Abstract
The aim of this study was to develop a quantitative 16S rRNA assay for determination of bacterial nucleic acid load in cerebrospinal fluid (CSF) shunt infection and to compare quantitative 16S rRNA polymerase chain reaction (PCR) findings to those of conventional bacterial culture in patients treated for CSF shunt infection. We developed a quantitative 16S rRNA PCR assay that detected bacterial load across a range of 2.5 × 109 down to 2.5 × 104 16S copies/mL CSF under experimental conditions for numerous Gram-positive and Gram-negative organisms. However, when applied to archived CSF samples from 25 shunt infection episodes, correlations between positive bacterial culture and 16S rRNA levels were seen in only half of infections, and 16S rRNA levels dropped precipitously after an initial peak on the first day of sample collection. Bacterial load measured using 16S rRNA PCR does not provide sufficient information beyond bacterial culture to inform CSF shunt infection treatment.
Keywords: Cerebrospinal, Shunt, Infection, Children, Bacterial load
1. Introduction
Cerebrospinal fluid (CSF) shunt placement has been the mainstay of treatment for hydrocephalus for over 50 years (Kestle, 2003). While CSF shunts allow patients to survive and avoid further brain injury, they can cause new and often chronic surgical and medical problems. CSF shunt infections are one such complication, causing 2200–2400 pediatric hospital admissions each year in the U.S. (Simon et al., 2008).
CSF shunt infections are usually caused by bacterial pathogens (Fan-Havard and Nahata, 1987; Nelson, 1984; Odio et al., 1984; Sells et al., 1977) and are notoriously difficult to treat. Because the bacteria adhere to the shunt itself, treatment requires both surgical management to remove and replace the shunt and medical therapy to kill the bacteria. Medical treatment generally involves prolonged intravenous (IV) antibiotic administration and occasionally intrathecal antibiotics (Fan-Havard and Nahata, 1987; Kestle et al., 2006). While IV antibiotics are a mainstay of clinical practice, duration of IV antibiotic use varies widely—from 4 to 47 days in 1 study (Kestle et al., 2006)—and may depend on both the surgical approach used (Nelson, 1984; Vinchon and Dhellemmes, 2006) and the pathogen involved (Fan-Havard and Nahata, 1987; Younger et al., 1987). Available data on optimal length of therapy are sparse. For children with first CSF shunt infection, the prognosis is poor with reinfection rates ranging from 12 to 26% (Kestle et al., 2006; Kulkarni et al., 2001; Vinchon and Dhellemmes, 2006). Rates of reinfection are higher still for children with second infections (Tuan et al., 2011).
Risk factors for CSF shunt reinfection are not well-understood and may be related to the management of the prior infection. Diagnosis and management of CSF shunt infection currently rely on conventional microbiologic cultures. The initial diagnosis is based on the recovery of a pathogen from culture, and length of therapy is often based on the number of days that cultures are positive. Conventional cultures can require 24–48 hours for organism identification, and the chances of recovering a bacterium are significantly reduced if the patient has received prior antimicrobial therapy. CSF indices (e.g., glucose, protein, cell count and differential, and Gram stain) are not always reliable in identifying infection or tracking its progression (Lan et al., 2003); however, these parameters are often followed by clinicians to determine duration of antimicrobial treatment and timing of shunt replacement. CSF shunt replacement generally does not occur until negative CSF cultures are obtained and the treatment course is completed.
Clinicians, patients, and their families clearly need better evidence to guide the treatment of CSF shunt infection in order to avoid reinfection. We recognized the increasing use of real-time polymerase chain reaction (RT-PCR) technology targeting conserved regions of the 16S rRNA gene was capable of detecting thousands of species of bacteria, including the most frequent CSF shunt infection pathogens (Banks et al., 2005; Blaschke and Voelkerding, 2006; Deutch et al., 2007). The potential utility of 16S rRNA PCR for the detection of bacterial DNA has been shown in otherwise sterile body fluids including amniotic fluid, neonates with bacteremia, and neutropenic cancer patients with fever as well as CSF (Banks et al., 2005; Won et al., 2012), but its utility in the detection of infection has been limited. We wondered if 16S rRNA PCR might be more beneficial for guiding therapy. We developed a quantitative 16S rRNA assay to quantify bacterial load and evaluate changes in bacterial load in response to antibiotic therapy and surgical management. The objectives of this study were to 1) develop a quantitative 16S rRNA assay for the determination of bacterial nucleic acid load in CSF shunt infection and 2) compare the findings of quantitative 16S rRNA PCR to those of conventional bacterial culture in patients treated for CSF shunt infection.
2. Materials and methods
2.1. Primer selection
A panel of target organisms for assay development was generated from those observed in CSF shunt infections at Primary Children’s Medical Center (PCMC) during a retrospective observational cohort study conducted from 1997 to 2006 (Simon et al., 2012) (Table 1). Sequences were aligned using Vector NTI Align × (Life Technologies, Grand Island, NY, USA), and primers were designed to target conserved sequences between organisms and to optimize primer characteristics. We selected a 17 base pair (bp) forward primer and a 21 bp reverse primer to amplify a 107 bp amplicon spanning bases 1149–1256 of the Escherichia coli 16S rRNA gene. The numbers of copies of 16S rRNA for each bacterial species were determined from the literature (Lee and Bussema, 2009). After we detected only low amounts of 16S rRNA using this primer set, we tested samples using a second set of previously published primers targeting the V6 region of the 16S rRNA gene (Chakravorty et al., 2007) that also targets all organisms shown in Table 1.
