Abstract
Purpose
Tuberculosis treatments need to be shorter and overcome drug resistance. Our previous large scale phenotypic high-throughput screening against Mycobacterium tuberculosis (Mtb) has identified 737 active compounds and thousands that are inactive. We have used this data for building computational models as an approach to minimize the number of compounds tested.
Methods
A cheminformatics clustering approach followed by Bayesian machine learning models (based on publicly available Mtb screening data) was used to illustrate that application of these models for screening set selections can enrich the hit rate.
Results
In order to explore chemical diversity around active cluster scaffolds of the dose-response hits obtained from our previous Mtb screens a set of 1924 commercially available molecules have been selected and evaluated for antitubercular activity and cytotoxicity using Vero, THP-1 and HepG2 cell lines with 4.3%, 4.2% and 2.7% hit rates, respectively. We demonstrate that models incorporating antitubercular and cytotoxicity data in Vero cells can significantly enrich the selection of non-toxic actives compared to random selection. Across all cell lines, the Molecular Libraries Small Molecule Repository (MLSMR) and cytotoxicity model identified ~10% of the hits in the top 1% screened (>10 fold enrichment). We also showed that seven out of nine Mtb active compounds from different academic published studies and eight out of eleven Mtb active compounds from a pharmaceutical screen (GSK) would have been identified by these Bayesian models.
Conclusion
Combining clustering and Bayesian models represents a useful strategy for compound prioritization and hit-to lead optimization of antitubercular agents.
Keywords: Bayesian models, clustering, Collaborative Drug Discovery Tuberculosis database, Dual-event models, Function class fingerprints, Lead optimization, Mycobacterium tuberculosis, Tuberculosis
INTRODUCTION
Research targeted toward the identification of small molecule inhibitors of Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), has more recently focused on whole-cell phenotypic screening (1–6). Even though effective treatments have been approved for drug-sensitive infections, an urgent need exists for next generation drugs (7, 8) to address rising drug resistance for a disease that infects approximately one-third of the world’s population and kills 1.7–1.8 million people annually (9). Key to this effort has been research on new drugs that would significantly decrease treatment time of drug-sensitive TB from its current 6–9 month regimen (7, 8). Although many laboratories have screened libraries numbering 103 – 106 compounds (4, 5), the hit rate is usually below 1% (2, 3) as typically seen in many other high-throughput screening (HTS) (2, 3, 10, 11) campaigns for TB as well as other therapeutic indications (10–12). Occasionally the hit rate can reach the low single digits (~1.7–5%) (4–6). These TB HTS efforts are delivering interesting and potentially promising hits (albeit at great cost, Table 1), and in excess of a thousand actives may be deserving of follow-up. Hit-to-lead optimization must be properly balanced with continuing efforts to screen even larger compound libraries to more thoroughly cover chemical space and/or sample different experimental conditions to better mimic human TB infection (13).
Table 1.
Scoring recent Mtb hits from high-throughput screens with dual-event Bayesian Models (bold text = predicted active)
| Name | Mtb Activity in vitro | Notes | Reference | MLSMR dose response and cytotoxicity Bayesian score | TAACF-CB2 dose response and cytotoxicity Bayesian score | TAACF Kinase dose response and cytotoxicity Bayesian score | Maximum Tanimoto similarity to MLSMR set using MDL Keys |
|---|---|---|---|---|---|---|---|
Oxyphenbutazone
|
MIC 10μg/ml | Active against non-replicating TB Metabolite active against replicating TB-screened ~5600 compounds | (10) | −0.88 | 0.51 | 1.34 | 0.76 |
GNF-NITD 101
|
IC50 3.4μM | Active in hypoxic model, used to screen of 600,000 compounds | (2) | 0.51 | 1.55 | −4.93 | 0.68 |
GNF-NITD 82
|
IC50 9.3μM | Active in hypoxic model, used to screen of 600,000 compounds | (2) | −18.96 | 4.02 | −4.63 | 0.79 |
GNF-NITD 46
|
IC50 7.4μM | Active in hypoxic model, used to screen of 600,000 compounds | (2) | −12.49 | −1.32 | −3.92 | 0.75 |
377790
|
IC50 0.5μM | From a screen of 20,000 compounds – Target DprE1 | (3) | −3.69 | 2.77 | 3.87 | 0.81 |
A039
|
IC90 1.5μM | From a screen of 20,000 compounds | (3) | −6.02 | 1.72 | −3.91 | 0.82 |
Gliotoxin
|
IC90 0.14μM | From a screen of 20,000 compounds | (3) | 4.40 | −2.20 | −2.25 | 0.64 |
C215
|
IC90 16μM | From a screen of 20,000 compounds – target MmpL3 | (3) | −17.44 | 1.48 | −3.86 | 0.74 |
AU1235
|
MIC 0.1μg/ml | From a screen of 12,000 compounds – Target MmpL3 | (53) | −8.24 | −3.16 | −8.07 | 0.76 |
The large number of hits for hit-to-lead optimization coupled with limited resources can benefit from established and highly efficient computational methods to expedite evolution of novel antitubercular lead compounds for clinical development. We and others (14–22) have suggested that computational approaches can assist in identifying compounds with activity against Mtb (20) and, in particular, Bayesian classification models are valuable (16–19, 23). More recently, we have described dual-event models that combine Mtb growth inhibition activity and cytotoxicity data to improve selection of actives with antitubercular activity (measured by IC90 – the concentration of compound inhibiting bacterial growth by 90%) less than 10 μg/mL (or 10 μM depending on the original chemical library format and dataset) and a selectivity index (SI = CC50/IC90 where CC50 = concentration of compound inhibiting growth of a cultured mammalian cell line, Vero cells, by 50%) greater than ten (22). We demonstrated using data from multiple laboratories that there are clear benefits of this approach: computational screening of 82,403 commercially available small molecules predicted 550 actives, which were assayed to identify 124 hits (22.5% hit rate) in one study (24), while another study computationally screened >13,000 molecules, assayed seven predicted actives and found five hits empirically (71% hit rate) (22).
We hypothesized that the Bayesian model technology could also positively impact hit-to-lead optimization. This phase of drug discovery is a significant driver of both process time and cost, typically entailing the design, synthesis, and biological evaluation of hundreds to thousands of compounds (25). A computationally-enhanced approach would expand the chemical space explored given the potentially unrestricted querying of commercial libraries for follow-up compounds and/or hit substituents derived from reactive building blocks. At the same time, this in silico approach could enable the efficient selection of a significantly smaller set of compounds for testing through the prioritization of analogs by their Bayesian score, which, in general, scales with the likelihood of activity. The following study describes the benefits of implementing Bayesian dual-event models in conjunction with the commonly used approach of hit structure clustering followed by the expansion of chemical space around core cluster scaffolds through commercial analog selections (26, 27) (Fig. 1). We also note how different mammalian cell types utilized in cytotoxicity determination can impact the rate at which active analogs are found.
Figure 1.
Schematic illustrating the integrated in vitro and computational processes described in this study
MATERIALS AND METHODS
Small Molecules
Small molecules for biological assay were purchased from Life Chemicals (Ontario, Canada) and ChemBridge (San Diego, CA).
CDD Database and SRI datasets
The development of the CDD TB database (Collaborative Drug Discovery Inc. Burlingame, CA) has been previously described (17). The Tuberculosis Antimicrobial Acquisition and Coordinating Facility (TAACF) and Molecular Libraries Small Molecule Repository (MLSMR) screening datasets (4–6) were collected and uploaded in CDD TB from sdf files and mapped to custom protocols (28). All of these Mtb datasets used in model building are available for free public read-only access and mining upon registration in the CDD database (18, 28–30), making them a valuable molecule resource for researchers along with available contextual data on these samples from other non Mtb assays. These datasets used previously for modeling are also publically available in PubChem (31). All data generated in this study (TB: ARRA) is available in the CDD TB database (Collaborative Drug Discovery, Burlingame, CA) (28).
Compound Selection and Clustering
Active compounds from previous H37Rv screens of the MLSMR, TAACF datasets and the kinase library from LifeChemicals (totaling ~4000 dose-response hits) have been clustered to identify common core scaffolds and analog series present among actives, as described previously (4–6). For cluster analyses a hierarchical clustering method implemented in LeadScope (LeadScope, Inc. Columbus OH.) was used applying default parameters. Clusters were separated using the ‘Complete Linkage (Furthest Neighbor)’ method with the cluster threshold distance set to 0.7. Each cluster may be characterized by a cluster scaffold that is a common core structure shared by all of its members. Clusters were also prioritized based on an enrichment ratio computed for each cluster, defined as the ratio of the percentage of compounds containing the cluster scaffold within the active (clustered) set and the percentage of such compounds within the entire library. High enrichment ratios are associated with structural motifs preferred among actives compared to primary screened compounds. Clusters with enrichment ratios below a specified threshold were excluded from further consideration; for the MLSMR and Chembridge datasets we used the threshold of ten and for the kinase library the more permissive value of five. We obtained 22, 29 and 26 conformational clusters corresponding to MLSMR, Chembridge and kinase library dose-response datasets, respectively. Out of these clusters, 30 cluster scaffolds were selected for follow-up by expanding variation around the core scaffold structures and potential SAR versus Mtb through the selection of analog series from commercial sources. Out of cluster scaffolds obtained from clustering dose-response hits from all three screens (MLSMR, TAACF datasets and the kinase library) totaling close to 4000 compounds, we selected 30 cluster scaffolds that were well represented among commercially available compounds. Analog compounds exploring chemical diversity around these 30 cluster scaffolds were selected from the Chembridge and Life Chemicals commercial libraries. Commercial compounds identical to any primary screened compounds were excluded except those that show inhibition of > 80% in primary screens but were not tested in dose-response. The final selection included 1847 compounds is described by the selected 30 major clusters out of which large clusters may be further grouped into sub-clusters, totaling 55 sub-clusters/clusters (or 30 major clusters). We also chose to add an additional cluster of 12 analogs (based on a designed ‘hybrid’ cluster scaffold) and 65 diverse compounds, totaling 1924 purchased from the aforementioned vendors.
