Abstract
The integration of chemical ecology and bacterial genome mining can enhance the discovery of structurally diverse natural products in functional contexts. By examining bacterial secondary metabolism in the framework of its ecological niche, insights can be drawn for the upregulation of orphan biosynthetic pathways and the enhancement of enzyme substrate supply to illuminate new secondary metabolic pathways that would otherwise be silent or undetected under typical laboratory cultivation conditions. Access to these new natural products (i.e., the chemotypes) facilitates experimental genotype-to-phenotype linkages. Here, we describe select functional natural products produced by Xenorhabdus and Photorhabdus bacteria, with experimentally linked biosynthetic gene clusters, as illustrative examples of synergy between chemical ecology and bacterial genome mining in connecting genotypes to phenotypes through chemotype characterization. These Gammaproteobacteria share a mutualistic relationship with nematodes and a pathogenic relationship with insects, and in select cases, humans. The natural products encoded by these bacteria distinguish their interactions with animal hosts and other microorganisms in their multipartite symbiotic lifestyles. Though both genera have similar lifestyles, their genetic, chemical, and physiological attributes are distinct. Both undergo phenotypic variation and produce a profuse number of bioactive secondary metabolites. We provide further detail in the context of regulation, production, processing, and function of these genetically encoded small molecules with respect to their roles in mutualism and pathogenicity. These collective insights more widely promote the discovery of atypical orphan biosynthetic pathways encoding novel small molecules in symbiotic systems, which could open new avenues for investigating and exploiting microbial chemical signaling in host-bacteria interactions.
Keywords: chemical signaling, natural products, secondary metabolism, biosynthesis, structure elucidation, symbiosis
Introduction
Chemical ecology is an interdisciplinary field that seeks to identify and elucidate the functions of natural molecules in mediating biotic and abiotic interactions among organisms and their surrounding environments. Hartmann has detailed the field's lost origins, describing early pioneering work in the mid-19th century primarily by Ernst Stahl, whose field observations of plant-herbivore interactions led him to study the chemical defenses of plants in the laboratory [55]. But the concept of chemical ecology remained largely ignored until the 1950s mainly due to the primitive belief that the majority of secondary metabolites were waste products from primary metabolism. The landmark paper by the entomologist Gottfried Fraenkel (1949) played a major role in propelling chemical ecology to the forefront, highlighting plant-insect studies and the evolutionary adaptation of plant secondary metabolites to either deter or attract insects [39]. This ever-expanding field has come to encompass all dynamic small molecule functional interactions among organisms, including vertebrates [92, 110], invertebrates [5, 7, 23, 54, 58, 103, 117], and microorganisms [91, 106, 116, 121]. These genetically encoded small molecules often represent extracellular extensions of genomic instructions and the intercellular transfer of information, which are regulated accordingly and are frequently “silent” in common laboratory media. As such, chemical ecology does not need to be mutually exclusive from microbial genome mining and synthetic biology approaches for secondary metabolite stimulation and discovery. Rather, the combination of these disciplines places natural products in genomic, regulatory, functional, and ecological contexts, dramatically enhancing the productivity and efficiency of emerging genes to molecule discovery platforms.
Over the last decade, the continuous flood of microbial genome sequencing has revealed a much greater biosynthetic potential for the synthesis of diverse and novel secondary metabolites than previously appreciated [11, 51, 67, 95, 115, 135, 138, 141]. The majority of secondary metabolic pathways observed in genomic analysis are “orphan” and encode “cryptic” metabolites. In this context, cryptic simply means that an encoded molecule has not yet been correlated to its biosynthetic pathway. While unknown secondary metabolite pathways are occasionally referred to as being “silent,” this term implies some regulatory knowledge of the pathway and its usage should only be applied when supporting experimental regulatory data is available. Indeed, many expressed pathways producing molecules ranging from inert to highly unstable are simply undetected due to incompatible extraction, processing, and/or analytical techniques. Functional approaches, such as bioassay-guided fractionation, remain robust methods to empirically optimize the production, extraction, isolation, and structural characterization of unknown natural products with often-unpredictable chemical and biological properties. Here in lies a key advantage to focusing on symbiotic systems for natural product discovery, as experimental conditions and screening parameters can be leveraged within the physiological and ecological contexts of a given symbiosis.
To establish synergy among chemical ecology, natural product discovery, and genome mining, fundamental evolutionary mechanisms in microbial functional adaptation should be considered. Microbial genomes are dynamic and major phenotypic differences among related species are often associated with the evolutionarily malleable secondary metabolic pathways. In bacteria and fungi, the genes responsible for the biosynthesis, regulation, resistance, and transport of a given metabolite are frequently encoded on a contiguous stretch of the genome (i.e., “gene cluster”), facilitating experimental genotype-to-phenotype correlations and the discovery of functional gene cluster-specific small molecules (also known as a chemotype). These secondary metabolic gene clusters, which are often not essential for cellular growth in ideal growth conditions, represent highly adaptable genetic elements that evolve through genetic mutation, gene duplication, gene deletion, sequence migration, and genome rearrangements through periods of genetic drift and natural selection [15, 38, 63, 88, 137].
While gene duplication and functional divergence substantially contribute to natural product structural diversification within a given species (e.g., the genetic expansion of polyketide synthases in fungal species) [72], the exchange of genetic information between organisms can be a central player in mediating host-bacteria interactions. For example, the acquisition of a new encoded trait via the horizontal gene transfer of chromosomal, plasmid, or phage sequences can rapidly redefine the landscape available to a microorganism [98]. The resulting genomic islands can contribute to functional adaptation by enhancing pathogenicity, symbiosis (mutualistic or parasitic traits), fitness, or drug resistance [32, 53]. Functional attributes that can migrate on genomic islands are wide ranging and include virulence factors, adhesins, invasins, modulins, effectors, secretion systems, iron acquisition systems, protein toxins, and in particular, bioactive small molecules. An important aspect of chemical ecology focuses on the microbial acquisition of orphan biosynthetic pathways involved in overcoming new challenges (e.g., functional small molecule contributions to host-bacteria interactions). These secondary metabolic pathways vary widely across species and even individual strains. In this regard, natural products themselves can promote bacterial niche differentiation, and ultimately, contribute to speciation [64, 73, 104].
To highlight our points, natural products from the two well-studied Gammaproteobacteria genera, Xenorhabdus and Photorhabdus, will be reviewed in greater detail. These bacteria produce a diverse array of small molecules that play an assortment of biological roles in regulating their multipartite symbioses [5, 7], in which the bacteria serve as both mutualists of specific nematode hosts and pathogens of a variety of insect hosts [17, 36, 48, 49, 59, 93, 112, 130]. Interestingly, Photorhabdus asymbiotica also made the evolutionary leap from invertebrate to human pathogenesis. In addition to infecting insects, P. asymbiotica represents an emerging human pathogen that causes soft tissue and blood infections [20, 45, 47, 124, 130, 134, 136]. Descriptions of the genomes from several Xenorhabdus and Photorhabdus species have been published [14, 34, 44, 56, 75, 99, 136], revealing an enormous potential for secondary metabolite synthesis. In part, the natural products encoded by these bacteria distinguish their interactions with animals and other microbes during their variable life histories. Here, we describe select natural products, with experimentally linked gene clusters, and highlight their likely functional aspects in the Xenorhabdus and Photorhabdus lifecycles. The examples are used to illustrate the synergy between chemical ecology and bacterial genome mining approaches for small molecule discovery in functional contexts, which in practice, could be more widely applied across diverse symbiotic systems. We then conclude by positing that such collective insights could facilitate the discovery of atypical orphan biosynthetic pathways encoding novel functional small molecule structural classes and/or features, which could open new avenues for investigating and exploiting microbial chemical signaling in host-bacteria interactions.
Xenorhabdus and Photorhabdus multipartite lifestyles
Xenorhabdus species are prolific antibiotic-producing Gammaproteobacteria that share a mutualistic relationship with nematodes in the Steinernema genus, whereas the related bioluminescent Photorhabdus species share a relationship with Heterorhabditis nematodes [17, 36, 48, 49, 59, 93, 112, 130]. The bacterium-nematode complexes are highly efficient at infecting and killing a wide range of insect larvae in the soil, leading to their commercialization as biological control agents of agricultural pests. In addition to producing immunomodulators to circumvent the insect's innate immune system and insecticides that assist in the insect's demise, the bacteria encode an armament of bioactive molecules and antibiotics to modulate their own development utilizing the insect's biomass as an energy source, influence the development of the nematode, and protect their decomposing insect prey from competing bacteria and fungi in the combative soil microenvironment. For P. asymbiotica, additional molecules are likely produced to circumvent the human immune system and establish niches in the soft tissue and bloodstream [20, 45, 47, 124, 130, 134, 136].
Despite their overall similarities, physiological, genetic, and genomic comparisons across genera indicate that Xenorhabdus and Photorhabdus underwent divergent evolution leading to convergent lifestyles [14, 34, 44, 49, 56, 75, 99, 136]. Consequently, the bacteria collectively produce a treasure trove of molecules for at least five biomedically-relevant objectives – immunomodulators, insecticides, developmental modulators, antibacterials, and antifungals – and are strikingly comparable to Streptomyces in regards to their secondary metabolic repertoire. Currently, there are 22 recognized species of Xenorhabdus and three species of Photorhabdus [93]. Combined with the intraspecies and interspecies chemical variability observed to date, they continue to represent a rich source of small molecules with potential commercial value. Unlike the poorly understood interactions that occur in complex microenvironments, such as in bulk soil, an understanding of the simpler animal-bacteria systems facilitates the upregulation of molecules for discovery in the lab and the functional connection(s) for which the molecules evolved to fulfill.
