Abstract
Genome sequencing is rapidly changing the field of natural products research by providing opportunities to assess the biosynthetic potential of strains prior to chemical analysis or biological testing. Ready access to sequence data is driving the development of new bioinformatic tools and methods to identify the products of silent or cryptic pathways. While genome mining has fast become a useful approach to natural product discovery, it has also become clear that identifying pathways of interest is much easier than finding the associated products. This has led to bottlenecks in the discovery process that must be overcome for the potential of genomics-based natural product discovery to be fully realized. In this perspective, we address some of these challenges in the context of our work with the marine actinomycete genus Salinispora, which is proving to be a useful model with which to apply genome mining as an approach to natural product discovery.
Keywords: genomics, natural product biosynthesis, genome mining, Salinispora
Introduction
Bacterial natural product discovery once relied heavily upon luck. Thousands of strains were typically cultured in a limited number of fermentation conditions in the hope that a minimum number would yield compounds of interest. The odds could be improved by creative fermentation techniques and targeting poorly studied taxa, however the process remained deeply invested in serendipity. While this strategy initially proved successful, it became less tenable by the close of the twentieth century as the rates of new compound discovery dropped to levels that could not be supported by the pharmaceutical industry. These diminishing returns, coupled with advances in combinatorial chemistry and high throughput screening, led major pharmaceutical companies worldwide to move en masse away from natural products as a resource for drug discovery [16], leaving this area of research largely in the realm of academia and small biotechnology companies.
Ready access to microbial genome sequencing has now changed the playing field for natural product discovery. Genome sequencing provides a highly informed approach by which strains can be prioritized based on a bioinformatic assessment of their biosynthetic potential. This potential can be used to infer the production of known compounds (de-replication), to make generalized predictions about the types of compounds that can be expected (e.g., polyketides, terpenes, etc.), and in some cases to make precise structural predictions. These capabilities provide opportunities to identify strains with the potential to produce compounds of interest, which once identified can be subjected to detailed fermentation studies, biological screening, and chemical analysis. Key genes in a targeted pathway can also be monitored for expression to help ensure that the appropriate fermentation conditions have been selected. Alternatively, entire pathways can be targeted for heterologous expression thereby bypassing regulatory hurdles in the native host.
Genome sequencing has fundamentally changed the way we think about natural product discovery. Nonetheless, it is far from a panacea, as many technical challenges remain. These challenges include the bioinformatic expertise required to handle large data sets and the lack of comprehensive pathway databases that can be used for rapid comparative analysis. One major challenge arises from the fact that genome sequences are rarely closed, with the number of contigs dependent upon the depth of sequencing and the efficiency of the assembly process. In the case of secondary metabolism, where biosynthetic gene clusters can exceed 100 kb, it is uncommon to capture large pathways on a single contig. In addition, the highly repetitive sequence motifs associated with many biosynthetic genes create assembly challenges that are not readily surmountable regardless of sequencing depth. Despite these challenges, it has become widely recognized that bacterial genomes harbor many more biosynthetic pathways than the number of compounds discovered from them would predict [23]. While pathways can be readily identified from sequence data using tools such as antiSMASH [22] and SBSPKS [2], it has become increasingly clear that the identification and structure elucidation of the compounds they produce, and the establishment of formal links between pathways and products, remain major bottlenecks.
The recognition that even well-studied species such as S. coelicolor can harbor a large number of pathways for which the products remain unknown came as something of a surprise [4]. This observation implies that the associated compounds are either not being produced or are not being detected using the techniques employed. Both of these issues can be addressed but not without significant effort. An alternative is heterologous expression, which may ultimately provide the most effective approach, but currently remains limited in application. Here we provide perspectives on these various topics derived from our experience with the marine actinomycete genus Salinispora. While it remains unclear how broadly applicable the results obtained for this model organism will be to other bacteria, the challenges are similar to those faced with better known secondary metabolite producing taxa such as the genus Streptomyces.
Salinispora genomics
The marine actinomycete genus Salinispora is comprised of only three species [20; 1] yet has yielded an impressive array of structurally diverse secondary metabolites [10]. Most significant among these is salinosporamide A [9], which has advanced to clinical trials for the treatment of cancer [11]. The first Salinispora genome to be sequenced revealed a surprisingly large number of biosynthetic pathways relative to the compounds that had been discovered [28]. The second genome sequence provided clear evidence that these pathways were clustered in genomic islands [24] and additional support for the observation that secondary metabolites were produced in species-specific patterns [14]. The analysis of additional genome sequences is providing new insight into the biosynthetic diversity within this taxon and information about the processes driving secondary metabolite gene evolution. These efforts are being made possible through the acquisition of more than 100 Salinispora genome sequences through the Joint Genome Institute Community Sequencing Program (www.jgi.doe.gov/CSP/overview). This program provides high quality, annotated draft genomes and is linked to a variety of tools that can be used to assist in genome analyses (http://img.jgi.doe.gov/).