Bacterial strain, growth conditions and media
Mtb H37Rv (ATCC 27294) was obtained from the American Type Culture Collection (Manassas, VA). To prepare permanent frozen stocks, H37Rv was grown as five mL subcultures (50 mL conical tubes, 36–37°C) in Middlebrook 7H9 broth (Becton Dickinson) supplemented with 0.2% glycerol (Becton Dickinson), 0.05% Tween 80 (Becton Dickinson), and 10% ADC enrichment (albumin, dextrose, catalase; Becton Dickinson). The subculture was mixed periodically and used to inoculate (5% inoculum) a second subculture (30 mL in 250 mL screw cap flask) when the turbidity reached a density similar to a #1 McFarland turbidity standard (A600 nm ~0.2). The subcultures were incubated with periodic mixing for 18–21 days until the turbidity reached a #3–#4 McFarland turbidity standard (A600nm ~0.6–0.8, 4–8×107 CFU/mL). The caps on both the conical tubes and flasks were loosened and wrapped in parafilm to allow for adequate gas exchange and to prevent evaporation during incubation. Prior to harvest, samples from all cultures were spotted onto Trypticase Soy Agar (TSA) plates and incubated for 3–4 days to check for contamination. Mtb grows poorly on TSA which supports the growth of most potential contaminating microorganisms. Each culture was then transferred to a 50 mL tube and allowed to settle at ambient temperature for one h. The upper half of each culture was aspirated and pooled in a flask. Aliquots of one mL were then transferred to two mL cryovials and frozen at −80°C. At least three frozen stocks were thawed and used to determine the viable count by plating dilutions, prepared in supplemented 7H9 broth, onto Middlebrook 7H11 Agar followed by incubation for up to 21 days. A contamination check on the thawed cultures was also performed as described above.
Mtb Assay
Primary screening against replicating cultures of Mtb were determined using modifications to the microplate Alamar Blue assay (MABA (32, 33)) as previously published (4). This assay is widely used for HTS screening by many laboratories as recently documented (34). Antitubercular activity was determined against Mtb H37Rv ATCC 27294 following 7 days incubation with test compounds. Compounds were evaluated initially in a stacked-plate dose response and final test concentrations for the compounds ranged from 100 μM to 0.0195 μM in two-fold dilutions with a final DMSO concentration of 1.0%.
Cytotoxicity in Vero Cells of compounds that inhibit Mtb
Cytotoxicity for Vero cells (ATCC CCL-81) was determined following 72 hours exposure (33). Cell viability was assessed using CellTiter-Glo reagent (Promega) according to the manufacturer’s protocol.
Cytotoxicity in THP-1 cells of compounds that Inhibit Mtb
This functional assay was developed for detection of compounds inhibiting THP-1 cells viability as a secondary screen to the Mtb bactericidal assay. The THP-1 cell line was chosen as a representative peripheral blood monocyte. In this assay, we treated THP-1 cells with compounds selected as “hits” in the Mtb assay over a 10 point 2-fold dilution series, ranging from 40 μM to 0.078 μM. Following 72 h of treatment, relative viable cell number was determined using Cell Titer Glo from Promega. Each plate contained 64 replicates of vehicle treated cells which served as controls. THP-1 cells were sub-cultured every 7 days in RPMI 1640 with 10% fetal bovine serum, incubated at 37 °C in 5% carbon dioxide. Cells were passaged as needed, harvested from flasks using 0.25% trypsin-EDTA and maintained for no more than 20 passages. Compounds and carrier controls were diluted in complete growth medium to prepare a 6× concentrated dosing solution which was dispensed into 384-well black clear-bottom tissue culture treated plates (5 μL volume). The final DMSO concentration for this assay was 0.4%. Cells were harvested as previously described. Twenty microliters of complete growth medium containing 3000 cells were dispensed per well. Plates were incubated at 37 °C, 5% CO2 for 72 h prior to endpoint detection. At the end of the treatment period, assay plates were removed from the incubator and equilibrated to room temperature for 10 min. Twenty-five μL of Cell Titer Glo reagent was added and plates were incubated for an additional 10 min in the dark. At the end of the incubation, assay plates were analyzed using a PerkinElmer Envision microplate reader in luminescence mode with an integration time of 0.1 s.
Cytotoxicity in HepG2 Cells of compounds that inhibit Mtb
This functional assay was developed for detection of compounds inhibiting HepG2 cells viability as a secondary screen to the Mtb bactericidal assay. In this assay, HepG2 cells were treated with compounds selected as “hits” in the Mtb assay for 72 h over a 10 point 2-fold dilution series, ranging from 20 μM to 0.39 μM. Following the incubation, the relative viable cell number was determined using Cell Titer Glo (Promega). Each plate contained 32 replicates of vehicle treated cells which served as negative controls and 32 wells of 100 μM hyamine-treated cells that represent a positive control. The maintenance of the HepG2 cells followed the recommendations of the ATCC. Cells were passaged as needed, harvested from flasks using 0.25% trypsin-EDTA and maintained for no more than 20 passages. On the day of the assay, compounds or carrier control (DMSO) were diluted to 6× in complete growth medium supplemented with 1% Penicillin/Streptomycin and 5 μL was dispensed into 384-well black clear-bottom tissue culture treated plates using a Biomek FX. The DMSO concentration was maintained at 0.2% final concentration. A 10 point 2-fold serial dilution was generated in the “stacked plate” method previously published by this group (4). The HepG2 cells were harvested as previously indicated and the concentration was adjusted to 1.5×105 cells/mL in complete growth medium supplemented with 1% Penicillin/Streptomycin. Using a Matrix WellMate in a certified biosafety cabinet, twenty microliters or approximately 3000 cells were dispensed to each well in the 384-well plate. The plates were then incubated at 37 °C, 5% CO2 for 72 h prior to endpoint detection. Following the 72 h incubation period, the assay plates were equilibrated to room temperature for 10 min and twenty-five microliters of Cell Titer Glo reagent (Promega) was added to each well using a WellMate (Matrix, Hudson, NH). The plates were then incubated for an additional 10 min at room temperature. At the end of the incubation, luminescence was measured using a Perkin Elmer Envision microplate reader with an integration time of 0.1 s.
Biological Data Analysis
All data were imported into ActivityBase (IDBS) data management system for analyses and calculation of IC50 and IC90 values. Percent Inhibition was calculated as: 100× (1-(Median of Test Compound – Median of Positive control)/Median of Negative control – Median of Positive control)). Selectivity Index (SI) was calculated as SI = CC50/IC90, where CC50 = concentration of compound inhibiting growth of cultured cells by 50%.
Using Dual Event Machine Learning Models with novel bioactivity and cytotoxicity data
We have previously described the generation and validation of the Laplacian-corrected Bayesian classifier models developed with cytotoxicity data to create Mtb dual-event models (22, 24) using Discovery Studio (16, 35–38). These models (22, 24) were developed based on: a. MLSMR dose response and cytotoxicity; b. CB2 dose response and cytotoxicity; and c. TAACF Kinase dose response and cytotoxicity, where cytotoxicity was determined in Vero cells for each set. All three models were generated using standard protocols using the following molecular descriptors: molecular function class fingerprints of maximum diameter 6 (FCFP_6) (39), AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area were calculated from input sdf files. Models were validated using leave-one-out cross-validation in which each sample was left out one at a time, a model was built using the remaining samples, and that model utilized to predict the left-out sample. Each model was internally validated and receiver operator characteristic (ROC) plots generated, and the cross-validated ROC area under the curve (XV ROC AUC) calculated. All models generated were additionally evaluated by leaving out 50% of the data and rebuilding the model 100 times using a custom protocol for validation, to generate the ROC AUC, concordance, specificity and selectivity as described previously (22, 24). The three models were used in this study to score a set of 1924 commercial analogs that expand the selected 30 major clusters (or 55 sub-clusters/clusters) obtained from cluster analyses of these screens. The set of 1924 compounds have been evaluated in dose-response in whole cell Mtb assay and Vero, THP-1 and HepG2 cytotoxicity assays. Defining non-toxic actives those that possess IC90 < 10 μg/ml and SI > 10, we obtained 82, 81 and 52 non-toxic Mtb actives based on Vero, THP-1 and HepG2 cytotoxicity data sets, respectively. The prediction data were evaluated using a ROC plot and also with standard statistics (sensitivity, specificity, prediction accuracy and Matthews correlation).
Further retrospective evaluation of Dual Event Machine Learning Models
The previously developed dual-event Mtb and cytotoxicity models (22, 24) were further evaluated using a set of nine molecules collated from recent academic Mtb HTS studies (Table 1) as well as eleven hit molecules from GSK (Table 2) (40). These molecules were sketched using the mobile application Mobile Molecular DataSheet (Molecular Materials Informatics, Montreal, CA) (41, 42) to create sdf files which were used in Discovery Studio for prediction with the Bayesian models.
Table 2.
Scoring Mtb hits from GSK high throughput screen with dual event Bayesian Models (bold text = predicted active)
| Molecule | Name | H37Rv MIC (μM) | MLSMR dose response and cytotoxicity Bayesian score | CB2 dose response and cytotoxicity Bayesian score | TAACF Kinase dose response and cytotoxicity Bayesian score |
|---|---|---|---|---|---|

