as a function of time. Data are presented as mean ± s.d. (n = 4). e, Organ to blood ratios (see data in Table S7) 6 h post-injection of cRGD-UPSi nanoprobe (n = 4). A549 tumor has 628-fold of signal amplification over blood by cRGD-UPSi. **P < 0.01, compared with other organs. f, Representative images of ex vivo tumors, muscles, and blood at 6 h post-injection of nanoprobes. g, Plasma concentration versus time curves (n = 4) for and UPSi nanoprobes. h, Biodistribution profiles (n = 4) of cRGD-UPSi and UPSi nanoprobes 6 h after intravenous injection. i, Correlation of nanoprobe activation with tumor vasculature (anti-CD31). The co-localization between nanoprobe and tumor vasculature is indicated by the yellow color in the merged images (green: blood vessels; red: nanoprobes; blue: nuclei. Scale bar = 100 µm).
For in vivo tumor imaging studies, cRGD-UPSi, UPSi, and cRGD-encoded always-ON nanoprobes were injected intravenously into A549 tumor-bearing mice (n = 4). As an additional control, a blocking dose of cRGD peptide (25 mg/kg) was injected 30 min before the cRGD-UPSi administration. As early as 30 min post-injection, the cRGD-UPSi group produced significantly higher fluorescence contrast in tumors (T/N ratio ~3) than the control groups (~1, P<0.05, Fig. 4d and Fig. S12). The fluorescence contrast reached a maximum at 6 h post-injection. UPSi, cRGD-always-ON and free cRGD competition controls had minimal fluorescence increase in the tumor over 6 h span (<2-fold). The OBR value was 628 for cRGD-UPSi, which is significantly higher than other organs/tissues (Fig. 4e). Despite comparable tumor accumulation percentages (e.g. 2.49 ± 0.44% vs. 2.11 ± 0.59% ID/g for cRGD-UPSi and UPSi 6h post-injection, respectively. Fig. 4h), the OBR value was significantly smaller for the UPSi control (78, P<0.01. Table S6 and Fig. S13). This difference (628 vs. 78) confirms that cell internalization is primarily responsible for the larger OBR for cRGD-UPSi. The always-ON nanoprobe control showed strong fluorescence background and low tumor/tissue contrast (<1.6-fold over 24 h, Fig. S14). The strategy of signal amplification by the cRGD-UPSi nanoprobes in angiogenic tumor vasculature was more markedly illustrated when compared to a small molecular cRGD-IRDye®800CW conjugate, where a maximum of 2-fold T/N ratio was observed in the 24 h span (Fig. S15).
We performed immunostaining of tumor sections (Fig. 4i and Fig. S16) to verify the locations of cRGD-UPSi activation. Tumor vessels were stained with Alexa Fluor®488-labeled anti-CD31. For cRGD-UPSi, majority of nanoprobe activation was found to colocalize with tumor vasculature (yellow color in Fig. 4i). In contrast, low levels of nanoprobe activation were observed in the UPSi and free cRGD blocking control groups. In these tumor sections, sporadic spots of nanoprobe activation were found outside the tumor vasculature, suggesting that non-vascular cells may also pick up a small population of nanoprobes through αvβ3-independent pathways.
To characterize the pharmacokinetic and biodistribution of UPSe/UPSi nanoprobes, we synthesized 3H-labeled nanoprobes through acetylation (-COCT3) of the free amino groups in the corresponding copolymers.3H-labeled UPSe, cRGD-UPSi and UPSi nanoprobes were injected at the same dose (10 mg/kg, or 2.0 mCi/kg) as in imaging studies (n = 5). For all three compositions, plasma concentration-time curves showed a two-phase behavior over 24 h (Fig. 4g and Fig. S17). The α-phase half-lives (t1/2,α) were 1.0±0.2, 2.3 ± 0.5 and 4.3 ± 0.7 h for UPSe, cRGD-UPSi and UPSi, respectively. The β-phase half-lives (t1/2,β) were 7.5±0.3, 17.0 ± 1.8 and 19.6 ± 2.1 h for UPSe, cRGD-UPSi and UPSi, respectively. The faster clearance of UPSe may be a result of higher CMC (2.38 µg/mL) than that of the UPSi (1.32 µg/mL). Between the two UPSi probes, cRGD surface functionalization resulted in decreased blood half-lives, consistent with other cRGD-encoded nanoparticle systems29.