Xenorhabdus and Photorhabdus phenotypic variation and regulation
In keeping with their alternative lifestyles in multiple animal hosts, Xenorhabdus and Photorhabdus species stochastically undergo phenotypic variation, which manifests in at least two highly distinct forms, primary and secondary. These forms, which are controlled by mechanistically distinct regulatory systems between genera, can be distinguished on Petri dishes simply by eye using dye supplements [1]. The primary form is associated with the secretion of a variety of extracellular products, including hemolysins, proteases, lipases, and antibiotics. Many of these factors are needed for both mutualistic nematode development and insect pathogenicity. Understanding the mechanisms of phenotypic variation and small molecule regulation are important for bridging the Xenorhabdus/Photorhabdus orphan biosynthetic gene clusters to the molecules they produce in a native context and the phenotypic effects they regulate in the symbiosis.
Early investigations of Xenorhabdus nematophila and Photorhabdus temperata revealed that the two genera exhibit remarkably dissimilar downstream regulatory mechanisms for antibiotic biosynthesis [21, 66]. In X. nematophila, a leucine-responsive regulatory protein (Lrp) is a positive regulator of phenotypes associated with the primary form (i.e., a Δlrp mutant lacks antibacterial zones of inhibition) [21]. The upstream regulatory mechanisms that control phenotypic variation leading to the primary form in Xenorhabdus are currently unclear, although it has been suggested that epigenetics could be an important player [122]. In P. temperata, a LysR-type transcriptional repressor (HexA) contributes to the regulation of primary form features, such as antibiotic activity (i.e., a ΔhexA mutant enhances antibacterial zones of inhibition) [66]. Despite the regulatory disparities – positive versus negative regulation – both Xenorhabdus and Photorhabdus primary forms produce relatively high titers of general insecticidal, antibacterial, and antifungal activities during insect infection [60, 61, 84], providing an advantage to the nematode-bacteria complex during pathogenicity.
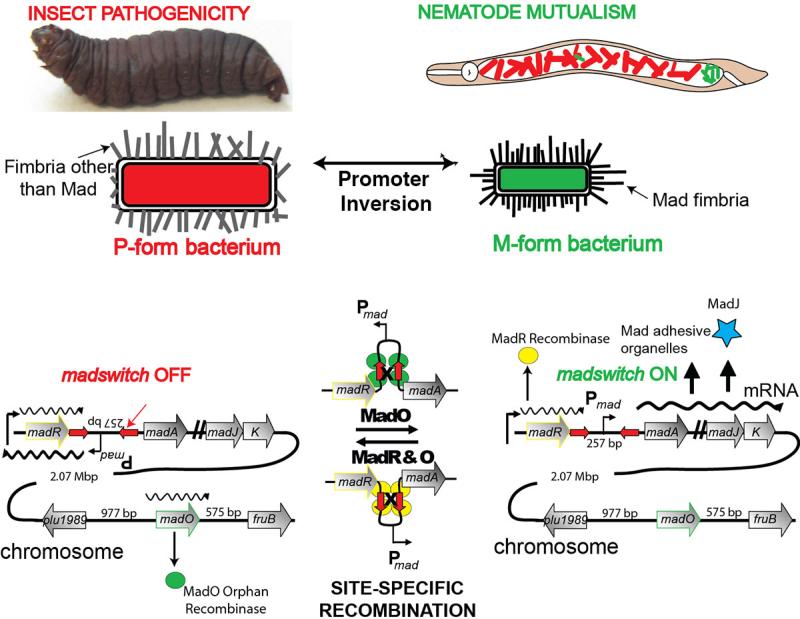
More recent genetic, pulse-chase, and microscopy experiments have provided further important insights into the mechanism of stochastic form switching in Photorhabdus [119]. A mutant library search for genes required in nematode colonization led to the identification of a fimbrial locus, maternal adhesion defective (mad), in Photorhabdus luminescens [118]. The locus was controlled by an ON/OFF invertible promoter switch, the madswitch (Fig. 1) [119]. Depending on the orientation of the 257-bp promoter region, which was flanked by 36-bp inverted repeats, the bacteria toggled between a pathogenic primary form (“P-form”) and a less prevalent small colony variant secondary form (“M-form”). The P-form robustly produces antibiotics, insect virulence factors, and mutualistic nematode factors. Strikingly, the small colony variant M-form, which was avirulent towards insects, selectively adhered to posterior intestinal cells of the maternal nematode to initiate mutualistic association (“M-form” bacteria). MadJ was identified in the locus as the transcriptional activator responsible for regulating the M-formation program. An understanding of the form switching mechanism enabled the bacteria to be engineered in either genetically-locked state [119]. Locked forms could be particularly helpful for novel bioactive small molecule discovery, as opposed to identifying molecules from minority cell variants in mixed bacterial populations. Locked forms also provide a unique opportunity to investigate the metabolic status between pathogenic and mutualistic initiation states of the infectious bacteria. Indeed, LC/MS analysis showed that the locked P-form robustly produced bioactive small molecules, such as bacterial stilbenes and anthraquinones, and microscopy confirmed that this engineered strain was incapable of generating M-form revertants during cultivation. Additionally, transcript microarray analysis revealed few upregulated orphan biosynthetic genes in the locked M-form during exponential growth relative to the P-form, suggesting that select bioactive small molecules could contribute not only in combative P-form infections, but perhaps also as currently unknown M-form colonization factors [119]. Small colony variants related to the M-form are common in chronic human infections [108], such as in cystic fibrosis, and it would be prudent to identify small molecules that might contribute to persistent bacterial colonization of animal hosts in general.
Figure 1.
Phenotypic variation in Photorhabdus. Promoter inversion toggles the bacterium between a pathogenic P-form and an M-form that initiates nematode mutualism. A new understanding of the genetic form switching mechanism enables engineered locked states to examine the metabolic status associated with phenotypic variation. Adapted from Somvanshi et al [62].
Parallel functional small molecule screening platforms
The innate immune systems of insects share similarities with mammalian innate immunity [70, 125]. Consequently, some bacterial virulence strategies utilized in invertebrate pathogenesis can be evolutionarily redeployed for vertebrates, increasing the utility of invertebrate model systems in the lab [132]. The dual insect-human pathogen P. asymbiotica represents an excellent example of bacterial virulence mechanisms being employed in insect and/or human hosts [20, 45, 47, 124, 130, 134, 136]. It is hypothesized that the Heterorhabditis nematode can actively penetrate intact and healthy human skin to deliver the pathogen [46]. Once delivered, P. asymbiotica effectively outmaneuvers the human immune system, leading to soft tissue and blood infections [20, 45, 47, 124, 130, 134, 136].
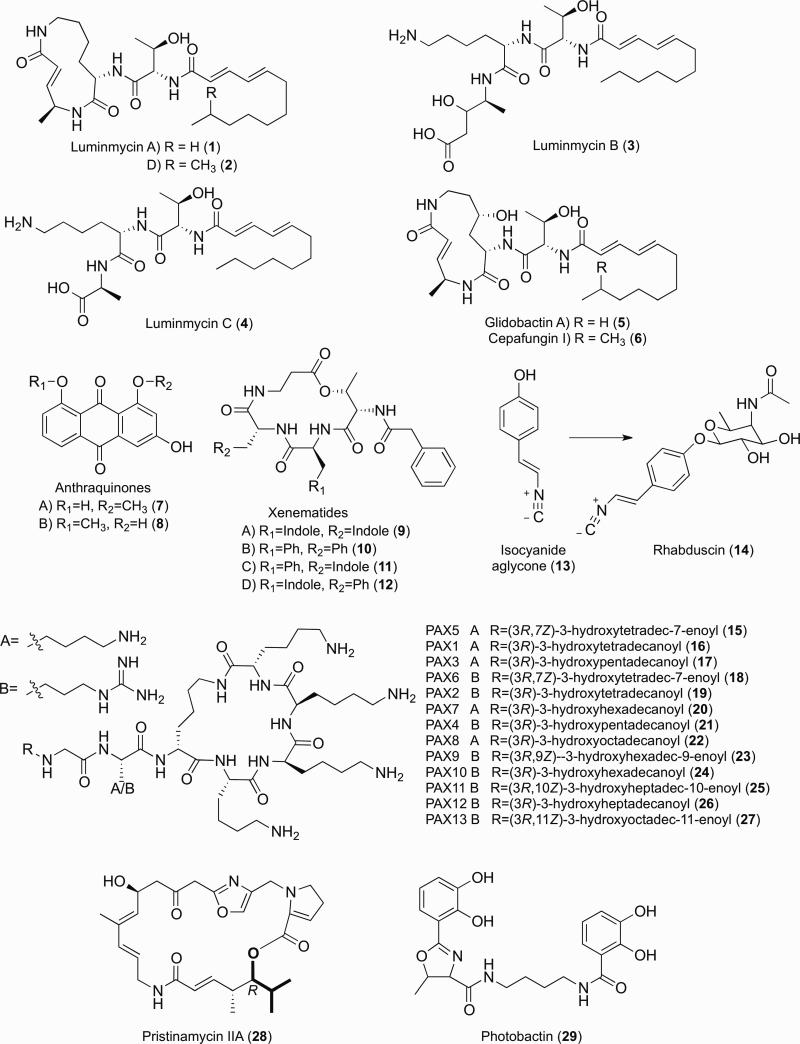
A parallel screening strategy, Rapid Virulence Annotation (RVA), was carried out to identify bacterial genomic islands in P. asymbiotica that contribute to host toxicity across diverse taxa – insects, nematodes, protozoa, and mammalian macrophage cell lines [131]. To identify small molecules toxic to select eukaryotes, RVA requires a heterologous bacterial host capable of expressing orphan biosynthetic pathways from genomic libraries in the presence of the eukaryote host. For P. asymbiotica, expression of a cosmid library in Escherichia coli led to the sequence identification of eight NRPS and two PKS orphan biosynthetic gene clusters that resulted in toxic phenotypes for the eukaryote hosts, suggesting that the orphan pathways could contribute to virulence. One of the orphan pathways toxic to insects and mammalian cells was predicted to synthesize antitumor glidobactin analogs based on bioinformatics [131]. Using a homologous recombination cloning strategy, a related biosynthetic pathway in P. luminescens was overexpressed in E. coli and experimentally shown to produce the glidobactin analogs, luminmycins A-C (Fig. 2, Structures 1, 3-4) [3, 41], where luminmycin A exhibited cytotoxicity against a colon cancer cell line in the nanomolar IC50-range [3]. Osmotic culture shock combined with a NMR-based proteasome inhibitory assay, led to the identification of glidobactin A (5) and the highly potent antitumor cepafungin 1 (6) in wildtype P. luminescens cultures [120]. Production of glidobactin A (5) and luminmycins A and D (1-2) were confirmed in P. asymbiotica cultures using a defined medium previously shown to induce secondary metabolism in some Streptomyces species [123]. These small molecule proteasome inhibitors were also expressed in bacterium-infected crickets, keeping with one of their regulatory niches and functional attributes in virulence. In principle, the manipulation of sequence inputs, engineered heterologous expression strains, and alternative functional screening outputs could readily be tailored to dramatically facilitate genotype to chemotype to phenotype correlations in any related parallel natural product discovery platform.