Pathway assembles
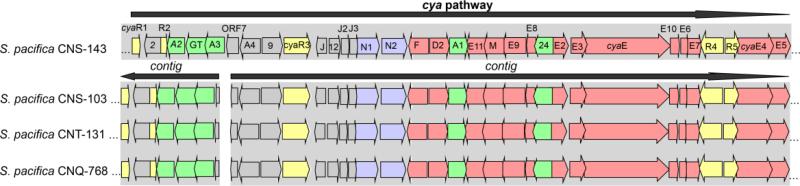
The poor assemblies observed for many secondary metabolite biosynthetic pathways creates challenges for bioinformatic-based structure predictions. However, the quality of the assembly can vary greatly depending not only upon the depth of sequencing but also on the type of biosynthetic pathway encountered. For example, of the 11 different type I modular PKS pathways (containing more than 3 modules) that have been detected to date in Salinsipora genomes, none were assembled. This was readily apparent from the detection of highly similar KS domains on different contigs and by the use of well-defined pathways, such as that for rifamycin biosynthesis [29], as templates for contig assembly. Type I modular PKSs are highly repetitive and thus it is not surprising that they create challenges for assembly algorithms. In some cases, modules are collapsed within the assemblies while in others they simply fail to assemble. Another interesting observation is that the same PKS pathway can be truncated in the identical location in different genome sequences. This is exemplified by the cya gene cluster, which is responsible for the biosynthesis of the cyanosporasides in S. pacifica strain CNS-143 [17]. In multiple strains that possess this pathway, the contigs truncate at orf7, which is annotated as a dihydrofolate reductase (figure 1). Further examination reveals that each contig ends at the same nucleotide, which suggests that the termination may be linked to the sequencing technology itself. Certainly new sequencing technologies that acquire longer read lengths, such as that marketed by Pacific Biosciences (http://www.pacificbiosciences.com/), will help solve this problem. Conversely, the majority of type II PKS pathways, which lack the highly repetitive structure of modular PKSs, are fully assembled in the Salinispora genomes.
Figure 1.
The cyanosporaside pathway cya. This pathway was originally characterized in S. pacifica strain CNS-143 [17] using a combination of fosmid sequencing and primer walking. The three S. pacifica draft genome sequences (strains CNS-103, CNT-131, and CNQ-768) all posses the pathway, which occurs on two contigs. These contigs all end at the same nucleotide position in ORF7.
Defining pathway boundaries
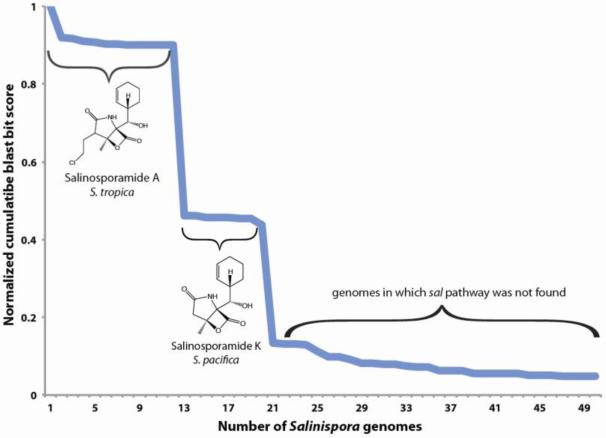
Identifying the boundaries of a biosynthetic gene cluster is a subjective process. Outside of the core biosynthetic genes and those associated with regulation and transport, there are often uncertainties about other genes in the cluster, especially in the flanking regions and for those with hypothetical annotations. Having access to multiple genomes representing strains that produce the same compound provides a useful method to predict the minimum pathway required for compound production. MultiGeneBlast [21] provides a useful tool for this type of analysis. The search output includes cumulative blast bit scores, which represent the sum of the BlastP bit scores for all genes in a genome that match the query sequence. This score provides a quantitative method to estimate the presence/absence of pathways in genome sequences as scores generally drop precipitously when a pathway is not present. Furthermore, this tool can be used to identify strains that contain variations of related pathways, which can be predictive of structural variations within a compound class. This is exemplified by the production of salinosporamides A and K by S. tropica and S. pacifica, respectively [8; 7]. In this case, plotting the normalized blast bit scores shows a clear stepwise decrease that corresponds to the distribution of the salinosporamide A and K biosynthetic pathways in the two Salinispora species (figure 2). A subsequent decrease in scores to <0.2 is then observed for genomes that do not possess the pathway. It’s particularly interesting to consider that the variations in the sal pathway correspond to a speciation event and to speculate on the potential ecological significance of the structural changes [12].