Biodistribution studies show that tumor uptake (2–3% ID/g tissue) of UPSe/UPSi nanoparticles was higher than most normal tissues (heart, lung, kidney, and muscle, Tables S5–7). Meanwhile, the RES system (i.e. liver and spleen) was responsible for the uptake of majority of nanoprobes. Interestingly, for both UPSi groups, higher uptake was observed in the spleen than the liver; however, probe activation was much greater in the liver than spleen (e.g. OBR >250 in liver while <50 in spleen 6 h post-injection). To help understand this discrepancy, we co-injected a mixture of always-ON nanoprobe (labeled with Cy3) and UPSi (Cy5.5) with the anticipation of using the always-ON probe to benchmark the tissue distribution of nanoparticles regardless of tissue environment, while the UPSi probe only shows signals upon cell uptake in these organs. The results were striking (Fig. S18): first, UPSi activation occurred only sporadically in cell punctuates (most likely spleen macrophages)30 in the red pulps of the spleen but was largely absent in white pulp, despite the wide-spread distribution in both pulps as indicated by the always-ON probes. In the liver, we observed UPSi activation in the majority of hepatocytes. This surprising combination (i.e. lack of nanoprobe activation in a major spleen component and unexpected activation in most liver hepatocytes) explains the divergent behaviors of UPSi in these two organs. Additional studies are warranted to further elucidate the mechanism of unexpected patterns of cell uptake in these organs.
To evaluate nanoprobe toxicity, we investigated the changes in animal body weight, liver and kidney functions, and histology of RES at 24 h and 7 d after nanoprobe injection (10 mg/kg). Results showed no weight loss, statistically insignificant changes of hepatic and kidney functions (e.g. aspartate transaminase and glutamic oxaloacetic transaminase, Fig. S19) and normal RES histology (data not shown), demonstrating the safety of these nanoparticles.
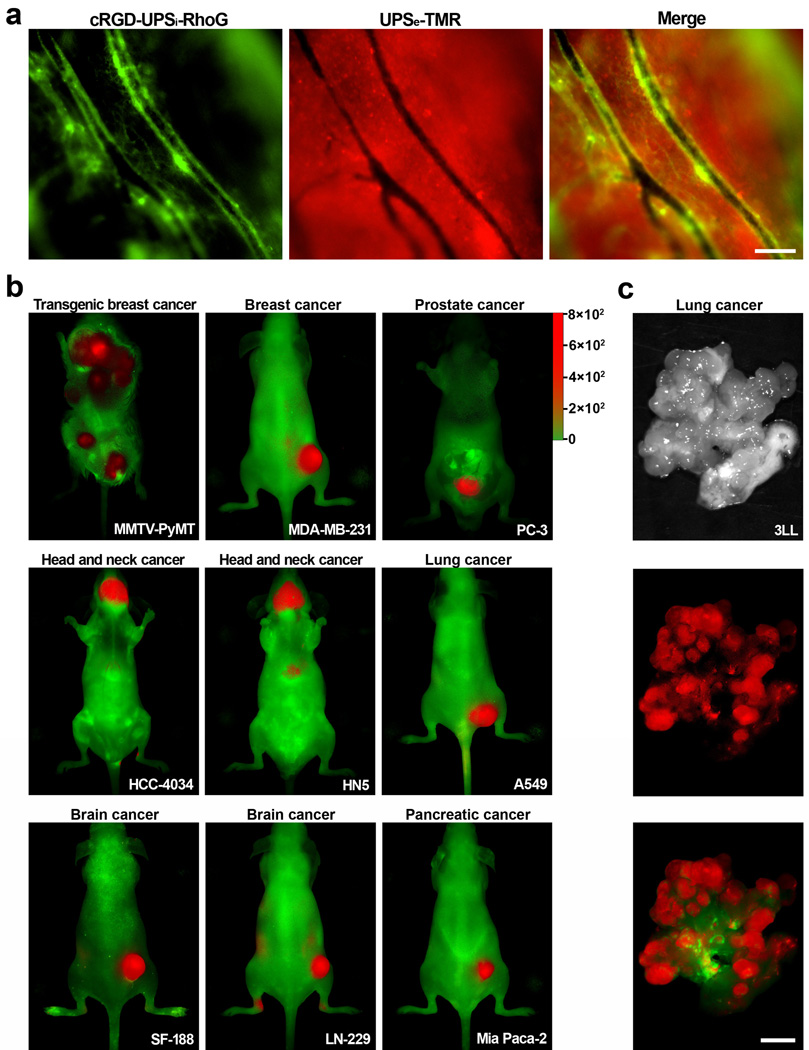
To explore the potential synergy in simultaneous imaging of tumor pHe and vasculature, we investigated the spatial pattern of UPSe and cRGD-UPSi activation in the tumor microenvironment. We employed intravital microscopy and subcutaneous A549 lung tumor xenograft in mice as our model system. To differentiate the two nanoprobes, we used tetramethyl rhodamine (TMR, λex/λem=550/580 nm) and rhodamine G (RhoG, λex/λem=502/527 nm) to label the UPSe and cRGD-UPSi, respectively. The dual nanoprobes were co-injected intravenously and imaged over time. Figure 5a shows complementary spatial activation patterns at 6 h post-injection: cRGD-UPSi-RhoG activation was mostly restricted to tumor vessels, whereas UPSe-TMR was illuminated in the interstitial space in the tumor parenchyma. Neither nanoprobe showed observable fluorescence inside tumor vasculature, demonstrating they remained ‘silent’ in blood.
Figure 5. iUPS nanoprobes target both acidic pHe and tumor vasculature with broad tumor specificity.