Figure 2.
Selected natural products from Xenorhabdus and Photorhabdus.
Functional small molecules from primary-form bacteria
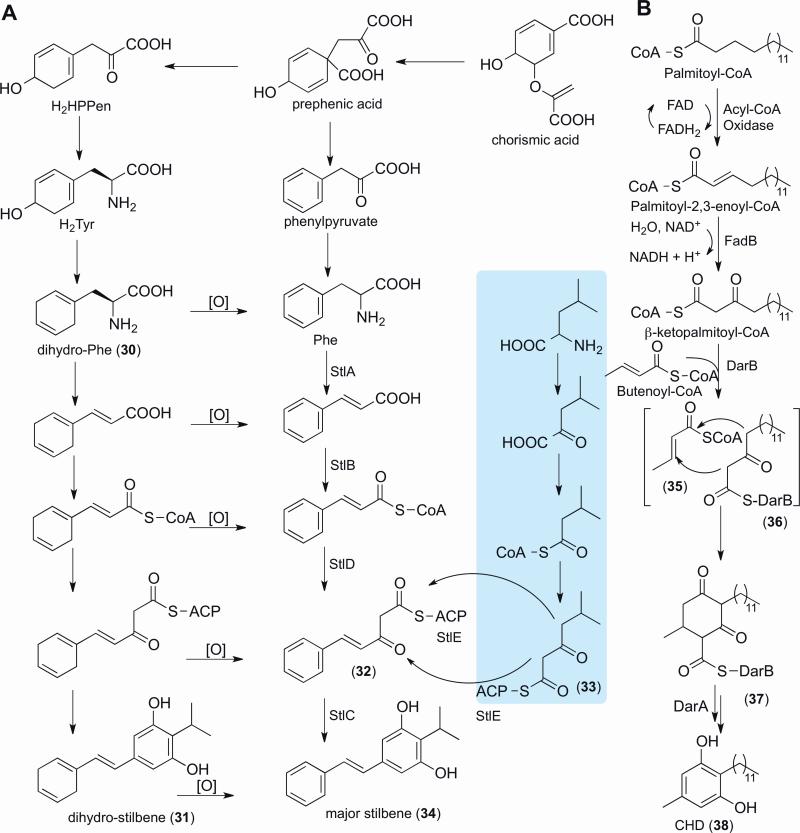
Earlier structural investigations of bioactive metabolites produced by Photorhabdus primary form cultures led to the discovery of anthraquinone polyketides (e.g., Fig. 2, Structures 7-8) and bacterial stilbenes (e.g., Fig. 3, Structure 34) [113]. The anthraquinone pigments, which are thought to provide anti-omnivory properties for the bacteria-nematode-insect complex [19], were linked to their discretely expressed (type II) polyketide synthase gene cluster by gene deletion, isotopic labeling, and product analysis [8]. The major antibiotic molecules present in laboratory cultures were identified as bacterial stilbenes [77, 113], which are disseminated throughout the Photorhabdus genera. The stilbenes are multipotent polyketide-phenylpropanoids, exhibiting antimicrobial activity against Gram-positive bacteria and fungi [61], harboring moderate inhibitory activity against the metalloenzyme phenoloxidase, an important terminal component of the insect's innate immune system [35], and crucially contributing to host nematode development [65]. A fragmented biosynthetic pathway (i.e., not on a contiguous stretch of the genome) was identified via genetics and product analysis. It was proposed that two β-keto-acyl-derivatives (32 and 33) were condensed in a head-to-head manner, leading to the stilbene scaffold (Fig. 3) [65]. Intriguingly, this biosynthetic strategy is much different than the type III PKS strategy common to plant stilbene biosynthesis, indicating convergent evolution to similar structural outcomes.
Figure 3.
Proposed stilbene and cyclohexanedione biosynthesis. Based on enzyme homology, it is plausible that stilbene biosynthesis could rather proceed as shown for cyclohexanedione biosynthesis.
Identification of regulators governing symbiotic interactions can also promote the discovery of small molecules involved in specific lifestyle transitions. Microbiologists have identified a panoply of regulatory elements in Xenorhabdus/Photorhabdus, and many other bacterial symbionts, capable of producing bioactive small molecules with pharmacological value. There are far fewer examples, however, in which natural product chemists examined the chemical potential of regulatory mutants directly connected to bacterial lifestyle decisions. In P. luminescens, for example, insertional inactivation of hexA, the repressor previously identified to be involved in regulating primary form characteristics [66], led to an upregulation of previously described and new bacterial stilbenes [71]. A newly identified dihydrostilbene derivative (31) was shown to undergo oxidation in aerobic conditions to form the major aromatic stilbene (34) (Fig. 3). Such easily oxidized metabolites could assist in overcoming oxidative stress from the insect's innate immune system [71]. L-2,5-Dihydrophenylalanine (30), an antimetabolite of phenylalanine biosynthesis, was identified in P. luminescens cultures and shown to be the substrate for dihydrostilbene in isotopic feeding studies [25]. A recently characterized nonaromatizing prephenate decarboxylase common to secondary metabolism [80, 81] was identified in a separate orphan biosynthetic pathway. Genetic, small molecule, and biochemical analyses determined that this previously orphan pathway initiated the biosynthetic process from prephenate to L-2,5-dihydrophenylalanine (Fig. 3) [25]. With over 500 other nonproteinogenic amino acid derivatives known, the continued elucidation of their underlying gene clusters will aid not only in the in vivo engineering of new nonribosomal peptides, but also increasingly in the ribosomal protein engineering field [127].
Related to the bacterial stilbene biosynthesis, two fatty acid precursors (Fig. 3, Structures 35-36) are condensed by a ketosynthase homolog to generate 2,5-dialkylcyclohexane-1,3-diones (CHDs, e.g., enzyme-tethered intermediate 37) [42]. Separate enzymes, including an aromatase, were required to oxidize the CHD scaffold to its corresponding resorcinol scaffold (38). The CHD natural product class was recently introduced, when they were discovered to serve as orchid sex pheromones [40]. Phylogenetic analysis indicated that the biosynthesis of CHDs is widespread and enriched in bacteria that interact with eukaryotes [42], supporting unresolved regulatory functions in various eukaryote-bacteria associations.
It has long been known that the alteration of growth conditions in the lab profoundly affects the production of secondary metabolites (e.g., as applied in the one strain-many compounds (OSMAC) approach [6]). Symbiotic systems provide immediate insights to guide cultivation parameters for secondary metabolite stimulation and discovery. Addition of hemolymph (insect blood) to Xenorhabdus and Photorhabdus laboratory cultures substantially stimulated the production of the previously noted stilbenes and anthraquinones in primary form cultures of P. luminescens TT01 and of a variety of new and previously described indole-containing antibiotics in X. nematophila [24]. Bioassay-guided fractionation of the hemolymph for the identification of antibiotic-induction factors identified L-proline and osmotic stress as principal induction factors. Addition of L-proline at low millimolar (mM) concentrations stimulated antibiotic production, while deletion of metabolic or osmotic proline transporters in P. luminescens dramatically reduced stilbene virulence factor production. Analysis of various insect hemolymph samples confirmed the presence of free amino acid L-proline between 3-73 mM, demonstrating a physiologically-relevant response [24]. While individual bacterial species and strains behave significantly different in regards to bioactive small molecule production, we regularly observe higher antibiotic titers in primary form Xenorhabdus/Photorhabdus cultures supplemented with free amino acids over typical peptide supplements (e.g., tryptone).
