Abstract
Many vaccines induce protective immunity via antibodies. Recent studies have used systems biological approaches to determine signatures that predict vaccine immunity in humans, but whether there is a ‘universal signature’ that can predict antibody responses to any vaccine, is unknown. Here we performed systems analyses of immune responses to the meningococcal polysaccharide and conjugate vaccines in healthy adults, in the broader context of our previous studies with the yellow fever and two influenza vaccines. To achieve this, we performed a large-scale network integration of public human blood transcriptomes, and systems-scale databases in specific biological contexts, and deduced a set of blood transcription modules. These modules revealed distinct transcriptional signatures of antibody responses to different classes of vaccines providing key insights into primary viral, protein recall and anti-polysaccharide responses. These results illuminate the early transcriptional programs orchestrating vaccine immunity in humans, and demonstrate the power of integrative network modeling.
Introduction
Recent studies have used systems biological approaches to identify molecular networks that orchestrate immunity to vaccination in humans 1–3. Analyses of the immune response to the yellow fever vaccine (YF-17D) have provided proof of concept that molecular signatures in the blood of humans, induced within a few days after vaccination, can be used to predict the magnitude of the later immune responses to a vaccine 4, and are beginning to yield novel insights about the nature of the innate and adaptive responses to vaccination 4, 5. Subsequently, systems biological approaches have been extended to identify predictive signatures to influenza vaccines 6, and are being used to study immune responses to other vaccines 7–11. The new field of systems vaccinology has emerged from this data and is poised to address the mechanisms that control immune responses to vaccination and identify predictors of vaccine efficacy 2, 12, 13.
A central question in systems vaccinology is whether there are ‘universal predictors’ of vaccine immunity. For example, given that many vaccines stimulate protective immunity through antibodies, are there molecular signatures that can be used to predict the antibody response to any vaccine? In studies of the yellow fever vaccine 4, 5, a robust but transient type I interferon response was seen in the blood transcriptomes of vaccinees. In studies of the trivalent inactivated influenza vaccine (TIV), a strong gene signature of antibody secreting cells was detectable seven days after vaccination 6. In both studies, expression of genes such as TNFRSF17 (BAFF receptor) was found to be highly predictive of the later antibody response 4, 6. How the early molecular and cellular events induced by vaccination impact the later antibody response remains a central question. Previous work on live attenuated virus vaccines (YF-17D and live attenuated influenza vaccine, LAIV) and an inactivated protein vaccine (TIV) suggest that different programs are induced by different vaccines 6. The question of whether there are common programs that drive antibody responses to different vaccines remains unanswered. For example, YF-17D triggers Toll-like receptors 2, 7, 8 and 9, as well as RIG-I and MDA-5 4, 14, and LAIV triggers TLR7 15, 16. However, bacterial polysaccharide do not trigger these receptors, which are involved in viral sensing. Do carbohydrate vaccines induce molecular signatures that reflect distinct molecular pathways that stimulate antibody production? To address this issue, we initiated a program aimed at comparing molecular signatures induced by different vaccines. As part of this effort, we performed a detailed analysis of the innate and adaptive responses to vaccination with the meningococcal polysaccharide vaccine or the meningococcal conjugate vaccine. The key questions that we addressed were: can the molecular signatures induced early after vaccination be used to predict the later magnitude of the antibody titers to these vaccines? Are these signatures similar to those elicited by other vaccines such as the influenza or yellow fever vaccines, and if so, to what extent are they capable of predicting antibody responses to these vaccines?
Neisseria meningitidis is a leading cause of meningitis and septicemia with 1.2 million cases per year worldwide 17. Two major classes of meningococcal vaccines available in the US are the polysaccharide vaccines, such as the quadrivalent polysaccharide vaccine (MPSV4) containing polysaccharides from serogroups A, C, Y and W-135, and the polysaccharide-protein conjugate vaccines, such as the quadrivalent conjugate vaccine (MCV4) that contains the same four polysaccharides conjugated to diphtheria toxoid. Vaccination induces anti-capsular antibodies with the ability to fix complement and trigger bacterial lysis, as measured in the serum bactericidal activity assay (SBA), which correlates with protection from clinical disease 18. Both classes of meningococcal vaccines induce high titers of functional antibodies one month after vaccination, however polysaccharide vaccines are believed to induce T-independent antibody responses, leading to waning humoral immunity and impaired memory, especially in infants 19. Moreover, repeated polysaccharide vaccination can result in hyporesponsiveness to serogroups C and W-135 20, 21. Despite the fact that these two vaccines contain the same polysaccharide antigens, the molecular mechanisms by which they elicit immunity may differ and are poorly understood.
In this study, we performed a detailed characterization of the innate and adaptive immune responses to vaccination with MPSV4 and MCV4 in healthy young adults. Comparative analysis was performed on five vaccines, combining the previous data on the yellow fever vaccine, and two influenza vaccines. A large-scale network integration of public human blood transcriptomes, with interactome, bibliome, and pathway databases and specific biological contexts was conducted to deduce a set of blood transcription modules, which were used to evaluate the correlation between the antibody response and the blood transcriptome. This approach revealed distinctive transcriptomic signatures that correlate with vaccine-specific antibody responses, providing key insights into primary viral, protein recall and anti-polysaccharide responses. Our results demonstrate the power of integrative network modeling, and show that immunological mechanisms can be successfully inferred from early blood transcriptomes.
Results
Antibody responses induced by meningococcal vaccines
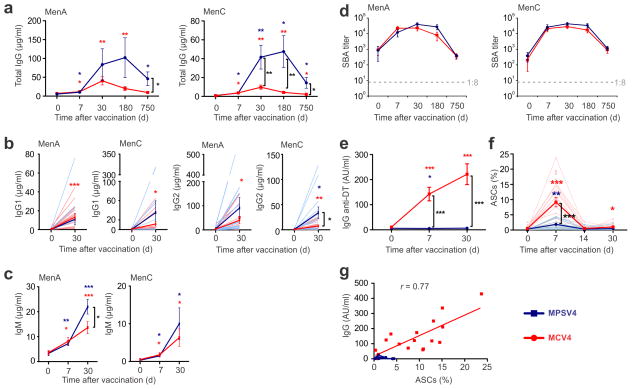
In a > 2-year longitudinal study, we immunized 30 healthy young adults with either MCV4 (n=17) or MPSV4 (n=13), and performed a comprehensive analysis of their innate and adaptive responses (Table 1). Serum antibody responses against meningococcal serogroups A and C were measured at days 0, 7, 30, 180 and 750 post-vaccination (Fig. 1a). Serogroup A is the cause of large pandemics, especially in Sub-Saharan Africa. Serogroup C is among the common types causing meningococcal infections in the United States, along with serogroups Y and B (http://www.cdc.gov/abcs/reports-findings/survreports/mening11.html). There is no licensed vaccine for serogroup B in the USA. Both vaccines induced robust polysaccharide-specific IgG binding antibody responses against serogroups A and C, as measured by ELISA. These responses peaked at day 30 (MCV4) or day 180 (MPSV4) (Fig. 1a). The magnitude and duration of the polysaccharide-specific IgG response was greater with MPSV4 compared to a single dose of MCV4 (Fig. 1a). Both MPSV4 and MCV4 induced polysaccharide-specific IgG2 and IgG1 antibody responses (Fig. 1b), as well as specific IgM antibody titers, which continued to rise until at least day 30 (Fig. 1c). Two years after immunization, there was still a significant concentration of polysaccharide-specific IgG response induced by MPSV4, whereas the response induced by the primary dose of MCV4 had declined, although it stayed substantially above baseline (Fig. 1a).
Table 1.
Study design of meningococcal vaccines, in healthy adult cohorts (18–45 yr, n=13 for MPSV4 and n=17 for MCV4). Blood samples were drawn at indicated time points. Whole blood cells were used for FACS and PBMCs were isolated for DNA microarray analyses. Plasma samples were used for the remaining assays.
| Time from vaccination (d) | 0 | 3 | 7 | 14 | 30 | 180 | 750 |
|---|---|---|---|---|---|---|---|
| Gene expression profile | x | x | x | ||||
| Serum bactericidal assay (SBA) | x | x | x | x | x | ||
| Anti-PS IgG (ELISA) | x | x | x | x | x | ||
| Anti-PS IgM (ELISA) | x | x | x | ||||
| Anti-DT IgG (Luminex) | x | x | x | ||||
| 27-plex cytokines (Luminex) | x | x | x | ||||
| Antibody secreting cells (FACS) | x | x | x | x |
Figure 1.
Antibody responses of meningococcal vaccinees. Serogroups A and C were measured for total IgG (a), IgG1 and IgG2 (b), IgM (c) and SBA titers (d). (e) A strong IgG response against diphtheria toxoid was detected in MCV4 vaccinees. (f) Antibody secreting cells (ASCs, selected as CD3− CD20− CD38hi CD27hi CD19+ cells) and (g) the number of ASCs at day 7 correlated with diphtheria toxoid-specific serum IgG at day 7. * p<0.05, ** p<0.01, *** p<0.001; unpaired t-test was used for comparisons between vaccine groups, paired t-test was used for comparisons to baseline within a group, Pearson’s correlation was used in (g). All plots show mean values (n=13 for MPSV4 and n=17 for MCV4) from one experiment, error bars as s.e.m.