a, Intravital fluorescent images show complimentary pattern of spatial activation of cRGD-UPSi-RhoG (green) and UPSe-TMR (red) inside tumor vasculature and parenchyma, respectively. The dual nanoprobes were co-injected intravenously and the images were taken 6 h post-injection. Scale bar = 100 µm. b–c, iUPS nanoprobes show broad tumor imaging specificity and efficacy in 10 different tumor models of different cancer types (breast, prostate, head and neck, lung, brain, and pancreatic cancers) and organ sites. In 3LL lung cancer model (c), explanted lung was shown to illustrate the effective detection of small metastatic nodules (<1 mm). Scale bar = 2 mm. In each model, high T/N ratios were observed demonstrating the success of targeting tumor microenvironment signals as a universal strategy to achieve broad tumor specificity (see Fig. S20a–j).
To exploit the synergy of pHe and tumor vasculature activation, we constructed an integrated cRGD-UPSe-Cy5.5 nanoprobe (iUPS) and investigated its tumor imaging efficacy in 10 different tumor models. These models include a transgenic MMTV-PyMT breast cancer (multi-foci along mammary glands), several orthotopic cancers (lung, head and neck, prostate) and various subcutaneous cancer models (brain, pancreatic cancers). In all 10 of the tumor models, we observed universal nanoprobe activation in the tumor microenvironment over surrounding normal tissues/organs (Fig. 5b–c and Supplementary Fig. 20a–j). In 3LL lung cancer (Fig. 5c), explanted tissues showed effective detection of small metastatic nodules (<1 mm). These data highlight the success of targeting tumor microenvironment as a more robust and universal strategy to achieve broad tumor specificity.
Cancer is a diverse set of diseases with vastly different genotypes and phenotypes. Cancer-specific biomarkers and their expression levels can vary considerably among cancer types and organ sites31. This heterogeneity makes it challenging to establish a universal strategy for tumor detection using cancer cell-centric approaches, despite the availability of many ligands with high affinity and specificity (e.g. Herceptin for Her2/neu). Compared to cancer cell targeted approaches, the tumor microenvironment contains biomarkers that are more consistent across a range of cancer types. Acidic pHe resulting from dysregulated glycolysis is a hallmark of cancer that has been associated with enhanced cell proliferation, evasion of apoptosis, invasion and metastasis17,32. Angiogenesis represents another hallmark of cancer, which is essential for sustained tumor growth and exchange of nutrients and metabolic waste. Although targeting the tumor microenvironment provides a promising strategy for broad tumor detection, the challenge resides in how to achieve sensitive visualization with high biological specificity. Imaging acidic tumor pHe (6.5–6.8) is difficult since it is not drastically different from blood (7.4). Small molecular pH sensors only yield <3-fold signal difference in this pH span (Fig. 2b). A considerably sharper pH response is necessary to differentiate the pH difference in tumor versus normal tissues. Meanwhile, tumor vasculature is comprised of a small volume fraction (~1.5%)33,34, which necessitates an amplification strategy for robust detection.
