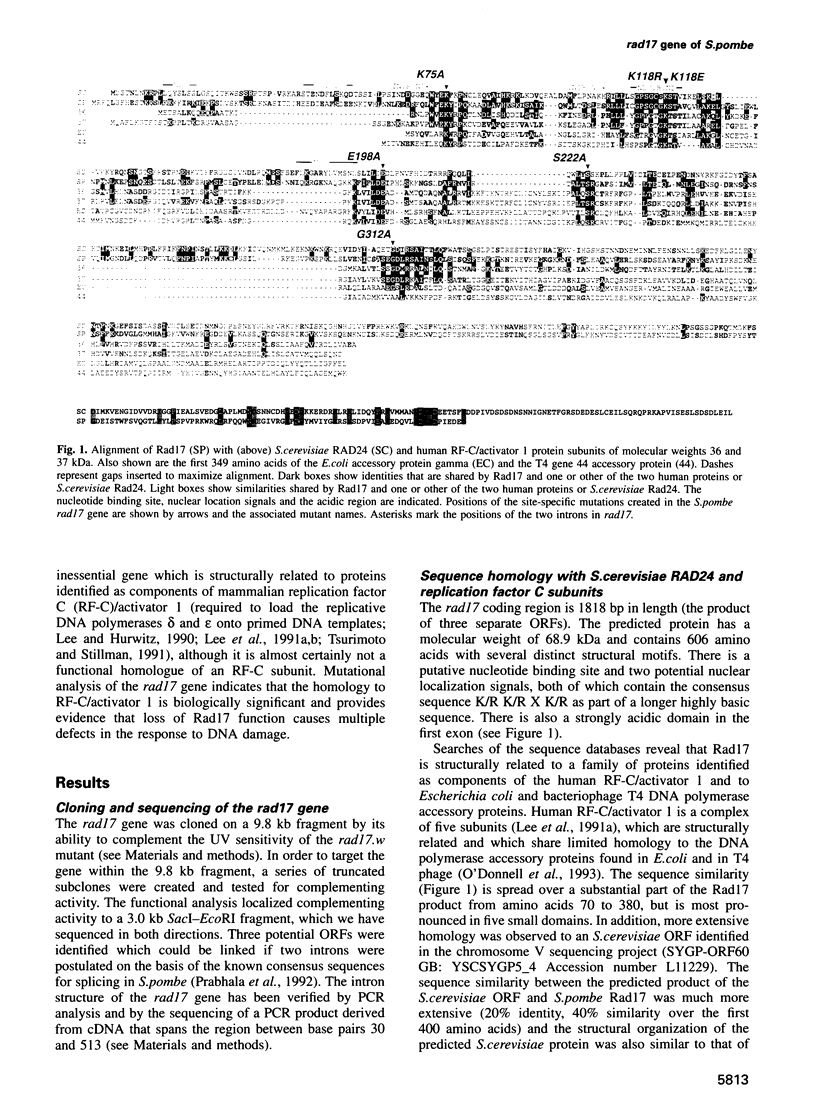
Abstract
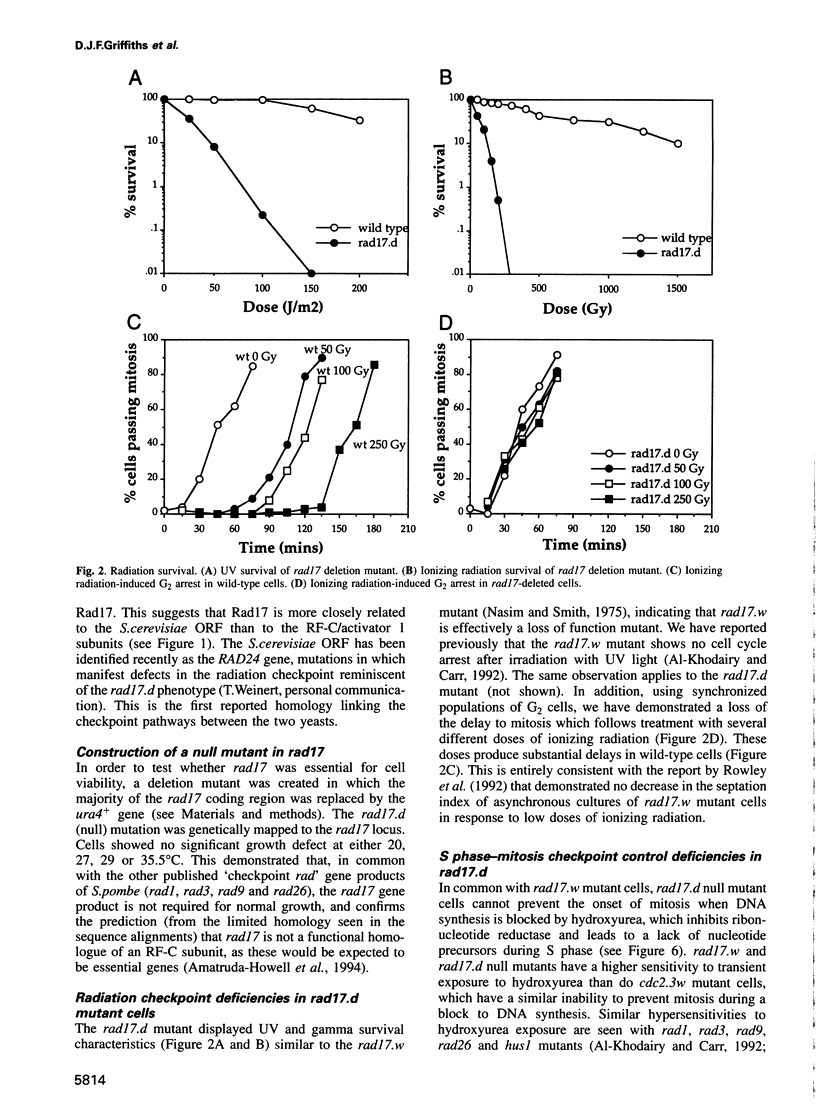
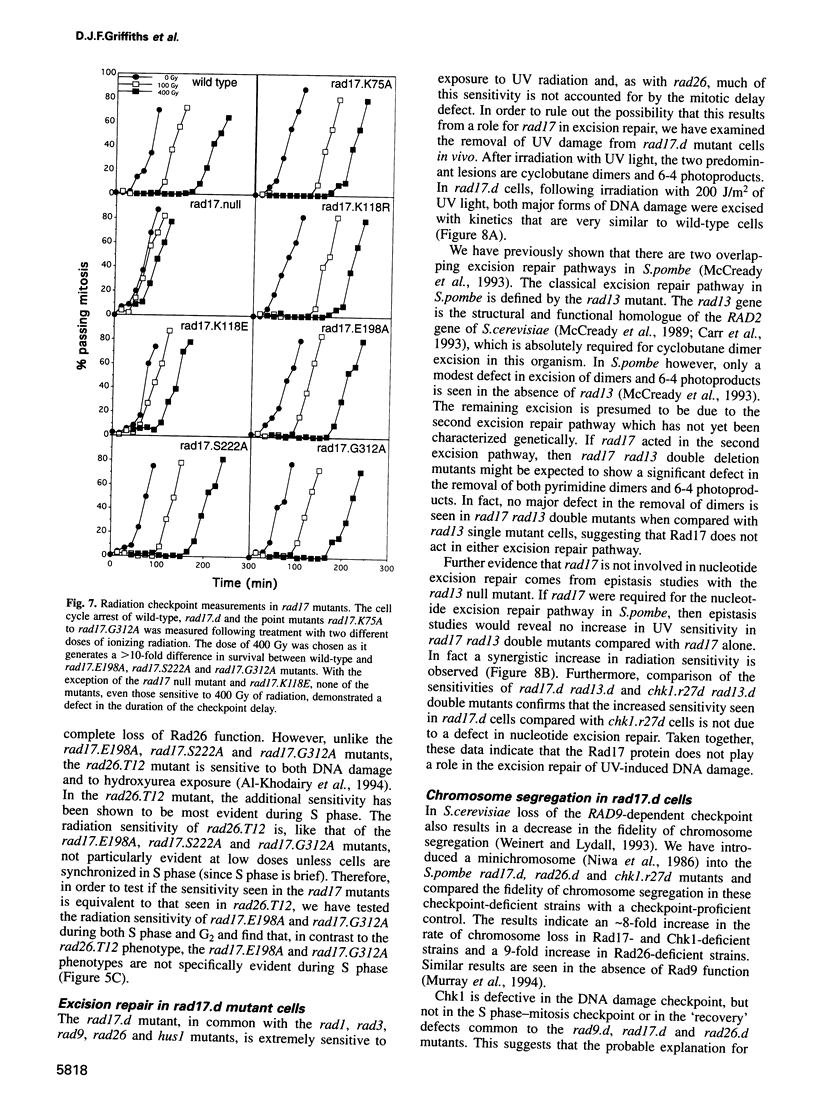
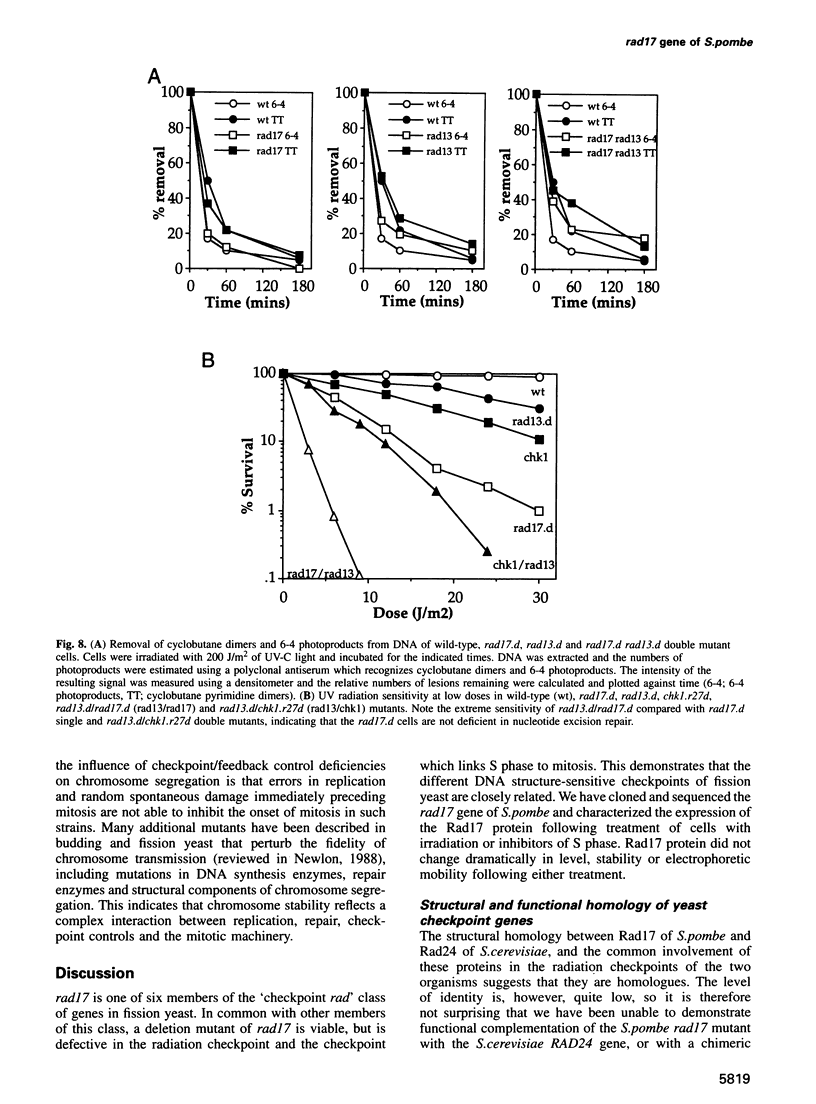
Following DNA damage or a block to DNA synthesis, checkpoint pathways act to arrest mitosis and prevent the attempted segregation of damaged or unreplicated DNA. The rad17 locus of Schizosaccharomyces pombe is one of seven known radiation-sensitive (rad) loci which are absolutely required to prevent mitosis following DNA damage in fission yeast. Six of these (rad1, rad3, rad9, rad17, rad26 and hus1) are also required for the checkpoint which prevents mitosis from occurring before DNA replication is complete. We report here that the predicted rad17 gene product is a basic hydrophilic protein of 606 amino acids which contains five domains with sequence homology to replication factor C (RF-C)/activator 1 subunits. Western analysis and fusion with Green Fluorescent Protein indicate that the abundance and electrophoretic mobility of Rad17 is not significantly modified following a block to DNA synthesis or following DNA damage, and that Rad17 is localized in the nucleus. Rad17 function is not essential for growth, but is required for the function of the DNA structure-dependent checkpoints. Site-directed mutagenesis has been used to demonstrate the biological significance of the RF-C/activator 1-related domains. These studies have also defined an element of the radiation sensitivity caused by loss of Rad17 function which is not associated with the radiation-induced G2 arrest defect seen in the rad17.d null mutant cells.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki H., Ropp P. A., Johnson A. L., Johnston L. H., Morrison A., Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992 Feb;11(2):733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Kremenstova E., Luo L. Complete transposition requires four active monomers in the mu transposase tetramer. Genes Dev. 1994 Oct 15;8(20):2416–2428. doi: 10.1101/gad.8.20.2416. [DOI] [PubMed] [Google Scholar]