Table 1.
CSF shunt infection organisms targeted during 16S rRNA PCR primer design.
| Gram positive | Gram negative |
|---|---|
| Enterococcus spp. | E. cloacae |
| Propionibacterium acnes | E. coli |
| S. aureus | Haemophilus spp. |
| Staphylococcus spp. (coagulase-negative) | K. oxytoca |
| Streptococcus pneumoniae | P. aeruginosa |
| Streptococcus spp. | Serratia marcescens |
| Streptococcus viridans | K. pneumoniae a |
| Corynebacterium spp.a | |
| Micrococcus spp.a |
Organism was not an original target during primer design, but primer is able to detect its 16S rRNA sequence based on alignment.
2.2. DNA extraction
We tested several conditions to extract bacterial DNA. Seven organisms were used to test DNA extraction from CSF: Staphylococcus aureus, Staphylococcus saprophyticus, Enterococcus faecalis, E. coli, Klebsiella oxytoca, Enterobacter cloacae, and Pseudomonas aeruginosa (ATCC, Manassas, VA, USA, and clinical isolates from Seattle Children’s Microbiology Lab). Cultures were grown overnight at 37° in LB broth and were pelleted and resuspended in uninfected donated CSF, which were then serially diluted using uninfected donated CSF to test extraction efficiency across a wide range, and particularly, for lower bacterial concentrations.
We initially tested approaches including Gentra Puregene (Qiagen, Valencia, CA, USA) and the column-based QIAmp (Qiagen) DNA mini kit, both using the body fluids protocol, with the addition of either lysozyme, cell lysis solution, proteinase K digestion, RNase digestion, and/or heat disruption protocols in various combinations. We also tested use of linear acrylamide, glycogen, and diluted glycogen to improve recovery when low concentrations of bacteria were expected, but bacterial contamination prevented the use of these approaches. Direct CSF lysis was also attempted to maximize recovery, but this proved ineffective.
After initial approaches demonstrated low yields with Gram-positive organisms, we switched to a more rigorous bead-beating cell disruption protocol using 0.1-mm zircona/silica beads. These smaller gauge beads were substituted into a standard MO BIO PowerSoil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) protocol showing dramatic improvement in extraction efficiency. The final approach to DNA extraction in CSF employed direct lysis of 100–μL CSF using MO BIO BiOstic bacteremia DNA isolation kit, a bead-beating protocol optimized for the purification of DNA (host and microbial) from body fluids. Volume for final elution of genomic DNA was 50 μL.
2.3. Assay refinement
Primers were synthesized by Integrated DNA Technologies (Iowa City, IA, USA). RT-PCR was performed with an Agilent Mx3005P QPCR System instrument using SYBR Green (Applied Biosystems; Life Technologies, Grand Island, NY, USA) for detection of PCR products. Each quantitative (qRT-PCR) reaction was performed in triplicate and included 0.4 μL each of 10 μmol/L forward and reverse primers, 5.0 μL 2× SYBRGreen PCR master mix (Applied Biosystems), 3.2 μL of ddH2O, as well as 1.0 μL of genomic DNA. Quantitative PCR cycling conditions were as follows: 95 °C for 10 minutes, and 40 cycles of 95 °C for 15 seconds, 60 °C for 1 minute. After each assay, a dissociation curve was run to confirm specificity of all PCR amplicons. Resulting crossing threshold values were converted to copy numbers and expressed as the average of triplicate samples ± 1 SD. If an individual well amplified poorly during qRT-PCR, the replicate was removed from triplicate measurements.