Both vaccines induced a rapid increase in serum bactericidal activity (SBA), which was detectable as early as day 7, peaked at day 30, began to decline by day 180, yet remained substantially above baseline at day 750 (Fig. 1d). Although there was a trend for MPVS4 to induce greater SBA titers than MCV4 (Fig. 1d), the difference in the magnitude of the SBA titers was not as pronounced as that observed with the magnitude of the ELISA binding titers (Fig. 1a). An SBA titer of 1:8 or higher, a correlate of protective immunity 22, was maintained in the majority of MPSV4 and MCV4 vaccinees two years after inoculation, with no difference observed between the two vaccine groups (Fig. 1d). MCV4, which contains the diphtheria toxoid (DT) protein as a conjugate, elicited a strong anti-DT IgG response at days 7 and 30, whereas MPSV4 containing polysaccharide alone did not (Fig. 1e).
The number of peripheral blood antibody secreting cells (ASCs) induced by vaccination was assessed. The ASCs showed a peak at day 7 in both vaccines (Fig. 1f). Surprisingly, even though a greater polysaccharide-specific antibody response was induced by MPSV4 (Fig. 1a, d), the magnitude of the ASC response induced by MPSV4 was lower than that induced by MCV4 (Fig. 1f). Polysaccharide- and DT-specific ELISpots in a subset of MCV4 vaccinees revealed that the majority of IgG ASCs detected in the blood at day 7 were specific for DT (Supplementary Fig. 1a, b). Indeed, the percentage of total ASCs in the blood at day 7 after MCV4 vaccination strongly correlated with the serum anti-DT IgG (Fig. 1g), and not with the anti-polysaccharide IgG response (data not shown).
Taken together, these data demonstrate that both the polysaccharide and conjugate vaccines induce robust polysaccharide-specific antibody titers in healthy young adults, but the magnitude and isotypes of the antibody responses vary substantially between the two vaccines. Interestingly, MPSV4 induced a greater magnitude of binding antibody titers than MCV4 (Fig. 1a), although the differences in SBA titers were more modest (Fig. 1d). This suggests that despite the noticeable difference in the binding antibody response, that MCV4 may be more efficient at inducing functional antibodies, perhaps as a result of increased antibody affinity. Importantly, MCV4 induced a robust antibody secreting cell response that was directed largely against the carrier protein, DT, whilst MPSV4 induced very little IgG antibody secreting cell response. These data prompt the question of what cells produce carbohydrate-specific antibodies to MCV4 and MPSV4. It is possible that such cells emerge more rapidly (or later) than the plasmablasts producing DT-specific antibodies, or such cells may not circulate in the blood. Further experimentation involving a more detailed kinetic analysis of the response in humans will provide additional insights into these issues.
Workflow to compare vaccine transcriptomic signatures
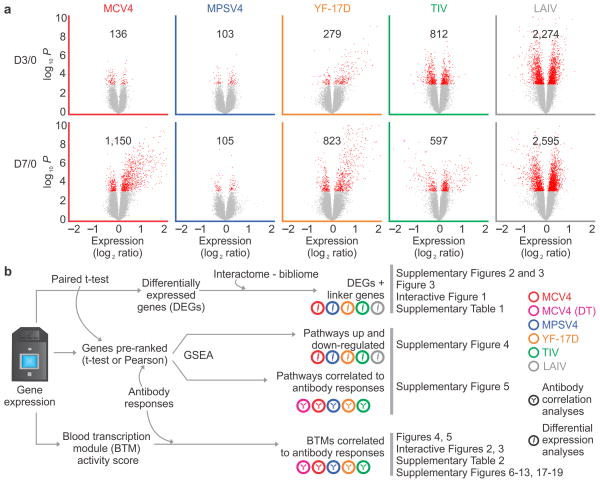
To gain insights into the molecular mechanisms underlying the responses to meningococcal vaccines, we analyzed the transcriptomic profiles in peripheral blood mononuclear cells (PBMC, days 0, 3 and 7) of the vaccinees using DNA microarrays (Table 1). At day 7 post-vaccination, we identified 1,150 differentially expressed genes (DEGs) in MCV4 vaccinees (Fig. 2a). These DEGs bear the hallmark of antibody secreting cells (ASC), which is consistent with the enhanced frequencies of these cells (Fig. 1f). In our previous study with influenza TIV, we observed that such an ‘ASC response’ is characterized by a transcriptional network regulated by XBP1, which is a key transcription factor associated with ER stress and plasma cell differentiation 6. This XBP1 network is also highly enriched for up-regulated genes at day 7 post-MCV4 vaccination (Supplementary Fig. 2).
Figure 2.
Analysis of blood transcriptomic data from five human vaccines. (a) Differential expression analysis was performed using paired t-test for each vaccine and each time point (day 3 or day 7 compared to baseline). The red dots in volcano plots show differentially expressed genes (DEGs, p < 0.001), with the numbers of DEGs. (b) A work flow to compare the transcriptomic signatures of five human vaccines.
However, only a small number of DEGs were identified at day 3 post-vaccination for both MPSV4 and MCV4, and at day 7 for MPSV4 (Fig. 2a). Consistent with this, analysis of plasma cytokines revealed little change induced by either vaccine (Supplementary Fig. 3). This is in contrast to what was observed with 3 other vaccines 4, 6: the yellow fever vaccine (YF-17D), the live attenuated influenza vaccine (LAIV) and the trivalent inactivated influenza vaccine (TIV), where the same differential expression analysis was performed (Fig. 2a). Similar results were obtained when we applied the additional gene filtering and false discovery rate as in our previous study 6 (Supplementary Table 1). Among these 5 vaccines, YF-17D and LAIV are live attenuated viruses, TIV is an inactivated vaccine that contains a viral coat protein, hemagglutinin (HA), MPSV4 contains meningococcal polysaccharides, and MCV4 contains the meningococcal polysaccharides conjugated to DT. With the exception of LAIV 6, all vaccines induce a robust antibody response in the blood, which is considered to be the principal correlate of protection for these vaccines 2, 18. Whether such transcriptomic differences reflect fundamental differences in the mechanisms of antibody response to these different vaccines is a key issue.
To address this issue, we devised a systematic framework for comparing the transcriptomic signatures of five human vaccines (Fig. 2b). Firstly, we applied to the lists of DEGs from the five studies a method that utilizes gene interaction data (interactome/bibliome), to put them into a biological context. Secondly, we tested molecular pathways for their association to vaccination and to antibody response, using a robust positional test (Gene Set Enrichment Analysis – GSEA 23). Lastly, we devised a novel approach using blood transcription modules (BTM), as an alternative to ‘conventional’ pathway analyses (Fig. 2b). Details of each step are described in the subsequent subsections.
Given the complexity and depth of these data and analyses, we provide access to an online data portal (http://www.immuneprofiling.org/papers/meni/). At this site, users can browse, filter and visualize the entire dataset through a web interface. Online interactive versions of major figures and details of our BTM module annotations are included as part of the website.
Comparative differential gene expression analysis
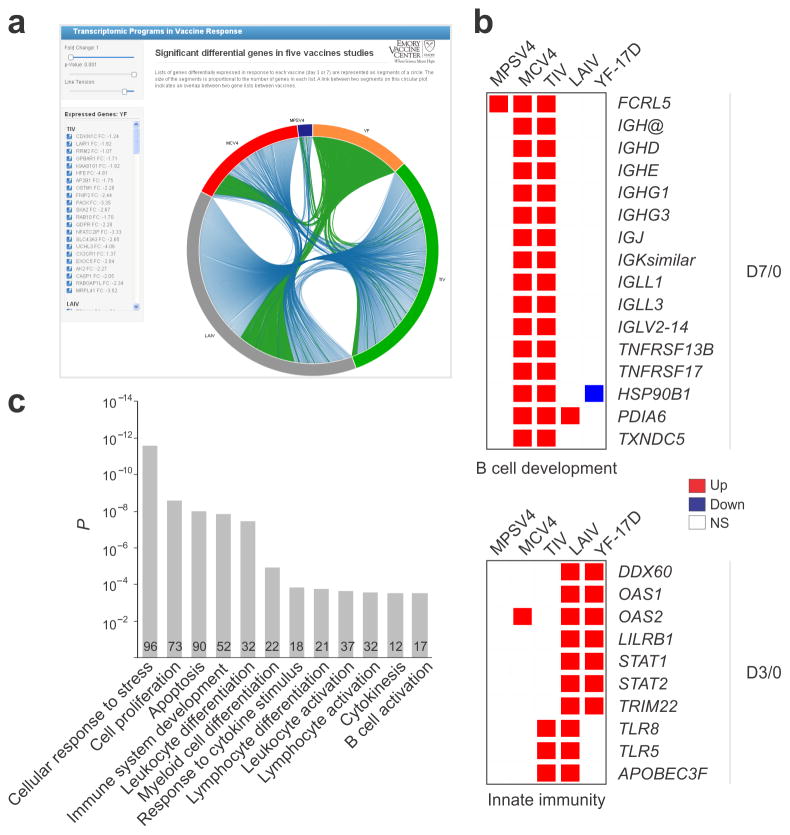
To find gene signatures shared by two or more vaccines, we cross-referenced the lists of DEGs (Fig. 2a) of all 5 vaccines. The DEGs common to any two vaccines are available online for interactive exploration, presented by blue curves linking two arcs on the circular plot (snapshot shown in Fig. 3a). Some of the DEGs across these vaccine datasets are shown (Fig. 3b). Several B cell genes were up-regulated at day 7 in MCV4 and TIV vaccinees (Fig. 3b), including TNFRSF17, a B cell differentiation gene that is predictive of antibody responses to the YF-17D and TIV vaccines 4, 6. Genes associated with innate immunity and interferon responses, such as OAS1, OAS2, STAT1, STAT2 and TRIM22 were up-regulated at day 3 post-vaccination with live attenuated virus vaccines (Fig. 3b).
