Abstract
The scaffolding protein NEDD9 is an established pro-metastatic marker in several cancers. Nevertheless, the molecular mechanisms of NEDD9 driven metastasis in cancers remain ill defined. Here, using a comprehensive breast cancer (BCa) tissue microarray, it was show that increased levels of NEDD9 protein significantly correlated with the transition from carcinoma in situ to invasive carcinoma. Similarly, it was shown that NEDD9 overexpression is a hallmark of highly invasive BCa cells. Moreover, NEDD9 expression is crucial for the protease-dependent mesenchymal invasion of cancer cells at the primary site but not at the metastatic site. Depletion of NEDD9 is sufficient to suppress invasion of tumor cells in vitro and in vivo, leading to decreased circulating tumor cells (CTCs) and lung metastases in xenograft models. Mechanistically, NEDD9 localized to invasive pseudopods and was required for local matrix degradation. Depletion of NEDD9 impaired invasion of cancer cells through inactivation of membrane-bound matrix metalloproteinase MMP14 by excess TIMP2 on the cell surface. Inactivation of MMP14 is accompanied by reduced collagenolytic activity of soluble metalloproteinases MMP2 and MMP9. Re-expression of NEDD9 is sufficient to restore the activity of MMP14 and the invasive properties of BCa cells in vitro and in vivo. Collectively, these findings uncover critical steps in NEDD9-dependent invasion of BCa cells.
Implications
This study provides a mechanistic basis for potential therapeutic interventions to prevent metastasis.
Keywords: NEDD9; invasion, metastasis; breast cancer; MMP14
Introduction
The cytoplasmic docking protein, NEDD9 has emerged as a biomarker of melanoma, pancreatic, and lung cancer metastasis and is required for the migration of tumor cells (1–6). The overexpression of NEDD9 leads to mesenchymal protease-dependent migration (4, 7). NEDD9 promotes epithelial-mesenchymal transition (EMT) and invasion of BCa cells (8).
Extracellular matrix (ECM) proteolysis is dependent upon soluble and membrane bound matrix metalloproteinases (MMPs) (9). Membrane bound MMP14 is a key enzyme involved in ECM degradation and invasion of tumor cells. The activity of MMP14 is regulated by phosphorylation, proteolysis, and endocytosis (10–11). MMP14 is activated by furin and is inhibited by tissue inhibitor of MMP, TIMP2, which makes a stable complex with the activated MMP14 (12–13). The tertiary complex of MMP14, TIMP2, and proMMP2 is required to form on the cell surface to activate MMP2 by a neighboring MMP14 free of TIMP2 (14). The presence of excess TIMP2 results in inhibition of MMP14 activity and decreased processing of proMMP2 to active MMP2 (15). In invasive cancer cells, MMP14 is localized at the membrane of invadopodia, ventral actin-based membrane protrusions (16–17). Local ECM degradation is initiated upon the assembly of: F-actin, cortactin, N-WASP, and the Arp2/3 complex (17–18). Functional mature invadopodia form when F-actin and cortactin-rich puncta initiate the accumulation of MMP14, thus forming proteolytically active protrusions (16). Activity of MMP14 is required for invasion of 3D collagen matrices in tissue culture and in nude mice (19–20).
While NEDD9 deficient cells are characterized by reduced migration accompanied with reduced levels of Src and FAK kinase activity (21–22) the role of NEDD9 in the function of invadopodia in BCa has been unclear. Although NEDD9 and another Cas family member, p130Cas, were previously identified in invadopodia of cancer cells (23–25), the differences between NEDD9 and p130Cas function in invasion are currently unknown.
In transgenic oncogene driven mammary tumor models such as MMTV-PyVmT or MMTV-Her2/Erb2 genetic ablation of NEDD9 leads to delays in tumor initiation and growth but has a limited effect on metastasis (21–22). The tumors arising in MMTV-Her2/NEDD9−/− or PyVmT/NEDD9−/− mice underwent a dramatic mutational selection that overcame the initial requirement for NEDD9. In this context it is difficult to determine the requirement for NEDD9 in metastasis since the selection of alternative signaling pathways happened prior to tumor formation. Also, it is currently unknown what stages of metastasis are affected by NEDD9.
Overexpression of NEDD9 is often mentioned in diverse cancers (1–4). However, as of now, a comprehensive analysis of NEDD9 expression with respect to BCa progression has not been conducted; To address these questions, we analyzed NEDD9 protein expression in a tissue microarray (TMA) consisting of 200 progressive BCa patient biopsies and in a panel of diverse BCa cell lines. We found that NEDD9 is overexpressed in invasive breast ductal carcinomas and invasive cell lines. We discovered that NEDD9 is involved in regulation of MMP14 activity on the cell surface, but not expression. Depletion of NEDD9 leads to decrease in proteolytic activity of soluble MMPs and transmembrane MMP14, resulting in the ablation of cancer cell invasion in vitro and in vivo. The decrease in activity was due to the association of MMP14 with its inhibitor TIMP2 on cell surface. Depletion of TIMP2 or MMP14 expression or addition of excess of recombinant TIMP2 to control cells led to a similar phenotype, suggesting that excessive TIMP2 bound to the MMP14 potentially is the primary reason for decreased invasion in carcinoma cells upon depletion of NEDD9.
Using inducible shRNAs against NEDD9 in xenograft models, we dissected particular stages of metastasis dependent upon NEDD9 expression and thus define the sensitive stages of tumor progression where anti-NEDD9 therapy could be applied to prevent metastasis. We found that reduction of NEDD9 expression in established tumors leads to a drastic decrease in MMPs activity and number of circulating tumor cells resulting in a decrease in the overall number and size of pulmonary metastases. Collectively, our findings suggest a novel mechanism for NEDD9 in accelerating cell invasion through regulation of MMP14 by TIMP2 thus defining the new therapeutic approach for anti-metastatic strategies via manipulation of NEDD9 expression.
Materials and methods
Plasmids and cell culture
Cell lines MDA-MB-231, MDA-MB-453, ZR-75-1, BT-549, MCF10A, MCF7, AU-565, BT-20, were purchased from and authenticated by American Type Culture Collection (ATCC), MDA-231-LN (Caliper Life Sci.) and grown based on manufacturer’s recommendations. shRNA expressing constructs against NEDD9, control (sequences available upon request) and smart pool siRNAs against MMP14 and TIMP2 and siControl were purchased from ThermoFisher Scientific as ready to use siRNAs or in pGIPZ, or in doxycycline-inducible pTRIPZ vectors. Lentiviral particles were prepared as previously described (26). For rescue experiments wild type cDNA of mouse NEDD9 was subcloned into pLUTZ lentiviral vector under doxycycline-inducible promoter (27). Cell medium and supplements were purchased from ATCC (Sigma).
MMP Antibody Array
MMP Antibody Arrays were purchased from RayBiotech, Inc. and assays were carried out according to the manufacturer’s protocol using whole cell lysate (WCL) and conditioned for 24h serum free medium (SFM).
Fluorescent-gelatin degradation assay
Foci of degraded matrix were visible as dark areas that lack fluorescence in the FITC-gelatin matrix (Life Technologies). Cells with pseudopods were identified by the presence of at least one actin/cortactin aggregate within the cell. Degradation per cell area was analyzed using ImageJ (NIH) as described (16). At least 100 cells were counted per each experimental condition. Data were pooled from multiple independent experiments.
DQ Collagen Assay
Cells expressing doxycycline-inducible shRNAs against NEDD9 and red fluorescent protein (RFP) embedded in DQcollagen I/IV combined with matrigel (BD Biosciences) to allow for cell tracing and dose-dependent manipulation of NEDD9 depletion. shRNA expression was induced for 72h. DQ collagen I/IV/matrigel assays were carried out according to a previously published protocol (28). Detailed protocol for data acquisition and analysis outlined in supplementary material.
EnzChek Gellatinase/Collagenese assay
Assay was purchased from Life Technologies and performed accordingly to manufacturer’s recommendations using shNEDD9 and shCon conditioned medium (24h). Fluorescence was measured at 485/545mn.
Gelatin zymography
The gelatin zymography protocol was performed as previously described (16, 24). Briefly, 4ml of 24h conditioned media was collected from cells, concentrated using AmiconUltra-4 columns and 1/20 of original volume loaded on the Zymogram Gelatin Gels (Life Technologies) and developed according to the manufacturer’s recommendations.