|
GSK153890A | 0.47 | −10.79 | −1.60 | −1.99 |
|
GSK163574A | 0.76 | −10.00 | −2.27 | −2.93 |

|
GSK1589673A | 0.25 | 26.39 | 3.33 | −10.81 |

|
GSK1829820A | 0.19 | 12.03 | 3.24 | 4.94 |

|
GSK1829731A | 0.19 | 3.51 | −0.46 | −3.85 |

|
GSK2200150A | 0.38 | −13.49 | 0.61 | −10.36 |

|
GSK353069A | 0.13 | −7.24 | 2.47 | −9.18 |

|
GSK358607A | 0.7 | −14.10 | 3.76 | −10.96 |

|
GSK749336A | 0.25 | −13.58 | 0.85 | −10.21 |

|
GSK888636A | 0.94 | 0.10 | 1.07 | −7.12 |

|
GW623128 | 0.47 | −9.78 | −2.56 | −2.66 |
RESULTS
Hierarchical clustering of actives from three previous antitubercular screens
Dose response hit compounds from three previous antitubercular screens (4–6) of the MLPCN (MLSMR), TAACF (Chembridge) and the Life Chemicals kinase libraries were pooled and clustered to identify common core scaffolds and analog series present among actives, as described previously (Fig. 2) (4–6). The pool of clustered compounds consisted of dose-response hits from the Chembridge and kinase libraries with IC90 < 10 μg/mL and additionally we chose to include all dose-response hits from the MLSMR screen, totaling close to 4000 hits that were included for cluster analysis (see Materials and Methods). Within these 4000 hits 737 compounds satisfy the criteria of IC90 < 10 μg/mL and SI (Vero cells) > 10, however we chose to use this larger, more inclusive set for the purposes of conformational clustering. The enrichment of cluster scaffolds among actives was assessed through the computation of cluster enrichment ratios, as described in detail under the ‘Materials and Methods’. Briefly, only cluster scaffolds represented as ‘enriched’ among dose-response hits compared to primary screening libraries were considered for the selection of commercial analogs. Based on the scaffolds shown in Fig. 2 a total of 1924 commercially available compounds were selected to explore the chemical space and Mtb SAR around these cluster scaffolds. As also shown in Fig. 2, large clusters have been grouped into sub-clusters, totaling 55 sub-clusters/clusters (or 30 major clusters, Table 3) and an additional cluster (Cluster 28) was also added based on a designed ‘hybrid’ substructure.
Figure 2.
Cluster scaffolds of MDR TB DR hits following hierarchical clustering as described in the Materials and Methods.
Table 3.
Number of cluster members listed for each cluster that contain at least one (or more) non-toxic active cluster member, defined as showing activity against Mtb. with IC90 (averaged over three runs) < 10 μg/mL while possessing a selectivity index (SI) > 10 in any one out of the three cytotoxicity assays (Vero, THP-1, HepG2 cell lines). For each cluster the numbers of compounds are listed as follows: total number per cluster (‘Total’), dose-response actives with IC90 < 10 μg/mL (‘DR-A’), and non-toxic actives as defined above (‘NT-A’). Cluster scaffold structures of the listed clusters are shown in Figure 2 Note, to avoid duplicate listings of the same compound, compounds that belong to multiple clusters are counted once, in the first listed cluster only.
| Cluster | Number of compounds | ||
|---|---|---|---|
| Total | DR-A | NT-A | |
| 1e | 204 | 69 | 1 |
| 1i | 98 | 17 | 1 |
| 2a | 53 | 13 | 1 |
| 2d | 20 | 5 | 1 |
| 2g | 49 | 5 | 1 |
| 4a | 9 | 2 | 1 |
| 4b | 51 | 25 | 13 |
| 4c | 34 | 13 | 9 |
| 4d | 9 | 3 | 2 |
| 5a | 16 | 9 | 4 |
| 5b | 13 | 9 | 5 |
| 5d | 9 | 3 | 3 |
| 5e | 52 | 11 | 6 |
| 7b | 17 | 2 | 2 |
| 7c | 9 | 1 | 1 |
| 7d | 55 | 8 | 7 |
| 8a | 41 | 7 | 1 |
| 9b | 61 | 9 | 2 |
| 10b | 17 | 2 | 1 |
| 10c | 42 | 12 | 2 |
| 11a | 15 | 3 | 1 |
| 15 | 45 | 2 | 2 |
| 17 | 43 | 14 | 7 |
| 20 | 33 | 15 | 5 |
| 21 | 30 | 7 | 1 |
| 22 | 27 | 3 | 1 |
| 23 | 25 | 16 | 13 |
| 24 | 24 | 11 | 1 |
| 25 | 32 | 5 | 2 |
| 26 | 10 | 2 | 1 |
In vitro Screens for growth inhibition of Mtb and cytotoxicity
The selected 1924 commercial compounds were tested in vitro for growth inhibition of Mtb and cytotoxicity versus three distinct mammalian cell lines: THP-1, Vero and HepG2. For each cell line used for cytotoxicity assessment, SI values have been calculated as the ratio of CC50 determined in each cytotoxicity assay and the antitubercular IC90 activity (averaged over three runs). Defining non-cytotoxic Mtb actives as possessing IC90 < 10 μg/ml and SI > 10, out of the 1924 commercial compounds 82, 81 and 52 molecules satisfy the criteria of non-cytotoxic actives in Vero, THP-1 and HepG2 cells, respectively (Table S1). These numbers represent ‘hit rates’ of 4.3%, 4.2% and 2.7% for the three cytotoxicity cell lines, retrospectively. The use of a more stringent efficacy and/or SI cutoff would naturally decrease the hit rate and be useful for exploring in hit evolution.
Bayesian Machine learning for hit-to-lead optimization
An alternate approach to expanding on the diversity of these ~4000 screening actives, while also seeking to enhance their antitubercular growth inhibition and SI values, relies on our recently published dual-event Bayesian machine learning. These models have been educated through learning which compound physiochemical and structural features are consistent with activity and promising SI. Importantly, the models have been validated through retrospective enrichment studies with published screening datasets as well as prospective prediction of actives from a GlaxoSmithKline antimalarial library (22). We have also utilized the dual-event models to score nine recently published hits from Mtb whole-cell screening campaigns (Table 1) that were derived after our dual-event models were built. Using the panel of three Bayesian models, we would have identified seven of nine of the molecules as actives. Interestingly, the maximal Tanimoto similarity using MDL keys and the MLSMR dose response and cytotoxicity model dataset was quite high (range 0.64–0.82) and yet the model only correctly identified two molecules. The TAACF-CB2 dose response and cytotoxicity model alone performed better with these molecules in this case and alone would have selected five of the nine compounds. Similarly, a second test dataset of 11 active molecules that were tabulated in a paper describing the HTS of two million compounds performed by GSK (40) against Mtb was analyzed, and at least one of the three models predicted eight of the compounds as hits (Table 2). These results highlight the need for further studies to comprehend what model factors influence predictive value, or whether the utilization of a consensus scoring approach with our dual-event Bayesian models could further enhance their ability to pick actives.
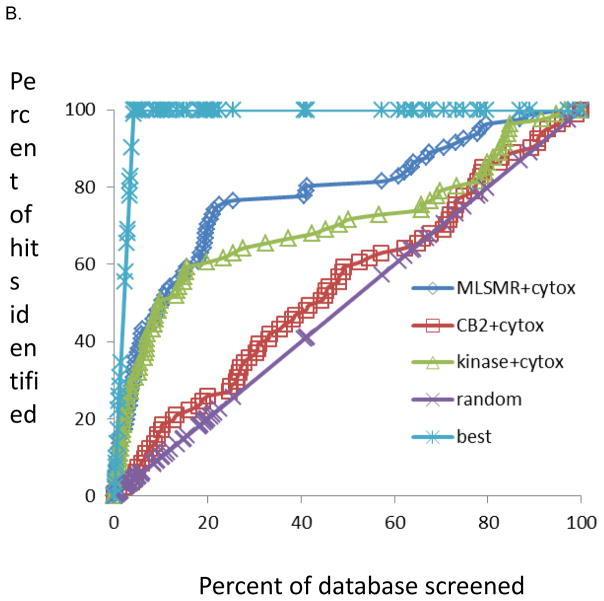
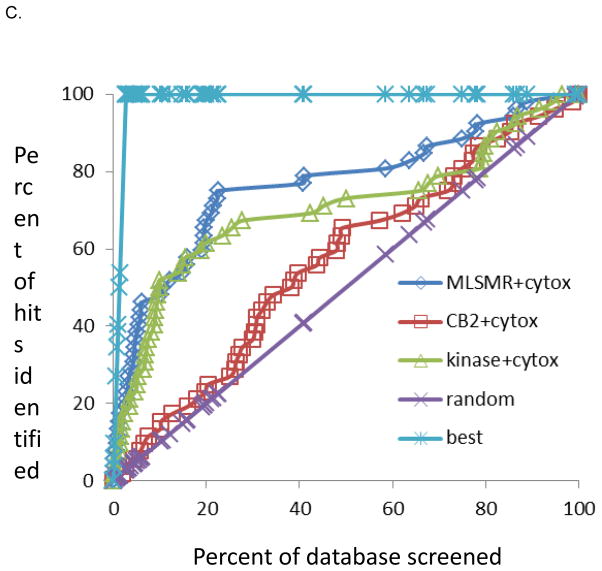