Figure 2.
Distribution of the sal pathway among Salinispora genomes. MultiGeneBlast [21] was used to BLAST the sal pathway from S. tropica (which is responsible for salinosporamide A production) against a database of Salinispora genomes. The resulting cumulative BLAST bit scores were normalized to the genome from which the query pathway was derived. A drop in scores to ca. 0.5 is observed for S. pacifica genomes that possess the version of the pathway responsible for salinosporamide K production. These strains lack the genes encoding the 26 kb chloroethylmalonyl-CoA portion of the pathway [7]. Scores then drop further to <0.2 in strains that do not possess the pathway.
Sequence tags
Given that many biosynthetic pathways are not fully assembled in most draft genome sequences, an alternative approach to predict the class of compounds that will be produced is through the use of sequence tags. NaPDoS was specifically developed using this concept for the classification of PKS and NRPS genes based on sequence tags corresponding to KS and C domains, respectively [31; 32]. Despite sequence lengths of only 200-300 amino acids, these tags can be used to make predictions about the pathway type (e.g., type I or type II PKS), the structural class of the product (e.g., an enediyne or a PKS-NRPS hybrid), and in the case of close matches (i.e., >90% nucleotide sequence identity), the structure of the product. The analysis of sequence tags can be a particularly useful strategy in the case of poorly assembled genome sequences and to make a quick assessment of the biosynthetic potential of individual strains including the de-replication of well-known compound classes. This approach can also be readily applied to environmental DNA in an effort to identify the best sample types from which to target cultivation efforts.
Iterative pathway-product analysis
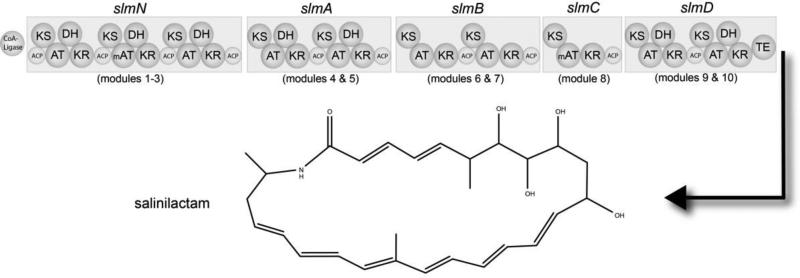
While genome sequences can be viewed as the ultimate predictors of secondary metabolite biosynthesis, there are numerous examples where generalized biosynthetic logic, such as the co-linearity rule, has been violated. These examples include the siderophore coelichelin, which included one more amino acid than predicted based on the associated tri-modular NRPS [18], and have led to an improved understanding of processes such as module skipping and stuttering [3]. Exceptions from traditional paradigms have helped expand our understanding of biosynthetic processes and emphasize the importance of having structurally characterized compounds that can be compared to the pathways responsible for their production [25]. Interestingly, structures can also be used to help resolve ambiguities in genome sequence data. Salinilactam A isolated from S. tropica provides one such example (figure 3). The initial characterization of this compound revealed a carbon skeleton that would require 10 extension modules. The candidate slm pathway however was found on separate contigs and, due to the high level of sequence similarity among modules (>99% in many regions), it was not clear how they should be assembled. An understanding of the basic structure of the compound provided the logic to assemble the pathway into 10 modules, which ultimately helped to close the S. tropica CNB-440 genome sequence [28]. The organization of the dehydratase domains could then be used to help assign the eight conjugated olefins in the compound, which were difficult to resolve by NMR, along with the position of a methyl group at C-18 based on the methyl-malonyl-CoA specificity of the associated AT domain. This type of iterative process between structure assignment and gene cluster assembly can be especially useful for large modular PKSs.
Figure 3.
Modular organization of the slm pathway as observed in S. tropica strain CNB-440 and its associated product salinilactam. ACP: acyl-carrier protein, KS: ketosynthase, AT: acyl-transferase, mAT: methyl-malonyl-CoA specific AT domain, DH: dehydratase, KR: ketoreductase, TE: thioesterase.