2.4. Assay testing
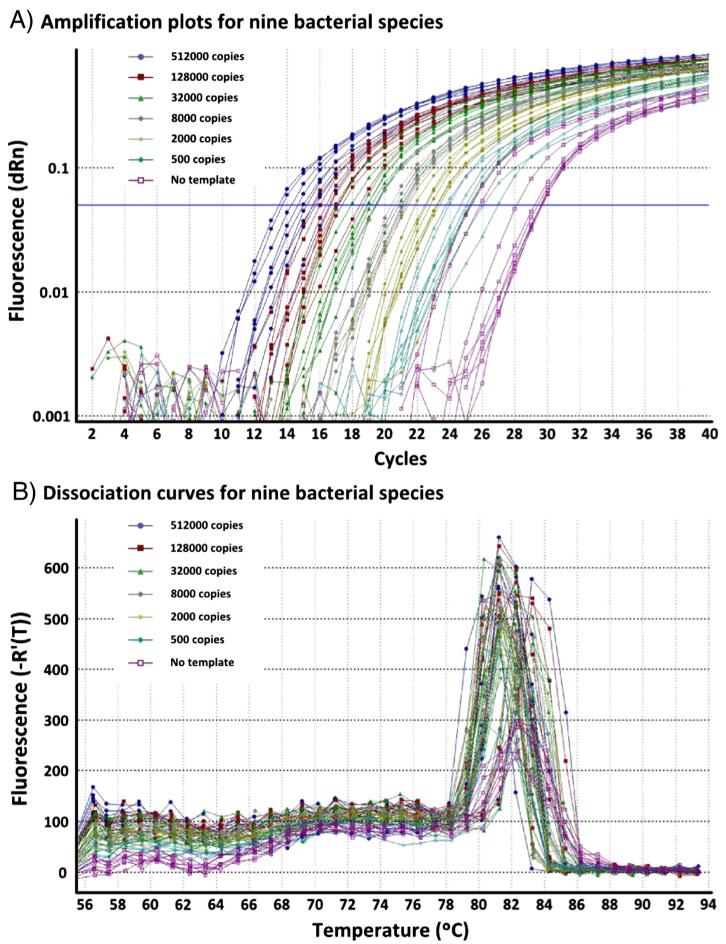
Assay efficiency was determined for both Gram-positive and Gram-negative CSF shunt infection organisms in dilution series of genomic DNA suspended in 10 mmol/L Tris-HCl. Nine organisms were provided by the Seattle Children’s Hospital (SCH) microbiology lab: S. aureus, S. saprophyticus, E. faecalis, S. pneumoniae, E. coli, K. oxytoca, E. cloacae, P. aeruginosa, and Haemophilus influenzae (ATCC); Staphylococcus epidermidis was provided by the Blaschke lab. Bacteria were harvested, and DNA was extracted using the Gentra Puregene (Qiagen) bacteria protocol including a lysozyme digest. Dilution series were created and qRT-PCR was performed (Fig. 1).
Fig. 1.
qRT-PCR for 16S rRNA showing dilution series of genomic DNA in water from 9 CSF shunt infection organisms (S. aureus, S. saprophyticus, E. faecalis, S. pneumoniae, E. coli, K. oxytoca, E. cloacae, P. aeruginosa, and H. influenzae). Caption: Nine species of bacteria were cultured, and genomic DNA was extracted. Dilution series for each bacterial species were created and tested using the 1149f and 1256r primers.
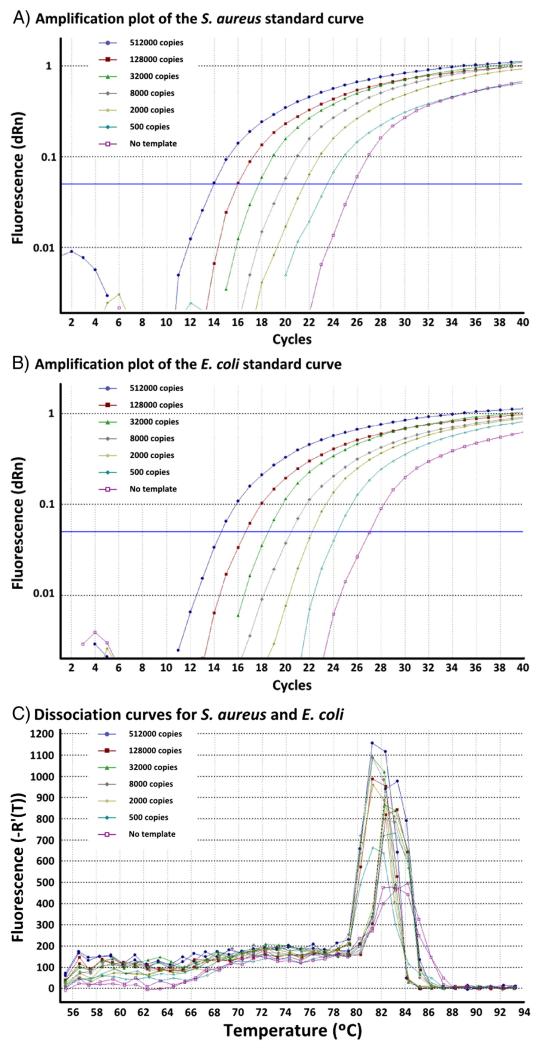
To generate quantification curves, purified DNA from S. aureus and E. coli bacteria in CSF was quantified using a PicoGreen DNA quantitation kit (Molecular Probes®; Life Technologies). This DNA was subsequently diluted serially by copy number (calculated by molecular weight) and amplified using the 16S rRNA qRT-PCR assay (Fig. 2).
Fig. 2.
RT-PCR for 16S rRNA showing standard copy number curves for S. aureus and E. coli bacteria in CSF. Panel A demonstrates a standard curve of S. aureus bacteria in CSF. Panel B demonstrates a standard curve of E. coli bacteria in CSF. Panel C demonstrates melt curves for both S. aureus (Tm 81.3 °C) and E. coli (Tm 82.8 °C) PCR amplicons.