MMP14-specific fluorogenic substrate degradation assay
2×104 cells were plated on 96w plate, for 16 hours, followed by 1 hour in L15 media (Life Technologies). MMP14-specific fluorogenic substrate (Millipore) was introduced in final concentration 10 µM. Fluorescence was measured at 328/400mn every 5 minutes for 12 hours, using Synergy H4 Plate Reader (BioTek). Averaged background signal was subtracted, and data plotted curves described by equation for one site binding kinetics and fitted to each data set with the R2 of 0.8 The curves were significantly different between control and NEDD9 deficient cells according to the Extra sum-of-squares F test, p<0.0001.
ECM coated Boyden chamber invasion assays
Invasion assays were performed using BD FluoroBlok™ insets coated with matrigel (BD Biosciences). Cells were added to the top chamber in serum free media, 10%FBS supplemented MEM was added to the bottom chamber, and incubated for 8h. Once cells migrated through the matrix, they were labeled with Calcein AM (Life Technologies) and detected by fluorescence plate reader (Genios) at 485/530nm excitation/emission.
Cell viability Trypan Blue exclusion assay
MDA-MB-231-shCon, -shNEDD9 cells were cultured in suspension using Ultra-Low attachment plates (Fisher Scientific) for 36 hours; 0.1 mL of a 0.4% solution of trypan blue in PBS was added to 1×106/mL of cells. The total number of cells and the number of blue staining cells were calculated by Countess automated cell counter (Life Technologies). Number of viable cells × 104 × 1.1 = cells/mL culture.
Fluorescence Activated Cell Sorting (FACS)
1×106 cells, with or without permeabilization by 0.5% Tween20 (Sigma), were stained for MMP14 (Novus International). Secondary fluorescence conjugated antibodies (BD Biosciences) were used at a concentration 0.5ug/1 ×106 cells. Cells were pre-blocked with 5mg/ml of Human IgG (Sigma). As a control, non-specific primary antibody, either Rabbit or mouse IgG were used (Sigma).
Western blotting (WB)
Western blotting was done using standard procedures (26, 30). Primary antibodies included mouse anti-NEDD9 mAb (2G9) (29), anti-phTyr, (Cell Signaling Technology Inc.), anti-p130Cas, -MMP2, -MMP9, -GAPDH, -RFP (ThermoFisher), anti-cortactin, -MMP14 (Novus), anti-β-actin, -α-tubulin, (Sigma) antibodies. Secondary HPR-conjugated antibodies and chemiluminescence-based detection and quantification methods were previously described (30).
Recombinant TIMP2 titration assay
Recombinant human TIMP2 was purchased from Life Technologies, diluted in cell culture medium and added to the cells in 0–100nM final concentration for 6–12h. After indicated times cells number of invaded cells was calculated as described in Boyden chamber invasion assay.
Immunofluorescence (IF)
IF was performed as previously described (30). Primary antibodies included: rabbit anti-phTyr, -Arp3, -N-WASP (Cell Signaling Technology), anti-cortactin, -MMP14 (Novus), anti-cortactin (4F11). The secondary antibodies included: Alexa Fluor 405, 488, 555 donkey anti-rabbit, and Alexa Fluor 647 goat anti-mouse and Rhodamine-540 or PacificBlue-405-conjugated phalloidin (Life Technologies). Images were captured by confocal microscope LSM 510 (Carl Zeiss). To analyze IF data, volumes of positively stained structures were evaluated per cell. Quantifications were performed on z stack projections, and collected using standard acquisition parameters with ImageJ (NIH) and LSM Image analysis software (Carl Zeiss).
Tissue Microarrays (TMA) and Immunohistochemistry (IHC)
The high density BCa tissue microarrays were purchased from US Biomax Inc., and content is outlined in Supplementary Table 1. All procedures were approved by the Institutional Biohazard Committee of West Virginia University. TMA sections were deparaffinized, endogenous peroxidase activity was quenched with 3% hydrogen peroxide, and slides were incubated with anti-NEDD9 antibodies (1:100 dilution) overnight at 4C; followed by staining with secondary peroxidase-labeled antibodies and developed with DAB (DAKO). Positive and negative controls were included with each run. Staining was done in duplicate. The IHC analysis and scoring protocol is available in Supplementary materials.
Lung metastasis analysis
Lungs were collected and fixed in 4%PFA for 24–48h embedded in paraffin by WVU Tissue Bank as previously described (30). Serial 6–10µm think sections were cut at each 40µm steps through the whole organ. Sections were subjected to hematoxylin and eosin (H&E) staining. Metastatic lesions were visually identified and images were acquired using Zeiss Axioplan microscope with ×4 and ×10 NA1.6 objective, followed by higher ×100 magnification evaluation of whole section. Multiple sections from each lung/mouse were analyzed in each cohort. Quantification of the metastases was done by WVU pathologist (Dr. Livengood) using serial sections of lungs and H&E staining. Results are plotted as mean number of metastases per mm2 +/− S.E.M, 3 independent experiments, n=6–10 per treatment group.
Animal Studies
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ/(NSG) immunodeficient mice were purchased from the Jackson Laboratory. Animals were housed in the WVU Animal Facility under pathogen-free conditions and approved by the Institutional Animal Care and Use Committee protocol. Primary tumors and lungs were collected, processed for histopathology, and analyzed by the WVU Tissue Bank Core Facility.
Animal Bioluminescence Imaging (BLI) mammary fat pad injections
For orthotopic injections luciferase-expressing MDA-MB-231LN and BT-549 cells expressing Control (Con) or NEDD9 (N1, N4) targeting shRNAs were grown, trypsinized, and resuspended in PBS (1×107 cells/ml), and 0.1 ml was injected into the 4th inguinal mammary gland of 6–8 weeks old NSG female mice. Tumor cells were injected in 6 mice and the experiments were repeated at least 3 times resulting in a total of 18 mice per cell line/per shRNA. Mice were imaged weekly for quantitative evaluation of tumor growth and dissemination for up to 6 weeks. For doxycycline-inducible shRNAs Dox diet (BioServ, S3888, 200mg/kg doxycycline) was introduced to start the shRNA expression when primary tumors in all experimental cohorts had reached similar size of 200mm3 (2–3 weeks post injection). shRNA expression was monitors by the increase in RFP fluorescence. 150 mg/kg D-Luciferin (Perkin Elmer) was injected into the peritoneum. Images were obtained using the IVIS® Lumina-II Imaging System and Living Image-4.0 software (Perkin Elmer).
Isolation and characterization of circulating tumor cells (CTC) from Peripheral Blood
Blood was collected from animals weekly via submandibular bleeding. About 200µl of blood was collected into centrifuge tubes containing anticoagulants (EDTA, heparin). Peripheral blood was depleted for erythrocytes using RBC Lysis buffer (eBiosciences) according to manufacturer’s guidelines. The presence of nucleated tumor cells was detected by co-staining with DAPI and quantified by direct fluorescence, of RFP-induced by Doxycycline only in tumor cells (red) was measured under fluorescence microscope.
Statistical Analysis
Statistical comparisons were made using two-tailed Student’s t test. When more than two groups were analyzed, one-way analysis of variance (ANOVA) was used. P≤0.05 was considered to be significant. Experimental values were reported as the means with +/− S.E.M. All calculations were made using the GraphPad InStat software. Random forests analysis provides an aggressive machine learning approach for classifying data into groups using ensembles of regression trees. Using this approach, we quantified the influence of NEDD9 IHC intensity staining in prediction of disease stage by the construction of the regression trees using a variable importance score averaged over all the trees.