The bottlenecks
Two major bottlenecks to genomics-based natural product research are readily apparent. One is the large number of biosynthetic pathways that appear to be silent. If in fact these pathways are not being expressed, it is clear that improved cultivation techniques, e.g., those that seek to better mimic natural conditions, must be sought. Alternatively, it is possible that many are being expressed yet the products are simply not being detected with the analytical methods employed. This could be due to low yield, the absence of a UV chromophore, or the complexity of the mixture within which they may reside. Alternatively, organic extraction methods, which frequently select for more lipophilic compounds, may not be appropriate for many of these products. A second bottleneck remains the establishment of formal links between specific pathways and structurally characterized compounds. In cases such as modular type I PKSs, bioinformatic predictions may correlate well with structures, however in others correlations are less apparent. Experimentally verified links are extremely important as once made they inform all future discovery efforts. Yet establishing these links requires considerable effort, such as knocking out key genes in the biosynthetic pathway, and thus remains a time consuming endeavor.
Salinispora genetics
While Salinispora genomics has provided considerable insight into the natural product biosynthetic diversity of the genus, the development of genetic protocols to work with these bacteria has been crucial to experimentally link biosynthetic pathways to specific metabolites. The first validated Salinispora biosynthetic pathway was the sal locus in S. tropica CNB-440, which is responsible for the construction of the -lactone proteasome inhibitor salinosporamide A [6]. Since then, numerous Salinispora biosynthetic gene clusters have been validated, including those associated with the production of the cyclic peptide cyclomarin (cym) in S. arenicola CNS-205 [26] and the enediyne polyketide cyanosporaside (cya) in S. pacifica CNS-143 [17]. The general methodology for interrogating the function of Salinispora genes involves PCR targeting via Red/ET recombineering, which is also useful in other actinomycetes [13]. In addition to facilitating gene deletions, -Red-mediated recombination has also been used to replace Salinispora genes with homologues in order to alter native pathway functions as in the genetic engineering of fluorosalinosporamide [5]. C31 phage-based vectors have also been employed to integrate DNA into Salinispora chromosomes at pseudo-attB sites [19]. These studies have shown that modifications to genetic methods commonly employed with terrestrial actinomycetes, such as the model organism S. coelicolor A3(2), are appropriate in Salinispora after taking into account its requirement for saline growth media. A recently constructed synthetic promoter library for actinomycetes based on -10 and -35 consensus sequences of native promoters proved equally effective in S. tropica CNB-440 as in several terrestrial actinomycete strains [27], thereby adding to the notion that Salinispora isolates are amenable to genetic protocols that are commonly employed in Streptomyces [15]. Thus, it comes at some surprise that a Salinispora biosynthetic pathway has yet to be heterologously expressed in a surrogate host, which is a well-established practice with terrestrial and more recently marine Streptomyces [30]. The development of an effective expression system for Salinispora strains will be needed if we are to effectively capture a greater percentage of the secondary metabolite biosynthetic potential of this marine actinomycete genus.
Natural products chemistry in the post-genomic era
The identification of diverse and abundant secondary metabolite biosynthetic pathways in bacterial strains has created considerable excitement about opportunities for the isolation of new metabolites. Of course, it is unclear how many of these pathways are expressed when strains are grown in the laboratory and, if they are, at what levels the associated compounds are produced. While bioinformatic analyses provide the opportunity to evaluate the biosynthetic potential of individual strains and predict, to varying degrees of accuracy, the structures and even stereochemical details of metabolites, a major difficulty remains the translation of this potential into purified molecules that can be evaluated using spectroscopic techniques and tested for biological activity. One major obstacle is compound yield. While sensitive techniques such as high-resolution mass spectroscopy can detect the presence of compounds that occur in very low yields, the isolation and structure elucidation of these compounds generally requires that they be obtained in milligram quantities. Given that laboratory cultures can produce compounds at the microgram per liter level, there remain significant challenges in the purification of sufficient quantities for identification. Although large-scale cultivation technologies can address this problem, these facilities are seldom available in academic settings. These issues contribute to the gap between the pathways observed in genome sequence data and the isolation and characterization of the associated compounds. The continued development of new isolation and spectroscopic methods that accommodate smaller sample sizes will surely facilitate the discovery of a greater percentage of these minor metabolites.