2.5. Samples from children with CSF shunt infection
Children <18 years old undergoing treatment for conventional culture-confirmed CSF shunt infection at either SCH or PCMC were eligible for enrollment in this study. A CSF shunt infection was defined as identification of organisms on microbiological culture of CSF fluid obtained from a partial or complete CSF shunt system. CSF shunt system(s) included ventriculoperitoneal, ventriculoatrial, ventriculopleural, arachnoid cyst shunts, subdural shunts, and lumboperitoneal shunts, while temporary devices only such as external ventricular drain(s), Ommaya reservoir(s), ventricular access devices (reservoirs), and subgaleal shunts were not included. For all study subjects, except those from PCMC prior to March 18, 2010, parental permission (and assent when appropriate) was obtained for additional CSF to be collected on each occasion that regular CSF samples were obtained during treatment for CSF shunt infection. Prior to March 18, 2010, at PCMC, we used CSF remaining after routine processing and testing in the PCMC Microbiology Laboratory. The first CSF sample for diagnosis of infection was usually obtained from needle aspiration of the shunt reservoir under sterile conditions outside the operating room in a bedside “shunt tap”. The initial CSF sample analyzed in this study either was leftover from this first diagnostic sample or was obtained in the operating room under sterile conditions from the system being removed during the first surgery to treat infection. Subsequent CSF samples, including those at the end of the infection, were generally obtained under sterile bedside conditions through a sampling port within sterile extension tubing attached to the external ventricular drain. The study received Institutional Review Board approval from the Seattle Children’s Research Institute and the University of Utah, as well as approval from the PCMC Privacy Board.
After CSF was obtained for the study, CSF was stored at 4 °C. For a minority of samples, CSF was stored in solution provided in the MO BIO BiOstic bacteremia DNA isolation kit; storage in this solution did not affect recovery of bacterial DNA. CSF was then aliquoted for the study and stored at −70 °C; PCMC samples were shipped overnight to Seattle on dry ice.
2.6. Analysis
DNA from 100 μL of CSF was extracted for each available sample over the course of infection treatment. BVY performed the 16S rRNA qRT-PCR assay in triplicate. CSF samples tested included a selection among children for whom consistent CSF sample collection was obtained over the course of infection. Negative controls included phosphate-buffered saline and uninfected donor CSF. We tested the original primers in 25 infection episodes total; for 13 of these infections, we also performed analysis using the V6 primers. We compared the results of each 16S rRNA PCR series to each other, as well as to conventional culture results.
3. Results
Testing serial dilutions of genomic DNA from Gram-positive and Gram-negative CSF shunt infection organisms in water demonstrated the ability of our qRT-PCR to detect bacterial DNA across a wide range of species (Fig. 1, data not shown for S. epidermidis). Some slight variation existed when comparing amplifications between species, but generally, the assay was highly efficient and linear regardless of bacterial template (average efficiency 97.4%, average RSq 0.996). Amplicon melting temperatures were similar for all organisms (81 °C) except for S. pneumoniae and P. aeruginosa, which had slightly lower (80 °C) and slightly higher (83 °C) melt temperatures, respectively.
The assay’s ability to detect quantified amounts of 16S rRNA DNA was demonstrated with linearity across a range of 2.5 × 109 copies/mL CSF down to a detection limit of approximately 2.5 × 104 16S copies/mL CSF using standard copy number curves of S. aureus (efficiency 107.1%, RSq 1.000) and E. coli (efficiency 105.4%, RSq 1.000) bacterial templates (Fig. 2).
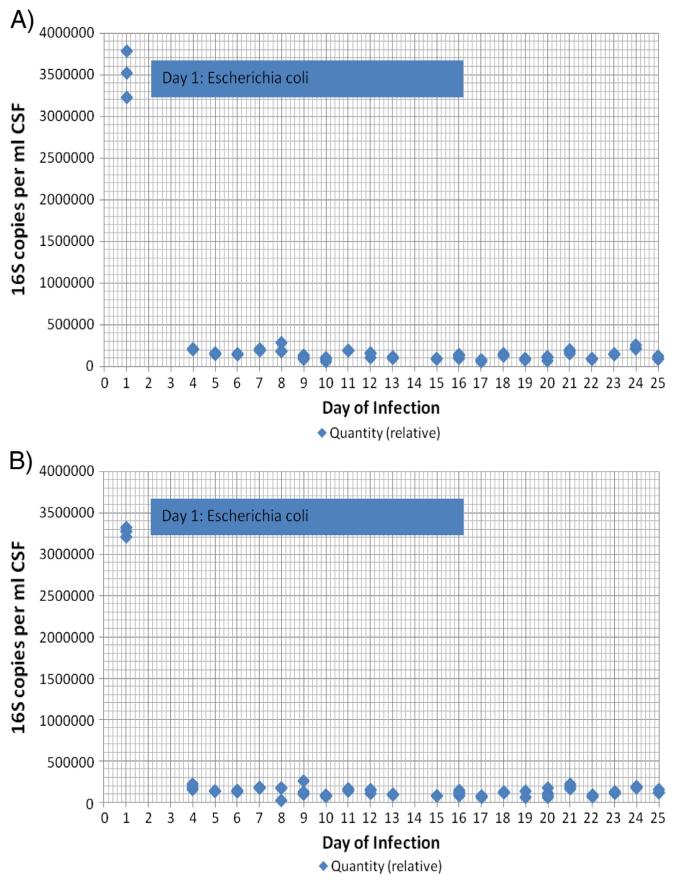
Data from 16S rRNA amplification in a representative infection episode are shown in Fig. 3. After an initial peak on the first day of sample collection, levels of quantitative 16S rRNA dropped precipitously and thereafter remained comparable to negative controls with both primer sets. The quantitative 16S rRNA PCR assays using the original and V6 primers were well correlated.