Results
Increase in NEDD9 protein correlates with an invasive state in BCa
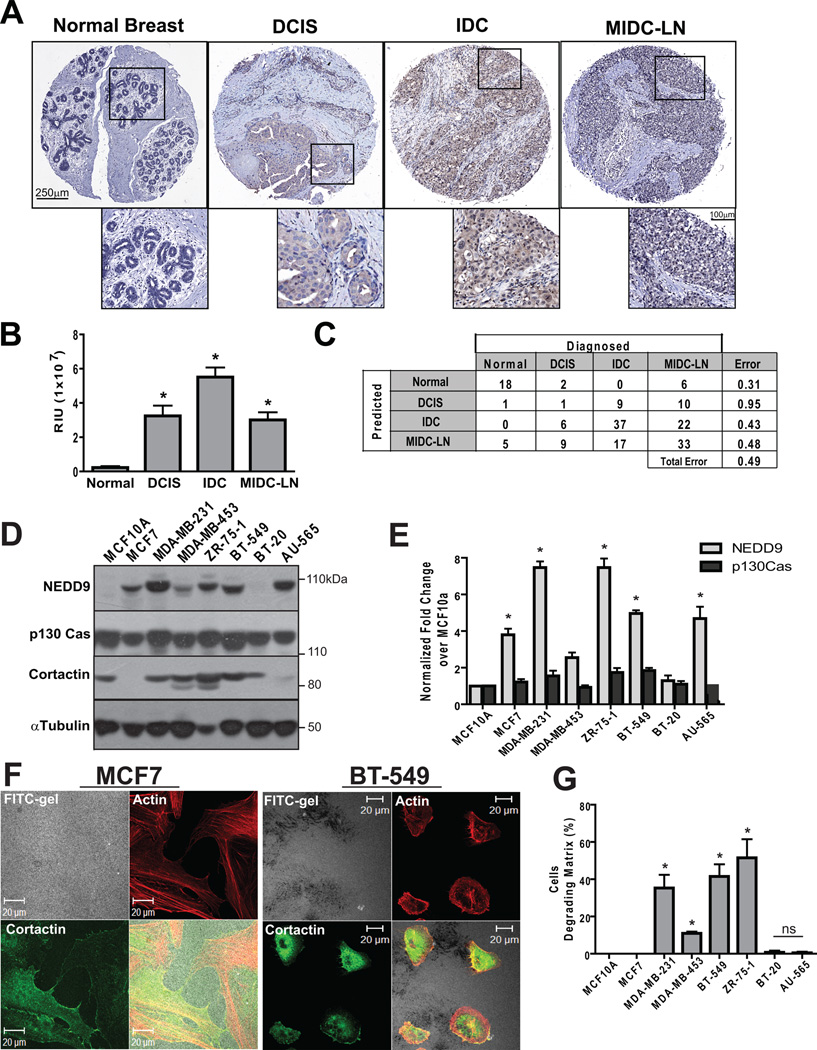
To assess the role of NEDD9 in cancer progression, we screened two independent TMAs using immunohistochemical staining (IHC) comprised of 200 BCa cases. Figure 1A shows representative images of NEDD9 staining in normal, DCIS (ductal carcinoma in situ), IDC (invasive ductal carcinoma), and MIDC-LN (metastatic invasive ductal carcinoma, lymph node). Statistical analysis of anti-NEDD9 staining intensity performed either by automated image capture system (Fig. 1B) or visual evaluation by pathologist (Suppl. Fig. 1A) suggests that NEDD9 levels positively correlate with disease progression. The lowest intensity was found in normal tissue, followed by a 10 fold increase in DCIS and a 30 fold increase in IDC, whereas only a 10 fold increase in expression was observed in MIDC-LN (Fig. 1B). Random forest statistical analysis of NEDD9 staining, which achieved an out-of-bag error rate of 0.49, indicates that by using NEDD9 protein levels, one can double the predictive power over chance of classifying tumor. The independent prognostic value of NEDD9 expression had 70% probability of identifying cases of IDC (Fig. 1C).
Figure 1. NEDD9 is overexpressed in invasive breast ductal carcinomas and BCa cell lines.
(A). Representative images of IHC staining with anti-NEDD9 antibodies in normal breast, DCIS, IDC MIDC-LN. (B). Statistical analysis of NEDD9 expression based on intensity of staining in (A), mean values +/− S.E.M, one-way ANOVA*, p<0.001 (Normal/IDC, Normal/MIDC-LN, Normal/DCIS), p<0.05 (DCIS/IDC, IDC/MIDC-LN), ns-non significant (DCIS/MIDC-LN) (C). The data (B) were analyzed by Random forest statistical software to assess the predictive value of NEDD9 expression. (D). Western Blot analysis (WB) of NEDD9, p130Cas, cortactin and tubulin in the indicated cell lines. (E). Quantification of NEDD9 and p130Cas as in (D), normalized to MCF10A, * p<0.001. (F). Immunofluorescent (IF) staining of MCF7 and BT-549 cells with Rhodamine-Phalloidin (red) and cortactin (green), FITC-gelatin (white). (G). Quantification of matrix degradation, 100 cells/cell line n=3, % of cells degrading matrix, +/− S.E.M, * p<0.0001 one-way ANOVA.
NEDD9 expression in BCa cells correlates with the capability to degrade ECM
We used a panel of characterized human BCa cell lines to determine if NEDD9 expression profiles identified in BCa patient samples correlate with invasion potential of cells,. We found that NEDD9 protein levels varied across the different cell lines in contrast to the p130Cas/BCAR1 protein (Fig. 1D–E). Several BCa cell lines including MDA-MB-231, ZR-75-1, and BT-549 have 5 to 8 fold more NEDD9 when compared to the non-transformed MCF10A line. The high NEDD9 expression levels correlate with the invasive capabilities of these cells (31–32). We confirmed these findings using FITC-gelatin degradation assays. Representative images and quantification of degradation by MCF7, BT-549 and ZR-75-1 shown in Figure 1F–G and Suppl. Fig. 1B. In contrast to the highly invasive MDA-MB-231 and BT-549, the AU-565 and MCF7 BCa cells were not proficient in gelatin degradation. We and others previously noticed that these cells expressed low levels of cortactin (Fig. 1D), MMP14, and MMP2 (33) which are required for invasion (16). This indicates that NEDD9 is required to support ECM degradation, although other components of the invasion machinery are also required for these processes.
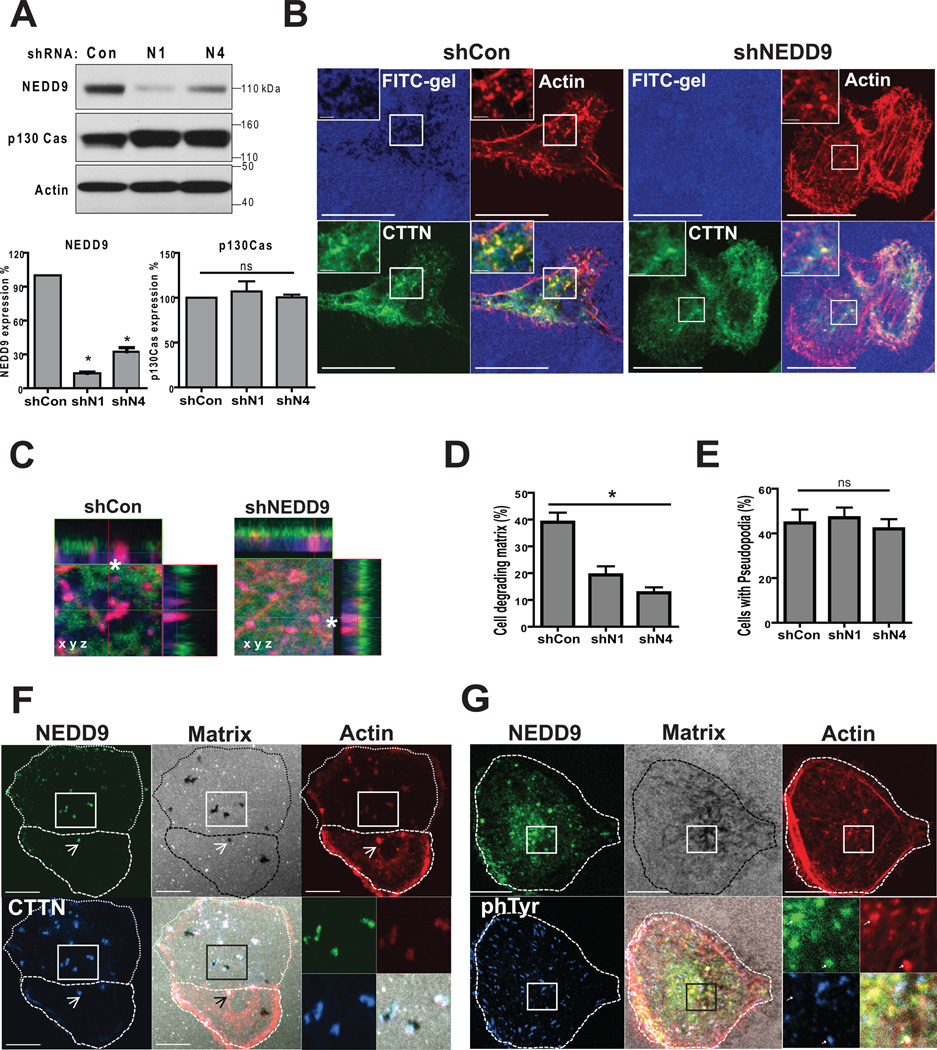
NEDD9 localizes to and promotes invadopodia maturation
To define the requirement of NEDD9 in formation and function of invadopodia, we infected MDA-MB-231, BT-549, and ZR-75-1 cells with two different shRNAs targeting NEDD9, which did not effect the expression of p130Cas (Fig. 2A). Cells were plated on FITC-gelatin coated coverslips and co-stained with invadopodia markers F-actin and cortactin to visualize the area of gelatin degradation and pseudopods (Fig. 2B–C). Depletion of NEDD9 led to a two- threefold reduction in the number of cells degrading matrix, while viability of cells was not affected (Fig. 2D). Interestingly, NEDD9 depleted cells had a similar number of F-actin/cortactin rich puncta (Fig. 2E) which are found during the initial stages of invadopodia formation (16) but show an absence of degradation beneath the puncta (Fig. 2C). p130Cas, which is highly expressed in BCa cells (Fig 2A), was not able to compensate for the decrease in NEDD9 under current experimental settings suggesting a critical role of NEDD9 in invadopodia based invasion in BCa. The localization of other major components of invadopodia, such as: Arp3 and N-WASP, were not affected by depletion of NEDD9 (Suppl. Fig. 1C).