Our iUPS design exploits amplification of tumor microenvironmental signals and background suppression to impact tumor imaging outcome. Compared to many small molecular imaging tracers (e.g. folate-FITC, one dye per tracer or cancer target), the nanoprobe design allows for a dramatic increase of dye payloads (e.g. 1,600 dye per particle) and thus deliver more imaging beacons for signal amplification (as demonstrated by the superior imaging outcome by cRGD-UPSi over cRGD-IRDye®800CW). Most nanoprobes have high background in the blood and low tumor contrast due to slow clearance compared to small molecular tracers. Our current UPS design solved this problem through homoFRET-induced fluorescence quenching where the blood signal is abolished, leading to large OBR values (>300-fold). Ultra-pH response is essential for the achieved imaging efficacy. Many previous studies on pH-sensitive nanoparticles showed improved antitumor response in drug delivery applications19–23. Most of these approaches were based on the breakings of pH-sensitive covalent bonds to induce nanoparticle degradation, which is time-consuming (e.g. 24 h) and not very pH responsive. The current UPS design utilizes proton transfer and non-covalent self-assembly that renders fast (<5 ms) and tunable response18. Consequently, UPS pertains an exquisite pH sensitivity over existing systems, as demonstrated by >100-fold ON/OFF activation within 0.25 pH unit with signal suppression in blood. When an appropriate pH threshold is selected (e.g. 6.9), the UPS platform illustrated a powerful differentiation of subtle pH transitions between blood and tumor microenvironment. This approach appears to be robust and universal, as demonstrated in 10 different tumor models with diverse cancer phenotypes and organ sites. Preliminary animal studies show negligible toxicities, and the approach avoids the exposure to radioactivity (e.g. 18FDG) required for positron emission tomography. Potentially, the UPS platform and fluorescence imaging can provide high resolution delineation of primary and metastatic tumors to achieve complete tumor resections during surgery. Finally, this nanoplatform can also serve as a valuable research tool to investigate cancer metabolism, to monitor acute perturbations of the microenvironment induced by metabolic inhibitors, and to illuminate the spatio-temporal dynamics of tumor hypoxia and acidic pH for the development of novel therapies.
Methods
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. Pharmacokinetic experiments involving radioactive materials (3H) were approved by the Radiation Safety Committee at the University of Texas Southwestern Medical Center.