- Barbet N. C., Carr A. M. Fission yeast wee1 protein kinase is not required for DNA damage-dependent mitotic arrest. Nature. 1993 Aug 26;364(6440):824–827. doi: 10.1038/364824a0. [DOI] [PubMed] [Google Scholar]
- Barbet N., Muriel W. J., Carr A. M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992 May 1;114(1):59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Carr A. M. Radiation checkpoints in model systems. Int J Radiat Biol. 1994 Dec;66(6 Suppl):S133–S139. [PubMed] [Google Scholar]
- Carr A. M., Sheldrick K. S., Murray J. M., al-Harithy R., Watts F. Z., Lehmann A. R. Evolutionary conservation of excision repair in Schizosaccharomyces pombe: evidence for a family of sequences related to the Saccharomyces cerevisiae RAD2 gene. Nucleic Acids Res. 1993 Mar 25;21(6):1345–1349. doi: 10.1093/nar/21.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T., Carr A. M., Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992 Nov;6(11):2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Enoch T., Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990 Feb 23;60(4):665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- Fikes J. D., Becker D. M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990 Jul 19;346(6281):291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- Ford J. C., al-Khodairy F., Fotou E., Sheldrick K. S., Griffiths D. J., Carr A. M. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science. 1994 Jul 22;265(5171):533–535. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- Grimm C., Kohli J., Murray J., Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988 Dec;215(1):81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Kastan M. B. Cell cycle control and cancer. Science. 