Figure 2. NEDD9 localizes to and promotes invadopodia maturation.
(A). Top: WB of NEDD9, p130Cas, actin in MDA-MB-231-shNEDD9 or -shCon cells, Bottom: quantification of NEDD9 and p130Cas expression normalized to shCon; one-way ANOVA, p<0.001. (B). IF staining of MDA-MB-231-shCon and -shNEDD9 cells with anti-cortactin (green), -actin (red) antibodies and FITC-gelatin (blue), scale bar 10µm. Inserts are enlarged areas indicated in the main panel. (C). Enlarged XYZ projections of IF as in (B). (D). Quantification of number of cells degrading matrix and F-actin/cortactin positive foci (E) in MDA-MB-231-shCon, -shNEDD9 (N1, N4) cells. Student’s t-test for (D): *p<0.001(shCon/shN1, shCon/shN4), ns -shN1/N4; and ns (E) +/− S.E.M. (F). IF of MDA-MB-231 cells with cortactin (blue), actin (red) and NEDD9 (green), and FITC-gelatin (white). (G). IF staining with anti-phTyr (blue), actin (red) and NEDD9 (green) antibodies, and FITC-gelatin (white). Scale bar −10 µm. Inserts are enlarged areas indicated in the main panel.
To determine if NEDD9 localized to invadopodia in BCa cells, we co-stained cells with anti-NEDD9 antibodies (29) and invadopodia markers (16). We found that NEDD9 was consistently co-localized with cortactin, actin, and phTyrosine (Fig. 2F–G) in all cells with invadopodia.
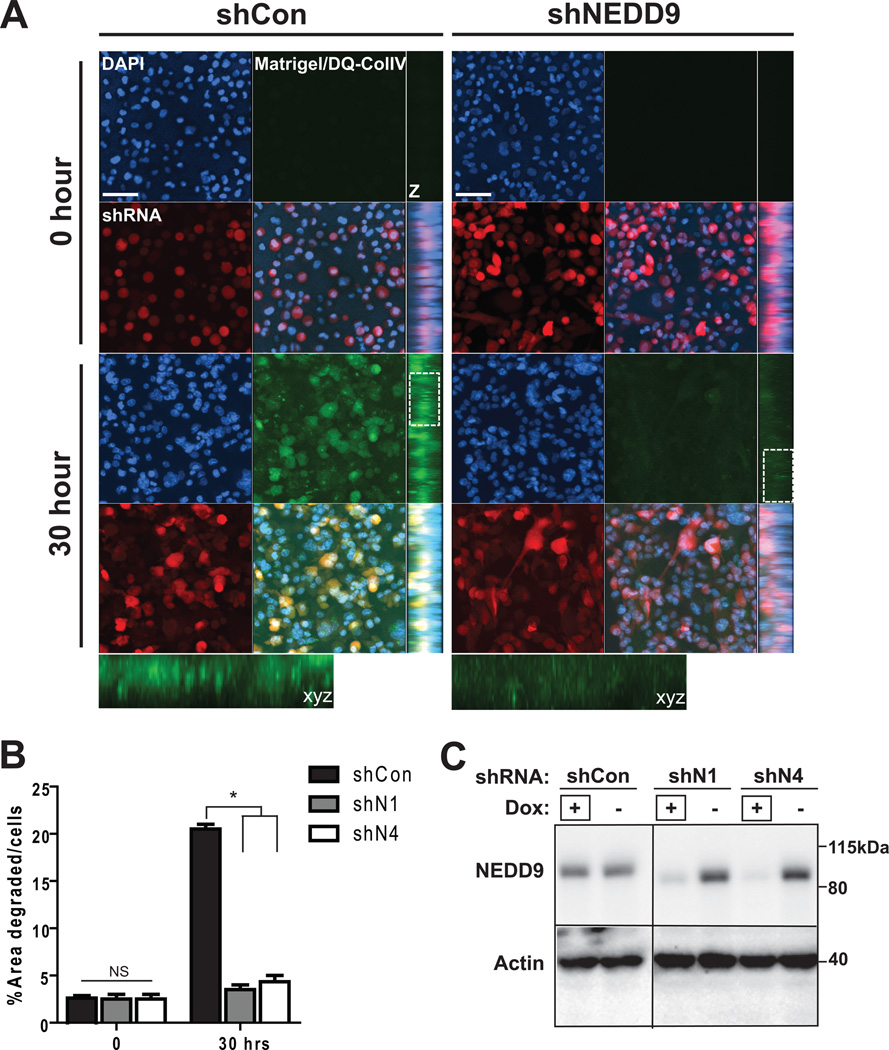
NEDD9 depletion in BCa cells leads to decrease in 3D invasion
To assess the impact of NEDD9 depletion on ECM degradation under more physiological conditions we used matrigel-DQcollagen I/IV invasion assay to trace invasion and pericellular ECM degradation in 3D matrix. BCa cells treated with shRNAs against NEDD9 were deficient in matrix degradation when embedded in 3D matrix (Fig. 3A–C). The levels of FITC-fluorescence as a result of proteolytical cleavage of DQcollagen I/IV were calculated (Fig. 3B) in shRNA expressing RFP-positive cells. The results were also evaluated in BT-549 and ZR-75-1 cell lines (Suppl. Fig. 2A–B). shNEDD9 cells demonstrated severe deficiency in collagen I/IV degradation indicated by a six fold decrease in fluorescence. We observed that the area of degradation in control cells had either a pericellular or dotted pattern (insert, Fig. 3A).
Figure 3. NEDD9 depletion ablates matrix degradation and invasion in 3D matrices.
(A). Representative images of 3D projections live cell imaging of MDA-MB-231-shCon, -shNEDD9 cells embedded in the DQ-CollagenI/IV/matrigel matrix, shRNA (red), DNA (blue) at indicated time points; xyz projections (z), scale bar −50µm. Insets are enlarged areas indicated in white rectangle. (B). Quantification of FITC-fluorescence intensity as in (A), % of area degraded per cell, +/− S.E.M, *p<0.0001 one-way ANOVA. (C). WB of cells in (A–B) with anti-NEDD9 and actin antibodies. +Dox indicates the lines used in A–B.
NEDD9 knockdown decreases MMPs activity, but does not affect MMPs expression or secretion
MMPs are the key enzymes responsible for matrix degradation. To determine the potential impact of NEDD9 on the function of MMPs, we analyzed protein levels of MMPs in shNEDD9 and control cells using a comprehensive MMP antibody array. Serum-free medium (SFM) conditioned by control and shNEDD9 cells was collected to measure the levels of secreted MMPs. The whole cell lysates (WCL) were used to define the total protein level of various expressed MMPs. No significant differences in the total amount of produced or secreted MMP and tissue specific inhibitor of MMP- TIMPs, were detected (Fig. 4A–B), except MMP13 which was slightly elevated in shNEDD9 cells. Nevertheless, analysis of SFM conditioned by shNEDD9, but not control, using EnzChek Gelatinase/Collagenase assay shows a severe deficiency in the activity of secreted collagenases (Fig. 4C). Next, we tested the activity of secreted MMPs by gel zymography. The activity of MMPs, particularly MMP2 and MMP9, was reduced by 60–80% upon NEDD9 depletion (Fig. 4D–E), indicated by the decrease in the amount of active 68kDa form of MMP2 (Fig. 4D, quantification of 68kDa form Fig. 4E) and 92-86kDa forms of MMP9 (Fig. 4E). We then tested the activity of MMP14 using a MMP14-specific fluorogenic substrate and show that NEDD9 depletion results in a significant decrease in MMP14 activity (Fig. 4F).