Syntheses and characterization of UPS nanoprobes
Dye-conjugated MeO-PEG-b-PDPA and MeO-PEG-b-PC7A, and maleimide-terminated block copolymers were synthesized by atom transfer radical polymerization method (Supplementary method). cRGD-UPSi nanoprobes were prepared following a previously published procedure35. In a typical procedure, 2 mg of MAL-PEG-b-PDPA and 18 mg of MeO-PEG-b-PDPA-Cy5.5 were dissolved in 1 mL THF. Then, the mixture was added into 4 mL of Milli-Q water under sonication. The mixture was filtered 4 times to remove THF using the micro-ultrafiltration system. After micelle formation, an excess amount of c(RGDf(ε-acetylthiol)K) and 0.05 M hydroxylamine in 0.05 M HEPES/0.01 M EDTA aqueous solution were added. The conjugation was allowed to occur for 4 h followed by filtration to remove any precipitates in micelle solution. The cRGD-UPSi nanoprobe was filtered for 6 times to remove free peptide. To prepare the UPSi or UPSe nanoprobes, 20 mg of MeO-PEG-b-PDPA-Cy5.5 or MeO-PEG-b-PC7A-Cy5.5 were dissolved in THF. The solution was added into water under sonication. Then, the mixture was filtered for 4 times to remove THF, followed by filtration. To prepare the always-ON PEG-b-PC7A nanoprobes, 19 mg of MeO-PEG-b-PC7A and 1 mg of MeO-PEG-b-PC7A-Cy5.5 were dissolved in THF, added in water, and filtered by ultrafiltration. To prepare the always-ON cRGD-encoded PEG-b-PDPA nanoprobes, 2 mg of MAL-PEG-b-PDPA, 17 mg of MeO-PEG-b-PDPA and 1 mg of MeO-PEG-b-PDPA-Cy5.5 were dissolved in THF, and same procedure shown above was used for cRGD conjugation. 1H-NMR was used to confirm the formation of core-shell structure and conjugation of cRGD peptide to micelle surface. The successful conjugation of cRGD on the surface of micelles was validated by the appearance of phenyl protons of cRGD at 7.4 ppm. Transmission electron microscopy was carried out with 1% phosphotungstic acid negative staining and visualized on a JEOL 1200EX electron microscope (JEOL 1200EX).
Fluorescence activation of UPS nanoprobes
Fluorescence emission spectra of UPS nanoprobes in different pH buffer solutions were obtained on a Hitachi fluorometer (F-7500 model). For each UPS nanoprobe, the sample (5 mg/mL) was prepared in Milli-Q water. Then, the solution was diluted in 50 mM PBS buffer with different pH values. The final polymer concentration was controlled at 0.1 mg/mL. The nanoprobe was excited at 675 nm, and the emission spectra were collected from 690 to 770 nm. The emission intensity at 710 nm was used to quantify the signal amplification for UPS nanoprobes. Fluorescent images of UPSi and UPSe nanoprobe solutions (0.1 mg/mL) at different pH were captured on Maestro in vivo imaging system (CRI. Inc. Woburn, MA) using the “orange” filter (645–820 nm).
In vitro serum stability
Fresh mouse serum was collected and filtered through 0.22 µm syringe filters. Then, 0.2 mL of cRGD-UPSi or UPSe nanoprobe (2 mg/mL) was added into 2 mL of serum. The mixture was incubated at 37°C in a humidified chamber. At each designated time point, 100 µL aliquots of serum mixture were collected and immediately imaged by Maestro in vivo imaging system under identical settings to quantify the fluorescence intensity.
Cell culture
The tumor cell lines used for in vivo implantation include A549 lung carcinoma, MDA-MB-231 breast cancer, HN5 and HCC4034 head-neck cancer, SF-188 glioma, LN-229 glioma, 3LL lung carcinoma, Mia Paca-2 pancreatic cancer and PC-3 prostate cancer cells. Cells were cultured in DMEM with 10% fetal bovine serum and antibiotics.
Animal models
Female athymic nu/nu mice (18–22 g) were purchased from Charles River (Wilmington, MA). Mice were inoculated s.c. on the right flank with A549 cells (5×106/mouse). Three to four weeks after implantation, animals with tumor size of 200–300 mm3 were used for pharmacokinetic, biodistribution, and imaging studies. To demonstrate the universal imaging applications of the integrated nanoprobe, orthotopic tumor models, including HN5 and HCC4034 head-neck cancers, PC-3 prostate cancer and 3LL Lewis lung carcinoma were developed. MMTV-PyMT transgenic mice bearing multifocal mammary tumors were established by Dr. DeBerardinis lab. Subcutaneous tumor models, including MDA-MB-231 breast cancer, SF-188 glioma, LN-229 glioma, and Mia Paca-2 pancreatic carcinoma were established.