Fig. 3.
A representative example of 16S rRNA PCR over the course of infection treatment for a single infection episode (P) using the original primer set (A) and the primers targeting V6 (B). For A, Concurrent negative controls of CSF alone produced a relative quantity of 232,500–265,000 copies of 16S per milliliter CSF. For B, Concurrent negative controls of CSF alone produced a relative quantity of 238,500–272,000 copies of 16S per milliliter CSF.
A summary of findings for all 25 infection episodes is shown in Table 2. Relative quantities of 16S above baseline, which might be expected in the presence of CSF that grew organisms in culture, were only observed intermittently. As demonstrated for the representative infection shown in Fig. 3, levels of quantitative 16S rRNA dropped precipitously after the first day of sample collection across all samples (data not shown). Correlations between positive bacterial culture and quantitative 16S rRNA levels did not differ by infecting organism and were seen in only 4 of 9 (44%) infections with coagulase-negative Staphylococcus, 4 of 7 (57%) infections with other single organisms, and 4 of 5 (80%) infections with mixed organisms (Table 2). Results using the original and V6 primers again correlated well. For 3 of 4 infections in children who went on to develop reinfections (H, M, T), we also examined levels of 16S rRNA PCR at the end of the initial infection episode. None of these levels were above that of the negative controls at the end of the first infection treatment (data not shown).
Table 2.
Determination of bacterial load with quantitative 16S rRNA DNA PCR using original and V6 primers, compared conventional bacterial culture findings only when cultures demonstrated organisms.
| Patient | Day | Bacterial culture | Original primer 16S copy number per ml CSF | V6 primer 16S copy number per mL CSF (if tested) |
|---|---|---|---|---|
| A. Coagulase-negative Staphylococcus only (n = 11). We had insufficient CSF to obtain PCR data for J and N on day 1, as well as B on day 1 and K on day 2. | ||||
| A | 1 | S. epidermidis | 477,000–835,000 | n/a |
| B | 5 | S. epidermidis | 253,000–328,500 | 227,500–235,500 |
| C | 1 | Staphylococcus lugdunensis | 194,000–280,500 | n/a |
| 2 | S. lugdunensis | 188,500–281,000 | n/a | |
| E | 1 | Coagulase-negative Staphylococcus | 211,850–247,150a and 384,200–453,950 and 344,050–358,200 | n/a |
| 3 | Coagulase-negative Staphylococcus | 201,050–242,750a | n/a | |
| F | 1 | Coagulase-negative Staphylococcus | 3,627,000–3,600,050 | n/a |
| G | 1 | S. epidermidis | 52,500–70,000 | n/a |
| I | 1 | S. epidermidis | 26,850–90,500 | n/a |
| 2 | S. epidermidis | 211,500–232,000 | n/a | |
| 3 | S. epidermidis | 173,000–244,500 | n/a | |
| 4 | S. epidermidis | 171,000–187,000 | n/a | |
| K | 1 | S. epidermidis | 435,500–483,000 | n/a |
| M (#2) | 1 | S. epidermidis | 206,050–220,300 | 193,000–216,000 |
| 2 | S. epidermidis | 129,100–164,050 | 150,500–171,000 | |
| 3 | S. epidermidis | 129,050–145,750 | 142,000–150,500 | |
| 4 | S. epidermidis | 106,500–147,250 and 133,000–197,850 | 103,500–117,500 and 156,500–177,500 | |
| B. Other single infecting organisms (n = 8). We had insufficient CSF to obtain PCR data for H (#2) on day 1 with S. aureus, as well as Ton day 3 with Serratia marcescens, Ron day 2 with E. faecalis and L on day 7 with Propionibacterium acnes. | ||||
| D | 1 | S. aureus | 153,500–165,000a | n/a |
| L | 1 | P. acnes | 2,355,000–2,765,000 | n/a |
| 2 | P. acnes | 168,500–225,000 | n/a | |
| M (#1) | 1 | Group A beta-hemolytic Streptococcus | 406,400–565,000 | 445,500–610,000 |
| O | 1 | P. acnes | 77,000–100,500 | 66,500–73,250 |
| P | 1 | E. coli | 3,225,000–3,785,000 | 3,205,000–3,325,000 |
| R | 1 | E. faecalis | 256,500–314,500 | 306,500–324,000 |
| T (#2) | 1 | S. marcescens | 950,000–1,150,000 | 1,125,500–1,331,500 |
| 2 | S. marcescens | 195,000–225,500 | 202,800–240,300 | |
| C. Infections with a several organisms (n = 6). We had insufficient CSF to obtain PCR data for S (#2) on day 1 with S. epidermidis, Staphylococcus warneri, and Diphtheroid bacilli, as well as for Q on day 1 with S. pneumoniae, S (#1) on day 17 and 18 with Micrococcus spp., and T (#1) on days 4 and 6 with S. aureus and day 11 with Staphylococcus hominis. | ||||
| H (#1) | 1 | S. epidermidis and S. aureus | 7,050,000–7,800,000 | n/a |
| 3 | S. aureus | 96,500–145,000a and 85,500–100,500a and 78,500–114,500a | n/a | |
| 4 | S. aureus | 21,550–164,000a and 4,670–27,550 | n/a | |
| 5 | S. aureus | 108,000–163,500a and 71,000–123,000a | n/a | |
| 6 | S. aureus | 110,000–131,000a and 42,850–60,500 | n/a | |
| Q | 3 | No growth | 3,140,000–3,775,000 | 3,440,000–4,080,000 |
| 5 | Staphylococcus capitis | 940,000–1,165,000 | 1,020,000–1,525,000 | |