Figure 3.
Differential expression analysis of five vaccines. (a) Online interactive figure showing DEGs shared between vaccine studies (online data portal http://www.immuneprofiling.org/papers/meni/, snapshot of Interactive Fig. 1). Links are shown in blue. When a given vaccine is selected, links are shown in green. (b) Selected DEGs involved in B cell development and innate immunity. (c) The DEGs plus “linker” genes identified by our interactome/bibliome integrative approach and shared by four or more vaccines are enriched with a number of gene ontology categories related to immune system. The genes found in ‘immune system development’ are shown in Supplementary Fig. 4f.
To better understand the interactions between these DEGs and to put them into biological context, we adopted an approach 24, 25 that utilizes gene connections extracted from interactome and bibliome data (Supplementary Fig. 4a; see online methods for details). The “interactome” refers to a collection of gene-gene interactions, including physical and causal interactions in pathways obtained from several public databases (e.g. HPRD, MINT, Reactome). The “bibliome” refers to pairs of genes that are associated to papers listed in PubMed. For each gene in the interactome or bibliome network, its association to DEGs in a vaccine response is assessed by statistical enrichment 24, 25. If such association is significant, this “linker” gene is added to the respective vaccine response network, even though it was not present in the original DEG list (Supplementary Fig. 4a). We note that these ‘linker’ genes are highly relevant to the study context. For example, among the 312 ‘linker’ genes added to the MCV4 network from Interactome, 115 have p-value under 0.05 in the gene expression data (enrichment p = 10−19). Using this integrative approach, the overlap of DEGs plus “linker” genes across the 5 vaccines were substantially increased (Supplementary Fig. 4b, c). The overlap network between MCV4 and TIV is enriched for genes expressed in antibody secreting cells (Supplementary Fig. 4d), whereas the overlap network between YF-17D and LAIV is enriched with TCR signaling genes and interferon-related genes (Supplementary Fig. 4e). The network of 1255 genes common to 4+ vaccines (Supplementary Fig. 4c) is highly enriched for a number of immunological-related processes, such as leukocyte differentiation and B cell activation (Fig. 3c, Supplementary Fig. 4f). Of these 1255 genes, 1231 are also found in the response network that we constructed from a published malaria RTS, S vaccine study 7 using the same method (data not shown), indicating that this common network may represent a “generic” signature of vaccine transcriptomic response.
Molecular pathways associated with vaccination and antibody responses
Beyond single gene level analysis, the expression of pathways can be also directly evaluated, providing more specific biological context and increasing statistical power 3, 26, 27. We adopted the positional test framework of GSEA 23 and Nature/NCI pathway interaction database 28 to perform pathway level analyses (see methods, Fig. 2b). Several pathways related to cell proliferation (ERBB1 downstream signaling, CDC42 signaling, E2F network, c-myc targets) were significantly induced by different vaccines, as well as innate pathways including interferon gamma pathway in YF-17D, mTOR pathway and TNFR signaling in MPSV4 and PDGFR-β signaling in YF-17D, MCV4 and MPSV4 (Supplementary Fig. 5).
We also identified pathways whose expression were correlated to antibody responses (Supplementary Fig. 6), such as “BCR signaling pathway” for TIV and MCV4 and “ATF-2 transcription factor network” for YF-17D. ATF-2 is a central transcription factor associated with tumorigenesis, DNA damage response 29 and cellular responses to amino acid starvation stress 30. We showed previously that another key sensor gene of amino acid starvation stress, EIF2AK4, predicts 4 and regulates (data not shown) the later CD8+ T cell response induced by YF-17D vaccination. Although further experimentation is necessary, our result suggests that the same amino acid starvation stress, through ATF-2 network, could be related to B cell responses.
Despite these interesting results, it should also be noted that pre-defined pathways are inherently limited in analyzing these blood transcriptome data 3, 27, 31, 32. Current pathway databases are biased to oncology and cover limited immunology 31. Also, many canonical pathways were defined in extreme or controlled experimental situations (e.g. knock-out experiments, pathological conditions, drug treatment, etc) 33, making them less suitable for describing how a healthy immune system responds under non-pathological conditions, such as vaccination. Additionally, genes in a pathway are not necessarily co-expressed. Furthermore, heterogeneous tissue like PBMCs, where expression signals from different cell types are mixed, can limit the sensitivity of pathway analysis. Therefore as described below, we undertook a novel approach of constructing gene transcription modules for blood tissues through large-scale data integration.
Blood transcription modules as a novel framework
Transcriptional networks can be computationally inferred from compendia of gene expression profiling data 34–37. Large volumes of human blood transcriptome data have already accumulated in public repositories, providing the opportunity to reconstruct gene networks that are specific to the context of immune response in the blood tissue. A special form of gene networks, the gene modules, has become a powerful tool of systems biology 34–37. This module approach was pioneered for human immunology using in-house microarray data38. However, one of the greatest challenges consists in finding modules that can be applied to the thousands of heterogeneous samples across the many different studies and platforms available (see Supplementary Note). To fully leverage the power embedded in these public data, we developed a large scale data integration approach, which reconstructed gene networks from publicly available microarray data, and identified context-specific gene modules from the reconstructed networks.
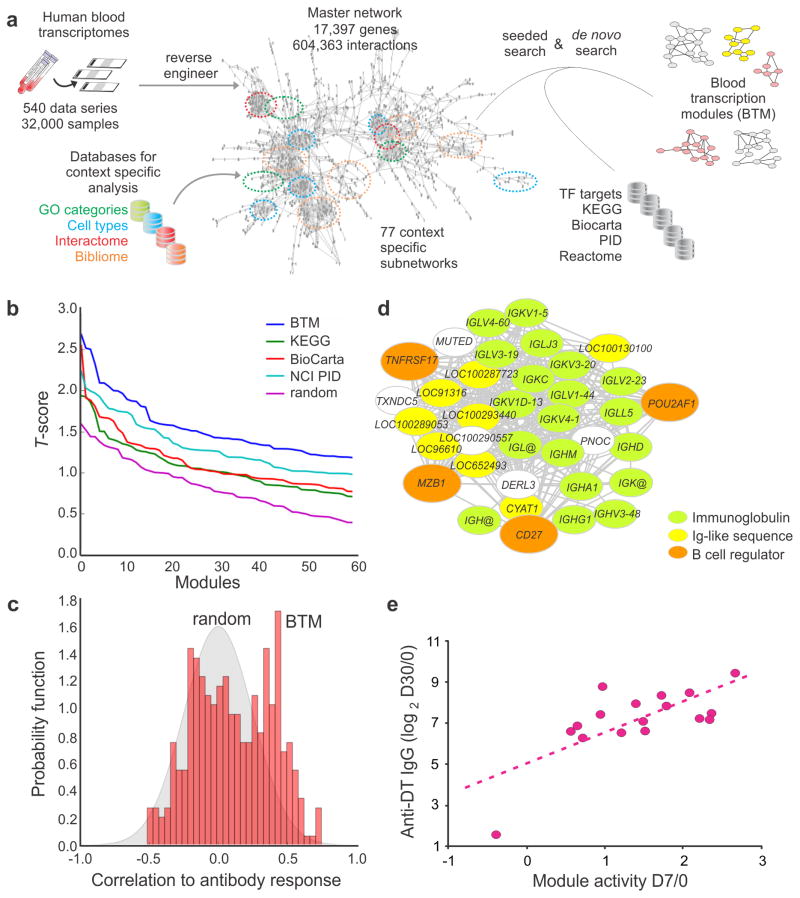
A detailed description of how we constructed the Blood Transcription Modules (BTM) is presented (Fig. 4a and full technical details are given in Supplementary Note). Over 30,000 human blood transcriptomes from more than 500 studies were compiled from public repositories. From each of these studies, we tested the dependency between genes using mutual information and extracted significant gene coexpression patterns 36, 39–42. Genes whose coexpression relationships were confirmed in three or more studies were connected to form a high-quality coexpression network, called the master network (Fig. 4a). Next, we obtained subnetworks of specific biological contexts by intersecting the master network with a gene ontology category, cell type-specific expression, interactome or bibliome. Finally, we implemented a massive parallel strategy to search through all networks for gene modules, which were defined as densely connected gene groups in a context-specific subnetwork. Current pathway databases and transcription factor binding data were also integrated into the search algorithm. This novel approach identified 334 BTMs that were annotated according to their biological functions and/or tissue-specific expression patterns. Only 37% of these BTMs have a greater than 25% overlap with current pathways, indicating that substantial new information is brought in. Full details of BTMs, including member genes and associated contexts, are provided in our online portal (http://www.immuneprofiling.org/papers/meni/) and in the Supplementary Tutorial, where a step-by-step demonstration of their applications is presented.
