Abstract
Kinase inhibitors are important cancer drugs, but they tend to display limited target specificity, and their target profiles are often challenging to rationalize in terms of molecular mechanism. Here we report that the clinical kinase inhibitor bosutinib recognizes its kinase targets by engaging a pair of conserved structured water molecules in the active site, and that many other kinase inhibitors share a similar recognition mechanism. Using the nitrile group of bosutinib as an infrared probe, we show that the gatekeeper residue and one other position in the ATP-binding site control access of the drug to the structured water molecules, and that the amino acids found at these positions account for the kinome-wide target spectrum of the drug. Our work highlights the importance of structured water molecules for inhibitor recognition, reveals a new role for the kinase gatekeeper, and showcases an effective approach for elucidating the molecular origins of selectivity patterns.
Protein kinases have long been prime targets for cancer therapy due to their central importance in controlling cellular growth pathways1. Today, numerous ATP-competitive kinase inhibitors are available that inhibit mutated or overexpressed kinases responsible for driving oncogenic signaling. The prototypical example, imatinib, targets BCR-Abl, a constitutively active form of the Abl tyrosine kinase that causes chronic myeloid leukemia (CML), and has transformed the treatment of this disease2.
Because of significant sequence conservation within the kinase ATP-binding site3, kinase inhibitors tend to have limited target specificity. Off-target effects can in some cases be beneficial, such as in the case of imatinib’s activity towards c-Kit, which contributes to the efficacy of the drug in CML4 and provides an effective treatment for gastrointestinal stromal tumors5. Nonetheless, kinase inhibitors with improved selectivity are in great demand, both as new cancer therapeutics with reduced toxicity, and as tools for studying signaling pathways6. Kinase inhibitors are now routinely profiled against the kinome (all ~500 human kinases), revealing that each compound has a unique and highly unpredictable target spectrum7. Understanding the origin of these complex patterns in terms of molecular mechanism is an important goal that would enhance the use of existing inhibitors and greatly benefit the process of inhibitor development.
Some kinase inhibitors obtain selectivity by recognizing particular inactive conformations favored by certain kinases8. Type II inhibitors, typified by imatinib, specifically recognize an inactive conformation in which a catalytically important Asp-Phe-Gly (DFG) motif is rotated by ~180° with respect to the active conformation (referred to as DFG-Out, in contrast to the active DFG-In conformation)9. The more numerous type I inhibitors bind to the DFG-In conformation, shared by all active kinases, and are usually less selective than type II inhibitors. An example is the 2nd generation BCR-Abl inhibitor bosutinib, developed to combat clinical resistance to imatinib in CML patients10, which also displays activity towards the Src-family kinases that is exploited in the treatment of other cancers11.
As with other kinase inhibitors, a single residue in the ATP-binding site, called the gatekeeper, appears to play an important role in determining bosutinib’s target profile12,13. The general importance of the gatekeeper is underscored by the fact that patients undergoing kinase inhibitor therapy frequently develop clinical resistance mediated by mutations at this position12,14. It is often argued that the gatekeeper exerts control over inhibitor binding by restricting access to a pocket deep inside the ATP-binding site, and compounds that extend into this region do tend to be selective for kinases with small gatekeeper residues15. However, while bosutinib is selective for threonine gatekeeper kinases16, and is ineffective against the common T315I gatekeeper mutation of BCR-Abl17, the structure of the drug bound to Abl revealed a cavity adjacent to the gatekeeper with ample room to accommodate larger gatekeeper residues18. The inability of a simple steric model to explain the preference for a threonine gatekeeper suggests that this residue may be mediating its effects through an as yet undiscovered mechanism. While pursuing this observation we discovered that most type I inhibitors leave a similar cavity next to the gatekeeper, that two structured water molecules typically occupy this space, and that these molecules form a network of hydrogen bonds in which the bound inhibitor often participates. Here we report a novel mechanism that explains bosutinib’s target spectrum in which the gatekeeper residue controls access of the drug to this conserved water-mediated hydrogen bond network.