1994 Dec 16;266(5192):1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Howell E. A., McAlear M. A., Rose D., Holm C. CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol Cell Biol. 1994 Jan;14(1):255–267. doi: 10.1128/mcb.14.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato R., Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994 Aug 11;22(15):3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993 Jul 30;74(2):371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Kwong A. D., Pan Z. Q., Hurwitz J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase delta. J Biol Chem. 1991 Jan 5;266(1):594–602. [PubMed] [Google Scholar]
- Lee S. H., Pan Z. Q., Kwong A. D., Burgers P. M., Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J Biol Chem. 1991 Nov 25;266(33):22707–22717. [PubMed] [Google Scholar]
- Lee S., Elenbaas B., Levine A., Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell. 1995 Jun 30;81(7):1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- McCready S. J., Burkill H., Evans S., Cox B. S. The Saccharomyces cerevisiae RAD2 gene complements a Schizosaccharomyces pombe repair mutation. Curr Genet. 1989 Jan;15(1):27–30. doi: 10.1007/BF00445748. [DOI] [PubMed] [Google Scholar]
- McCready S., Carr A. M., Lehmann A. R. Repair of cyclobutane pyrimidine dimers and 6-4 photoproducts in the fission yeast Schizosaccharomyces pombe. Mol Microbiol. 1993 Nov;10(4):885–890. doi: 10.1111/j.1365-2958.1993.tb00959.x. [DOI] [PubMed] [Google Scholar]
- McCready S., Cox B. Repair of 6-4 photoproducts in Saccharomyces cerevisiae. Mutat Res. 1993 Mar;293(3):233–240. doi: 10.1016/0921-8777(93)90074-q. [DOI] [PubMed] [Google Scholar]
- Mitchison J. M., Carter B. L. Cell cycle analysis. Methods Cell Biol. 1975;11:201–219. [PubMed] [Google Scholar]