Figure 4. NEDD9 depletion decreases MMPs activity, but does not affect MMPs expression or secretion.
(A). WB of total MMPs expressed (WCL) and secreted (SFM) by indicated cells, 48h post shRNA expression. Schematic outline the MMP microarray. (B). Quantification of MMPs and TIMPs expression, n=3, one-way ANOVA for all pairs p is non-significant; p<0.05 for MMP13 (shCon/shN4). (C). Quantification of gellatinase/collagenese activity in SFM conditioned by cells using EnzChek Gellatinase/Collagenese assay. Clostridium Collagenase, 0.1U/ml, is positive control, p<0.05 (shCon/shN1 or /N4), linear regression analysis. (D). Representative image of -gel zymography, SFM conditioned by MDA-MB-231-shCon, -shNEDD9 cells, WB of MMP2 and MMP9. (E). Quantification MMP2 and MMP9 activity as in (D) normalized to total level of MMP2 and MMP9, +/− S.E.M; MMP9: p=0.007 (*) shCon/N1, or /N4 and ns - shN1/N4, one-way ANOVA. MMP2: p=0.00031, shCon/N1, /N4, ns -shN1/N4, one-way ANOVA. (F). MMP14-specific fluorogenic substrate assay, at 328/400mn. Data plotted as curves described by equation for one site binding kinetics, fitted to each data set with the R2 of 0.8, Extra sum-of-squares F test, p<0.0001.
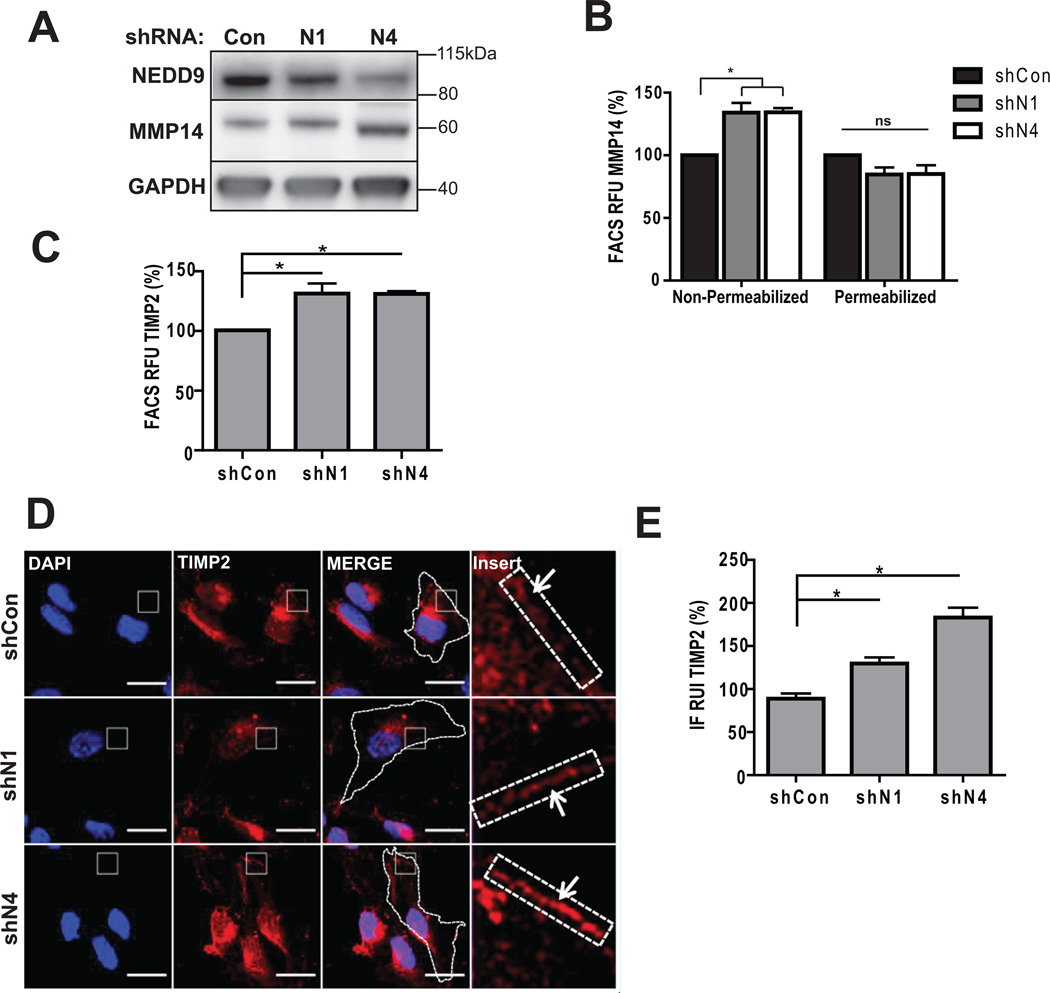
NEDD9 depletion leads to inactivation of MMP14 by TIMP2
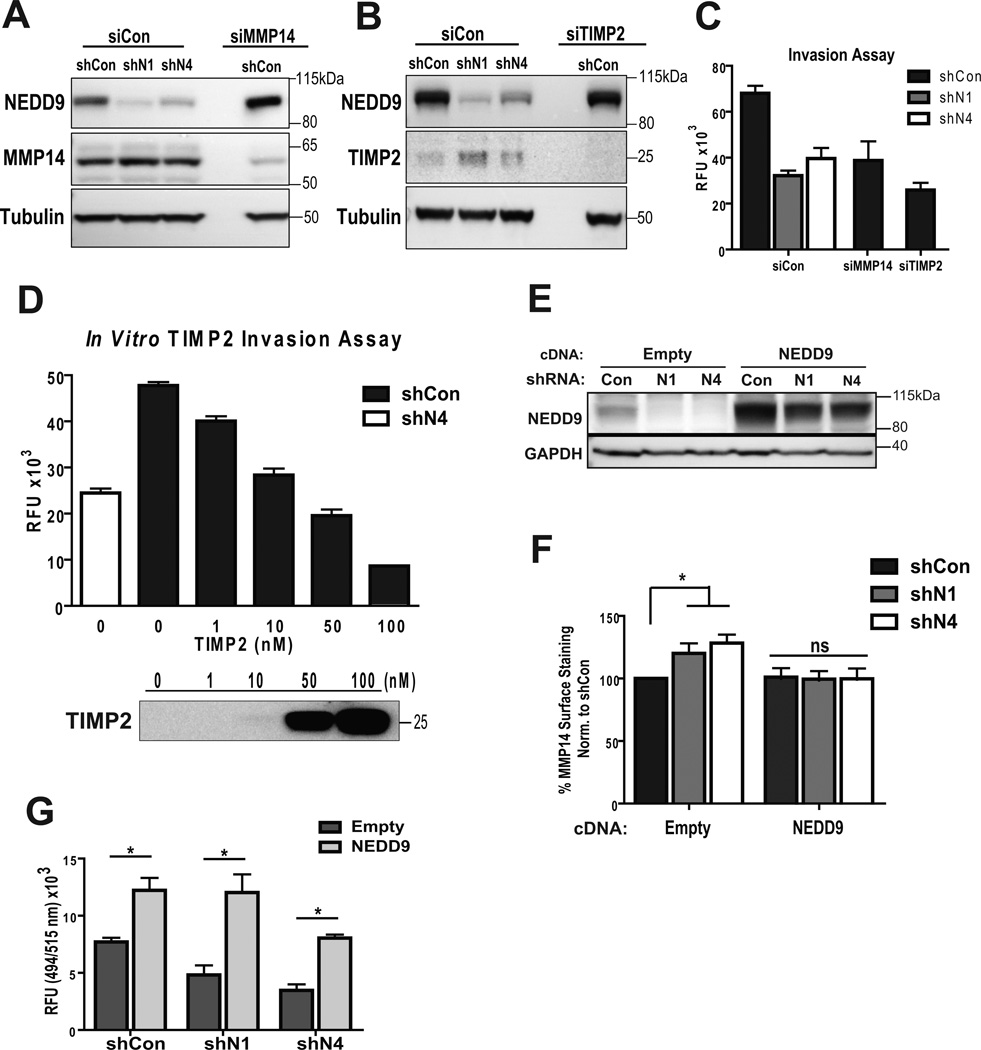
Membrane bound MMP14 is one of the key molecules involved in activation of soluble MMPs and pericellular degradation (34). Total MMP14 protein level (Fig. 5A) and localization patterns (Suppl. Fig. 2C) were not impacted by NEDD9 depletion. Interestingly, cell surface levels of MMP14 and TIMP2 proteins assessed by FACS (Fig. 5B–C) and IF (Fig. 5D–E) had increased up to 40% upon NEDD9 depletion. Note, that according to the MMP array analysis performed earlier depletion of NEDD9 did not affect total expression or secretion of TIMP2 (Fig. 4A–B).