Figure 4.
Blood transcription modules provide a sensitive and robust statistical framework. (a) Construction of blood transcription modules (BTM) through large scale data integration. Full details are given in the Supplementary Note. (b) BTMs show superior sensitivity (assessed by t-score) compared to canonical pathways in class comparison. MCV4 transcriptomic data at D3/0 are used as example. Additional examples can be found in Supplementary Fig. 9. (c) Assessing the statistical significance of BTM correlation to antibody data. Each module is collapsed to a single activity score (mean value of all member genes), and Pearson correlation to antibody data is calculated across all subjects (red bars). The gray area is the distribution of random data generated by permutations of module gene memberships and sample labels. Data shown is the D3/0 MCV4 transcriptome data. (d) BTM module M156.1 consists of mostly immunoglobulin genes. Each edge represents a coexpression relationship learned from public data. CD27, TNFRSF17 and MZB1 are known B cell regulators, and POU2AF1 is a regulator of TNFRSF17. (e) Correlation of module M156.1 activity and later antibody response (MCV4-DT).
The BTM construction process recovered many known protein complexes, demonstrating that our reverse engineering approach had great sensitivity (see Supplementary Note). For example, 79 human genes from the ribosome complex are presented in the master network and are highly interconnected by 2,418 edges (Supplementary Fig. 7a). Additional examples of protein complexes, such as splicesome and nuclear pore complexes, are represented as BTM modules (Supplementary Fig. 7b, c). A large majority of BTM modules represent specific biological processes. The genes in module M18 for example are fully supported by literature for their involvement in T cell differentiation (Supplementary Fig. 8a), yet no database described a pathway that consisted of these genes. Many modules are self-evident, e.g. M240 contains genes whose majority is specific to Y-chromosome (Supplementary Fig. 8b).
Many statistical methods commonly used in pathway analyses can be applied to our BTM modules. For this study, we combined the expression values of member genes into a single module activity score (the mean value). Then, standard t-test and Pearson correlation were used on these activity scores. Using the t-test, we benchmarked the discriminative power of BTMs in the vaccine datasets and additional test datasets, and showed a superior performance of BTM modules compared to canonical pathways (Fig. 4b, Supplementary Fig. 9). Using a pathway/module activity score is an established practice 43, 44. These methods are statistically powerful because random elements within a module are expected to have no net contribution to the score. We thus have a framework to assess the statistical significance of correlations to antibody data, where BTM modules can be distinguished from random data (Fig. 4c). The Plasma cells-Immunoglobulin module is illustrated (Fig. 4 d). This module consists of mostly immunoglobulin genes and genes that bear sequence similarity to immunoglobulins. It is noteworthy that the four genes out of this category, TNFRSF17, POU2AF1, MZB1 and CD27, are all known B cell regulators. Due to the inherent nature of this module, one can expect its correlation to antibody production. Indeed, the increased activity of this module at day 7 MCV4 data is well correlated to later DT-specific IgG response (Fig. 4e), and it is consistent with the increase of antibody secreting cells at day 7 (Fig. 1f). A similar result is seen for influenza TIV vaccine (Supplementary Fig. 10), which also induces an expansion of antibody secreting cells at day 7 6. These results indicate that gene interactions can be learned from public data, and BTMs provide a powerful tool to decrypt immune responses.
Distinctive transcriptional programs in vaccine antibody responses
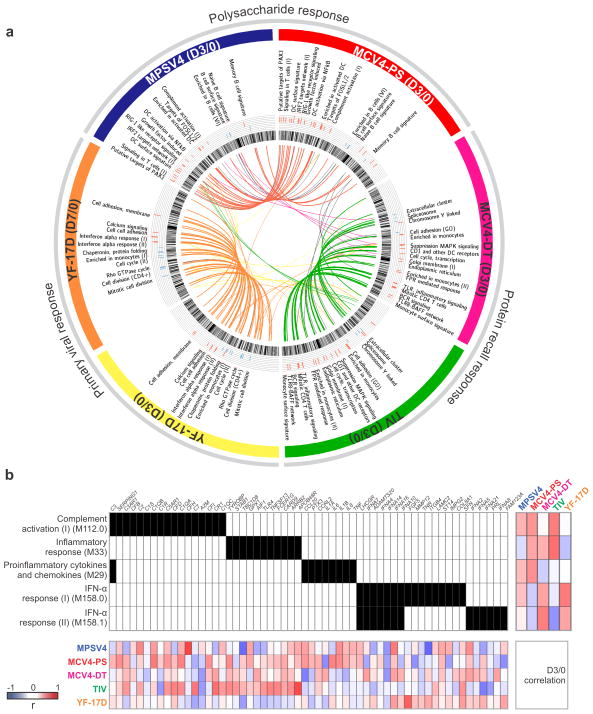
We applied our BTM framework to examine the transcriptomic programs that correlate with antibody responses to different vaccines. For each of the YF-17D (live attenuated virus), TIV (inactivated), MPSV4 (carbohydrate) and MCV4 (polysaccharide-conjugate) vaccines, the correlation to antibody response was computed for all BTM modules, and statistical significance was assessed by permuting gene memberships and sample labels (see on-line methods). The conjugate vaccine MCV4 yielded two sets of correlation profiles from the same blood transcriptome data, one for the Diphtheria Toxoid (DT) antibody response and one for the polysaccharide (PS) antibody response. With the distinct nature of these vaccines and variations between cohorts, one might have expected a unique profile for each vaccine, as suggested by the earlier pathway analysis (Supplementary Fig. 6). Yet to our surprise, the early BTM correlation profiles at day 3 displayed three distinctive patterns (Fig. 5a and Supplementary Fig. 11) that represent the protein recall response (shared by TIV and MCV4 anti-DT), the polysaccharide response (shared by MPSV4 and MCV4 anti-PS) and primary the viral response (YF-17D). The YF-17D profile at day 3 is very different from other vaccines, but closely resembles that of YF-17D at day 7. In this context, an important consideration is that differences in the signatures observed with MPSV4 versus MCV4 may be due to the fact that one vaccine is given intramuscularly and the other subcutaneously. However, we feel that this is unlikely, since the signatures that correlate with the anti-polysaccharide antibody responses to MPSV4 and MCV4 are similar (Fig. 5a).
Figure 5.
BTM analysis reveals distinct mechanisms of antibody response. (a) Each vaccine data set is shown as one of six segments on the circular plot. In each segment, the inner circular bands show an ordered list of all BTM modules, layered by histograms of modules significantly correlated to the antibody response, red for positive correlation and blue for negative correlation. Significant modules that are common between vaccines (as in Supplementary Fig. 12b, c, d) are linked by a color curve in the center (gray links for modules omitted in Supplementary Figure 11). An interactive version of this figure is available (online data portal, Interactive Figure 2). (b) Illustration of module activity. A filled unit in the center grid indicates the membership of the gene (top axis) in the corresponding module (left axis). The heat map on the right shows the Pearson correlation between module activity and antibody response in each study. The bottom heat map shows correlation between module member genes and the antibody response.
In the primary viral response (Fig. 5a, b, Supplementary Fig. 12), higher antibody response is associated with greater activity in interferon response and cell adhesion modules. Although we previously showed that interferon activity is induced after YF-17D vaccination, no correlation was found between the interferon response and antibody production 4, 5, 36. Using our novel module approach, we identified for the first time for YF-17D this robust correlation (Fig. 5b), which is consistent with the ability of YF-17D to induce robust type I IFNs production in plasmacytoid DCs 14. The cell adhesion molecules are a hallmark of leukocyte migration and activation, thus a sensitive indicator of vaccine immunogenicity. Inversely correlated to the antibody response are a number of cell division modules, including PLK1 signaling events. The PLK1 pathway is generally an antagonist of interferon signaling 45, 46, implicating a concerted immune modulation.
The anti-polysaccharide response at day 3 (Fig. 5a, top; Supplementary Fig. 12) contains a number of modules associated with antigen presenting cells and their activation, as well as complement and proinflammatory cytokines (Fig. 5b). This profile suggests the active involvement of myeloid dendritic cells (Supplementary Fig. 12,13), possibly through TNF-NF-κB signaling. Gene members of these modules include dendritic cell (DC) markers: CD83, HLA-DR and HLA-DQ (Supplementary Fig. 14), and inflammatory cytokines: CCL20 and IL1B (Supplementary Fig. 13). The correlation of DC surface signature module to anti-polysaccharide IgG in both MCV4 and MPSV4 is shown (Supplementary Fig. 14b).