- Murray J. M., Carr A. M., Lehmann A. R., Watts F. Z. Cloning and characterisation of the rad9 DNA repair gene from Schizosaccharomyces pombe. Nucleic Acids Res. 1991 Jul 11;19(13):3525–3531. doi: 10.1093/nar/19.13.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Doe C. L., Schenk P., Carr A. M., Lehmann A. R., Watts F. Z. Cloning and characterisation of the S. pombe rad15 gene, a homologue to the S. cerevisiae RAD3 and human ERCC2 genes. Nucleic Acids Res. 1992 Jun 11;20(11):2673–2678. doi: 10.1093/nar/20.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Tavassoli M., al-Harithy R., Sheldrick K. S., Lehmann A. R., Carr A. M., Watts F. Z. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol Cell Biol. 1994 Jul;14(7):4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim A., Smith B. P. Genetic control of radiation sensitivity in Schizosaccharomyces pombe. Genetics. 1975 Apr;79(4):573–582. doi: 10.1093/genetics/79.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas T. A., Zhou Z., Elledge S. J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995 Jan 13;80(1):29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Newlon C. S. Yeast chromosome replication and segregation. Microbiol Rev. 1988 Dec;52(4):568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M., Onrust R., Dean F. B., Chen M., Hurwitz J. Homology in accessory proteins of replicative polymerases--E. coli to humans. Nucleic Acids Res. 1993 Jan 11;21(1):1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhala G., Rosenberg G. H., Käufer N. F. Architectural features of pre-mRNA introns in the fission yeast Schizosaccharomyces pombe. Yeast. 1992 Mar;8(3):171–182. doi: 10.1002/yea.320080303. [DOI] [PubMed] [Google Scholar]
- Rowley R., Subramani S., Young P. G. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992 Apr;11(4):1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D. A., Smith S., Uziel T., Sfez S. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995 Jun 23;268(5218):1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Seaton B. L., Yucel J., Sunnerhagen P., Subramani S. Isolation and characterization of the Schizosaccharomyces pombe rad3 gene, involved in the DNA damage and DNA synthesis checkpoints. Gene. 1992 Sep 21;119(1):83–89. doi: 10.1016/0378-1119(92)90069-2. [DOI] [PubMed] [Google Scholar]