RESULTS
Bosutinib forms a water-mediated interaction with Src
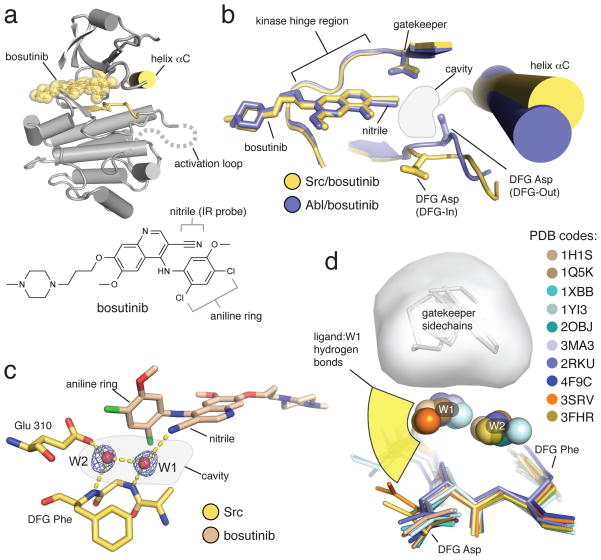
Type I inhibitors are usually visualized in crystal structures bound to the active DFG-In conformation, but in our structure of bosutinib bound to Abl the kinase is in the inactive DFG-Out conformation18. This was attributed to the propensity of Abl to adopt the DFG-Out conformation at the low pH value of the crystallization condition19, and complicated the analysis of the kinase/drug interaction. To see how the drug interacts with an active kinase we determined the x-ray structure of the drug bound to Src to 2.1 angstroms (Supplementary Results, Supplementary Table 1). The overall binding mode is very similar to that observed in the Abl/bosutinib structure, but the kinase is in the active, DFG-In conformation (Fig. 1a and 1b). As seen in the Abl structure, the drug is in van der Waals contact with the gatekeeper, but leaves a sizeable cavity directly adjacent it (Fig. 1b). The active conformation of the DFG motif, which forms the floor of this cavity, positions the backbone NH groups of the DFG Asp and Phe residues pointing into the cavity, where they serve to anchor two ordered water molecules, referred to as W1 and W2, respectively (Fig. 1c). The nitrile group of the drug points into the cavity, where it forms a hydrogen bond with water molecule W1 (Fig. 1c). This water-mediated interaction is one of only two hydrogen bonds formed by the drug; the other is an interaction with the kinase hinge region, a feature shared with most kinase inhibitors.
Figure 1. The structure of bosutinib bound to Src shows the drug participates in a water-mediated hydrogen bond network.
(a) Overview of the structure of bosutinib bound to Src, with the DFG motif and helix αC of Src colored yellow, and the drug shown as semi-transparent yellow spheres. The approximate path of the C-terminal portion of the activation loop, which is disordered in the structure, is shown as a dotted gray line. The P-loop is omitted for clarity. The chemical structure of bosutinib is shown underneath the x-ray structure. (b) Comparison of the structures of bosutinib bound to Src (yellow) and Abl (blue). The aniline ring of bosutinib, which forms one side of the water-filled cavity, is omitted for clarity. (c) View of the water-mediated hydrogen bond network, with hydrogen bonds shown as dashed yellow lines. The blue mesh represents a simulated annealing omit map contoured at 3 standard deviations above the mean electron density. (d) Structural alignment of 10 structures of kinases bound to a diverse set of type I inhibitors, in which both water molecules are observed and the ligand engages W1 in a hydrogen bond. The angular distribution of the ligand/water hydrogen bonds is represented schematically in yellow. The space occupied by the gatekeeper sidechains is shown as a gray surface.
Interestingly, we found that the water molecules W1 and W2 observed in the Src/bosutinib structure are conserved in many structures of active kinases bound to nucleotides20–23. These water molecules form hydrogen bond networks linking the nucleotide α-phosphate, as opposed to the bosutinib nitrile, to a conserved, catalytically important, glutamate residue (Glu 310 in Src, chicken c-Src numbering), which projects into the active site from helix αC of the kinase (Supplementary Fig. 1a). The precise positioning of helix αC is critical for productive catalysis24, and the water-mediated hydrogen bond network may contribute to kinase regulation by coupling the position of helix αC, the active conformation of the DFG motif, and the position of the nucleotide phosphates.
The prevalence of kinase inhibitor/W1 hydrogen bonds
Given the conservation of W1 and W2 in active kinase structures, we speculated that other type I inhibitors might recognize these water molecules in a similar manner to bosutinib. To quantify the frequency with which this occurs we used the web-based program PDBeMotif25 to search the protein databank for type I inhibitor complexes by constraining the backbone torsion angles of the DFG motif to the DFG-In conformation. Of 968 active kinase structures identified in our search, 630 contained non-nucleotide ligands in the ATP-binding site. In the vast majority of these structures the ligand does not extend into the water-filled cavity, and both water molecules W1 and W2 are commonly observed (Supplementary Fig. 1b). For structures determined to better than 2 Å, W1 is seen in ~75% and W2 in ~60% of cases. We found that hydrogen bonds between W1 and a donor or acceptor on the ligand are in fact very common (seen in the majority of structures where W1 is present), while interactions with W2 are found in ~5% of cases.