Pharmacokinetics and biodistribution studies
3H-labeled cRGD-UPSi were prepared from 90% MeO-PEG-b-PDPA-C(O)CT3 and 10% MAL-PEG-b-PDPA. UPSi and UPSe were prepared from MeO-PEG-b-PDPA-C(O)CT3 and MeO-PEG-b-PC7A-C(O)CT3, respectively. For pharmacokinetic experiment, mice bearing A549 tumors were randomly divided into three groups (n=4–5 for each group) for cRGD-UPSi, UPSi, and UPSe. The mice were injected intravenously with micelle solutions. Blood was collected at 2 min, 30 min, 1, 3, 6, 12, and 24 h after injection. Plasma (20 µL) was isolated by centrifugation at 5,000 g for 10 min. Plasma was subsequently mixed with a tissue solubilizer solution (1 mL, BTS-450; Beckman) at room temperature for 12 h followed by addition of a liquid scintillation mixture (10 mL, Ready Organic, Beckman) for 24 h. Amount of radioactive isotope was measured by a liquid scintillation counter (Beckman LS 6000 IC). Biodistribution of cRGD-UPSi, UPSi and UPSe nanoprobes in tumor and other organs was performed in a separate group of A549 tumor-bearing mice (n=4 for each group). Mice were perfused with PBS buffer (30 mL) at pre-designated time points (6 and 24 h). Dissected organs were weighed, homogenized, and treated with scintillation mixtures. The nanoprobes distribution in different organs/tissues was calculated as the percentage of injected dose per gram of tissue (%ID/g).
In vivo and ex vivo NIR fluorescence imaging
For tumor vasculature imaging, cRGD-UPSi or UPSi (10 mg/kg) was administrated intravenously into the A549 tumor-bearing mice (n=4 for each group). Time-course fluorescent images were captured on Maestro in vivo imaging system using the “orange” filter. To elucidate the role of αvβ3-mediated endocytosis, a group of mice were injected with cRGDfK (25 mg/kg) 30 min before cRGD-UPSi injection. To demonstrate EPR effect, always-ON nanoprobe was also used as a control.
For pHe imaging, UPSe (10 mg/kg) was injected into A549 tumor-bearing mice (n=4 for each group). Time-lapse NIR images were captured on Maestro system using the “orange” filter. As controls, 2-DG (250 mg/kg) or CHC (250 mg/kg) was injected 12 h before the UPSe nanoprobe administration. Then, the mice were monitored at pre-designated time points.
Ten tumor models described above were used to demonstrate the universal application of cRGD-UPSe-Cy5.5 integrated nanoprobe in tumor microenvironment imaging. Integrated nanoprobe (10 mg/kg) was administrated intravenously into tumor-bearing mice (n=4 for each tumor model). Fluorescent images were captured at 24 h post-injection.
Tumor/normal tissue (T/N) ratios were determined by comparing the average fluorescence intensities in the tumor and the whole body except the tumor site. After imaging, the mice were sacrificed. Excised tumor and organs were imaged by Maestro system. Fluorescence intensities of ex vivo tumors were quantified and normalized to the value of the muscle and blood.
Intravital imaging
Mice bearing A549 tumors were anesthetized with isoflurane and fixed under a Nikon ECLIPSE intravital microscope (Nikon, Japan) with a two-channel method in which one channel was used to image the activation of cRGD-UPSi nanoprobe in tumor vasculature and the other channel was used to probe the signal amplification of UPSe nanoprobe in acidic tumor microenvironment. Mixtures of cRGD-UPSi-RhoG (10 mg/kg, green) and UPSe-TMR (10 mg/kg, red) were intravenously injected into tumor bearing mice (n=4). Images were captured with a resolution of 1024×768 pixels with 10× Nikon objectives.
Immunofluorescence staining
In tumor vasculature imaging studies, the mice were sacrificed at 6 h post-injection. Tumors were snap frozen and cut into 8-µm sections. The slices were fixed in cold acetone and rinsed with PBS thrice, and blocked with 10% BSA for 1 h at room temperature. Subsequently, the slices were incubated with rat anti-mouse CD31 antibody (BD Biosciences) at 4°C overnight. Then, Alexa Flour®488-conjugated secondary antibody was added to stain the slices. The slides were mounted with DAPI-containing medium. The images were captured on a fluorescence microscope (Nikon ECLIPSE TE2000-E, Japan).