Summary
The increasing availability of DNA sequence data has brought natural products research into the genomic era. Genome sequences provide valuable blueprints that can speed the de-replication process and direct the selection of strains for detailed chemical and genetic studies. As the utility of genome sequence data becomes apparent, so do a number of challenges that need to be overcome for the potential of genomics-based natural product research to be fully realized. These challenges include the accurate assembly of highly repetitive sequence motifs, the isolation and structural characterization of compounds that are produced in low yields, and the creation of formal links between pathways and compounds, which requires tractable genetic approaches that are applicable to diverse organisms. While heterologous expression remains a particularly promising approach, considerable work remains before this technique will become broadly applicable to natural product discovery. Despite these challenges, genomics has taken center stage in the field of natural product discovery. The renewed interest in this field, coupled with increasingly cost effective genome sequencing, will undoubtedly continue to drive future discovery efforts and help realize the potential of microorganisms to yield new chemical scaffolds that can be explored for applications in medicine and biotechnology.
Acknowledgements
PJ and WF acknowledge financial support from the National Institutes of Health (NIH R37 CA 044848 and RO1-GM086261) and the Fogerty Center International Cooperative Biodiversity Groups program (grant U01-TW007401-01). PJ, WF, and BSM acknowledge support from the NIH (grant R01-GM085770).
References
- 1.Ahmed L, Jensen P, Freel K, Brown R, Jones A, Kim B-Y, Goodfellow M. Salinispora pacifica sp. nov., an actinomycete from marine sediments. Antonie Van Leeuwenhoek. 2013;103:1069–1078. doi: 10.1007/s10482-013-9886-4. [DOI] [PubMed] [Google Scholar]
- 2.Anand S, Prasad MVR, Yadav G, Kumar N, Shehara J, Ansari MZ, Mohanty D. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Research. 2010;38:487–496. doi: 10.1093/nar/gkq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann BO, Ravel J. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 4.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 5.Eustáquio AS, O'Hagan D, Moore BS. Engineering fluorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide. A Journal of Natural Products. 2010;73:378–382. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eustáquio AS, Pojer F, Noe JP, Moore BS. Discovery and characterization of a marine bacterial SAM-dependent chlorinase. Nat Chem Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eustáquio AS, Nam S-J, Penn K, Lechner A, Wilson MC, Fenical W, et al. The discovery of salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. ChemBioChem. 2011;12:61–64. doi: 10.1002/cbic.201000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Nat Acad Sci. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angewandte Chemie. 2003;115:369–371. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 10.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 11.Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorg Medicinal Chem. 2009;17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freel KC, Nam S-J, Fenical W, Jensen PR. Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol. 2011;77:7261–7270. doi: 10.1128/AEM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proceedings of the National Academy of Sciences. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. John Innes Foundation Norwich; 2000. [Google Scholar]
- 16.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 17.Lane AL, Nam S-J, Fukuda T, Yamanaka K, Kauffman CA, Jensen PR, et al. Structures and comparative characterization of biosynthetic gene clusters for cyanosporasides, enediyne-derived natural products from marine actinomycetes. Journal of the American Chemical Society. 2013;135:4171–4174. doi: 10.1021/ja311065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 19.Lechner A, Eustáquio A, Gulder TAM, Hafner M, Moore BS. Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem Biol. 2011;18:1527–1536. doi: 10.1016/j.chembiol.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, et al. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 21.Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Molecular Biology and Evolution. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Research. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Natural Product Reports. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross AC, Xu Y, Lu L, Kersten RD, Shao Z, Al-Suwailem AM, et al. Biosynthetic multitasking facilitates thalassospiramide structural diversity in marine bacteria. Journal of the American Chemical Society. 2012;135:1155–1162. doi: 10.1021/ja3119674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, et al. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Am Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 27.Siegl T, Tokovenko B, Myronovskyi M, Luzhetskyy A. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metabolic Engineering. 2013;19:98–106. doi: 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson MC, Gulder TAM, Mahmud T, Moore BS. Shared biosynthesis of the saliniketals and rifamycins in Salinispora arenicola is controlled by the sare1259-encoded cytochrome P450. Journal of the American Chemical Society. 2010;132:12757–12765. doi: 10.1021/ja105891a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamanaka K, Ryan KS, Gulder TAM, Hughes CC, Moore BS. Flavoenzyme-catalyzed atropo-selective N,C-bipyrrole homocoupling in marinopyrrole biosynthesis. Journal of the American Chemical Society. 2012;134:12434–12437. doi: 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziemert N, Jensen PR. Phylogenetic approaches to natural product structure prediction. Methods in Enzymology. 2012;517:161–182. doi: 10.1016/B978-0-12-404634-4.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7:e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]