All together, examples of a ligand engaging W1 encompass a significant pool of 40 different kinases and 164 different ligands spanning a broad range of chemical space, many of which are potent type I inhibitors in clinical or preclinical development (Supplementary Data Set)26–31. The ligand/W1 hydrogen bonds possess a length distribution typical of water-mediated hydrogen bonds (Supplementary Fig. 1c)32, and the interactions appear to be highly adaptable to the requirements of the ligand, as seen by the wide range of angles from which hydrogen bonding groups of the ligand approach W1 (Fig. 1d). One surprising example is the complex of Syk kinase with imatinib, in which the drug adopts a DFG-In binding mode that is distinct from the DFG-Out binding mode employed when binding to Abl and other high affinity imatinib targets33. Thus bosutinib’s engagement in the water-mediated hydrogen bond network is representative of a widespread feature of type I kinase inhibitors that distinguishes them from type II inhibitors, and to our knowledge, has not been previously noted. As described in the following, bosutinib provides a unique and highly local spectroscopic reporter of the presence or absence of a hydrogen bond to W1.
The bosutinib nitrile probes its interaction with W1
Water-mediated hydrogen bonds are a common feature of protein/ligand interactions34, but their importance for kinase inhibitor recognition has not been broadly established. We wondered whether the ability of a given inhibitor to engage the W1 water molecule might depend on the nature of the amino acid sidechains that line the water-filled cavity, and in particular on the gatekeeper residue, which is in van der Waals contact with W1 in the majority of the complexes discussed above (Fig. 1d). However, unlike the direct interaction between a ligand and an amino acid sidechain, the indirect nature of water-mediated hydrogen bonds makes it challenging to predict how they will be affected by nearby substitutions. In the case of bosutinib, the nitrile group of the drug fortuitously provides a facile experimental readout of its participation in the hydrogen bond network, since the stretching vibrations of nitriles appear in a background-free region of the infrared (IR) spectrum and exhibit pronounced blue-shifts (shifts to higher frequency) in response to hydrogen bonding35,36. We exploited this to investigate how nearby residue substitutions modulate the engagement of the bosutinib nitrile in the hydrogen bond network.
We used Src to generate a set of active site chimeras carrying substitutions in the ATP-binding site that mimic a diverse set of kinases. Substitutions were chosen by consulting a sequence alignment of the human kinome to determine the residues found at the cavity-lining positions, and we made mutations covering most of this sequence variation, including 9 residues at the gatekeeper position (Supplementary Fig. 2). The absorbance bands corresponding to the bosutinib nitrile probe are spread across a frequency range of 13 cm−1 in the IR spectra of this set of Src mutants, comparable to the largest frequency shifts seen for nitriles across protic and aprotic chemical solvents (Supplementary Table 2)35. This strongly suggests that the probe’s hydrogen bond status differs in the different mutants. Substitutions of the gatekeeper (T338) and one other position (A403) have the largest effect on the probe, producing red-shifts (shifts to lower frequency) as large as 11 cm−1, while substitutions at the two other positions result in considerably smaller shifts (Fig. 2a and 2b, Supplementary Fig. 3).
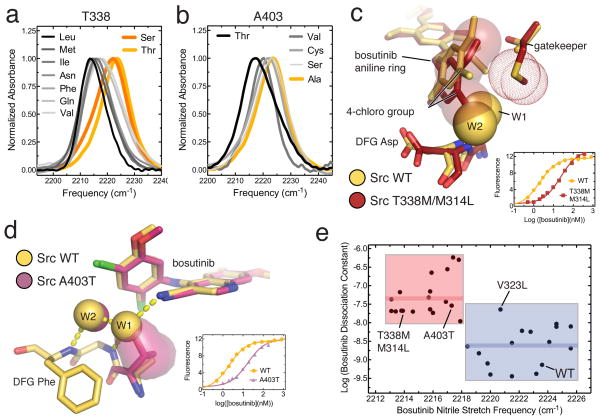
Figure 2. Substitutions of the cavity-lining residues modulate bosutinib’s engagement in the hydrogen bond network and affect binding.
(a) IR spectra of bosutinib bound to wildtype Src (yellow) and 8 gatekeeper mutants (black, gray and orange). (b) IR spectra of bosutinib bound to wildtype Src (yellow) and mutants of Src bearing substitutions at position 403 (black and gray). (c) Comparison of WT Src and the T338M/M314L mutant bound to bosutinib. For the mutant, the aniline ring of bosutinib is highlighted as transparent spheres, and the methionine gatekeeper sidechain as dots. The positions of W1 and W2 in the structure of WT Src are shown as yellow spheres. The inset shows fluorescence data for bosutinib binding to WT Src and Src T338M/M314L. (d) Comparison of WT Src and the A403T mutant bound to bosutinib. The sidechain of residue 403 in the mutant is show as a surface representation. The inset shows fluorescence data for bosutinib binding to WT Src and the A403T mutant. (e) Plot of the logarithm of the dissociation constant of bosutinib (in molar units) as a function of the nitrile stretch frequency for 34 Src mutants. Mutants in which the bosutinib nitrile is thought to be hydrogen bonded are highlighted by a blue box, mutants in which the hydrogen bond is lost are highlighted by a red box. The mean value of the dissociation constants for the red and blue sets are shown on the plot as thick red and blue lines, respectively.