L-Proline media supplementation enabled the upregulation and discovery of other bioactive metabolites and their assignment to previously orphaned pathways. Particularly in X. nematophila, it led to the identification of a diversified family of NRPS cyclodepsipeptides xenematides A-D (9-12) [26] and a new isocyanide- and amidoglycosyl-functionalized natural product, rhabduscin (14) [24]. The antibacterial xenematide A (Fig. 2, 9) had been previously characterized from primary form X. nematophila cultures [74], and its absolute configuration was determined by total synthesis [62]. Both Xenorhabdus and Photorhabdus species produce a variety of other nonribosomal peptides with a variety of biological activities. For example, a family of lysine-/arginine- rich cyclic peptides, the PAX peptides (Fig. 2, 15-27), were identified from X. nematophila, which harbor potent antifungal activity [43, 52].
The less prevalent isocyanide natural products have recently been identified from a number of cultured and environmental bacteria [10]. L-proline stimulated rhabduscin (14) production by over an order of magnitude in laboratory cultures of X. nematophila, facilitating its structural determination [24]. Rhabduscin exhibited potent nanomolar-level inhibition against the insect's innate immune enzyme, phenoloxidase, and its biosynthetic pathway was critical for infection [27]. At high bacterial inoculum, rhabduscin deletion mutants were markedly impaired in infectivity, and at lower inoculum, the mutants were avirulent. In contrast, wildtype bacterium killed all of the larvae at all cell inoculum concentrations, indicating that the small molecule pathway could represent a make-or-break point for pathogenicity. While rhabduscin was identified, in part, as a diffusible product in the spent culture medium, the molecule was also found localized to the outer bacterial lipopolysaccharide by stimulated Raman scattering microscopy. The inhibitor's location at the cell's periphery provides a spatially appropriate and high local concentration to defend the bacteria from the insect's phenoloxidase response. The rhabduscin gene cluster has been identified in genomes from both Xenorhabdus and Photorhabdus species, including the human pathogen P. asymbiotica. The isocyanide-aglycone intermediate (13) was also identified in Vibrio cholerae pathogens [27]. V. cholerae is thought to exploit arthropods as reservoirs to persist in endemic areas as well as vehicles to spread to alternative locales [109]. Consequently, isocyanides could potentially contribute in part to cholera spread through its potent inhibition of phenoloxidases in arthropod hosts and/or inhibition of other metalloenzymes in the human innate immune system [27].
Gene duplication of primary metabolic genes and functional divergence to secondary metabolic functions was likely an early evolutionary mechanism leading to the expansion of an enzyme class that utilizes α-keto acid substrates in secondary metabolism. The pyruvate dehydrogenase complex (E1, E2, and E3) in primary metabolism is an important connector of glycolysis with the TCA cycle, converting the simple α-keto acid pyruvate to acetyl-CoA [105]. Other α-keto acids, such as those processed by the related branched chain α-keto acid dehydrogenase complex (BCKAD) [100], similarly yield their related CoA-products. Secondary metabolic genes have been frequently observed in genome databases clustered with or fused to related pyruvate dehydrogenase-like subunits. In X. nematophila, rather than release the free acyl-CoA, the acyl-group is likely transferred directly to the CoA-derived phosphopantetheine arm of a PKS acyl-carrier protein to initiate synthesis of branched-chain fatty acids and the streptogramin-type antibiotic pristinamycin IIa (Fig. 2, Structure 28 with starter unit bolded) [9]. In this example, the Xenorhabdus gene cluster is flanked by transposase sequences supporting its acquisition from an unidentified extrachromosomal source and its repurposing for the Xenorhabdus lifestyle. Similar chemistry had also been observed from metagenomic DNA sources in the construction of branched-chain fatty acid substrates in select N-acyl-amino acid biosynthesis [22]. Pristinamycin IIa, a well-known Streptomyces antibiotic, is a precursor for the semi-synthesis of one active constituent of the two-component commercial antibiotic synercid [18], and the pristinamycin gene cluster in Streptomyces pristinaespiralis was recently reported [83].
Proteolytic Processing of Nonribosomal Peptides
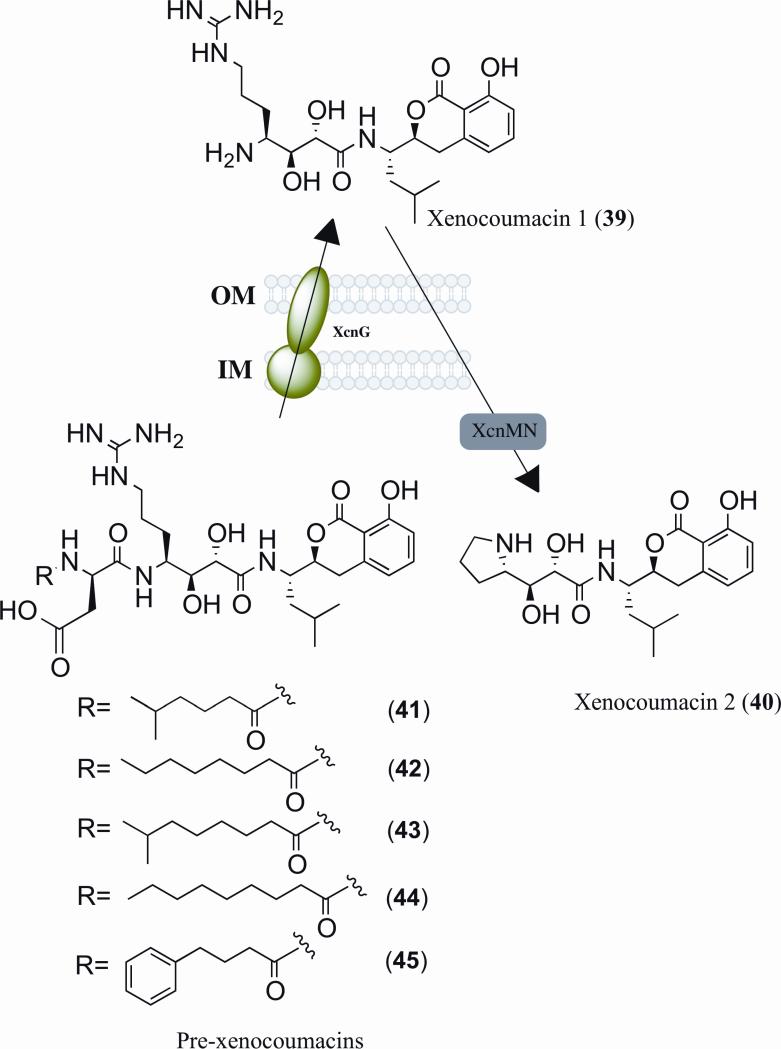
An X. nematophila compound thought to play a major role in insect cadaver sterility is the water-soluble benzopyran-1-one isocoumarin derivative xenocoumacin 1 (Fig. 4, Xcn 1, 39). Both Xcn 1 and xenocoumacin 2 (Xcn 2, 40) were first isolated from insect cadavers and as major compounds produced from the culture broth of Xenorhabdus stains [84]. Xcn 1 exhibits strong antibacterial activity against Gram-positive [140] and Gram-negative bacteria, antifungal activity, and potent antiulcer activity [85]. On the other hand, Xcn 2 shows substantially reduced bioactivity. Derivates of Xcn 1 and 2 have also been identified at trace amounts, though their bioactivity has not been reported [111].
Figure 4.
Proteolytic processing in xenocoumacin biosynthesis and activation. OM, outer membrane; IM, inner membrane.
The 34-kb biosynthetic gene cluster associated with the production of xenocoumacins (Xcn) contains 14 genes (xcnA-xcnN), which includes two NRPS (xcnAK), three PKS (xcnFHL), and nine accessory genes [33, 111]. The PKS and NRPS genes xcnAKFHL are necessary for Xcn 1 synthesis, and the accessory genes xcnBCDE are specifically involved in the biosynthesis of the extender unit hydroxymalonyl-ACP [13, 33, 111]. The conversion of Xcn 1 into Xcn 2 is due to xcnM and xcnN genes, which encode proteins homologous to saccharopine dehydrogenases and fatty acid desaturases, respectively [111]. When Xcn 2 production is attenuated, an increase in Xcn1 is observed along with a 20-fold reduction in cell viability, suggesting that conversion of Xcn 1 to Xcn 2 is a resistance mechanism utilized by the bacteria to avoid self-toxicity [33].