In pHe imaging studies, the tumor-bearing mice were intravenously injected with UPSe (10 mg/kg). At 5 h post-injection, the animals were injected with pimonidazole (60 mg/kg). One hour later, tumors were collected, frozen and cut into 8-µm sections. Adjacent tumor sections were exposed to primary antibody for 1 h at room temperature. Primary antibodies used were as follows: FITC-conjugated murine antipimonidazole monoclonal antibody (HPI Inc.); rat anti-mouse CD31 antibody; and rabbit anti-mouse Ki-67 antibody (Millipore). Sections were washed thrice with PBS and incubated with the appropriate secondary antibodies for 1 h. CD31 was detected with Alexa Flour®488-conjugated secondary antibody. Ki-67 was detected with Cy2-conjugated goat anti-rabbit antibody. The sections were scanned on an image analysis system consisting of Nikon fluorescence microscope using a computer-controlled motorized stage with a digital camera. All images were scanned at ×200 magnification. Composite images of sections were generated by the software from individual microscopic images.
Statistical analysis
Data were expressed as mean ± s.d.. Differences between groups were assessed using the paired, two-sided Student t-test. *P < 0.05 was considered significant, and **P < 0.01 was considered highly significant.
Supplementary Material
Acknowledgments
This work is supported by the NIH (R01EB013149 and R01CA129011) and Cancer Prevention and Research Institute of Texas (RP120094). Animal imaging work is supported by the UT Southwestern Small Animal Imaging Resource Grant (U24 CA126608) and Simmons Cancer Center Support Grant (P30 CA142543). We thank H. Zhou for help with the Maestro imaging, X. Luo for assistance with animal handling, J.T. Hsieh and L. Gandee for help with histology.
Footnotes
Author contributions
Y.W. and J.G. are responsible for all phases of the research; K.Z., G.H., X.H., X.M., and T.Z. helped with syntheses of different dye-conjugated polymers and characterization of UPS nanoprobes; C.H. and R.J.D. designed metabolic inhibition experiments and performed in vitro cell studies; R.J.D. supplied the transgenic MMTV-PyMT breast tumor model; B.D.S. guided the preclinical development of the experiments. Y.W. and G.H. wrote the initial draft. R.J.D., B.D.S. and J.G. revised the final draft.
The authors declare no competing financial interests.
Supplementary information accompanies this paper on www.nature.com/naturematerials.
References
- 1.Stuart MA, et al. Emerging applications of stimuli-responsive polymer materials. Nat. Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 2.de Las Heras Alarcon C, Pennadam S, Alexander C. Stimuli responsive polymers for biomedical applications. Chem. Soc. Rev. 2005;34:276–285. doi: 10.1039/b406727d. [DOI] [PubMed] [Google Scholar]
- 3.von Maltzahn G, et al. Nanoparticles that communicate in vivo to amplify tumour targeting. Nat. Mater. 2011;10:545–552. doi: 10.1038/nmat3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellomo EG, Wyrsta MD, Pakstis L, Pochan DJ, Deming TJ. Stimuli-responsive polypeptide vesicles by conformation-specific assembly. Nat. Mater. 2004;3:244–248. doi: 10.1038/nmat1093. [DOI] [PubMed] [Google Scholar]
- 5.Welsher K, et al. A route to brightly fluorescent carbon nanotubes for near-infrared imaging in mice. Nat. Nanotechnol. 2009;4:773–780. doi: 10.1038/nnano.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.So MK, Xu C, Loening AM, Gambhir SS, Rao J. Self-illuminating quantum dot conjugates for in vivo imaging. Nat. Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 7.Kircher MF, et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat. Med. 2012;18:829–834. doi: 10.1038/nm.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian X, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 9.Olson ES, et al. Activatable cell penetrating peptides linked to nanoparticles as dual probes for in vivo fluorescence and MR imaging of proteases. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urano Y, et al. Selective molecular imaging of viable cancer cells with pH-activatable fluorescence probes. Nat. Med. 2009;15:104–109. doi: 10.1038/nm.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dam GM, et al. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-alpha targeting: first in-human results. Nat. Med. 2011;17:1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 12.Ke S, et al. Near-infrared optical imaging of epidermal growth factor receptor in breast cancer xenografts. Cancer Res. 2003;63:7870–7875. [PubMed] [Google Scholar]