Figure 5. NEDD9 depletion leads to increase in amount of surface-bound TIMP2 and MMP14.
(A). WB of WCL with anti-NEDD9, -MMP14, -GAPDH antibodies. (B). FACS of MMP14: surface (non-permeabilized) and total (permeabilized) normalized to shCon; one-way ANOVA *p<0.005 (shCon/N1 or /N4) -surface cells, ns, -total. (C). FACS of TIMP2-surface, normalized to shCon; t-test *p=0.039 (shCon/N1), *p=0.0029 (shCon/N4). (D). Representative images of IF, MDA-MB-231-shCon, -shNEDD9 cells with anti-TIMP2 antibody (red), DNA (blue). Scale bar, 20µm. Inserts are ×100 magnified areas identified on the image. (E). Quantification of TIMP2 fluorescence intensity at the membrane as in (D) normalized to shCon; t-test *p<0.001 (shCon/shN1 or /N4), ns - shN1/shN4.
TIMP2 is a well-known regulator of MMP14 and MMP2 activity. An increased amount of TIMP2-bound MMP14 is indicative of its decreased activity (35–36). Depletion of MMP14 (Fig. 6A) and TIMP2 in BCa cells (Fig. 6B), using specific siRNAs, resulted in a similar to shNEDD9 invasion deficiency (Fig. 6C) suggesting that MMP14 and TIMP2 are potentially the downstream targets affected by NEDD9. Moreover, addition of increasing amounts of recombinant TIMP2 to the medium of control cells leads to a similar inhibition of invasion at the higher concentrations of TIMP2 (Fig. 6D). Importantly, re-expression of NEDD9 (Fig. 6E) restored the amount of surface-bound MMP14 (Fig. 6F) and invasion proficiency of shNEDD9 cells (Fig. 6G).
Figure 6. Inhibition or depletion of MMP14 or TIMP2 recapitulates invasion deficiency of shNEDD9 cells.
(A–B). WB of WCL of MDA-MB-231-shCon, -shNEDD9 cells treated with siCon, siMMP14 or siTIMP2, using anti-NEDD9, -TIMP2 and -a-tubulin antibodies. (C). Boyden Chamber invasion assay, cells as in (A–B); plotted as relative fluorescence units (RFU), one-way ANOVA *p<0.0001 (shCon/siCon, shN1/siCon, or shN4/siCon), ns -shN1/N4-siCon and shCon/siMMP14 or shCon/siTIMP2; p<0.005 shCon/siCon, shCon/siMMP14 or shCon/siTIMP2; (D). Boyden Chamber invasion assay of MDA-MB-231-shCon cells with increased amount of TIMP2. MDA-MB-231-shN4 cells used as a reference control. Results plotted as RFU, n=3. One-way ANOVA *p<0.0001 shCon-0nM/shCon-10nM or more, ns: shN4/shCon-10nM. (E). WB of NEDD9 rescue in MDA-MB-231-shCon, -shNEDD9 cells with empty vector or cDNA-NEDD9 construct, using anti-NEDD9, -GAPDH antibodies. (F). FACS of MMP14 surface staining as in (E); one-way ANOVA *p<0.0001 empty vector-shCon/shN1 or /N4; ns cDNANEDD9-shCon/shN1 or /N4). (G). Boyden-Chamber assay, cells as in (E–F). Data plotted as RFU, one-way ANOVA *p<0.0001 (shCon-empty/shCon-NEDD9, shN1-empty/shN1-NEDD9, shN4-empty/shN4-NEDD9); p<0.005 (shCon-empty/shN1-empty or -shN4-empty); ns (shCon-NEDD9/shN1-NEDD9 or /shN4-NEDD9).
NEDD9 depletion inhibits invasion and intravasation of tumor cells from primary site and growth of metastasis in xenograft models
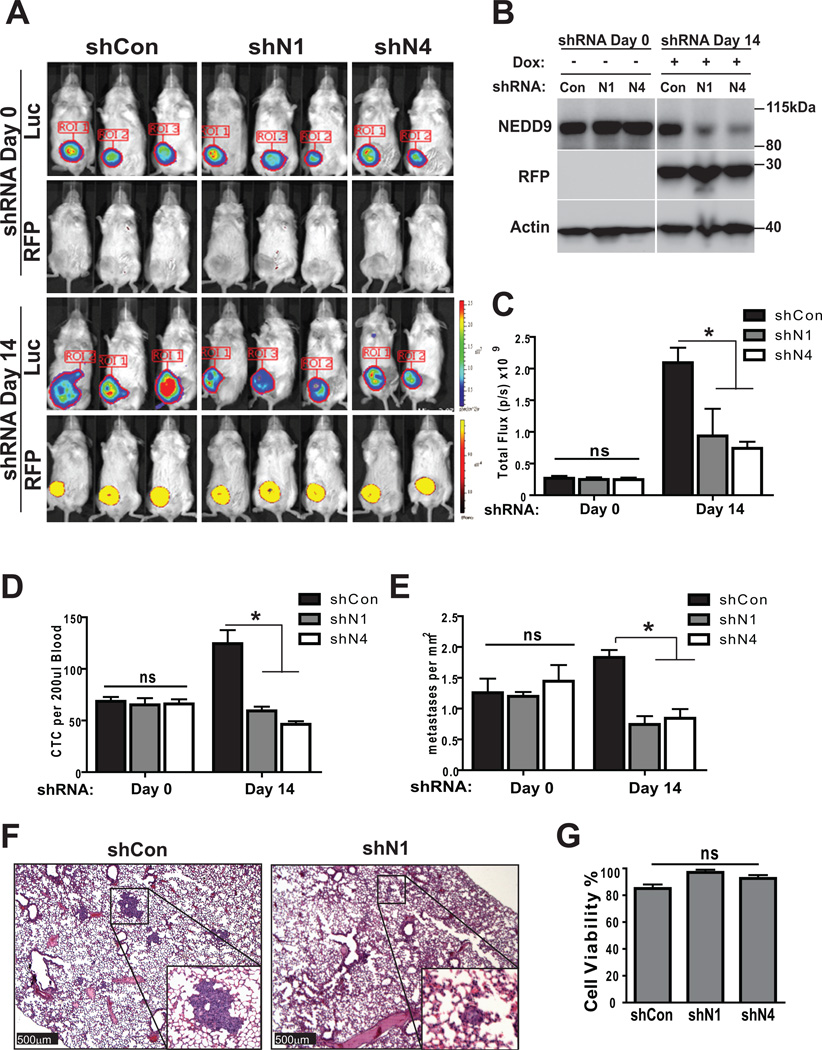
To determine the requirements of NEDD9 for invasion and metastasis in vivo, we utilized human tumor xenografts combined with bioluminescence imaging (BLI) (27, 30). MDA-MB-231-shCon and shNEDD9 (N1, N4) cells were injected into the mammary fat pad and allowed to develop primary tumors without induction of shRNA. When tumor volume reached 150–200 mm3 (Fig. 7A-shRNA-Day0), animals were placed on doxycycline-containing food. shRNA expression was monitored by RFP fluorescence, along with BLI (Fig. 7A-shRNA-Day14) and western blotting (Fig. 7B). Animals were euthanized at shRNA Day0 or Day14 which corresponded to 3, and 5 weeks post injection of tumor cells to assess for the number of CTCs in the peripheral blood, number and size of metastases in the lungs and gross pathology of primary tumor. At the shRNA Day0, all animals had similar primary tumor size and volume based on caliper measurements and BLI (Fig. 7C-shRNA-Day0). Analysis of peripheral blood samples for CTCs reveals similar number in all three cohorts of animals (shCon/shN1/shN4) (Fig. 7D-shRNA-Day0), as well as similar number of metastases seeded in the lungs (Fig. 7E-shRNA-Day0). NEDD9 depletion during the following 14 days in established primary tumors and metastases led to a drastic decrease in the number of CTCs (Fig. 7D-shRNA-Day14) and number of metastases in lungs (Fig. 7E–F-shRNA-Day14). The primary tumors had continued to grow during the shRNA induction based on BLI data (Fig. 7C-shRNA-Day0/Day14); thus, the decrease in the number of CTCs could be either due to a decrease in the ability of tumor cells to invade the surrounding matrix and intravasate in blood stream or due to decrease in overall survival. We have established that NEDD9 depletion did not affect the survival of MDA-MB-231 cells (Fig. 7G). Similar results were obtained with another metastatic BCa cell line BT-549 supporting the general phenomenon of NEDD9 function in invasion and metastasis (Suppl. Fig. 3A–D). Collectively, our findings suggest that the application of shRNAs against NEDD9 to already established tumors could drastically reduce the number of tumor cells released into circulation due to an invasion deficiency, thereby, eliminating further seeding and decrease the total number of metastasis.