A prodrug activation mechanism was recently identified in the biosynthesis of Xcn 1 from X. nematophila [30]. An in-frame deletion of peptidase gene xcnG produced five xenocoumacin-related compounds. The five identified pre-xenocoumacins (Fig. 4, PXCN, 41-45) contained the Xcn 1 chemical core structure bound to a unit at the N-terminus comprising of a D-asparagine amino acid attached to one of five different acyl chains. Interestingly, these PXCN compounds did not exhibit antimicrobial activity. The xcnG gene encodes a protein with an N-terminal periplasmic peptidase domain that contains a signal peptide sequence and a C-terminal transmembrane domain. In this prodrug activation model (Fig. 4), X. nematophila produces the inactive PXCN in the cytoplasm, which is then translocated to the periplasmic membrane. Once there, PXCN is cleaved into the potent antimicrobial Xcn 1 and is thought to be pumped out of the outer membrane by XcnG-ABC transporter-TolC protein complex. When present in the insect gut, Xcn 1 inhibits growth of other bacteria, allowing for a competitive advantage for X. nematophila. To reduce self-toxicity, Xcn 1 is cleaved by xcnM and xcnN into the less potent antibiotic Xcn 2 through the formation of the pyrrolidine ring [30].
XcnG homolog proteins have been identified in Bacillus pumilus, Bacillus thuringiensis, and E. coli [30], which produce amicoumacin [107], zwittermicin [79], and colibactin [97], respectively. The zwittermicin protein ZmaM is larger than the XcnG protein, containing six additional transmembrane helices and a C-terminal ABC-transporter nucleotide-binding domain. In a similar manner, ZmaM transports the pre-zwittermicin compound to the periplasm, where pre-zwittermicin is cleaved at the N-terminus into zwittermicin before being transported out of the cell [69]. Colibactin is a small genotoxin of unknown structure, biosynthesized by an NRPS-PKS gene cluster present in E. coli [97]. Recent studies have suggested a similar prodrug strategy in the production of colibactin. Biochemical analysis [12] and structural characterization [2] have identified an N-myristoyl-D-Asn product from the colibactin gene cluster. It is postulated that the ClbP peptidase protein cleaves a yet uncharacterized pre-colibactin molecule into the active cytotoxin(s), releasing N-myristoyl-D-Asn. Proteolytic cleavage of nonribosomal peptides is emerging as a more prominent small molecule processing mechanism in a variety of bacterial genera, implying an evolutionary preservation and likely importance in their symbiotic associations.
Secondary Metabolites and Iron Acquisition
The functional aspects of siderophores are clear for their roles in iron metabolism [89, 94, 128]. Most organisms require iron for growth and have evolved multiple mechanisms for its acquisition. In the majority of microbial habitats, bioavailable Fe(II) is oxidized to Fe(III), which leads to ferric oxide hydrate complexes (Fe2O3 × nH2O) and a resulting free Fe(III) concentration in the range of 10−9 to 10−18 M [89]. Siderophores, iron-chelating small molecules produced by NRPS and NRPS-independent pathways, scavenge Fe(III) to support cellular growth and serve as virulence factors in iron-limiting pathogen-host associations [28, 89, 102]. More recently, siderophores were identified as growth promoting factors that enabled previously uncultured bacteria from sediment communities to grow in the lab [29]. Such expanded cultivation of previously uncultured bacteria could significantly increase the available sources of new bioactive natural products, particularly when an understanding of the regulatory interactions between the natural products and their producers is desired [29, 76]. For microorganisms that encode siderophore-related biosynthetic pathways, biosynthesis is most often upregulated under iron-limiting conditions, and the resulting small molecule siderophores are actively secreted to scavenge Fe(III) from the extracellular environment. Consequently, growth in iron-limiting conditions in the lab has facilitated the discovery of many siderophores in a wide variety of structural classes [94]. In contrast and in keeping with the often idiosyncratic nature of secondary metabolite regulation, media supplementation with metals, such as bioavailable Fe(II), can enhance antibiotic production particularly for pathways containing common Fe(II)/α-ketoglutarate-dependent oxygenases [78, 82, 114, 129].
Siderophore chemistry plays a role in iron acquisition and antibiosis in the Photorhabdus lifecycle. In P. luminescens, a catecholate siderophore photobactin (Fig. 2, Structure 29) contributes to iron acquisition and enables the bacterium to grow under iron limiting conditions in vitro [16]. Photobactin is biosynthetically and structurally related to known catecholate siderophores, such as vibriobactin [50] from the human pathogen V. cholerae and agrobactin [101] from the plant pathogen Agrobacterium tumefaciens. The photobactin gene cluster contains a transposase-like element, and like many of the other secondary metabolic pathways in Photorhabdus, hints at its nomadic origin and its bacterial benefit in the multipartite symbiosis [16]. Unlike V. cholerae, in which a mutant defective in vibriobactin synthesis exhibited attenuated virulence in a mouse model [57, 139], P. luminescens mutants defective in photobactin synthesis did not lead to a detectable defect in supporting nematode mutualism or insect pathogenicity [16]. Rather, the TonB complex and YfeB transporter were required for metal transport in the insect and nematode environments [133]. Photobactin harbors antimicrobial activity and likely contributes to iron acquisition and antibiosis during the bacterium's competition with saprophytic bacteria near the decaying insect carcass [16]. Siderophore-related chemistry can promote antimicrobial activities against microbes lacking their cognate small molecule receptors, or in other cases, siderophore-antibiotic conjugates (sideromycins) can exploit siderophore uptake for antibiotic delivery. For Xenorhabdus, genome sequence analysis and secreted iron-binding activity under iron-limiting growth conditions suggest that both X. nematophila and X. bovienii are also capable of producing siderophores [14], and their roles remain unclear in the context of the symbiosis.
An approach for the discovery of novel secondary metabolism
Past and current bioinformatics strategies used to mine genomic information for natural product discovery largely focus on homology-based searches using previously described enzymes involved in antibiotic synthesis as inputs [37]. That is, you get what you search for and potentially miss biosynthetic systems that are currently unknown. For example, programs like antiSMASH utilize a collection of known antibiotic biosynthetic proteins as search criteria, which dramatically enhances initial secondary metabolite evaluations of genomic content [4, 86]. In cases of reasonably high sequence similarity, approximations of core structures can be predicted. However, many microbial genes in genome databases are annotated as encoding hypothetical proteins, and their functions cannot be predicted by sequence homology. Nor do they necessarily cluster with enzymes of perceived relevance to secondary metabolism. For some hypotheticals, functional insights can be loosely assessed through structural topology predictions that then guide subsequent experimental evaluations [68]. Intriguingly, many of these hypotheticals fall within genomic islands, suggesting that they might contribute to microbial functional adaptations. Some of these proteins will undoubtedly represent new biosynthetic catalysts that synthesize structurally diverse and currently unknown functional small molecules.
In addition to the homology-based searches, genome synteny (colocalization of genetic loci) among related organisms can provide a complementary view of gene content and genomic islands. Bioinformatics platforms, such as MicroScope, provide an excellent online source for visualizing genome synteny in annotated bacterial genomes [126]. Biosynthetic gene clusters, which are frequently transferred between organisms via horizontal gene transfer, often appear as acquired or lost sequence regions relative to closely related organisms in the comparative genome meta-analysis. Because many of the classical-type biosynthetic systems are “non-syntenic” across species, based on core evolutionary principles, we propose that atypical and currently unrecognized biosynthetic pathways should behave similarly. In this context, atypical biosynthetic pathways are multigenic clusters lacking genes similar to well-known biosynthetic systems and often containing multiple hypothetical enzymes. These gene clusters may encode proteins related to functionally recognizable enzymes, but they would not be exclusive to secondary metabolism. That is, these protein systems would be missed in the traditional homology-based searches for “antibiotic-like” biosynthetic proteins. As highlighted above, gene duplication of primary metabolic enzymes and functional divergence to secondary metabolic pathways is one evolutionary mechanism for bioactive small molecule synthesis. Putative atypical biosynthetic pathways can be observed in Xenorhabdus and Photorhabdus genomes, as well as other antibiotic-producing bacteria. We expect that this mixed chemical ecology-microbial genomics approach could provide a forward avenue for discovering and characterizing these potentially new biosynthetic enzyme systems, their resulting novel small molecule products and structural features, and the new biological functions for which they were evolutionarily selected to fulfill.
Conclusion
Microbial natural products have played and continue to play a major role in the drug development process [96], as their privileged scaffolds have been crafted by functional evolutionary selection. The reader is directed to an accompanying review by Demain in this Special Issue on Genome Mining dedicated to Sir David Hopwood for an overview of pharmacologically-relevant natural products [31]. The development of high-throughput and low cost microbial genome sequencing combined with the investigative exploitation of complex host-microbe interactions and the design of newly emerging synthetic biology platforms (e.g., [87, 90]) have ushered in a new era of natural products discovery that require fresh perspectives and interdisciplinary research programs. A revitalization of natural product discovery platforms in the post-genomic era shows great promise for the discovery of new functional small molecule classes and/or structural features. Chemical ecology can dramatically enhance these discovery platforms by placing orphan biosynthetic pathways and the cryptic molecules they produce in regulatory, genomic, functional, and ecological contexts.
We highlighted some illustrative natural product examples in the Xenorhabdus and Photorhabdus lifecycles, in which a greater understanding of the regulatory roles in their symbiotic lifestyle enhanced the discovery of new bioactive molecules in the lab. Additionally, their ecological contexts facilitated functional assignments that these molecules likely evolved to fulfill. With the numerous secondary metabolic genes emerging in the ever-expanding microbial genome databases, notwithstanding the increasing number of hypothetical genes providing sparse functional insights, interdisciplinary platforms to decode orphan secondary metabolic pathways and genomic islands will certainly reveal new biosynthetic catalysts involved in natural product synthesis and diversification, new evolutionarily privileged and functional small molecule structural features, and new chemical signaling events with potentially quite profound biomedical and commercial implications.