| S(#1) | 1 | Corynebacterium spp. | 406,400–565,000 | 148,000–181,500 |
| 10 | Corynebacterium spp. | 138,450–151,050 | 204,000–241,000 | |
| 11 | Corynebacterium spp. | 289,350–347,150 | 211,500–267,000 | |
| 12 | Corynebacterium spp. | 201,750–366,900 | 177,500–187,500 | |
| T (#1) | 1 | S. aureus | 133,500–202,500 | 96,000–116,500 |
| 2 | S. aureus | 234,500–256,500a | 199,000–205,500 | |
| 3 | S. aureus | 175,000–219,500a | 103,500–111,500 | |
| 5 | S. aureus | 226,500–293,500a | 224,000–239,000a | |
| 16 | Coagulase-negative Staphylococcus | 291,000–336,000 | 258,000–293,500 | |
| U | 1 | K. pneumoniae and Streptococcus mitis/oralis | 318,350–383,650 | 349,650–471,950 |
| 3 | K. pneumoniae | 137,850–159,900 | 151,400–177,300 | |
| 5 | K. pneumoniae | 177,050–192,950 | 181,850–265,100 | |
| 16 | S. epidermidis and S. aureus | 92,200–124,450 | 91,100–153,500 | |
Subsequent days of infection treatment with negative CSF cultures are not provided here. Bolded primer counts are significantly different from negative CSF controls and later (culture-negative) samples drawn during the infection episode; these examples resemble the representative infection episodes shown in Figs. 3A and 3B.
Above CSF negative controls, but not different from other samples drawn during the infection episode.
4. Discussion
We hypothesized that amplification of bacterial DNA using 16S rRNA PCR could be used as a marker of CSF bacterial load and might be useful to inform antibiotic management and timing of shunt replacement to avoid reinfection. We developed a quantitative 16S rRNA PCR assay that demonstrated an ability to detect genomic DNA from both Gram-positive and Gram-negative CSF shunt infection organisms across a range of 2.5 × 109 copies/mL CSF down to a detection limit of approximately 2.5 × 104 16S copies/mL CSF under experimental conditions. However, when this assay was applied to clinical samples, there was inconsistent detection of 16S rRNA over background levels when compared to bacterial culture. Levels of 16S rRNA at the end of infection did not correlate with reinfection. This quantitative 16S rRNA PCR assay did not augment treatment information provided by conventional CSF cultures among children undergoing treatment for CSF shunt infection.
Previous work has examined the use of 16S PCR for the rapid detection of CSF shunt infection (Banks et al., 2005) or in external ventricular drain infections (Deutch et al., 2007). The use of broad-range 16S rRNA PCR for the detection of pathogens has the potential advantage of unbiased amplification of any bacterial species present (Nadkarni et al., 2002) and thus could theoretically be used for infections caused by a range of organisms. While initial data for broad range PCR were promising, (Banks et al., 2005; Blaschke and Voelkerding, 2006; Deutch et al., 2007; Won et al., 2012), more recent studies have demonstrated significant limitations due to the ubiquitous nature of bacterial DNA and the high rate of false-positive detection (Corless et al., 2000; Millar et al., 2002). Due to high background levels of bacterial DNA, the extended PCR cycling required for optimal detection of organisms present in low concentrations cannot be performed (Sontakke et al., 2009).
Because of the well-documented sensitivity issues with broad-range bacterial amplification, we were seeking to use 16S PCR not to diagnose infection but to quantify bacterial load in culture-documented infection as a method to guide antibiotic management. While our experimental quantification curves showed good correlation between bacterial load and 16S copy number, this did not translate to practical data with utility for guiding management of CSF infection. Some authors have advised use of both 16S PCR and conventional culture in clinical scenarios (Deutch et al., 2007; Sarookhani et al., 2010). In our hands, the PCR did not add sufficient information to justify its routine clinical use. It is likely that the ability of our assay to detect bacterial load was complicated by low levels of bacterial DNA present in samples, particularly after the removal of CSF shunt and initiation of antibiotic treatment.
Measurement of viral load using quantitative real-time PCR is an established method to guide therapy in viral infections such as those caused by cytomegalovirus and human immunodeficiency virus (Berger and Preiser, 2002). The use of quantitative bacterial PCR in clinical care is less well-established. Literature supports its potential benefit in the diagnosis and evaluation of disease severity in pneumococcal pneumonia and pneumococcal sepsis as well as in meningococcal disease (Darton et al., 2009; Peters et al., 2004; Rello et al., 2009). Studies that use bacterial load to guide therapeutic management are rarer still.