The dataset of 1924 molecules selected by clustering was virtually screened with our current three previously generated dual-event Mtb and cytotoxicity models. The molecules were ranked using the classification from all three models, and the receiver operator curve plot was generated (Fig. 3). The MLSMR dose response and cytotoxicity model appeared to perform the best at identifying the active compounds and scoring them highly. This is exemplified by the MLSMR and cytotoxicity model identifying ten active molecules (~12%) in the top ranked 20 molecules (~1% of the entire dataset) when using the Vero cell cytotoxicity dataset. With random screening of the molecules, we would have expected less than one active (at the 4.3% hit rate empirical hit rate with the Mtb and Vero cell screens) (11.8 fold enrichment). For THP cells nine actives were in the top 20 molecules (10.7 fold enrichment). For HepG2 cells six actives were in the top 20 molecules (11.1 fold enrichment), however there were also fewer actives in this cell line. The TAACF Kinase dose response and cytotoxicity model has shown enrichments from 6.7 to 11.1 fold in the top 1% while the CB2 dose response and cytotoxicity model consistently performed poorly in all cell lines (Figure 3, Tables 4–6). Based on earlier studies (17, 18) we have focused on how the models enrich the top ranked molecules (top 1%) as this would suggest that we could screen a much smaller fraction of a library. In other applications it is advantageous to consider a larger percentage e.g. the top 10%, as well as use multiple models for compound selection. We would still observe a considerable enrichment over random as approximately 50% of the actives are identified by the MLSMR and cytotoxicity and TAACF Kinase dose response and cytotoxicity models, while random screening would have delivered only 10% of the hits (5 fold enrichment, Figure 3). Retrospectively, out of the three Bayesian models developed based on the MLSMR, CB2 and kinase library dose-response screens and cytotoxicity counter screens we found that the MLSMR-based model predicted the identity of actives out of the 1924 compound set most accurately, outperforming the kinase library based model, while the CB2 dataset-based model did not perform much better than random selection. These findings are illustrated in Figure 3 and also reflected in the number of true positives and prediction accuracy (Tables 4–6). These findings also underline the usefulness of applying multiple computational models to predict activity/toxicity since in typical applications it is not known a priori which model may perform better.
Figure 3.
Results for the 1924 compounds tested from the ChemBridge and Life Chemicals libraries screened for whole-cell TB activity and predicted with dual-event Bayesian models shown as receiver operator characteristic curves. The random rate is based on the empirical HTS hit rate; MLSMR+cytotox is based on the MLSMR dose response and cytotoxicity model; CB2+cytotox is based on the CB2 dose response and cytotoxicity model. Kinase+cytotox is based on the MLSMR dose response and cytotoxicity model. The best curve is based on a 100% hit rate. A. Vero cells, B. THP cells, HepG2 cells.
Table 4.
Model statistics for predictions made with the set of 1924 follow up compounds using Vero cells for cytotoxicity.
| Model | True Positive | True Negative | False Positive | False Negative | Sensitivity | Specificity | Prediction Accuracy | Matthews Correlation |
|---|---|---|---|---|---|---|---|---|
| MLSMR dose response and cytotoxicity | 66 | 1294 | 548 | 16 | 80 | 70 | 70.7 | 0.2 |
| CB2 dose response and cytotoxicity | 56 | 695 | 1147 | 26 | 68 | 38 | 38.7 | 0.02 |
| TAACF Kinase dose response and cytotoxicity | 59 | 1208 | 636 | 21 | 74 | 66 | 65.8 | 0.16 |
Table 6.
Model statistics for predictions made with the set of 1924 follow up compounds using HepG2 cells for cytotoxicity.
| Model | True Positive | True Negative | False Positive | False Negative | Sensitivity | Specificity | Prediction Accuracy | Matthews Correlation |
|---|---|---|---|---|---|---|---|---|
| MLSMR dose response and cytotoxicity | 39 | 1297 | 575 | 13 | 75 | 69 | 69.4 | 0.15 |
| CB2 dose response and cytotoxicity | 36 | 705 | 1167 | 16 | 69 | 38 | 38.5 | 0.02 |
| TAACF Kinase dose response and cytotoxicity | 35 | 1211 | 661 | 17 | 67 | 65 | 64.76 | 0.11 |
DISCUSSION
With the advent of antibacterial screening and chemotherapy in the early twentieth century, rapid advances led to a variety of new antibacterial agents. Research from the 1940’s – 1960’s led to current tuberculosis treatments (e.g., streptomycin, isoniazid and rifampicin) via the design and synthesis of small numbers of compounds (100’s per program) and their assessment in in vitro and in vivo models. Unfortunately, disease-focused research often occurs in spurts depending on perceived public health threat, pharmaceutical market size, and available funding. This phenomenon is best exemplified by the hunt for a cure for tuberculosis, caused by one of humankind’s oldest pathogens – Mtb. With the rapid development of effective antitubercular agents, the notion developed that tuberculosis would be eradicated worldwide, and this perception led to a reduction in efforts to maintain the tuberculosis research infrastructure, particularly the capabilities needed to drive new drug discovery. Beyond the incredible magnitude of eradicating this disease worldwide, the realities of treating latent disease and more recently resistant forms of tuberculosis have strained the public health infrastructure and led to the realization that new sources of drugs will continually be needed in order to simply contain the disease. The acute need for new faster acting therapies not subject to current drug-resistant strains is being partially addressed through large-scale renewed screening efforts much like those established in the 1940’s. Due to the advent of modern technology and HTS, millions of compounds can and have been screened for antitubercular efficacy under different metabolic conditions representing models of various states of human infection. Specific target-based screening of large synthetic libraries was found to be a relatively ineffective approach to antibacterial drug discovery due to a variety of reasons including bacterial permeability (43). On the other hand, whole-cell phenotypic screens suffer from the disadvantage of being target agnostic making compound optimization and selectivity problematic. In spite of these issues, recent large-scale phenotypic (4–6) HTS and computational- assisted HTS (22, 24) screens against Mtb have identified thousands of potential hit compounds. Our challenge is now to follow up on these data in a timely and efficient manner as described herein.
Our prior studies have demonstrated that dual-event Bayesian machine learning models can enrich hit discovery (22, 24). Using public Mtb screening data as a whole (actives and inactives) can enable us to make more effective decisions to identify active compounds. Our Bayesian models also indirectly take into account both uptake and activity against a growth-relevant target, making use of positive and negative information. This empirical, activity-based approach derived from large sets of screening results may be a useful and rapid alternative to other methods for predicting bacterial permeability, such as MycPermCheck which requires five molecular descriptors to be calculated (44).
As we see an increase in academic-industry collaborations around HTS such as the TB Drug Accelerator (45) screening for compounds active against Mtb, it is likely that the number of hits in the literature will only increase. Efforts to follow up on these compounds will create a bottleneck, perhaps similar to what we have seen with the wealth of antimalarial screening data (46). Therefore, the approach (Fig. 1) we propose of using the dual-event Bayesian classifiers to assist in selection of follow-up compounds would seem a natural progression, learning from all the data generated previously. Considering the tight research budgets and likely reductions in government supported tuberculosis drug discovery, it may be in the best interests of the academic research community to more widely employ these proven computational methods that are used in pharmaceutical company drug discovery programs, in order to accelerate progress. The potential for sharing the Mtb models derived from published literature (as used in this and previous studies (22, 24)) could quickly impact these efforts.