Figure 7. NEDD9 depletion inhibits metastasis in xenograft models.
(A). Representative images of bioluminescence and RFP fluorescence of mice orthotopically injected with MDA-MB-231LN-shCon or –shNEDD9 cells. shRNA-Day0–3 weeks post injection, regular chow; shRNA-Day14–2 weeks doxycycline chow; 6 mice/shRNA/3 independent experiments. (B). WB of primary tumor tissue at (shRNA-Day0, -Day14) with anti-NEDD9, -RFP and -actin antibodies. (C). Quantification of tumor growth based on bioluminescence in 3 independent experiments, n=6/group, mean photon flux, +/− S.E.M, one-way ANOVA, p is ns for shRNA-Day0 (shCon/shN1 or /N4), shRNA-Day14 (shCon/shN1, shN1/shN4); *p=0.032 for shRNA-Day14-shCon/shN4. (D). Quantification of RFP/luc2/DNA positive tumor cells in peripheral blood of shCon and shNEDD9 mice at indicated times, n=6/group/3 independent experiments; plotted as mean number of cells per 200µl of blood, +/− S.E.M, two-way ANOVA, ns (shRNA-Day0: shCon/shN1 or /N4); p<0.001 (shRNA-Day14: shCon/shN1 or /N4). (E). Quantification of metastases in lungs of orthotopically injected animals at indicated times by pathologist using serial sections of lungs and H&E staining, plotted as mean number of metastases per mm2 +/− S.E.M, n=6/group/3 independent experiments; two-way ANOVA p -ns (shRNA-Day0: shCon/shN1 or /N4); p<0.001 (shRNA-Day14: shCon/shN1 or /N4). (F). Representative images of H&E staining as in (E). Scale bar-500µm, main panel, 50µm -insert. (G). Quantification of number of trypan blue negative vs. positive shCon or shNEDD9 cells, 72 in suspension. One-way ANOVA, p=0.0946.
Discussion
While recent studies have established NEDD9 as an essential factor for invasion (2–4) and metastasis (1, 37) the precise mechanism of how NEDD9 functions and its contribution to various stages of metastasis have been poorly defined. In our current study we show that NEDD9 is upregulated in breast tumors, and specifically increases in IDC, giving NEDD9 the potential to serve as a strong diagnostic marker for invasive tumors.
Interestingly, NEDD9 is decreased in metastatic samples suggesting that high level of NEDD9 is required during the invasion from the primary site, most probably to enable protease-dependent degradation of the basal membrane and EMT re-programming. While still increased compared to normal tissue, lesser amounts of NEDD9 are sufficient during growth at distant metastatic sites. Alternatively, upon arrival at distant sites, a decrease in NEDD9 expression might be beneficial in order to undergo mesenchymal to epithelial transition to establish the colony (8). This data is in accordance with the previously published studies showing that established metastasis in lungs had less NEDD9 when compared to primary tumor (6–7, 21–22). It is possible that decrease in NEDD9 might be beneficial for migration through filamentous collagen rich matrices; while in the basal membrane like matrix, the presence of NEDD9 was an absolute requirement for efficient invasion.
Decreases in NEDD9 expression, induced by constitutive or inducible shRNAs against NEDD9, result in a dramatic decrease in matrix degradation and invasion of BCa cells in 2D, 3D environments, and limits number of CTCs and metastases, thus establishing NEDD9 as an important factor for invasion and metastasis in BCa.
In a search for a molecular mechanisms underlying NEDD9 dependent invasion, we found that depletion of NEDD9 does not affect formation of nascent invadopodia, characterized by aggregation of F-actin, cortactin, and MMP14 (17), but prevents further maturation of invadopodia into proteolytically active structures (16). These findings are in agreement with previous report in SCC-9 cells where NEDD9 expression was required for matrix degradation, but disagree with regard of NEDD9 requirement for invadopodia formation (23). The differences might be associated with artificial exogenous overexpression of NEDD9 in SCC9 cells. The source of differences could also reside in tissue specific behavior of breast versus head and neck cells. It is also needs to be noted that another NEDD9 family member protein p130Cas was previously identified in invadopodia (24). In the cell lines used in our study depletion of NEDD9 did not affect the formation of invadopodia, indicating the different roles of these adaptor proteins in invasion.
The maturation of invadopodia in BCa cells and the capacity of tumor cells to invade surrounding tissue are directly linked to accumulation and activation of MMP14 and soluble MMPs (10, 34). We found that depletion of NEDD9 leads to a 60–80% decrease in activity of secreted MMP2 and MMP9 and pericellular proteolysis. Activity of MMP14 is tightly regulated by TIMP2 which plays dual roles as both inhibitor and activator of MMP14, depending upon concentration (35–36). While there is no difference in produced or secreted protein levels of MMPs and TIMPs in shNEDD9 and shCon cells, we observed that the concentration of cell surface bound TIMP2 and MMP14 is up to 50% higher in shNEDD9 cells, suggesting that the decrease in activity of MMP14 and other MMPs is potentially due to an increased amount of TIMP2 associated with MMP14 on the cell surface. Interestingly, depletion of MMP14 or TIMP2 results in a similar decrease in MMP2 activity and invasion deficiency recapitulating the shNEDD9 phenotype. TIMP2 is a potent inhibitor of most soluble MMPs (39), thus TIMP2 depletion could also relieve the inhibition of other soluble MMPs produced by cells. Nevertheless, our findings indicate that depletion of TIMP2 alone is not sufficient to restore the activity of soluble MMPs most likely due to the presence of other TIMPs such as TIMP1, 3 and 4 (39–40). Re-expression of NEDD9 restores the MMPs activity and invasion proficiency of shNEDD9 cells.
Binding of TIMP2 results in increased endocytosis of MMP14 (10, 38) followed by dissociation of MMP14/TIMP2 complexes within the endocytic vesicles and regenerate an active protease (34). Accumulation of MMP14/TIMP2 complexes on the cell surface is indicative of endocytotic deficiency in NEDD9 depleted cells. Interestingly, MMP14 can be phosphorylated by Src kinase and this phosphorylation is required for invasion and endocytosis of MMP14 (19), thus directly linking the NEDD9 dependent decrease in Src and FAK activity (4, 21) with inactivation of MMP14. Further work is required to uncover the intricate link between NEDD9 and MMP14 trafficking.
The decrease in number of metastases and CTCs upon NEDD9 depletion in cancer cells could advocate for development of shRNA based NEDD9-targeted therapy against BCa invasion. Importantly, even established, late stage, cancers with seeded metastases based on our in vivo inducible xenograft studies might still benefit from shNEDD9 treatment and prevent further spreading of disease. Collectively, our findings establish a critical role for NEDD9 as a regulator of invasion in BCa through modulation of MMP14 activity, thus delineating a new signaling pathway for targeting NEDD9 dependent invasion.
Supplementary Material
Acknowledgements
We thank Erica Peterson and Dr. Michael Schaller for critically reading the manuscript.
Financial support: This work was supported by NIH/NCI grant CA148671, Susan G. Komen for Cure Foundation KG100539 and in part by NIH/NCRR P20 RR016440-09 to ENP. WVU MIF and AIMF, supported by the MBRCC and NIH grants P20-RR016440, P30-RR032138/GM103488, P20-RR016477, and S10-RR026378. WVU Flow Cytometry Facility was supported by NIH grants P30GM103488, P30RR032138 and RCP1101809.
Footnotes
Author Contributions. S.L.M, R.I.J, S.A.W and E.N.P conceived the project and wrote the manuscript. TMA statistical analysis was performed by R.I.J and M.V.C. Animal pathology and tumor analysis was done by R.H.L. All remaining experiments and data analysis were performed by S.L.M, R.I.J, A.R, P.Y.K, Y.V.L, V.K.K, A.V.I and E.N.P.