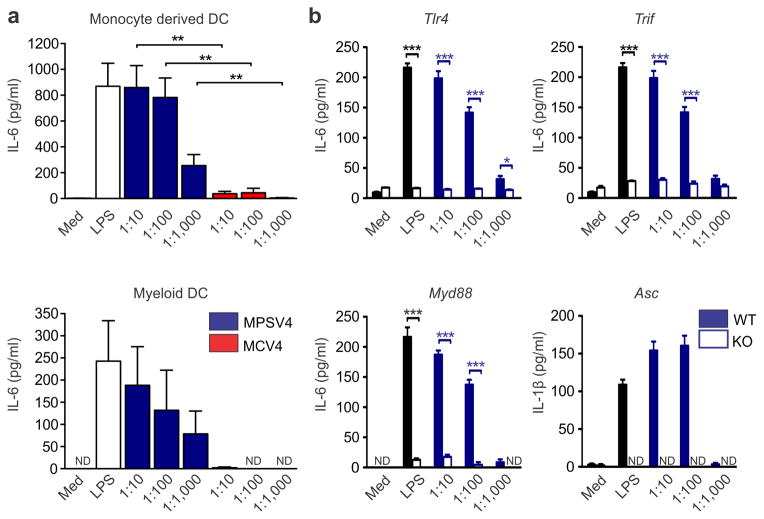
We therefore further investigated the involvement of DCs in polysaccharide vaccination. The polysaccharide vaccine MPSV4 was able to induce the maturation of blood myeloid DC isolated from human PBMC (Supplementary Fig. 15). Up-regulation of co-stimulatory molecules and secretion of IL-6, TNF and IL-12p40 was detected in human DCs stimulated by MPSV4 (Fig. 6a). A similar result was seen in mouse splenic CD11c+ DCs. Using these cells from the appropriate knockout mice, we demonstrated that DC maturation induced by MPSV4 is dependent on Myd88, Trif and Tlr4, as well as inflammasome activation (Fig. 6b, Supplementary Fig. 16). These data suggest that MPSV4 vaccination involves pattern recognition receptors, a finding that is in line with previous reports 47. To determine whether these molecules were essential for the immunogenicity of MPSV4, we vaccinated mice deficient in these molecules with MPSV4 or MCV4. Vaccintation with MPSV4 did not reliably induce antibody titers (Supplementary Fig. 17), even after two successive immunizations (Supplementary Fig. 17d), thus posing a challenge to further investigation using animal models. However, despite there being a correlation of the DC surface signature module to anti-polysaccharide IgG (Supplementary Figure 14b), MCV4 did not induce robust activation of DCs in vitro (Fig. 6a & Supplementary Fig. 16). This suggests that the MCV4 may activate DCs in vivo through an indirect mechanism, and highlight the challenges in extrapolating results in vitro to in vivo studies.
Figure 6.
The meningococcal polysaccharide vaccine activates myeloid dendritic cells. (a) Induction of cytokines by MPSV4 from human myeloid DC (4 independent replicates) and monocyte derived DC (6 independent replicates). Mean values of independent replicates are shown, error bars as s.e.m. Secreted IL-6 protein levels are shown but the same result was observed for TNF and IL-12p40 levels. (b) Mouse knockout DCs show that the MPSV4 stimulation is dependent on Myd88, Trif, Tlr4 and Asc in vitro. CD11c+ DCs were isolated by MACS from spleens of C57BL/6 mice (WT) or various knock-out mice and stimulated for 24 h; *p<0.05, ***p<0.0001, unpaired t-test was used to compare WT vs. KO. Four independent replicates were performed for Myd88, Trif and Tlr4 knockouts, six for Asc knockouts. One representative experiment is shown, with mean values and s.e.m. from technical replicates.
The modules whose activity on day 3 correlate with HAI titers (TIV) and the DT-specific antibody titers (MCV4) are likely be part of a transcriptomic program associated with protein recall responses (Fig. 5a, lower right; Supplementary Fig. 12). This program displays many monocyte features, especially of Toll-like receptor (TLR) signaling. The FPR (formyl peptide receptor)-mediated response module is also highly expressed in monocytes. Linked to TLRs are TNFSF13B (also known as BAFF, M25) and TNFSF13 (also known as APRIL, M48) (Supplementary Fig. 12d, Supplementary Table 2). Together with BCR signaling, these modules indicate the downstream activation of B cells. Plasma and memory B cell modules at day 7 are indeed positively correlated with the antibody response in both vaccines (Supplementary Fig. 18).
The distinctive early patterns of the protein recall response and the polysaccharide response become less apparent by day 7 (Supplementary Fig. 19). At this time point, several BTM modules were in fact common to 3 out of the 5 vaccines. For instance, cell cycle related modules are shared among MPSV4, TIV and MCV4 anti-DT responses (Supplementary Fig. 19). The TIV antibody response and the MCV4 DT-specific response still follow their day 3 pattern (protein recall response), sharing among the top correlated BTMs plasma cells and immunoglobulin modules (Supplementary Fig. 19,20). TNFRSF17, the receptor for B cell growth factor BAFF and a top antibody predictor gene from previous studies 4, 6, is a member of these modules. Interestingly, these plasma cells and immunoglobulin modules, as well as memory B cell modules, are inversely correlated to antibody titers in YF-17D (Supplementary Fig. 12e, Supplementary Table 2), which is the opposite of what was observed with TIV and MCV4, where there is a positive correlation with the antibody responses (Supplementary Fig. 20). This difference may reflect some intriguing biology in the putative mechanisms that differentially regulate the generation of short-lived plasmablasts during recall responses (TIV, MCV4) versus long-lived plasma cells (YF-17D), and remains to be explored mechanistically.
In summary, our BTM analysis revealed rich information underlying blood transcriptomes of vaccinated individuals. Besides the most pronounced features discussed above, more details are reported (Supplementary Fig. 12 and Supplementary Table 2). For instance, T cell activation appears to be negatively correlated with antibody responses in MPSV4 (D7) and TIV (D3 and D7), consistent with our previous report 6. A male-specific module (M240) is negatively correlated to antibody response in all these vaccines with varying statistical significance, suggesting a gender bias in antibody production. This is in agreement with several epidemiological studies that show that vaccinated females have a greater immune response than males 48.
Discussion
The major goals of systems vaccinology are to study the molecular mechanisms of action of vaccines and to identify signatures that predict vaccine immunity 2, 49. These goals can be more easily achieved when applied to vaccines such as YF-17D and TIV, which induce robust cellular responses and changes in gene expression in the blood. However, vaccination with MPSV4 and MCV4 induced more subtle changes in the blood transcriptome of vaccinees. Thus, deciphering the molecular mechanisms underlying immunity to different types of vaccines poses a great challenge. An additional problem inherent to the field 27, 49 is the relatively limited size of many study cohorts, which poses challenges in achieving statistical robustness in data analysis. Another challenge stems from the usage of canonical pre-defined pathways, which lack specificity in the context of blood transcriptomic analysis. Using these pre-defined pathways introduces bias and limits the information that can be obtained from antibody correlation analysis. To overcome these challenges, we developed a large-scale data integration approach, which leverages prior knowledge and data in the public domain, to construct blood transcription modules and to develop them into a robust yet interpretable statistical framework. We chose to use a straightforward module activity score, and compute its Pearson correlation to antibody data with permutation tests. However, it is important to mention that BTM modules can be used as gene sets for GSEA analyses and for other pathway methods, making it easier for any researcher to study BTM activity in their own experiment.
Two sets of antibody correlation profiles were observed for the conjugate vaccine MCV4: one specific to anti-PS and another for anti-DT antibodies. Such resolution of ‘dual profiles’ was only achieved by our BTM approach – and not by gene-level analysis – and, therefore represents a considerable increase in sensitivity. The result also indicates that distinct molecular mechanisms are induced by different components of the same vaccine, and these may impact the study and development of conjugate vaccines in general.
A surprising result from this study is that although both MPSV4 and MCV4 induced robust polysaccharide-specific antibody responses, the vast majority of plasmablasts induced by MCV4 secreted antibodies against the carrier protein, rather than against the polysaccharides. This raises the possibility that carbohydrate-specific antibodies are produced by a population of B cells, which are not present in the blood.
Our BTM framework identified three distinctive patterns in blood transcriptomes at day 3 post-vaccination that are related to the nature of the vaccines studied. Among the BTMs whose activity correlates to antibody response at one month or later, we found type I interferon modules for primary viral response (YF-17D), modules associated with DC and complement activation for polysaccharide responses (MCV4-PS and MPSV4) and modules such as BCR signaling and plasma cell Igs for protein recall response (TIV and MCV4-DT). Several additional BTMs were correlated to antibody responses, indicating potentially novel networks of innate immunity that should be further characterized. These data suggest that gene expression predictors of antibody response are unlikely to be “universal”, but are dependent on the type of vaccine. This is consistent with our previous proposal that there would be different classes of vaccines that induces similar signatures of immunogenicity 1.