- Sheldrick K. S., Carr A. M. Feedback controls and G2 checkpoints: fission yeast as a model system. Bioessays. 1993 Dec;15(12):775–782. doi: 10.1002/bies.950151202. [DOI] [PubMed] [Google Scholar]
- Sunnerhagen P., Seaton B. L., Nasim A., Subramani S. Cloning and analysis of a gene involved in DNA repair and recombination, the rad1 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1990 Jul;10(7):3750–3760. doi: 10.1128/mcb.10.7.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen M. P., Onel K., Holloman W. K. The REC1 gene of Ustilago maydis involved in the cellular response to DNA damage encodes an exonuclease. J Biol Chem. 1994 Jan 7;269(1):747–754. [PubMed] [Google Scholar]
- Traut T. W. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur J Biochem. 1994 May 15;222(1):9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N., Davey S., Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993 May 27;363(6427):368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993 May;134(1):63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994 Mar 15;8(6):652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Weinert T., Lydall D. Cell cycle checkpoints, genetic instability and cancer. Semin Cancer Biol. 1993 Apr;4(2):129–140. [PubMed] [Google Scholar]
- al-Khodairy F., Carr A. M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992 Apr;11(4):1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Khodairy F., Fotou E., Sheldrick K. S., Griffiths D. J., Lehmann A. R., Carr A. M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994 Feb;5(2):147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]