As an example we have demonstrated that assessment of compounds suggested by four academic groups and GSK from the literature as Mtb hits represents (in the absence of their entire screening libraries) one way to determine whether the three Mtb Bayesian dual-event models would have classified them as actives (Table 1, Table 2). Seven out of the nine (78%) academic screening derived compounds were identified by at least one model (Table 1) and eight out of eleven (73%) compounds in the dataset from GSK (Table 2). While we do not have access to the complete screening libraries used by these groups (ranging from tens of thousands to two million compounds) to do a complete assessment, predictions on their published hits may be instructive. Extension of such retrospective analysis is likely optimistic but it does suggest the benefits of using multiple models likely to cover a broader chemical space. Frequently, we have seen multiple Bayesian models perform differently with different datasets (17–19, 22, 24) and the current study using nearly 2000 compounds selected by clustering, is no exception. The CB2 and cytotoxicity model performed better with the literature compounds (Table 1 and 2) than with the 1924 compounds derived from clustering (Table 4–6, Fig. 3). This result may be a reflection of the diversity of the respective training sets for each model (compared to the test compounds), and, as we have seen previously, one of the models performed well in selecting Mtb active compounds from a library of antimalarial compounds (22). At the very least this result suggests that large libraries of compounds screened against Mtb can be used to generate Bayesian models (that incorporate activity and cytotoxicity information obtained in previous screens) to improve the selection of compounds for subsequent screening sets that are enriched in non-toxic actives. Full release of the large GSK dataset of two million compounds should allow significant improvement of these computational models, but results for the currently available 11 compounds prominently described in the paper suggest the models are performing well for hit to lead optimization and in line with our own data from previous studies (22, 24).
Bayesian classification models have been applied for identifying antibacterials in retrospective testing with 1–2 fold enrichments (23) and thus could have broader applicability than just finding compounds active against Mtb. In addition, Bayesian classification methods have also been used for ADME/Tox models (36, 47–49). Thus, using Bayesian models for hit follow-up outside of Mtb is worthy of further exploration. Limitations of using such models based on whole cell data are that there is of course no information on a target or SAR for a target, although this may not be necessary for further pursuit of a lead.
Interestingly, data derived with different cell types for cytotoxicity does suggest the benefits of using more than one cell line, as different cells appear to have different sensitivities based on the variation in hit rates observed. Fewer actives were present in HepG2 cells than Vero and THP-1, respectively. We are not aware of any discussion of such differences with or without mechanistic underpinning, although others have used many cell types to derive general cytotoxicity models (49). It could reflect expression of metabolizing enzymes involved in molecule activation versus detoxification, transport differences (import/export) or other possibilities. Some have compared the use of cardiac, hepatic and kidney derived cell lines at predicting compounds specific to each organ and found similar cytotoxicity across all cell types (50). It should also be noted in this study that we have used models incorporating only Mtb activity and cytotoxicity and have not tried to directly account for absorption, distribution, metabolism and excretion properties. A panel of models for different bacteria using different cell types for cytotoxicity could also be helpful for scoring potential compounds for follow up, to understand selectivity versus broad spectrum action and multi-targeting. As illustrated in Figure 3 and also reflected in model statistics (Tables 4–6) the MLSMR and Vero cytotoxicity based model performs well in the selection of non-toxic actives out of the set of 1924 compounds, with a prediction accuracy close to 70% using each of the three cytotoxicity datasets. The model developed based on the kinase library performed close to 65% with each of the three cytotoxicity datasets. The CB2 model performed relatively poorly as applied to the set of 1924 compounds. Out of clusters represented in the 1924 compound set cluster scaffolds containing at least one or more non-cytotoxic active hits are listed in Table 3. While the CB2 compound set utilized in the development of the CB2 model was lacking cluster 1 members (and its sub-clusters), all other clusters were represented equivalently or better compared to the MLSMR or the kinase set. Out of the three sets, the kinase library set was most under-represented in non-cytotoxic active scaffolds listed in Table 3 and yet performed well for the prediction of non-cytotoxic active compounds out of the set of 1924. In the case of a new (unknown) set of compounds, it is likely the best results may be achieved through the application of all three models followed by pooling top scoring compounds from each model. Non-cytotoxic actives identified in this study are distributed over a number of clusters as shown in Table 1. Core scaffolds shared among cluster members are related among sub-clusters. Among the less desirable are clusters that contain many evaluated or active compounds but only one (or few) non-cytotoxic active(s) such as sub-clusters 1e, 1i or 2a, d, g.
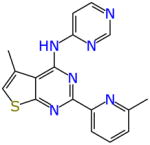
From the screen of 1924 compounds there were 33 compounds that met the bioactivity and selectivity criteria for all three cell lines (Table 7). Twenty seven of these compounds had been predicted as active with the MLSMR dose response and cytotoxicity Bayesian model. Twenty two were predicted active with the CB2 dose response and cytotoxicity Bayesian model while 23 were predicted active with the TAACF kinase dose response and cytotoxicity Bayesian score. AB00952642 is the most active compound out of these based on the IC90 (0.63 μM and 0.2 μg/ml). Noteworthy is the observation that AB00953420 and AB00953487 share the tetrahydropyrazolopyrimidine carboxamide common to potent antitubercular agents recently disclosed by both GlaxoSmithKline (51) and the Novartis Institute for Tropical Diseases (52) as well as resembling previous active compounds identified in our own laboratories (22, 24). Many of the other compounds in Table 7 also represent promising starting points for drug discovery optimization.
Table 7.
Compounds that were active and with selectivity index (SI) > 10 across all three cell lines along with dual-event Bayesian Model predictions (bold text = predicted active)
| Molecule | Structure | H37Rv IC90 (μg/ml) | H37Rv IC90 (μM) | SI Vero | SI HepG2 | SI THP-1 | MLSM R dose response and cytotoxicity Bayesian score | CB2 dose response and cytotoxicity Bayesian score | TAACF Kinase dose response and cytotoxicity Bayesian score |
|---|---|---|---|---|---|---|---|---|---|
| AB00077575 |