These analyses also enable the formulation of hypotheses about some potentially novel mechanisms of vaccine immunity. For example, the ‘signaling in T cells (I)’ module is strongly correlated to the carbohydrate-specific antibody response to MPSV4 (carbohydrate vaccine). Contrary to the prevailing belief that immune responses to MPSV4 is T cell-independent, our results raise the possibility that CD4+ T cells may be involved. This hypothesis is consistent with a recent report describing polysaccharide-specific T cells 50. Furthermore, surprisingly, modules for RIG-1-like signaling and IRF-2 strongly correlated with carbohydrate-specific antibody responses to MPSV4 and MCV4. This raises the possibility that these vaccines may be using the RIG-1 pathway to induce antibody responses – something that remains to be tested. Finally, the observation that the ATF-2 network correlates with antibody responses suggest a potential role for the integrated stress response in mediating antibody responses. This is consistent with our previous work showing that expression of EIF2AK4,, which encodes a key sensor gene of amino acid starvation, predicts 4 and regulates (data not shown) the later CD8+ T cell response induced by YF-17D vaccination.
Finally, while our BTMs offer a valuable tool to analyze blood transcriptomic data, further research is needed to better model the system and understand it. Thus the fine kinetics and potential regulatory mechanisms of genes contained within the BTMs could be elucidated in studies with more frequent sampling of the blood. Given the relatively modest strength of the early transcriptional signatures induced by MPSV4 and MCV4, a key question is the extent to which such changes represent variations in environmental confounding factors such as external temperature, time of collection of blood and atmospheric pressure. However, the biological relevance of many of these signatures to vaccine immunity, renders it likely that these results are indeed due to vaccination. Nevertheless, future studies could benefit from including placebo groups of subjects. Furthermore, this analysis presents several hypotheses about the functional relevance of the networks identified. In summary, these findings highlight the utility of systems biological approaches in discovering novel correlates of immunogenicity to vaccination. The identification of a compendium of signatures that correlate with antibody responses to different classes of vaccines represents a first step in addressing the question of whether there are universal correlates of antibody responses to vaccination. Extending this analysis to a broader range of vaccines will pave the way towards the development of a ‘vaccine chip,’ which could be used to predict vaccine immunity against a broad range of vaccines.
On-line Methods
Clinical study organization
Healthy adult volunteers (18–45 years of age) were randomized to be vaccinated subcutaneously with MPSV4 (Menomune®, n=13) or intramuscularly with MCV4 (Menactra™, n=17) (Sanofi Pasteur, Swiftwater, PA). Whole blood samples were collected using CPT tubes at days 0, 3, 7, 14, 30, 180 and 750 post-vaccination. Randomization was done by the pharmacist using Research Randomizer Form. The trial was double blinded to participants and laboratory staff during data collection. Informed consent was obtained on all participants prior to any study procedure. This research was approved by the Emory University Institutional Review Board and by the CDC Human Subject Office. The trial was designed as an exploratory study, without pre-specified effect size. All samples from the subjects that completed the study were included for analysis.
Cell and plasma isolation
Peripheral blood mononuclear cells (PBMC) and plasma were isolated from fresh blood (CPTs; Vacutainer® with Sodium Citrate; BD), as described in 6. PBMCs were frozen in DMSO with 10% FBS and stored at −80 °C and then transferred on the next day to liquid nitrogen freezers (−210 °C). Plasma samples from CPTs were stored at −80 °C. Trizol (Invitrogen) was used to lyse fresh PBMCs (1 ml of Trizol to ~1.5 × 106 cells) and to protect RNA from degradation. Trizol samples were stored at −80 °C.
Blood transcriptome analysis
Total RNA was extracted from PBMCs stored in Trizol according to the manufacturer’s recommendation. The quality of RNA was evaluated by a NanoDrop spectrophotometer and BioAnalyzer (Agilent Technologies). DNA microarray experiment was performed as previously described 6, on Affymetrix human U133 Plus 2.0 arrays (GeneTitan platform). Hybridization data of all samples passed quality control and expression data were normalized by RMA R package.
Measurement of anti-Meningococcal antibodies by ELISA
Immulon 2 HB plates (Thermo, Milford, MA) were coated with a final concentration of 5 μg/ml for N. meningitidis serogroup A or 2.5 μg/ml for serogroup C purified polysaccharide (NIBSC, England) co-mixed for a final concentration of 2.5 μg/ml methylated serum (mHSA) albumin of serogroup A of 5 μg/ml mHSA for serogroup C. A blocking step with 0.01M PBS buffer (pH 7.2) containing 5% new born bovine serum (NBBS, GIBCO-Invitrogen, Grand Island, NY) was performed for 1 h and decanted. Serum samples were pre-diluted 1:25 and then serially diluted (2-fold) seven times in SC buffer (PBS with 5% NBBS and 0.1% Brij35). Samples were incubated over night at 4°C along with quality control serum CDC900385 and the reference standard CDC1992 51, as previously described 52. Following overnight incubation samples were removed by washing and horse radish peroxidase (HRP) conjugate serum (mouse anti-human IgG, monoclonal antibody HP6043, Hybridoma, Baltimore, MD) was added to detect the bound serum IgG in samples collected at days 0, 7, 30, 180 and year 2. For detection of IgM antibodies mouse anti-IgM Fc monoclonal antibody (HP6083, Hybridoma) conjugated to HRP were used instead in samples from days 0, 7 and 30. Following a 2.5 h incubation, excess conjugate was removed by washing. The amount of bound conjugate was detected by addition of liquid SureBlue™ TMB (KPL, Gaithersburg, MD) and incubated for 30 minutes. The reaction was stopped by adding 100 μl of 1N HCl. Plates were read at 450 nm with a 630 nm background wave length in an ELx808 plate reader (BioTek, Winooski, VT). Data was captured with Gen5™ (BioTek) and analyzed using ELISA for Windows (CDC, Atlanta, GA). Data files were stored in a secured network folder. Subclass IgG1 antibodies were measured using mouse anti-human IgG1 (Alpha Diagnostics, San Antonio, TX) conjugated to HRP. IgG2 antibodies were measured using mouse anti-human IgG2 (Invitrogen, Frederick, MA) conjugated to alkaline phosphatase. This later conjugate was developed using BluePhos® kit from KPL (Gaithersburg, MD). IgG1 and IgG2 antibodies were only measured in samples obtained at days 0, 7 and 30.
Measurement of anti-Diphtheria IgG by Luminex
Immune response to Diphtheria toxoid (DT) carrier protein was assessed at Days 0, 7 and 30 using anti- DT IgG capture microsphere assay. Fluorescent microspheres (Luminex Corp, Austin, TX) were covalently conjugated to DT (List Biologicals, Campbell, CA) and used to capture and quantify DT specific IgG in sera based on previous method with minor modifications 53. An in-house DT reference serum was generated with a human Intravenous immunoglobulin (IVIG C911, Gamunex, Telecris, NC) and anti-DT IgG titer (relationship between the median fluorescence intensity (MFI) and dilution factor) was expressed as arbitrary units per milliliter (AU/ml). Data reduction and analysis was performed with Masterplex CT/QT (MiraiBio, San Francisco, CA).
Measurement of serum bactericidal titers (SBA)
Serum samples collected at days 0, 7, 30, 180 and year 2 were added undiluted to microtiter plates (Costar) and then serially diluted (3-fold) in Hank’s balanced salt solution (HBSS, GIBCO/Invitrogen). A pre-determined dilution of meningococcal reference bacterial stock (serogroup A, strain F8238 or serogroup C, strain C11) from a 4 h active culture grown in brain heart infusion (BHI) agar supplemented with 1% horse serum that yielded ~400 to 750 bacteria per well was added to each reaction plate. A qualified source of complement from 3 to 4 week-old rabbit complement (Pel Freez, AR) was added at a concentration of 25% v/v. Plates were incubated for 1 h (T60), 37°C in the absence of CO2. Following the incubation period, a 10 μl aliquot per well was tilted onto supplemented BHI agar. Viability counts were read with an automated colony counter (Synbiosis Protocol, United Kingdom) after 16 to 18 h incubation. Continuous titers were determined by interpolation of the serum dilution yielding 50% killing as compared to the T60 growth control plate average. Reaction plates also included a serum growth control to check for any inhibitory factors in the serum sample and complement controls as described 22, 54. A titer of 1.33 was assigned to sera with no activity in the initial serum dilution of 1:4. Sera were also tested with N. meningitidis serogroup A strain 3125, L10 immunotype as described by Poolman et al. 55.
Measurement of antibody secreting cells by FACS and ELISpot
Blood ASC were identified by flow cytometry as CD3neg, CD20neg, CD38hi, CD27hi, CD19+ cells as described previously 56. Antigen specific and total IgG, IgM, and IgA secreting cells were quantified by ELISpot. Goat anti-human IgG, IgM, and IgA capture antibody (Jackson Immunoresearch; 100 ng per well) was used for total Ig capture. For capture of antibodies specific for meningitis polysaccharides, the tetravalent meningococcal polysaccharide vaccine (Menomune; Sanofi Pasteur) was conjugated to poly-L-lysine as previously described 57. One poly-L-lysine conjugated vaccine dose was coated per ELISpot plate (500 ng of each polysaccharide per well). For capture of antibodies specific for the diphtheria toxoid component of the conjugate meningitis vaccine, ELISpot plates were coated with CRM mutant diphtheria toxin (Calbiochem; 100 ng per well). Captured antibodies were detected with biotinylated anti human IgG, IgA, or IgM (Jackson Immunoresearch) and avidin HRP (Vector Labs).