Conventional culture techniques examine free-floating clonal populations of a single species during logarithmic growth phase (Rhoads et al., 2012). In contrast, biofilms are surface-associated groups, often composed of multiple genera, of organisms held together by an extracellular matrix, and both the growth conditions and population within the biofilm can be spatially and temporally variable (Rickard et al., 2003). These complex communities attach to surfaces in a largely dormant mode (Bayston et al., 2012) that promotes resistance to antibiotics. Biofilms have been implicated in several chronic infections (Costerton et al., 1999). Some work has suggested the presence of biofilms on CSF shunts (Fux et al., 2006; Guevara et al., 1987). One possible explanation of our findings is that biofilms are present in CSF shunt infections, and bacterial DNA is only periodically detected by either 16S PCR or conventional culture in CSF due to the highly dormant state of bacteria in these communities. Nonetheless, reinfection occurs at high rates. This apparent paradox indicates a need to consider different models for understanding the microbial environment in CSF shunt infection.
This pilot study took the first step to examine the utility of molecular quantification of CSF pathogens for patients undergoing treatment for CSF shunt infection. Based on our data, we conclude that molecular quantification of bacterial load using broad-range 16S rRNA PCR does not appear useful as a novel adjunct to optimize treatment of CSF shunt infection. It is possible that other approaches, such as examination of biofilm, species-specific amplification, and/or markers of inflammation may be more suitable, and these approaches deserve further study. For the moment, clinicians continue to require an optimal treatment strategy for CSF shunt infections that accurately determines when bacteria have been eradicated and reinfection after shunt replacement is unlikely.
Acknowledgments
We would like to thank the children and families who participated in the study at SCH and PCMC. We also thank those who made CSF sample collection at SCH and PCMC possible, including the pediatric neurosurgeons and neurosurgical staff at SCH and PCMC; Hydrocephalus Clinical Research Network coordinators Amy Anderson and Tracey Habrock-Bach who assisted our study staff with tracking of eligible children; and study staff who enrolled patients and ensured appropriate collection and storage of CSF specimens including Robert Johnson, Courtney Dethlefs, and Marshal Werfelman at SCH and Priscilla Cowan, Trenda Barney, and Abby Phillips at PCMC. We thank Jennifer Forsyth for early work on assay development; members of SCH Microbiology lab for assistance with primer design and provision of bacteria, including Xuan Qin and Shannon Rich; as well as Chris Pope for a suggestion to add a primer set targeting V6. We thank members of TS’ advisory committee, including Danielle Zerr, Bonnie Ramsey, Nino Ramirez, Jeff Ojemann, and Rita Mangione-Smith; and last but most importantly Stephan Nemeth IV and Gabriel Finn Nemeth.
Footnotes
Financial Disclosure: This publication was supported by STARS-kids; Clinical and Translational Science Award (CTSA) pilot funds from the University of Utah [Grant UL1RR025764 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH)]; the Department of Neurosurgery at University of Utah; Seattle Children’s Center for Clinical and Translational Research (CCTR); CTSA Grant Number ULI RR025014 from the NCRR; and Diabetes and Endocrinology Research Center grant number P30DK017047. TDS is supported by Award K23NS062900 from the National Institute of Neurological Disorders And Stroke, the Child Health Corporation of America via the Pediatric Research in Inpatient Setting Network Executive Council, and Seattle Children’s CCTR. AJB is supported by 1K23AI079401 from the National Institute of Allergy and Infectious Diseases. None of the sponsors participated in design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or NIH.
Conflicts of Interest: AJB collaborates with BioFire Diagnostics, Inc., on NIH and CDC-funded projects. AJB has intellectual property in BioFire Diagnostics, Inc.
References
- Banks JT, Bharara S, et al. Polymerase chain reaction for the rapid detection of cerebrospinal fluid shunt or ventriculostomy infections. Neurosurgery. 2005;57(6):1237–43. doi: 10.1227/01.neu.0000186038.98817.72. [discussion 1237-1243] [DOI] [PubMed] [Google Scholar]
- Bayston R, Ullas G, et al. Action of linezolid or vancomycin on biofilms in ventriculoperitoneal shunts in vitro. Antimicrob Agents Chemother. 2012;56(6):2842–5. doi: 10.1128/AAC.06326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Preiser W. Viral genome quantification as a tool for improving patient management: the example of HIV, HBV, HCV and CMV. J Antimicrob Chemother. 2002;49(5):713–21. doi: 10.1093/jac/dkf050. [DOI] [PubMed] [Google Scholar]
- Blaschke AJ, Voelkerding K, et al. Broad-Range Bacterial PCR of CSF in Infants Evaluated for Sepsis; 44th Annual Meeting of the Infectious Diseases Society of America.2006. [Google Scholar]
- Chakravorty S, Helb D, et al. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2):330–9. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless CE, Guiver M, et al. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38(5):1747–52. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, et al. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Darton T, Guiver M, et al. Severity of meningococcal disease associated with genomic bacterial load. Clin Infect Dis. 2009;48(5):587–94. doi: 10.1086/596707. [DOI] [PubMed] [Google Scholar]