|
0.58 | 1.54 | 68.85 | 34.42 | 65.63 | 27.55 | −12.19 | −9.52 |
| AB00110214 |

|
0.32 | 1.07 | 126.05 | 63.09 | 113.31 | −16.67 | 4.61 | −9.40 |
| AB00143367 |

|
1.77 | 4.14 | 22.62 | 11.31 | 22.62 | 6.18 | −1.20 | 1.96 |
| AB00145154 |

|
1.65 | 3.99 | 24.29 | 12.14 | 24.29 | 42.50 | 4.87 | 8.47 |
| AB00285526 |

|
0.37 | 0.85 | 41.41 | 54.20 | 80.80 | 6.72 | −3.88 | 8.86 |
| AB00297140 |

|
1.46 | 3.85 | 27.46 | 13.73 | 11.83 | 28.99 | 6.43 | 6.64 |
| AB00305092 |

|
0.58 | 1.35 | 15.11 | 34.78 | 45.34 | 7.08 | −0.63 | 9.07 |
| AB00310530 |

|
0.86 | 2.33 | 46.51 | 23.26 | 46.51 | 3.47 | −1.17 | 2.53 |
| AB00315662 |

|
0.87 | 2.07 | 18.51 | 22.94 | 40.09 | 11.92 | 1.96 | 13.48 |
| AB00316014 |

|
0.20 | 0.60 | 27.02 | 102.56 | 35.83 | −5.19 | −5.51 | −6.85 |
| AB00370800 |

|
1.41 | 3.40 | 28.47 | 14.23 | 28.47 | −10.68 | −1.36 | −17.21 |
| AB00372473 |

|
1.36 | 3.42 | 15.13 | 10.24 | 29.33 | −12.32 | 2.59 | −18.66 |
| AB00434956 |

|
1.40 | 3.46 | 28.59 | 14.30 | 28.59 | 36.74 | 1.97 | 3.17 |
| AB00952522 |
|
1.66 | 4.01 | 13.36 | 11.43 | 14.07 | 25.22 | 3.18 | 23.26 |
| AB00952642 |

|
0.20 | 0.63 | 20.69 | 22.86 | 15.64 | −5.25 | 3.54 | −2.56 |
| AB00952825 |

|
0.56 | 1.47 | 70.96 | 35.46 | 70.92 | 22.70 | 4.06 | 16.70 |
| AB00952828 |

|
0.91 | 2.34 | 11.45 | 11.07 | 31.47 | 24.98 | 5.21 | 15.53 |
| AB00952834 |

|
0.93 | 2.52 | 13.37 | 21.55 | 34.55 | 29.02 | 5.43 | 17.76 |
| AB00952835 |

|
1.18 | 2.73 | 33.82 | 16.91 | 33.81 | 22.72 | 3.01 | 16.36 |
| AB00952911 |

|
1.88 | 5.52 | 21.27 | 10.64 | 21.28 | 24.79 | 0.21 | 18.01 |
| AB00953015 |

|
1.73 | 4.24 | 23.14 | 11.57 | 23.14 | 35.05 | −9.14 | −10.05 |
| AB00953018 |

|
1.10 | 2.55 | 36.53 | 18.26 | 25.26 | −10.48 | 1.89 | −10.40 |
| AB00953098 |

|
1.62 | 3.90 | 24.77 | 12.38 | 24.77 | 10.87 | 0.55 | 6.15 |
| AB00953127 |

|
1.77 | 4.90 | 22.62 | 11.31 | 19.12 | 24.04 | −0.62 | 13.46 |
| AB00953133 |
|
1.03 | 3.53 | 38.70 | 19.34 | 21.89 | 33.48 | 3.39 | 9.90 |
| AB00953146 |
|
1.48 | 4.80 | 27.11 | 13.55 | 13.65 | 35.82 | 4.27 | 12.49 |
| AB00953181 |

|
1.06 | 2.83 | 18.98 | 16.95 | 15.10 | 16.80 | 5.69 | 8.26 |
| AB00953202 |
|
1.90 | 5.54 | 21.08 | 10.54 | 19.59 | 27.53 | 2.24 | 8.73 |
| AB00953319 |