Integration of Interactome and Bibliome to vaccine response networks
The interactome data used in this study was retrieved from Pathway Commons (Oct. 26, 2011,58), including data from several databases: HPRD 59, BioGRID 60, IntAct 61, MINT 62, Reactome 63, NCI/Nature PID 28 and HumanCyc 64. The Bibliome network was built using gene keywords in Pubmed entries (retrieved Aug. 26, 2010), where concurrence of two genes in the same paper will constitute an edge between two genes. Papers with 10 or more keyword genes were excluded, as they are likely to be based on high throughput assays.
From a vaccination blood transcriptome dataset, differentially expressed genes (DEGs) were identified (using p < 0.001 in Supplementary Table 1, no gene filtering). A DEG network was first built by connecting the upregulated DEGs with the edges from reference network (Interactome or Bibliome). Next, a “linker” gene is added into the DEG network if its connecting neighbors are significantly enriched within the DEG network compared to its neighbors in the interactome/bibliome network (Fisher exact test, p < 0.05 after Benjamini Hochberg correction). Its validity here can be evaluated by examining the “linker” genes. For instance, among the 312 “linker” genes that were added to the MCV4 network from interactome, 115 have p-value under 0.05 in the gene expression data (enrichment p-value 1.60E–19). Malaria RTS, S vaccine data 7 were used for further validation, where the top 1000 genes in paired t-test between the third vaccination and 24 hours post-vaccination were used as input DEGs. The same integration method yielded a RTS, S response network of 2951 genes, including 1231 genes that were found in our common network with 4+ vaccines.
Pathway and Gene Ontology analyses
We retrieved the XML version of NCI/Nature PID database (Sept. 14, 2011, 28), and parsed them into plain text gene sets. These PID pathways were used as external file in the GSEA program 23. Genes were ranked by t-score (comparing the transcriptome after and before vaccination) or Pearson correlation (correlating gene expression with antibody response), and were input to GSEA as externally supplied pre-ranked list. This approach offers good sensitivity and robustness, without relying on gene selection as in other over-representation methods. The significance of pathway enrichment in either scenario was assessed by permutation in GSEA program. Gene Ontology enrichment test (Fig. 3c) was performed using NIAID DAVID bioinformatics server 65.
Blood transcription module construction and correlation to antibody response
The construction and evaluation of blood transcription modules (BTM) is fully described in the Supplementary Note. The expression values of member genes in a module are combined into a single module activity score (the mean value). The module activity scores are used for subsequent analysis, such as Student t-test or Pearson correlation. All vaccine data were excluded from the BTM construction process. In antibody correlation analysis, the statistical significance of BTM modules was estimated by comparing to random permutation data, i.e., module memberships and sample labels were permutated to estimate the null distribution. Such significance is further enhanced when the same module is seen in two independent vaccine datasets. The analysis result with GSEA using BTM modules is comparable to that presented in this paper (Supplementary Table 3).
Day 30 polysaccharide specific IgG (sum of serogroup A and C) data subtracted by day 0 baseline were used for the antibody responses in MCV4 and MPSV4, as they were robust indicators throughout the study (Supplementary Fig. 1c). Day 30/0 anti-DT IgG data was used as DT specific response in MCV4. As previously reported, day 60 neutralizing antibody titers were used for the yellow fever vaccine 4, and maximum fold change of hemagglutination inhibition (HAI) titers (day 28/0) were used for the influenza TIV vaccine 6.
In vitro DC activation
Human PBMCs were obtained from buffy coat donors by centrifugation over Lymphoprep, and washed twice in MACS buffer (PBS, 2%FBS, 2mM EDTA). BDCA1+ blood myeloid DCs (mDC) and CD14+ monocytes were isolated with magnetic beads according to manufacturer’s instructions (Miltenyi Biotec, Auburn, CA, catalog numbers 130-090-506, 130-050-201). Monocytes were cultured for 6 days in complete RPMI medium-10% FBS with 20 ng/ml GM-CSF and 40ng/ml IL-4 (Peprotech, Rocky Hill, NJ) to generate monocyte-derived DCs (moDC). mDC and moDC were stimulated overnight (106/ml) in 96-well plates with dilutions of the MPSV4 or MCV4 vaccines (final concentration 0.1x, 0.01x, 0.001x), or LPS (10ug/ml Invivogen, San Diego, CA). Cells were stained with CD80-FITC, CD83-PE, HLA-DR-PerCP and CD86-APC (BD Biosciences, San Jose, CA), acquired on an LSR and analysed using FlowJo. The clones used for FACS staining were CD80-FITC (L307.4), CD83-PE (HB15e), HLA-DR-PerCP (L243) and CD86-APC (2331) (BD Biosciences). Cell supernatants were harvested and assessed for IL-6, TNF and IL-12p40 using ELISA kits, according to manufacturer’s instructions (BD Biosciences, catalog numbers 555220, 555212, 555171).
Spleens were isolated from C57BL/6 wild-type or knock-out mice, digested with collagenase (Worthington, Lakewood, NJ) for 30min at 37°C, and passed through a 40um cell strainer. The splenocytes were washed twice with MACS buffer, and CD11c+ DCs were isolated with magnetic beads (Miltenyi Biotec, catalog number 130-052-001). DCs were stimulated overnight (106/ml in 96-well plates) with dilutions of the MPSV4 or MCV4 vaccines (final concentration 0.1x, 0.01x, 0.001x) or TLR-ligand controls (Invivogen): Pam3Cys (10ug/ml), polyIC (100ng/ml), LPS (10ug/ml), FSL-1 (100ng/ml), R848 (10ug/ml) or CpG (10ug/ml). Cell supernatants were harvested and assessed for IL-6, TNF and IL-1b by ELISA (BD Biosciences, catalog numbers 555240, 555268, 559603). All mice were 12–16 weeks old, gender and age matched in all experiments. Mice were housed and handled according to local, state and federal regulations. All experimental procedures were carried out according to protocols approved by the Institutional Animal Care and Use Committee at Emory University.
Supplementary Material
Antibody responses induced by Meningococcal vaccines.
Day 7 activity of Plasma cells/Immunoglobulin module (M156.1, Fig. 4d) in blood transcriptome is correlated to day 30 antibody response in TIV.
Hierarchical clustering of antibody correlations of BTM modules.
Distinctive early transcription programs correlated to vaccine antibody response.
Dendritic cell related BTM modules and genes.
The DC surface signature module is positively correlated to polysaccharide response.
Polysaccharide vaccine stimulates dendritic cells.
MPSV4 stimulation of DC is dependent on MyD88, TRIF, TLR4 and ASC in vitro
Antigen-specific serum antibody and SBA titers induced by MCV4 and MPSV4 in mice in vivo.
Cell type patterns in BTM-antibody analysis.
BTM modules whose activity on day 7 is correlated to later antibody response in 2 or more datasets.
The XBP-1 network is up-regulated in MCV4 transcriptomic response.
B cell related BTM modules and genes.
Differential expression in five vaccine studies.
Correlation of BTM activity to vaccine antibody responses.
GSEA analysis using the BTM modules.
Supplementary Note. Construction of Blood Transcription Modules.
Supplementary Tutorial. Data and tutorial package of Blood Transcription Modules. This package provides BTMs in reusable data formats, and a step-by-step demonstration of their applications.
Cytokines and chemokines measured by Luminex assay for meningococcal vaccines.
Integration of DEGs with Interactome and Bibliome data.
Molecular pathways induced by different vaccines.
Pathways associated with antibody response.
Examples of protein complexes detected during BTM construction.
Examples of BTM modules.
Discriminative power of BTM modules compared to canonical pathways.
Acknowledgments
We thank Beverly Weaver and the Hope clinic Staff for their assistance with the clinical study. This study was supported by funding from the US National Institutes of Health (U19AI090023, U54AI057157, R37AI48638, R37DK057665, U19AI057266, AI100663-02) to B.P. laboratory; by the Georgia research Alliance GRA and Emory University Research Committee and from the Clinical and Translational Science Award Program, NIH/NCRR to Nadine Rouphael.
Footnotes
Data accession
Access to Online data portal (Firefox and Safari are recommended for best results): http://www.immuneprofiling.org/papers/meni/. The accession numbers for microarray data at Gene Expression Omnibus are MPSV4 and MCV4: GSE52245, YF-17D: GSE13485, Flu TIV: GSE29617, Flu LAIV: GSE29615.
Contributions
S.L. did analyses in Figures 4 and 5, Supplementary Figures 4–14 and 18–20. S.L. and H.I.N. did analyses in Figures 2 and 3, Supplementary Figure 2. N.R. organized the clinical study. S.D. did experiments and analyses in Figure 6, Supplementary Figures 15–17. S.L. and S.D. did analysis in Supplementary Figure 3. S. R-S., D.S.S., S.E.J., A.M., G.R. and G.M.C. did experiments in Figure 1. S.P., C.Q., D.C. and A.K.P. prepared online data portal and Figure 3a. C.D. did experiments in Supplementary Figure 1a, b. S.K. assisted with experiments in Figure 6 and Supplementary Figure 16. M.J.M. supervised the clinical study. R.A. supervised the study in Supplementary Figure 1a, b. D.S.S. helped conceive of and design the study. H.I.N. helped with study design and presentation. B.P. conceived of the study and designed and supervised the experiments and analyses. S.L., H.I.N. and B.P. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC).