Publisher's Disclaimer: Disclaimers:
This manuscript contains original work only and has not been published nor submitted elsewhere. All authors have directly participated in the planning, execution and analysis of this study and approved the submitted version of this manuscript.
Conflicts of interest: The authors declare that there are no conflicts of interest.
References
- 1.Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Kondo S, Iwata S, Yamada T, Inoue Y, Ichihara H, Kichikawa Y, et al. Impact of the integrin signaling adaptor protein NEDD9 on prognosis and metastatic behavior of human lung cancer. Clin Cancer Res. 2012;18:6326–6338. doi: 10.1158/1078-0432.CCR-11-2162. [DOI] [PubMed] [Google Scholar]
- 3.Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, Cox BD, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–1732. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- 4.Ahn J, Sanz-Moreno V, Marshall CJ. The metastasis gene NEDD9 product acts through integrin beta3 and Src to promote mesenchymal motility and inhibit amoeboid motility. J Cell Sci. 2012;125:1814–1826. doi: 10.1242/jcs.101444. [DOI] [PubMed] [Google Scholar]
- 5.Fashena SJ, Einarson MB, O'Neill GM, Patriotis C, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002;115:99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- 6.Guerrero MS, Parsons JT, Bouton AH. Cas and NEDD9 Contribute to Tumor Progression through Dynamic Regulation of the Cytoskeleton. Genes Cancer. 2012;3:371–381. doi: 10.1177/1947601912458585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 8.Kong C, Wang C, Wang L, Ma M, Niu C, Sun X, et al. NEDD9 is a positive regulator of epithelial-mesenchymal transition and promotes invasion in aggressive breast cancer. PLoS One. 2011;6(7):e22666. doi: 10.1371/journal.pone.0022666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 10.Poincloux R, Lizarraga F, Chavrier P. Matrix invasion by tumour cells: a focus on MMP14 trafficking to invadopodia. J Cell Sci. 2009;122:3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- 11.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MMP14) J Cell Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 12.Yana I and Weiss SJ. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol Biol Cell. 2000;11(7):2387–2401. doi: 10.1091/mbc.11.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato H, Kinoshita T, Takino T, Nakayama K, Seiki M. Activation of a recombinant membrane type 1-matrix metalloproteinase (MT1-MMP) by furin and its interaction with tissue inhibitor of metalloproteinases (TIMP)-2. FEBS Lett. 1996;393(1):101–104. doi: 10.1016/0014-5793(96)00861-7. [DOI] [PubMed] [Google Scholar]
- 14.Shofuda K, Moriyama K, Nishihashi A, Higashi S, Mizushima H, Yasumitsu H, et al. Role of tissue inhibitor of metalloproteinases-2 (TIMP-2) in regulation of pro-gelatinase A activation catalyzed by membrane-type matrix metalloproteinase-1 (MT1-MMP) in human cancer cells. J Biochem. 1998;124(2):462–470. doi: 10.1093/oxfordjournals.jbchem.a022136. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Barrantes S, Toth M, Bernardo MM, Yurkova M, Gervasi DC, Raz Y, et al. Binding of active (57 kDa) membrane type 1-matrix metalloproteinase (MMP14) to tissue inhibitor of metalloproteinase (TIMP)-2 regulates MMP14 processing and pro-MMP-2 activation. J Biol Chem. 2000;275:12080–12089. doi: 10.1074/jbc.275.16.12080. [DOI] [PubMed] [Google Scholar]
- 16.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- 17.Ayala I, Baldassarre M, Giacchetti G, Caldieri G, Tete S, Luini A, et al. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J Cell Sci. 2008;121:369–378. doi: 10.1242/jcs.008037. [DOI] [PubMed] [Google Scholar]
- 18.Bowden ET, Onikoyi E, Slack R, Myoui A, Yoneda T, Yamada KM, et al. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp Cell Res. 2006;312:1240–1253. doi: 10.1016/j.yexcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Nyalendo C, Beaulieu E, Sartelet H, Michaud M, Fontaine N, Gingras D, et al. Impaired tyrosine phosphorylation of membrane type 1-matrix metalloproteinase reduces tumor cell proliferation in three-dimensional matrices and abrogates tumor growth in mice. Carcinogenesis. 2008;29:1655–1664. doi: 10.1093/carcin/bgn159. [DOI] [PubMed] [Google Scholar]
- 20.Wu X, Gan B, Yoo Y, Guan JL. FAK-mediated src phosphorylation of endophilin A2 inhibits endocytosis of MMP14 and promotes ECM degradation. Dev Cell. 2005;9:185–196. doi: 10.1016/j.devcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69:7198–7206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little JL, Serzhanova V, Izumchenko E, Egleston BL, Parise E, Klein-Szanto AJ, et al. A requirement for Nedd9 in luminal progenitor cells prior to mammary tumorigenesis in MMTV-HER2/ErbB2 mice. Oncogene. 2013 doi: 10.1038/onc.2012.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas JT, Jr, Salimath BP, Slomiany MG, Rosenzweig SA. Regulation of invasive behavior by vascular endothelial growth factor is HEF1-dependent. Oncogene. 2010;29:449–4459. doi: 10.1038/onc.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brabek J, Constancio SS, Siesser PF, Shin NY, Pozzi A, Hanks SK. Crk-associated substrate tyrosine phosphorylation sites are critical for invasion and metastasis of SRC-transformed cells. Mol Cancer Res. 2005;3:307–315. doi: 10.1158/1541-7786.MCR-05-0015. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, McNiven MA. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J Cell Biol. 2012;196(3):375–385. doi: 10.1083/jcb.201105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinn KR, Chaudhuri TR, Szafran AA, O'Quinn D, Weaver C, Dugger K, et al. Noninvasive bioluminescence imaging in small animals. ILAR J. 2008;49:103–115. doi: 10.1093/ilar.49.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedeszko C, Sameni M, Olive MB, Moin K, Sloane BF. Visualizing protease activity in living cells: from two dimensions to four dimensions. Curr Protoc Cell Biol. 2008;Chapter 4(Unit 20) doi: 10.1002/0471143030.cb0420s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–946. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ice RJ, McLaughlin SM, Culp MV, Eddy ER, Livengood RH, Ivanov AV, et al. NEDD9 depletion destabilizes Aurora A kinase and heightens the efficacy of Aurora A inhibitors: implications for treatment of metastatic solid tumors. Cancer Res. 2013;73(10):3168–3180. doi: 10.1158/0008-5472.CAN-12-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madsen MW, Briand P. Relationship between tumorigenicity, in vitro invasiveness, and plasminogen activator production of human breast cell lines. Eur J Cancer. 1990;26:793–797. doi: 10.1016/0277-5379(90)90154-l. [DOI] [PubMed] [Google Scholar]
- 32.Ratnikov BI, Deryugina EI, Strongin AY. Gelatin zymography and substrate cleavage assays of matrix metalloproteinase-2 in breast carcinoma cells overexpressing membrane type-1 matrix metalloproteinase. Lab Invest. 2002;82:1583–1590. doi: 10.1097/01.lab.0000038555.67772.db. [DOI] [PubMed] [Google Scholar]
- 33.Sounni NE, Devy L, Hajitou A, Frankenne F, Munaut C, Gilles C, et al. MMP14 expression promotes tumor growth and angiogenesis through an up-regulation of vascular endothelial growth factor expression. FASEB J. 2002;16:555–564. doi: 10.1096/fj.01-0790com. [DOI] [PubMed] [Google Scholar]
- 34.Li XY, Ota I, Yana I, Sabeh F, Weiss SJ. Molecular dissection of the structuralmachinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol Biol Cell. 2008;19:3221–3233. doi: 10.1091/mbc.E08-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Catalan C, Bode W, Huber R, Turk D, Calvete JJ, Lichte A, et al. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 1998;17:5238–5248. doi: 10.1093/emboj/17.17.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67:8975–8979. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remacle A, Murphy G, Roghi C. Membrane type I-matrix metalloproteinase (MMP14) is internalised by two different pathways and is recycled to the cell surface. J Cell Sci. 2003;116:3905–3916. doi: 10.1242/jcs.00710. [DOI] [PubMed] [Google Scholar]
- 39.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochimica Biophysica Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Liu Y, Feng H, Bian X, Zhao W, Yang Z, et al. CD133 affects the invasive ability of HCT116 cells by regulating TIMP-2. Am. J. Pathol. 2013;182(2):565–576. doi: 10.1016/j.ajpath.2012.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.