- Deutch S, Dahlberg D, et al. Diagnosis of ventricular drainage-related bacterial meningitis by broad-range real-time polymerase chain reaction. Neurosurgery. 2007;61(2):306–11. doi: 10.1227/01.NEU.0000255526.34956.E4. [discussion 311-302] [DOI] [PubMed] [Google Scholar]
- Fan-Havard P, Nahata MC. Treatment and prevention of infections of cerebrospinal fluid shunts. Clin Pharm. 1987;6(11):866–80. [PubMed] [Google Scholar]
- Fux CA, Quigley M, et al. Biofilm-related infections of cerebrospinal fluid shunts. Clin Microbiol Infect. 2006;12(4):331–7. doi: 10.1111/j.1469-0691.2006.01361.x. [DOI] [PubMed] [Google Scholar]
- Guevara JA, Zuccaro G, et al. Bacterial adhesion to cerebrospinal fluid shunts. J Neurosurg. 1987;67(3):438–45. doi: 10.3171/jns.1987.67.3.0438. [DOI] [PubMed] [Google Scholar]
- Kestle JR. Pediatric hydrocephalus: current management. Neurol Clin. 2003;21(4):883–95. doi: 10.1016/s0733-8619(03)00016-1. vii. [DOI] [PubMed] [Google Scholar]
- Kestle JR, Garton HJ, et al. Management of shunt infections: a multicenter pilot study. J Neurosurg. 2006;105(3 Suppl):177–81. doi: 10.3171/ped.2006.105.3.177. [DOI] [PubMed] [Google Scholar]
- Kulkarni AV, Rabin D, et al. Repeat cerebrospinal fluid shunt infection in children. Pediatr Neurosurg. 2001;35(2):66–71. doi: 10.1159/000050393. [DOI] [PubMed] [Google Scholar]
- Lan CC, Wong TT, et al. Early diagnosis of ventriculoperitoneal shunt infections and malfunctions in children with hydrocephalus. J Microbiol Immunol Infect. 2003;36(1):47–50. [PubMed] [Google Scholar]
- Lee ZM, Bussema C., 3rd rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–93. doi: 10.1093/nar/gkn689. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar BC, Xu J, et al. Risk assessment models and contamination management: implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol. 2002;40(5):1575–80. doi: 10.1128/JCM.40.5.1575-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni MA, Martin FE, et al. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(Pt 1):257–66. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Nelson JD. Cerebrospinal fluid shunt infections. Pediatr Infect Dis. 1984;3(3 Suppl):S30–2. doi: 10.1097/00006454-198405001-00011. [DOI] [PubMed] [Google Scholar]
- Odio C, McCracken GH, Jr, et al. CSF shunt infections in pediatrics. A seven-year experience. Am J Dis Child. 1984;138(12):1103–8. doi: 10.1001/archpedi.1984.02140500009004. [DOI] [PubMed] [Google Scholar]
- Peters RP, van Agtmael MA, et al. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis. 2004;4(12):751–60. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- Rello J, Lisboa T, et al. Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest. 2009;136(3):832–40. doi: 10.1378/chest.09-0258. [DOI] [PubMed] [Google Scholar]
- Rhoads DD, Wolcott RD, et al. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci. 2012;13(3):2535–50. doi: 10.3390/ijms13032535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard AH, Gilbert P, et al. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11(2):94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- Sarookhani MR, Ayazi P, et al. Comparison of 16S rDNA-PCR amplification and culture of cerebrospinal fluid for diagnosis of bacterial meningitis. Iran J Pediatr. 2010;20(4):471–5. [PMC free article] [PubMed] [Google Scholar]
- Sells CJ, Shurtleff DB, et al. Gram-negative cerebrospinal fluid shunt-associated infections. Pediatrics. 1977;59(4):614–8. [PubMed] [Google Scholar]
- Simon TD, Riva-Cambrin J, et al. Hospital care for children with hydrocephalus in the United States: utilization, charges, comorbidities, and deaths. J Neurosurg Pediatr. 2008;1(2):131–7. doi: 10.3171/PED/2008/1/2/131. [DOI] [PubMed] [Google Scholar]
- Simon TD, Whitlock KB, et al. Revision surgeries are associated with significant increased risk of subsequent cerebrospinal fluid shunt infection. Pediatr Infect Dis J. 2012;31(6):551–6. doi: 10.1097/INF.0b013e31824da5bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontakke S, Cadenas MB, et al. Use of broad range16S rDNA PCR in clinical microbiology. J Microbiol Methods. 2009;76(3):217–25. doi: 10.1016/j.mimet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Tuan TJ, Thorell EA, et al. Treatment and microbiology of repeated cerebrospinal fluid shunt infections in children. Pediatr Infect Dis J. 2011;30(9):731–5. doi: 10.1097/INF.0b013e318218ac0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinchon M, Dhellemmes P. Cerebrospinal fluid shunt infection: risk factors and long-term follow-up. Childs Nerv Syst. 2006;22(7):692–7. doi: 10.1007/s00381-005-0037-8. [DOI] [PubMed] [Google Scholar]
- Won H, Yang S, et al. A broad range assay for rapid detection and etiologic characterization of bacterial meningitis: performance testing in samples from sub-Sahara. Diagn Microbiol Infect Dis. 2012;74(1):22–7. doi: 10.1016/j.diagmicrobio.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JJ, Christensen GD, et al. Coagulase-negative staphylococci isolated from cerebrospinal fluid shunts: importance of slime production, species identification, and shunt removal to clinical outcome. J Infect Dis. 1987;156(4):548–54. doi: 10.1093/infdis/156.4.548. [DOI] [PubMed] [Google Scholar]