References and Notes
- 1.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nature Reviews Immunology. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 2.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakaya HI, Pulendran B. Systems vaccinology: its promise and challenge for HIV vaccine development. Current Opinion in HIV and AIDS. 2012;7:24. doi: 10.1097/COH.0b013e32834dc37b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology. 2008;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. The Journal of experimental medicine. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vahey MT, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS, S malaria vaccine. J Infect Dis. 2010;201:580–589. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- 8.Bucasas KL, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zak DE, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Natl Acad Sci U S A. 2012;109:E3503–3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberg AL, Kennedy RB, Li P, Ovsyannikova IG, Poland GA. Systems biology approaches to new vaccine development. Current opinion in immunology. 2011;23:436–443. doi: 10.1016/j.coi.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trautmann L, Sekaly R. Solving vaccine mysteries: a systems biology perspective. Nature immunology. 2011;12:729. doi: 10.1038/ni.2078. [DOI] [PubMed] [Google Scholar]
- 14.Querec T, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. The Journal of experimental medicine. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geeraedts F, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 2008;4:e1000138. doi: 10.1371/journal.ppat.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koyama S, et al. Plasmacytoid dendritic cells delineate immunogenicity of influenza vaccine subtypes. Sci Transl Med. 2010;2:25ra24. doi: 10.1126/scitranslmed.3000759. [DOI] [PubMed] [Google Scholar]
- 17.Rouphael NG, Stephens DS. Neisseria meningitidis: Biology, Microbiology, and Epidemiology. Neisseria Meningitidis: Advanced Methods and Protocols. 2012;799:1–20. doi: 10.1007/978-1-61779-346-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969;129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayhty H, Karanko V, Peltola H, Sarna S, Makela PH. Serum antibodies to capsular polysaccharide vaccine of group A Neissera meningitidis followed for three years in infants and children. J Infect Dis. 1980;142:861–868. doi: 10.1093/infdis/142.6.861. [DOI] [PubMed] [Google Scholar]
- 20.Jokhdar H, et al. Immunologic hyporesponsiveness to serogroup C but not serogroup A following repeated meningococcal A/C polysaccharide vaccination in Saudi Arabia. Clin Diagn Lab Immunol. 2004;11:83–88. doi: 10.1128/CDLI.11.1.83-88.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Findlow H, et al. Immunoglobulin G subclass response to a meningococcal quadrivalent polysaccharide-diphtheria toxoid conjugate vaccine. Clin Vaccine Immunol. 2006;13:507–510. doi: 10.1128/CVI.13.4.507-510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrow R, Balmer P, Miller E. Meningococcal surrogates of protection--serum bactericidal antibody activity. Vaccine. 2005;23:2222–2227. doi: 10.1016/j.vaccine.2005.01.051. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PloS one. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nibbe RK, Koyuturk M, Chance MR. An integrative -omics approach to identify functional subnetworks in human colorectal cancer. PLoS Comput Biol. 2010;6:e1000639. doi: 10.1371/journal.pcbi.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Nakaya HI, Kazmin DA, Oh JZ, Pulendran B. Systems biological approaches to measure and understand vaccine immunity in humans. Semin Immunol. 2013 doi: 10.1016/j.smim.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau E, Ronai ZA. ATF2-at the crossroad of nuclear and cytosolic functions. Journal of Cell Science. 2012;125:2815–2824. doi: 10.1242/jcs.095000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaveroux C, et al. Identification of a Novel Amino Acid Response Pathway Triggering ATF2 Phosphorylation in Mammals. Molecular and Cellular Biology. 2009;29:6515–6526. doi: 10.1128/MCB.00489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haining WN, Pulendran B. Identifying gnostic predictors of the vaccine response. Current opinion in immunology. 2012 doi: 10.1016/j.coi.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaussabel D, Pascual V, Banchereau J. Assessing the human immune system through blood transcriptomics. BMC biology. 2010;8:84. doi: 10.1186/1741-7007-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser IDC, Germain RN. Navigating the network: signaling cross-talk in hematopoietic cells. Nature Immunology. 2009;10:327–331. doi: 10.1038/ni.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 35.Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 36.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 37.Lee I, et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 38.Chaussabel D, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC bioinformatics. 2006;7 (Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lefebvre C, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre C, Rieckhof G, Califano A. Reverse-engineering human regulatory networks. Wiley Interdiscip Rev Syst Biol Med. 2012;4:311–325. doi: 10.1002/wsbm.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sales G, Romualdi C. parmigene--a parallel R package for mutual information estimation and gene network reconstruction. Bioinformatics. 2011;27:1876–1877. doi: 10.1093/bioinformatics/btr274. [DOI] [PubMed] [Google Scholar]
- 43.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular Systems Biology. 2007;3 doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fredlund E, Ringner M, Maris JM, Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vitour D, et al. Polo-like kinase 1 (PLK1) regulates interferon (IFN) induction by MAVS. J Biol Chem. 2009;284:21797–21809. doi: 10.1074/jbc.M109.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, et al. The scaffold protein TANK/I-TRAF inhibits NF-kappaB activation by recruiting polo-like kinase 1. Mol Biol Cell. 2010;21:2500–2513. doi: 10.1091/mbc.E09-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zughaier SM. Neisseria meningitidis capsular polysaccharides induce inflammatory responses via TLR2 and TLR4-MD-2. J Leukoc Biol. 2011;89:469–480. doi: 10.1189/jlb.0610369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein SL, Poland GA. Personalized vaccinology: One size and dose might not fit both sexes. Vaccine. 2013;31:2599–2600. doi: 10.1016/j.vaccine.2013.02.070. [DOI] [PubMed] [Google Scholar]
- 49.Nakaya HI, Li SZ, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdisciplinary Reviews-Systems Biology and Medicine. 2012;4:193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avci FY, Li XM, Tsuji M, Kasper DL. A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design. Nature Medicine. 2011;17:1602–U1115. doi: 10.1038/nm.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holder PK, et al. Assignment of Neisseria meningitidis serogroup A and C class-specific anticapsular antibody concentrations to the new standard reference serum CDC1992. Clin Diagn Lab Immunol. 1995;2:132–137. doi: 10.1128/cdli.2.2.132-137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gheesling LL, et al. Multicenter comparison of Neisseria meningitidis serogroup C anti-capsular polysaccharide antibody levels measured by a standardized enzyme-linked immunosorbent assay. J Clin Microbiol. 1994;32:1475–1482. doi: 10.1128/jcm.32.6.1475-1482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Gageldonk PG, van Schaijk FG, van der Klis FR, Berbers GA. Development and validation of a multiplex immunoassay for the simultaneous determination of serum antibodies to Bordetella pertussis, diphtheria and tetanus. J Immunol Methods. 2008;335:79–89. doi: 10.1016/j.jim.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Maslanka SE, et al. Standardization and a multilaboratory comparison of Neisseria meningitidis serogroup A and C serum bactericidal assays. The Multilaboratory Study Group. Clin Diagn Lab Immunol. 1997;4:156–167. doi: 10.1128/cdli.4.2.156-167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poolman JT, et al. Measurement of functional anti-meningococcal serogroup a activity using strain 3125 as the target strain for serum bactericidal assay. Clin Vaccine Immunol. 2011;18:1108–1117. doi: 10.1128/CVI.00549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreno RL, et al. A murine model for the study of immune memory in response to pneumococcal conjugate vaccination. Vaccine. 2004;22:3069–3079. doi: 10.1016/j.vaccine.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aranda B, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ceol A, et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romero P, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibody responses induced by Meningococcal vaccines.
Day 7 activity of Plasma cells/Immunoglobulin module (M156.1, Fig. 4d) in blood transcriptome is correlated to day 30 antibody response in TIV.
Hierarchical clustering of antibody correlations of BTM modules.
Distinctive early transcription programs correlated to vaccine antibody response.
Dendritic cell related BTM modules and genes.
The DC surface signature module is positively correlated to polysaccharide response.
Polysaccharide vaccine stimulates dendritic cells.
MPSV4 stimulation of DC is dependent on MyD88, TRIF, TLR4 and ASC in vitro
Antigen-specific serum antibody and SBA titers induced by MCV4 and MPSV4 in mice in vivo.
Cell type patterns in BTM-antibody analysis.
BTM modules whose activity on day 7 is correlated to later antibody response in 2 or more datasets.
The XBP-1 network is up-regulated in MCV4 transcriptomic response.
B cell related BTM modules and genes.
Differential expression in five vaccine studies.
Correlation of BTM activity to vaccine antibody responses.
GSEA analysis using the BTM modules.
Supplementary Note. Construction of Blood Transcription Modules.
Supplementary Tutorial. Data and tutorial package of Blood Transcription Modules. This package provides BTMs in reusable data formats, and a step-by-step demonstration of their applications.
Cytokines and chemokines measured by Luminex assay for meningococcal vaccines.
Integration of DEGs with Interactome and Bibliome data.
Molecular pathways induced by different vaccines.
Pathways associated with antibody response.
Examples of protein complexes detected during BTM construction.
Examples of BTM modules.
Discriminative power of BTM modules compared to canonical pathways.