Acknowledgments
Our work on the discovery of bacterial natural product pathways in functional contexts is supported by the National Institutes of Health (R00-GM097096), the Searle Scholars Program (13-SSP-210), and the Damon Runyon Cancer Research Foundation (DFS:05-12). We thank Todd Ciche (Monsanto Company) for a preliminary version of Figure 1.
Footnotes
This manuscript is dedicated to Sir David Hopwood on the occasion of his 80th birthday.
References
- 1.Akhurst RJ. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Micro. 1980;121:303–309. [Google Scholar]
- 2.Bian X, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart AF, Zhang Y, Muller R. In vivo evidence for a prodrug activation mechanism during colibactin maturation. ChemBioChem. 2013;14(10):1194–1197. doi: 10.1002/cbic.201300208. [DOI] [PubMed] [Google Scholar]
- 3.Bian X, Plaza A, Zhang Y, Muller R. Luminmycins A-C, cryptic natural products from Photorhabdus luminescens identified by heterologous expression in Escherichia coli. J Nat Prod. 2012;75(9):1652–1655. doi: 10.1021/np300444e. [DOI] [PubMed] [Google Scholar]
- 4.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13(2):224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 6.Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 2002;3(7):619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann AO, Bode HB. Identification and bioanalysis of natural products from insect symbionts and pathogens. Adv Biochem Eng Biot. 2013 doi: 10.1007/10_2013_192. doi:10.1007/10_2013_192. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann AO, Joyce SA, Jenke-Kodama H, Schwar G, Clarke DJ, Bode HB. A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. ChemBioChem. 2007;8(14):1721–1728. doi: 10.1002/cbic.200700300. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann AO, Reimer D, Lorenzen W, Augusto Alonso E, Kopp Y, Piel J, Bode HB. Reciprocal cross talk between fatty acid and antibiotic biosynthesis in a nematode symbiont. Angew Chem Int Edit. 2012;51(48):12086–12089. doi: 10.1002/anie.201205384. [DOI] [PubMed] [Google Scholar]
- 10.Brady SF, Bauer JD, Clarke-Pearson MF, Daniels R. Natural products from isnA-containing biosynthetic gene clusters recovered from the genomes of cultured and uncultured bacteria. J Am Chem Soc. 2007;129(40):12102–12103. doi: 10.1021/ja075492v. [DOI] [PubMed] [Google Scholar]
- 11.Brady SF, Simmons L, Kim JH, Schmidt EW. Metagenomic approaches to natural products from free-living and symbiotic organisms. Nat Prod Rep. 2009;26(11):1488–1503. doi: 10.1039/b817078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brotherton CA, Balskus EP. A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J Am Chem Soc. 2013;135(9):3359–3362. doi: 10.1021/ja312154m. [DOI] [PubMed] [Google Scholar]
- 13.Chan YA, Boyne MT, 2nd, Podevels AM, Klimowicz AK, Handelsman J, Kelleher NL, Thomas MG. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc Natl Acad Sci U S A. 2006;103(39):14349–14354. doi: 10.1073/pnas.0603748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, et al. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PloS One. 2011;6(11):e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choulet F, Aigle B, Gallois A, Mangenot S, Gerbaud C, Truong C, Francou FX, Fourrier C, Guerineau M, Decaris B, Barbe V, Pernodet JL, Leblond P. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol Biol Evol. 2006;23(12):2361–2369. doi: 10.1093/molbev/msl108. [DOI] [PubMed] [Google Scholar]
- 16.Ciche TA, Blackburn M, Carney JR, Ensign JC. Photobactin: a catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl Environ Microbiol. 2003;69(8):4706–4713. doi: 10.1128/AEM.69.8.4706-4713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke DJ. Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cell Microbiol. 2008;10(11):2159–2167. doi: 10.1111/j.1462-5822.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- 18.Cocito C. Antibiotics of the virginiamycin family, inhibitors which contain synergistic components. Microbiol Rev. 1979;43(2):145–192. doi: 10.1128/mr.43.2.145-192.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copping LG, Duke SO. Natural products that have been used commercially as crop protection agents. Pest Manag Sci. 2007;63(6):524–554. doi: 10.1002/ps.1378. [DOI] [PubMed] [Google Scholar]
- 20.Costa SC, Girard PA, Brehelin M, Zumbihl R. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect Immun. 2009;77(3):1022–1030. doi: 10.1128/IAI.01064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol. 2007;9(5):1311–1323. doi: 10.1111/j.1462-5822.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- 22.Craig JW, Brady SF. Discovery of a metagenome-derived enzyme that produces branched-chain acyl-(acyl-carrier-protein)s from branched-chain alpha- keto acids. ChemBioChem. 2011;12(12):1849–1853. doi: 10.1002/cbic.201100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford JM, Clardy J. Bacterial symbionts and natural products. Chem Commun. 2011;47(27):7559–7566. doi: 10.1039/c1cc11574j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford JM, Kontnik R, Clardy J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr Biol. 2010;20(1):69–74. doi: 10.1016/j.cub.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford JM, Mahlstedt SA, Malcolmson SJ, Clardy J, Walsh CT. Dihydrophenylalanine: a prephenate-derived Photorhabdus luminescens antibiotic and intermediate in dihydrostilbene biosynthesis. Chem Biol. 2011;18(9):1102–1112. doi: 10.1016/j.chembiol.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford JM, Portmann C, Kontnik R, Walsh CT, Clardy J. NRPS substrate promiscuity diversifies the xenematides. Org Lett. 2011;13(19):5144–5147. doi: 10.1021/ol2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crawford JM, Portmann C, Zhang X, Roeffaers MB, Clardy J. Small molecule perimeter defense in entomopathogenic bacteria. Proc Natl Acad Sci U S A. 2012;109(27):10821–10826. doi: 10.1073/pnas.1201160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17(3):254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniela R, Klaas MP, Marco T, Peter G, Helge BB. A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat Chem Biol. 2011;7(12):888–890. doi: 10.1038/nchembio.688. [DOI] [PubMed] [Google Scholar]
- 31.Demain AL. Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biot. 2013;xx(xx):xx–xx. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- 32.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2(5):414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 33.Dongjin P. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol Microbiol. 2009;73(5):938–949. doi: 10.1111/j.1365-2958.2009.06817.x. [DOI] [PubMed] [Google Scholar]
- 34.Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Medigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21(11):1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 35.Eleftherianos I, Boundy S, Joyce SA, Aslam S, Marshall JW, Cox RJ, Simpson TJ, Clarke DJ, ffrench-Constant RH, Reynolds SE. An antibiotic produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Proc Natl Acad Sci U S A. 2007;104(7):2419–2424. doi: 10.1073/pnas.0610525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 2010;18(12):552–560. doi: 10.1016/j.tim.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Fedorova ND, Moktali V, Medema MH. Bioinformatics approaches and software for detection of secondary metabolic gene clusters. Method Mol Biol. 2012;944:23–45. doi: 10.1007/978-1-62703-122-6_2. [DOI] [PubMed] [Google Scholar]
- 38.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci U S A. 2008;105(12):4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraenkel GS. The Raison d'Être of Secondary Plant Substances: These odd chemicals arose as a means of protecting plants from insects and now guide insects to food. Science. 1959;129(3361):1466–1470. doi: 10.1126/science.129.3361.1466. [DOI] [PubMed] [Google Scholar]
- 40.Franke S, Ibarra F, Schulz CM, Twele R, Poldy J, Barrow RA, Peakall R, Schiestl FP, Francke W. The discovery of 2,5-dialkylcyclohexan-1,3-diones as a new class of natural products. Proc Natl Acad Sci U S A. 2009;106(22):8877–8882. doi: 10.1073/pnas.0900646106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30(5):440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 42.Fuchs SW, Bozhuyuk KA, Kresovic D, Grundmann F, Dill V, Brachmann AO, Waterfield NR, Bode HB. Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occuring ketosynthase. Angew Chemie. 2013;52(15):4108–4112. doi: 10.1002/anie.201210116. [DOI] [PubMed] [Google Scholar]
- 43.Fuchs SW, Proschak A, Jaskolla TW, Karas M, Bode HB. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org Biomol Chem. 2011;9(9):3130–3132. doi: 10.1039/c1ob05097d. [DOI] [PubMed] [Google Scholar]
- 44.Gaudriault S, Duchaud E, Lanois A, Canoy AS, Bourot S, Derose R, Kunst F, Boemare N, Givaudan A. Whole-genome comparison between Photorhabdus strains to identify genomic regions involved in the specificity of nematode interaction. J Bacteriol. 2006;188(2):809–814. doi: 10.1128/JB.188.2.809-814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerrard J, Waterfield N, Vohra R, ffrench-Constant R. Human infection with Photorhabdus asymbiotica: an emerging bacterial pathogen. Microbes Infect. 2004;6(2):229–237. doi: 10.1016/j.micinf.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Gerrard JG, Joyce SA, Clarke DJ, ffrench-Constant RH, Nimmo GR, Looke DF, Feil EJ, Pearce L, Waterfield NR. Nematode symbiont for Photorhabdus asymbiotica. Emerg Infect Dis. 2006;12(10):1562–1564. doi: 10.3201/eid1210.060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerrard JG, Waterfield NR, Sanchez-Contreeras M. Photorhabdus asymbiotica: Shedding light on a human pathogenic bioluminescent bacterium. Clin Microbiol Newsletter. 2011;33(14):103–110. [Google Scholar]