|
1.29 | 2.87 | 31.13 | 15.56 | 31.13 | 4.14 | −0.49 | 6.27 |
| AB00953326 |

|
1.26 | 3.89 | 31.67 | 15.84 | 13.96 | 43.38 | 3.47 | 11.73 |
| AB00953420 |

|
0.40 | 0.91 | 99.34 | 49.63 | 95.50 | 28.98 | 3.16 | −14.71 |
| AB00953487 |

|
0.45 | 1.00 | 21.26 | 19.48 | 22.86 | 26.51 | 3.35 | −6.36 |
| AB00953603 |
|
0.75 | 2.46 | 50.90 | 26.53 | 28.24 | 35.19 | 1.18 | 15.42 |
In summary, we have shown how computational approaches such as hierarchical clustering and Bayesian models could be used to assist human decision making in hit follow up for Mtb. Three Bayesian models have been developed based on Mtb dose-response activity and cytotoxicity datasets obtained for three previously screened libraries. We applied these models retrospectively for the prediction of actives out of a set of 1924 commercial compounds. The latter set consists of commercial analogs exploring chemical diversity around cluster scaffolds obtained from conformational clustering of the three previously screened libraries. The set of 1924 compounds was evaluated for antitubercular activity and cytotoxicity in three cell lines resulting in the identification of 82, 81 and 52 non-cytotoxic active compounds (IC90 < 10 μg/ml and SI > 10) using Vero, THP-1 and HepG2 cytotoxicity results, respectively. The selection of the 1924 commercial compounds was based on cluster scaffolds of dose-response hits from previous screens, followed by chemical diversity selection for clusters with large numbers of commercially available compounds. Compared to this strategy the current study demonstrates that the selection of such new sets of compounds may be achieved more effectively through the application of Bayesian models incorporating available antitubercular activity and cytotoxicity datasets. Multiple dual-event Bayesian models can increase the enrichment of non-cytotoxic actives in the top 1% of compounds to greater than tenfold and thus decrease the number of compounds purchased and tested. For example the application of our MLSMR model onto the 1924 compounds chosen by standard clustering (using the Vero cytotoxicity dataset) achieved an 11.8-fold enrichment of non-cytotoxic actives in the top 1% compared to random selection. Ideally the Bayesian models should be used prior to purchasing and testing of compounds to maximize the number of active compounds selected. Considering the likely limited budgets for purchasing follow up active samples for screening, this approach also allows virtual screening of even larger commercial databases and the purchase of a small, select set of compounds for follow-up that will be enriched in active compounds potentially leading to larger numbers of active compounds for mechanism of action studies than chemical diversity selection following conformational clustering alone. As further examples, the computational models can also be used to score compounds already identified by others and may be useful to triage the overwhelming number of hits and follow up screening set samples which themselves would consume valuable testing resources if they were all to be followed up.
Supplementary Material
Table 5.
Model statistics for predictions made with the set of 1924 follow up compounds using THP cells for cytotoxicity.
| Model | True Positive | True Negative | False Positive | False Negative | Sensitivity | Specificity | Prediction Accuracy | Matthews Correlation |
|---|---|---|---|---|---|---|---|---|
| MLSMR dose response and cytotoxicity | 62 | 1291 | 552 | 19 | 77 | 70 | 70.3 | 0.20 |
| CB2 dose response and cytotoxicity | 52 | 692 | 1151 | 29 | 64 | 38 | 38.7 | 0 |
| TAACF Kinase dose response and cytotoxicity | 53 | 1201 | 642 | 28 | 65 | 65 | 65.2 | 0.13 |
Acknowledgments
S.E. acknowledges colleagues at CDD. Accelrys are kindly acknowledged for providing Discovery Studio. The Bayesian models created in Discovery Studio are available from the authors upon written request
The CDD TB has been developed thanks to funding from the Bill and Melinda Gates Foundation (Grant#49852 “Collaborative drug discovery for TB through a novel database of SAR data optimized to promote data archiving and sharing”)
R.C.R. acknowledges the American Reinvestment and Recovery Act Grant 1RC1AI086677-01 that provided support for the presented study (National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID)) – “Targeting MDR-Tuberculosis.”
S.E. acknowledges that the Bayesian models described were developed with support from Award Number R43 LM011152-01 “Biocomputation across distributed private datasets to enhance drug discovery” from the National Library of Medicine
J.S.F. acknowledges funding from UMDNJ NJMS and the Foundation of UMDNJ.
Footnotes
Conflicts of Interest
SE is a consultant for Collaborative Drug Discovery, Inc.
Supplemental material for this article may be found at:
Table S1 – 1924 molecules with data used in this study with Bayesian model predictions
The complete dataset created under this grant is available as a public dataset TB: ARRA which is available upon registration http://web.collaborativedrug.com/pages/signup
References
- 1.Ballel L, Field RA, Duncan K, Young RJ. New small-molecule synthetic antimycobacterials. Antimicrobial agents and chemotherapy. 2005;49:2153–2163. doi: 10.1128/AAC.49.6.2153-2163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak PA, Rao SP, Ping Tan M, Lin X, Chyba J, Tay J, Ng SH, Tan BH, Cherian J, Duraiswamy J, Bifani P, Lim V, Lee BH, Ling Ma N, Beer D, Thayalan P, Kuhen K, Chatterjee A, Supek F, Glynne R, Zheng J, Boshoff HI, Barry CE, 3rd, Dick T, Pethe K, Camacho LR. A High-Throughput Screen To Identify Inhibitors of ATP Homeostasis in Non-replicating Mycobacterium tuberculosis. ACS Chem Biol. 2012;7:1190–1197. doi: 10.1021/cb2004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai H, Zhang L, Hung DT. Identification of Novel Inhibitors of M. tuberculosis Growth Using Whole Cell Based High-Throughput Screening. ACS Chem Biol. 2012;7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddry JA, Ananthan S, Goldman RC, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Sosa MI, White EL, Zhang W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinburgh, Scotland) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananthan S, Faaleolea ER, Goldman RC, Hobrath JV, Kwong CD, Laughon BE, Maddry JA, Mehta A, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Shindo N, Showe DN, Sosa MI, Suling WJ, White EL. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinburgh, Scotland) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds RC, Ananthan S, Faaleolea E, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Sosa MI, Thammasuvimol E, White EL, Zhang W, Secrist JA., 3rd High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinburgh, Scotland) 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko T, Cooper C, Mdluli K. Challenges and opportunities in developing novel drugs for TB. Future Med Chem. 2011;3:1373–1400. doi: 10.4155/fmc.11.115. [DOI] [PubMed] [Google Scholar]
- 9.Balganesh TS, Alzari PM, Cole ST. Rising standards for tuberculosis drug development. Trends Pharmacol Sci. 2008;29:576–581. doi: 10.1016/j.tips.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Gold B, Pingle M, Brickner SJ, Shah N, Roberts J, Rundell M, Bracken WC, Warrier T, Somersan S, Venugopal A, Darby C, Jiang X, Warren JD, Fernandez J, Ouerfelli O, Nuermberger EL, Cunningham-Bussel A, Rath P, Chidawanyika T, Deng H, Realubit R, Glickman JF, Nathan CF. Nonsteroidal anti-inflammatory drug sensitizes Mycobacterium tuberculosis to endogenous and exogenous antimicrobials. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16004–16011. doi: 10.1073/pnas.1214188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnet S, Hartkoorn RC, Szekely R, Pato J, Triccas JA, Schneider P, Szantai-Kis C, Orfi L, Chambon M, Banfi D, Bueno M, Turcatti G, Keri G, Cole ST. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis (Edinburgh, Scotland) 2010;90:354–360. doi: 10.1016/j.tube.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Impact of high-throughput screening in biomedical research. Nature reviews. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 13.Nathan C. Making space for anti-infective drug discovery. Cell host & microbe. 2011;9:343–348. doi: 10.1016/j.chom.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Periwal V, Rajappan JK, Jaleel AU, Scaria V. Predictive models for anti-tubercular molecules using machine learning on high-throughput biological screening datasets. BMC Res Notes. 2011;4:504. doi: 10.1186/1756-0500-4-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Periwal V, Kishtapuram S, Consortium OS, Scaria V. Computational models for in-vitro anti-tubercular activity of molecules based on high-throughput chemical biology screening datasets. BMC Pharmacol. 2012;12:1. doi: 10.1186/1471-2210-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prathipati P, Ma NL, Keller TH. Global Bayesian models for the prioritization of antitubercular agents. Journal of chemical information and modeling. 2008;48:2362–2370. doi: 10.1021/ci800143n. [DOI] [PubMed] [Google Scholar]