- 13.Paik S, et al. HER2 and choice of adjuvant chemotherapy for invasive breast cancer: national surgical adjuvant breast and bowel project protocol B-15. J. Natl. Cancer Inst. 2000;92:1991–1998. doi: 10.1093/jnci/92.24.1991. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ. HER-2/neu protein expression in breast cancer evaluated by immunohistochemistry. A study of interlaboratory agreement. Am. J. Clin. Pathol. 2000;113:251–258. doi: 10.1309/980M-E24R-V19K-595D. [DOI] [PubMed] [Google Scholar]
- 15.Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat. Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 16.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat. Rev. Drug Discovery. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 17.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 18.Zhou K, et al. Tunable, ultrasensitive pH-responsive nanoparticles targeting specific endocytic organelles in living cells. Angew. Chem. Int. Ed. 2011;50:6109–6114. doi: 10.1002/anie.201100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachelder EM, Beaudette TT, Broaders KE, Dashe J, Frechet JM. Acetal-derivatized dextran: an acid-responsive biodegradable material for therapeutic applications. J. Am. Chem. Soc. 2008;130:10494–10495. doi: 10.1021/ja803947s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae Y, Fukushima S, Harada A, Kataoka K. Design of environment-sensitive supramolecular assemblies for intracellular drug delivery: polymeric micelles that are responsive to intracellular pH change. Angew. Chem. Int. Ed. 2003;42:4640–4643. doi: 10.1002/anie.200250653. [DOI] [PubMed] [Google Scholar]
- 21.Griset AP, et al. Expansile Nanoparticles: Synthesis, Characterization, and in Vivo Efficacy of an Acid-Responsive Polymeric Drug Delivery System. J. Am. Chem. Soc. 2009;131:2469–2471. doi: 10.1021/ja807416t. [DOI] [PubMed] [Google Scholar]
- 22.Lee ES, Na K, Bae YH. Super pH-sensitive multifunctional polymeric micelle. Nano Lett. 2005;5:325–329. doi: 10.1021/nl0479987. [DOI] [PubMed] [Google Scholar]
- 23.Potineni A, Lynn DM, Langer R, Amiji MM. Poly(ethylene oxide)-modified poly(beta-amino ester) nanoparticles as a pH-sensitive biodegradable system for paclitaxel delivery. J. Control. Release. 2003;86:223–234. doi: 10.1016/s0168-3659(02)00374-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhou K, et al. Multicolored pH-tunable and activatable fluorescence nanoplatform responsive to physiologic pH stimuli. J. Am. Chem. Soc. 2012;134:7803–7811. doi: 10.1021/ja300176w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonveaux P, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J. Control. Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 27.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 28.Kleiter MM, et al. A comparison of oral and intravenous pimonidazole in canine tumors using intravenous CCI-103F as a control hypoxia marker. Int. J. Radiat. Oncol. Biol. Phys. 2006;64:592–602. doi: 10.1016/j.ijrobp.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, et al. A reexamination of active and passive tumor targeting by using rod-shaped gold nanocrystals and covalently conjugated peptide ligands. ACS Nano. 2010;4:5887–5896. doi: 10.1021/nn102055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moghimi SM, Hedeman H, Muir IS, Illum L, Davis SS. An Investigation of the Filtration Capacity and the Fate of Large Filtered Sterically-Stabilized Microspheres in Rat Spleen. Biochim. Biophys. Acta. 1993;1157:233–240. doi: 10.1016/0304-4165(93)90105-h. [DOI] [PubMed] [Google Scholar]
- 31.Polyak K. Heterogeneity in breast cancer. J. Clin. Invest. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhanabal M, et al. Endostatin induces endothelial cell apoptosis. J. Biol. Chem. 1999;274:11721–11726. doi: 10.1074/jbc.274.17.11721. [DOI] [PubMed] [Google Scholar]
- 34.Folkman J. What is the evidence that tumors are angiogenesis dependent? J. Natl. Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 35.Nasongkla N, et al. Multifunctional polymeric micelles as cancer-targeted, MRI-ultrasensitive drug delivery systems. Nano Lett. 2006;6:2427–2430. doi: 10.1021/nl061412u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.