- 48.Goodrich-Blair H. They've got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr Opin Microbiol. 2007;10(3):225–230. doi: 10.1016/j.mib.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol. 2007;64(2):260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 50.Griffiths GL, Sigel SP, Payne SM, Neilands JB. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259(1):383–385. [PubMed] [Google Scholar]
- 51.Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26(11):1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 52.Gualtieri M, Aumelas A, Thaler JO. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J Antibiot. 2009;62(6):295–302. doi: 10.1038/ja.2009.31. [DOI] [PubMed] [Google Scholar]
- 53.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 54.Hartmann T. Plant-derived secondary metabolites as defensive chemicals in herbivorous insects: a case study in chemical ecology. Planta. 2004;219(1):1–4. doi: 10.1007/s00425-004-1249-y. [DOI] [PubMed] [Google Scholar]
- 55.Hartmann T. The lost origin of chemical ecology in the late 19th century. Proc Natl Acad Sci U S A. 2008;105(12):4541–4546. doi: 10.1073/pnas.0709231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heermann R, Fuchs TM. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics. 2008;9:40. doi: 10.1186/1471-2164-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henderson DP, Payne SM. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun. 1994;62(11):5120–5125. doi: 10.1128/iai.62.11.5120-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev. 2012;10(9):641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- 59.Herbert EE, Goodrich-Blair H. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev. 2007;5(8):634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- 60.Hu K, Li J, Webster JM. Quantitative analysis of a bacteria-derived antibiotic in nematode-infected insects using HPLC-UV and TLC-UV methods. J Chromatogr. 1997;703(1-2):177–183. doi: 10.1016/s0378-4347(97)00398-8. [DOI] [PubMed] [Google Scholar]
- 61.Hu K, Webster JM. Antibiotic production in relation to bacterial growth and nematode development in Photorhabdus--Heterorhabditis infected Galleria mellonella larvae. FEMS Microb Lett. 2000;189(2):219–223. doi: 10.1111/j.1574-6968.2000.tb09234.x. [DOI] [PubMed] [Google Scholar]
- 62.Hung KY, Harris PW, Heapy AM, Brimble MA. Synthesis and assignment of stereochemistry of the antibacterial cyclic peptide xenematide. Org Biomol Chem. 2011;9(1):236–242. doi: 10.1039/c0ob00315h. [DOI] [PubMed] [Google Scholar]
- 63.Jenke-Kodama H, Muller R, Dittmann E. Evolutionary mechanisms underlying secondary metabolite diversity. Prog Drug Res. 2008;65(119):121–140. doi: 10.1007/978-3-7643-8117-2_3. [DOI] [PubMed] [Google Scholar]
- 64.Jensen PR. Linking species concepts to natural product discovery in the post-genomic era. J Ind Microbiol. 2010;37(3):219–224. doi: 10.1007/s10295-009-0683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joyce SA, Brachmann AO, Glazer I, Lango L, Schwar G, Clarke DJ, Bode HB. Bacterial biosynthesis of a multipotent stilbene. Angew Chem Int Edit. 2008;47(10):1942–1945. doi: 10.1002/anie.200705148. [DOI] [PubMed] [Google Scholar]
- 66.Joyce SA, Clarke DJ. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol Microbiol. 2003;47(5):1445–1457. doi: 10.1046/j.1365-2958.2003.03389.x. [DOI] [PubMed] [Google Scholar]
- 67.Kalaitzis JA, Lauro FM, Neilan BA. Mining cyanobacterial genomes for genes encoding complex biosynthetic pathways. Nat Prod Rep. 2009;26(11):1447–1465. doi: 10.1039/b817074f. [DOI] [PubMed] [Google Scholar]
- 68.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 69.Kevany BM, Rasko DA, Thomas MG. Characterization of the complete zwittermicin A biosynthesis gene cluster from Bacillus cereus. Appl Environ Microbiol. 2009;75(4):1144–1155. doi: 10.1128/AEM.02518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khush RS, Lemaitre B. Genes that fight infection: what the Drosophila genome says about animal immunity. Trends Genet. 2000;16(10):442–449. doi: 10.1016/s0168-9525(00)02095-3. [DOI] [PubMed] [Google Scholar]
- 71.Kontnik R, Crawford JM, Clardy J. Exploiting a global regulator for small molecule discovery in Photorhabdus luminescens. ACS Chem Bio. 2010;5(7):659–665. doi: 10.1021/cb100117k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100(26):15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci U S A. 2012;109(50):20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lang G, Kalvelage T, Peters A, Wiese J, Imhoff JF. Linear and Cyclic Peptides from the Entomopathogenic Bacterium Xenorhabdus nematophilus. J Nat Prod. 2008;71(6):1074–1077. doi: 10.1021/np800053n. [DOI] [PubMed] [Google Scholar]
- 75.Lanois A, Ogier JC, Gouzy J, Laroui C, Rouy Z, Givaudan A, Gaudriault S. Draft genome sequence and annotation of the entomopathogenic bacterium Xenorhabdus nematophila Strain F1. Genome Announcements. 2013;1(3) doi: 10.1128/genomeA.00342-13. doi:10.1128/genomeA.00342-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis K, Epstein S, D'Onofrio A, Ling LL. Uncultured microorganisms as a source of secondary metabolites. J Antibiot. 2010;63(8):468–476. doi: 10.1038/ja.2010.87. [DOI] [PubMed] [Google Scholar]
- 77.Li J, Chen G, Wu H, Webster JM. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl Environ Microbiol. 1995;61(12):4329–4333. doi: 10.1128/aem.61.12.4329-4333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin H-Y, Rao YK, Wu W-S, Tzeng Y-M. Ferrous ion enhanced lipopeptide antibiotic Iturin A production from Bacillus amyloliquefaciens B128. Int J Appl Sci En. 2007;5(2):123–132. [Google Scholar]
- 79.Luo Y, Ruan LF, Zhao CM, Wang CX, Peng DH, Sun M. Validation of the intact zwittermicin A biosynthetic gene cluster and discovery of a complementary resistance mechanism in Bacillus thuringiensis. Antimicrob Agents Chemother. 2011;55(9):4161–4169. doi: 10.1128/AAC.00111-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahlstedt S, Fielding EN, Moore BS, Walsh CT. Prephenate decarboxylases: a new prephenate-utilizing enzyme family that performs nonaromatizing decarboxylation en route to diverse secondary metabolites. Biochemistry. 2010;49(42):9021–9023. doi: 10.1021/bi101457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahlstedt SA, Walsh CT. Investigation of anticapsin biosynthesis reveals a four-enzyme pathway to tetrahydrotyrosine in Bacillus subtilis. Biochemistry. 2010;49(5):912–923. doi: 10.1021/bi9021186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martinez-Molina E, Del Rio LA, Olivares J. Copper and iron as determinant factors of antibiotic production by Pseudomonas reptilivora. J Appl Bacteriol. 1976;41(1):69–74. doi: 10.1111/j.1365-2672.1976.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 83.Mast Y, Weber T, Golz M, Ort-Winklbauer R, Gondran A, Wohlleben W, Schinko E. Characterization of the 'pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb Biotechnol. 2011;4(2):192–206. doi: 10.1111/j.1751-7915.2010.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maxwell PW, Chen G, Webster JM, Dunphy GB. Stability and activities of antibiotics produced during infection of the insect Galleria mellonella by two isolates of Xenorhabdus nematophilus. Appl Environ Microbiol. 1994;60(2):715–721. doi: 10.1128/aem.60.2.715-721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McInerney BV, Gregson RP, Lacey MJ, Akhurst RJ, Lyons GR, Rhodes SH, Smith DR, Engelhardt LM, White AH. Biologically active metabolites from Xenorhabdus spp., Part 1. Dithiolopyrrolone derivatives with antibiotic activity. J Nat Prod. 1991;54(3):774–784. doi: 10.1021/np50075a005. [DOI] [PubMed] [Google Scholar]
- 86.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Medema MH, Breitling R, Bovenberg R, Takano E. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol. 2011;9(2):131–137. doi: 10.1038/nrmicro2478. [DOI] [PubMed] [Google Scholar]
- 88.Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Muller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RA, Breitling R, Takano E. The sequence of a 1.8-mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol. 2010;2:212–224. doi: 10.1093/gbe/evq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mitchell W. Natural products from synthetic biology. Curr Opin Chem Biol. 2011;15(4):505–515. doi: 10.1016/j.cbpa.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 91.Molnar K, Farkas E. Current results on biological activities of lichen secondary metabolites: a review. Zeitschrift fur Naturforschung. 2010;65(3-4):157–173. doi: 10.1515/znc-2010-3-401. [DOI] [PubMed] [Google Scholar]
- 92.Müller-Schwarze D. Chemical ecology of vertebrates. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- 93.Murfin KE, Dillman AR, Foster JM, Bulgheresi S, Slatko BE, Sternberg PW, Goodrich-Blair H. Nematode-bacterium symbioses--cooperation and conflict revealed in the “omics” age. Biol Bull. 2012;223(1):85–102. doi: 10.1086/BBLv223n1p85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 95.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26(11):1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 98.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 99.Ogier JC, Calteau A, Forst S, Goodrich-Blair H, Roche D, Rouy Z, Suen G, Zumbihl R, Givaudan A, Tailliez P, Medigue C, Gaudriault S. Units of plasticity in bacterial genomes: new insight from the comparative genomics of two bacteria interacting with invertebrates, Photorhabdus and Xenorhabdus. BMC Genomics. 2010;11:568. doi: 10.1186/1471-2164-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oku H, Kaneda T. Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J Biol Chem. 1988;263(34):18386–18396. [PubMed] [Google Scholar]