- 17.Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 18.Ekins S, Kaneko T, Lipinksi CA, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Ernst S, Yang J, Goncharoff N, Hohman M, Bunin B. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis. Molecular bioSystems. 2010;6:2316–2324. doi: 10.1039/c0mb00104j. [DOI] [PubMed] [Google Scholar]
- 19.Ekins S, Freundlich JS. Validating new tuberculosis computational models with public whole cell screening aerobic activity datasets. Pharm Res. 2011;28:1859–1869. doi: 10.1007/s11095-011-0413-x. [DOI] [PubMed] [Google Scholar]
- 20.Ekins S, Freundlich JS, Choi I, Sarker M, Talcott C. Computational Databases, Pathway and Cheminformatics Tools for Tuberculosis Drug Discovery. Trends in Microbiology. 2011;19:65–74. doi: 10.1016/j.tim.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarker M, Talcott C, Madrid P, Chopra S, Bunin BA, Lamichhane G, Freundlich JS, Ekins S. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium tuberculosis. Pharm Res. 2012;29:2115–2127. doi: 10.1007/s11095-012-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekins S, Reynolds R, Kim H, Koo MS, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chem Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh N, Chaudhury S, Liu R, Abdulhameed MD, Tawa G, Wallqvist A. QSAR Classification Model for Antibacterial Compounds and Its Use in Virtual Screening. Journal of chemical information and modeling. 2012;52:2559–2569. doi: 10.1021/ci300336v. [DOI] [PubMed] [Google Scholar]
- 24.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PLOSONE. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature reviews. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 26.Wilkens SJ, Janes J, Su AI. HierS: hierarchical scaffold clustering using topological chemical graphs. Journal of medicinal chemistry. 2005;48:3182–3193. doi: 10.1021/jm049032d. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira RS, Simeonov A, Jadhav A, Eidam O, Mott BT, Keiser MJ, McKerrow JH, Maloney DJ, Irwin JJ, Shoichet BK. Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors. Journal of medicinal chemistry. 2010;53:4891–4905. doi: 10.1021/jm100488w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collaborative Drug Discovery, Inc. http://www.collaborativedrug.com/register.
- 29.Ekins S, Gupta RR, Gifford E, Bunin BA, Waller CL. Chemical space: missing pieces in cheminformatics. Pharm Res. 2010;27:2035–2039. doi: 10.1007/s11095-010-0229-0. [DOI] [PubMed] [Google Scholar]
- 30.Hohman M, Gregory K, Chibale K, Smith PJ, Ekins S, Bunin B. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Disc Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 31.The PubChem Database. http://pubchem.ncbi.nlm.nih.gov/
- 32.Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrobial agents and chemotherapy. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falzari K, Zhu Z, Pan D, Liu H, Hongmanee P, Franzblau SG. In vitro and in vivo activities of macrolide derivatives against Mycobacterium tuberculosis. Antimicrobial agents and chemotherapy. 2005;49:1447–1454. doi: 10.1128/AAC.49.4.1447-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franzblau SG, DeGroote MA, Cho SH, Andries K, Nuermberger E, Orme IM, Mdluli K, Angulo-Barturen I, Dick T, Dartois V, Lenaerts AJ. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis (Edinburgh, Scotland) 2012;92:453–488. doi: 10.1016/j.tube.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL. Analysis of Pharmacology Data and the Prediction of Adverse Drug Reactions and Off-Target Effects from Chemical Structure. ChemMedChem. 2007;2:861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 36.Klon AE, Lowrie JF, Diller DJ. Improved naive Bayesian modeling of numerical data for absorption, distribution, metabolism and excretion (ADME) property prediction. Journal of chemical information and modeling. 2006;46:1945–1956. doi: 10.1021/ci0601315. [DOI] [PubMed] [Google Scholar]
- 37.Hassan M, Brown RD, Varma-O’brien S, Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol Divers. 2006;10:283–299. doi: 10.1007/s11030-006-9041-5. [DOI] [PubMed] [Google Scholar]
- 38.Rogers D, Brown RD, Hahn M. Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J Biomol Screen. 2005;10:682–686. doi: 10.1177/1087057105281365. [DOI] [PubMed] [Google Scholar]
- 39.Jones DR, Ekins S, Li L, Hall SD. Computational approaches that predict metabolic intermediate complex formation with CYP3A4 (+b5) Drug Metab Dispos. 2007;35:1466–1475. doi: 10.1124/dmd.106.014613. [DOI] [PubMed] [Google Scholar]
- 40.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, Gonzalez R, Lozano S, Huss S, Santos-Villarejo A, Martin-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuinan-Blanco M, Lavandera JL, Perez-Herran E, Gamo-Benito FJ, Garcia-Bustos JF, Barros D, Castro JP, Cammack N. Fueling Open-Source Drug Discovery: 177 Small-Molecule Leads against Tuberculosis. ChemMedChem. 2013 doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark AM. Basic primitives for molecular diagram sketching. J Cheminform. 2010;2:8. doi: 10.1186/1758-2946-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark AM, Williams AJ, Ekins S. Cheminformatics workflows using mobile apps. Chem-Bio Informatics J. 2013;13:1–18. [Google Scholar]
- 43.Payne DA, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Disc. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 44.Merget B, Zilian D, Muller T, Sotriffer CA. MycPermCheck: The Mycobacterium tuberculosis permeability prediction tool for small molecules. Bioinformatics (Oxford, England) 2012;29:62–68. doi: 10.1093/bioinformatics/bts641. [DOI] [PubMed] [Google Scholar]
- 45.Nathan C. Fresh approaches to anti-infective therapies. Science translational medicine. 2012;4:140sr142. doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 47.Pan Y, Li L, Kim G, Ekins S, Wang H, Swaan PW. Identification and Validation of Novel hPXR Activators Amongst Prescribed Drugs via Ligand-Based Virtual Screening. Drug metabolism and disposition: the biological fate of chemicals. 2011;39:337–344. doi: 10.1124/dmd.110.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zientek M, Stoner C, Ayscue R, Klug-McLeod J, Jiang Y, West M, Collins C, Ekins S. Integrated in silico-in vitro strategy for addressing cytochrome P450 3A4 time-dependent inhibition. Chemical research in toxicology. 2010;23:664–676. doi: 10.1021/tx900417f. [DOI] [PubMed] [Google Scholar]
- 49.Langdon SR, Mulgrew J, Paolini GV, van Hoorn WP. Predicting cytotoxicity from heterogeneous data sources with Bayesian learning. J Cheminform. 2010;2:11. doi: 10.1186/1758-2946-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Z, Will Y. Evaluation of drugs with specific organ toxicities in organ-specific cell lines. Toxicol Sci. 2012;126:114–127. doi: 10.1093/toxsci/kfr339. [DOI] [PubMed] [Google Scholar]
- 51.Remuinan MJ, Perez-Herran E, Rullas J, Alemparte C, Martinez-Hoyos M, Dow DJ, Afari J, Mehta N, Esquivias J, Jimenez E, Ortega-Muro F, Fraile-Gabaldon MT, Spivey VL, Loman NJ, Pallen MJ, Constantinidou C, Minick DJ, Cacho M, Rebollo-Lopez MJ, Gonzalez C, Sousa V, Angulo-Barturen I, Mendoza-Losana A, Barros D, Besra GS, Ballell L, Cammack N. Tetrahydropyrazolo[1,5-a]Pyrimidine-3-Carboxamide and N-Benzyl-6′,7′-Dihydrospiro[Piperidine-4,4′-Thieno[3,2-c]Pyran] Analogues with Bactericidal Efficacy against Mycobacterium tuberculosis Targeting MmpL3. PloS one. 2013;8:e60933. doi: 10.1371/journal.pone.0060933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokokawa F, Wang G, Chan WL, Ang SH, Wong J, Ma I, Rao SPS, Manjunatha U, Lakshminarayana SB, Herve M, Kounde C, Tan BH, Thayalan P, Ng SH, Nanjundappa M, Ravindran S, Gee P, Tan M, Wei L, Goh A, Chen P-Y, Lee KS, Zhong C, Wagner T, DI, CAK, Pethe K, Kuhen K, Glynne R, Smith P, Bifani P, Jiricek J. Discovery of tetrahydropyrazolopyrimidine carboxamide derivatives as potent and orally active antitubercular agents. ACS Med Chem Lett. 2013 doi: 10.1021/ml400071a. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nature chemical biology. 2012;8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.