- 101.Ong SA, Peterson T, Neilands JB. Agrobactin, a siderophore from Agrobacterium tumefaciens. J Biol Chem. 1979;254(6):1860–1865. [PubMed] [Google Scholar]
- 102.Oves-Costales D, Kadi N, Challis GL. The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem Commun. 2009;(43):6530–6541. doi: 10.1039/b913092f. [DOI] [PubMed] [Google Scholar]
- 103.Pawlik JR. The chemical ecology of sponges on Caribbean reefs: Natural products shape natural systems. BioScience. 2011;61(11):888–898. [Google Scholar]
- 104.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3(10):1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perham RN. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30(35):8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 106.Phelan VV, Liu WT, Pogliano K, Dorrestein PC. Microbial metabolic exchange--the chemotype-to-phenotype link. Nat Chem Biol. 2012;8(1):26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pinchuk IV, Bressollier P, Sorokulova IB, Verneuil B, Urdaci MC. Amicoumacin antibiotic production and genetic diversity of Bacillus subtilis strains isolated from different habitats. Res Microbiol. 2002;153(5):269–276. doi: 10.1016/s0923-2508(02)01320-7. [DOI] [PubMed] [Google Scholar]
- 108.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev. 2006;4(4):295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 109.Purdy AE, Watnick PI. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc Natl Acad Sci U S A. 2011;108(49):19737–19742. doi: 10.1073/pnas.1111530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rath CM, Dorrestein PC. The bacterial chemical repertoire mediates metabolic exchange within gut microbiomes. Curr Opin Microbiol. 2012;15(2):147–154. doi: 10.1016/j.mib.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reimer D. A new type of pyrrolidine biosynthesis is involved in the late steps of xenocoumacin production in Xenorhabdus nematophila. ChemBioChem. 2009;10(12):1997–2001. doi: 10.1002/cbic.200900187. [DOI] [PubMed] [Google Scholar]
- 112.Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol. 2009;11:1025–33. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Richardson WH, Schmidt TM, Nealson KH. Identification of an anthraquinone pigment and a hydroxystilbene antibiotic from Xenorhabdus luminescens. Appl Environ Microbiol. 1988;54(6):1602–1605. doi: 10.1128/aem.54.6.1602-1605.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rollins MJ, Jensen SE, Westlake DWS. Regulation of antibiotic production by iron and oxygen during defined medium fermentations of Streptomyces clavuligerus. Appl Environ Microbiol. 1989;31:390–396. doi: 10.1139/m89-186. [DOI] [PubMed] [Google Scholar]
- 115.Sanchez JF, Somoza AD, Keller NP, Wang CC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012;29(3):351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scherlach K, Graupner K, Hertweck C. Molecular bacterial-fungal interactions with wmpact on the environment, food and medicine. Annu Rev Microbiol. 2013 doi: 10.1146/annurev-micro-092412-155702. doi:10.1146/annurev-micro-092412-155702. [DOI] [PubMed] [Google Scholar]
- 117.Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J. Origin and variation of tunicate secondary metabolites. J Nat Prod. 2012;75(2):295–304. doi: 10.1021/np200665k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Somvanshi VS, Kaufmann-Daszczuk B, Kim KS, Mallon S, Ciche TA. Photorhabdus phase variants express a novel fimbrial locus, mad, essential for symbiosis. Mol Microbiol. 2010;77:1021–1038. doi: 10.1111/j.1365-2958.2010.07270.x. [DOI] [PubMed] [Google Scholar]
- 119.Somvanshi VS, Sloup RE, Crawford JM, Martin AR, Heidt AJ, Kim KS, Clardy J, Ciche TA. A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science. 2012;337(6090):88–93. doi: 10.1126/science.1216641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stein ML, Beck P, Kaiser M, Dudler R, Becker CF, Groll M. One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor. Proc Natl Acad Sci U S A. 2012;109(45):18367–18371. doi: 10.1073/pnas.1211423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stocker-Worgotter E. Metabolic diversity of lichen-forming ascomycetous fungi: culturing, polyketide and shikimate metabolite production, and PKS genes. Nat Prod Rep. 2008;25(1):188–200. doi: 10.1039/b606983p. [DOI] [PubMed] [Google Scholar]
- 122.Sugar DR, Murfin KE, Chaston JM, Andersen AW, Richards GR, deLeon L, Baum JA, Clinton WP, Forst S, Goldman BS, Krasomil-Osterfeld KC, Slater S, Stock SP, Goodrich-Blair H. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ Microbiol. 2012;14(4):924–939. doi: 10.1111/j.1462-2920.2011.02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Theodore CM, King JB, You J, Cichewicz RH. Production of cytotoxic glidobactins/luminmycins by Photorhabdus asymbiotica in liquid media and live crickets. J Nat Prod. 2012;75(11):2007–2011. doi: 10.1021/np300623x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tounsi S, Blight M, Jaoua S, de Lima Pimenta A. From insects to human hosts: Identification of major genomic differences between entomopathogenic strains of Photorhabdus and the emerging human pathogen Photorhabdus asymbiotica. IJMM. 2006;296(8):521–530. doi: 10.1016/j.ijmm.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 125.Uvell H, Engstrom Y. A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 2007;23(7):342–349. doi: 10.1016/j.tig.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 126.Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, Rouy Z, Roche D, Salvignol G, Scarpelli C, Medigue C. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009 doi: 10.1093/database/bap021. 2009:bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walsh CT, O'Brien RV, Khosla C. Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew Chem Int Edit. 2013;52(28):7098–7124. doi: 10.1002/anie.201208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 129.Wang G, Jia S, Wang T, Chen L, Song Q, Li W. Effect of ferrous ion on epsilon-poly-L-lysine biosynthesis by Streptomyces diastatochromogenes CGMCC3145. Curr Microbiol. 2011;62(3):1062–1067. doi: 10.1007/s00284-010-9828-6. [DOI] [PubMed] [Google Scholar]
- 130.Waterfield NR, Ciche T, Clarke D. Photorhabdus and a host of hosts. Annu Rev Microbiol. 2009;63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- 131.Waterfield NR, Sanchez-Contreras M, Eleftherianos I, Dowling A, Wilkinson P, Parkhill J, Thomson N, Reynolds SE, Bode HB, Dorus S, Ffrench-Constant RH. Rapid Virulence Annotation (RVA): identification of virulence factors using a bacterial genome library and multiple invertebrate hosts. Proc Natl Acad Sci U S A. 2008;105(41):15967–15972. doi: 10.1073/pnas.0711114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Waterfield NR, Wren BW, Ffrench-Constant RH. Invertebrates as a source of emerging human pathogens. Nat Rev Microbiol. 2004;2(10):833–841. doi: 10.1038/nrmicro1008. [DOI] [PubMed] [Google Scholar]
- 133.Watson RJ, Millichap P, Joyce SA, Reynolds S, Clarke DJ. The role of iron uptake in pathogenicity and symbiosis in Photorhabdus luminescens TT01. BMC Microbiol. 2010;10:177. doi: 10.1186/1471-2180-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weissfeld AS, Halliday RJ, Simmons DE, Trevino EA, Vance PH, O'Hara CM, Sowers EG, Kern R, Koy RD, Hodde K, Bing M, Lo C, Gerrard J, Vohra R, Harper J. Photorhabdus asymbiotica, a pathogen emerging on two continents that proves that there is no substitute for a well-trained clinical microbiologist. J Clin Microbiol. 2005;43(8):4152–4155. doi: 10.1128/JCM.43.8.4152-4155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wenzel SC, Muller R. The impact of genomics on the exploitation of the myxobacterial secondary metabolome. Nat Prod Rep. 2009;26(11):1385–1407. doi: 10.1039/b817073h. [DOI] [PubMed] [Google Scholar]
- 136.Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez-Contreras M, Vlisidou I, Barron A, Bignell A, Clark L, Ormond D, Mayho M, Bason N, Smith F, Simmonds M, Churcher C, Harris D, Thompson NR, Quail M, Parkhill J, Ffrench-Constant RH. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics. 2009;10:302. doi: 10.1186/1471-2164-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wink M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry. 2003;64(1):3–19. doi: 10.1016/s0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 138.Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15(1):22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 139.Wyckoff EE, Mey AR, Payne SM. Iron acquisition in Vibrio cholerae. Biometals. 2007;20(3-4):405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 140.Zhou T. Global transcriptional responses of Bacillus subtilis to xenocoumacin 1. J Appl Microbiol. 2011;111(3):652–662. doi: 10.1111/j.1365-2672.2011.05086.x. [DOI] [PubMed] [Google Scholar]
- 141.Zotchev SB, Sekurova ON, Katz L. Genome-based bioprospecting of microbes for new therapeutics. Curr Opin Biotech. 2012;23(6):941–947. doi: 10.1016/j.copbio.2012.04.002. [DOI] [PubMed] [Google Scholar]