To verify that these pronounced frequency shifts of the probe report on the loss of the hydrogen bond to W1, we solved x-ray structures of bosutinib bound to Src bearing mutations at either T338 or A403. Methionine is the most common gatekeeper residue in human kinases, but the T338M mutant of Src yielded small crystals that diffracted x-rays poorly. However, when the T338M mutation was combined with the M314L substitution of another cavity-lining residue, which is found in most methionine gatekeeper kinases, larger crystals were obtained that diffracted well. We also determined the structure of the Src A403T mutant, which produces the largest red-shift of the probe among substitutions at this position (Fig. 2b). Both structures, which were obtained in the same crystal form as WT Src, and are very similar in overall conformation, were determined to 2.6 Å resolution (Supplementary Table 1).
In the T338M/M314L structure, a slight closure of the N- and C-terminal lobes of the kinase domain is observed, concomitant with a rotation of the aniline ring of bosutinib, which is wedged between the two lobes. This results in improved packing of the aniline ring against the hydrophobic methionine gatekeeper, but the 4-chloro group on the aniline ring occludes the binding sites for both water molecules, and the nitrile group of the drug is consequently not hydrogen bonded (Fig. 2c). In the Src A403T structure, the A403T sidechain hydroxyl directly occludes the binding site of W1, but is itself too far from the nitrile (3.5 angstroms) to form its own hydrogen bond (Fig. 2d). These structures confirm that substitutions of the gatekeeper and A403 can interfere with the access of bosutinib to the hydrogen bond network, and that the nitrile probe faithfully reports on this process.
Loss of the bosutinib/W1 interaction weakens binding
To test whether the participation of the drug in the hydrogen bond network affects binding we measured the dissociation constants of bosutinib for all Src mutants using a fluorescence assay18 (Supplementary Fig. 4, Supplementary Table 2). Substitutions that produce pronounced red-shifts of the probe do typically result in weaker binding. For instance, the T338M/M314L double mutant, which produces a 10 cm−1 red-shift of the probe, results in a 30-fold reduction in binding affinity compared to WT Src (Fig. 2c, inset), whereas the A403T mutation produces a 6 cm−1 red-shift accompanied by a 40-fold reduction in binding affinity (Fig. 2d, inset).
For individual substitutions an assessment of the energetic contribution of the hydrogen bond is confounded by other energetic factors. However, the large shifts in probe frequency report primarily on hydrogen bonding, and so by correlating binding with hydrogen bonding for a large number of mutants other energetic factors should be averaged out. This is apparent when the binding affinity is plotted as a function of the probe vibrational frequency for all mutants (Fig. 2e), which clusters the mutants into two groups: a tight binding group in which the drug is engaged in the hydrogen bond network (highlighted in blue) and a weak binding group in which the nitrile/W1 hydrogen bond is lost (highlighted in red). On average, participation in the hydrogen bond network is associated with a ~25 fold increase in affinity, corresponding to a significant energetic contribution of ~2 kcal/mol.
A surface representation of the cavity is shown in Fig 3a, colored as in Fig 2e according to how substitutions affect the water-mediated interaction with bosutinib. Since the mutants highlighted in red in Fig. 2e possess substitutions only at positions T338 and A403, these positions appear to have exclusive control. Interestingly, the A403 position has been previously implicated in controlling selectivity of the kinase inhibitor PP137. While substitutions of the other two cavity-lining positions (M314 and V323) do not disrupt hydrogen bonding, the V323L mutation, which is associated with resistance to kinase inhibitors including bosutinib17, does weaken bosutinib binding considerably and constitutes an outlying data point in Fig. 2e.
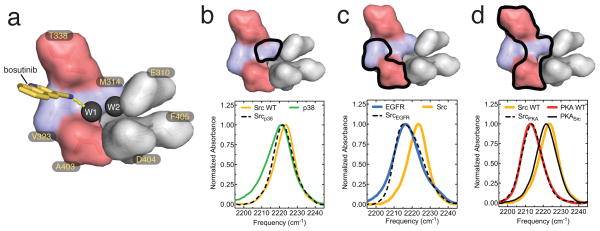
Figure 3. The probe environment is determined exclusively by the residues that comprise the water-filled cavity.
(a) Surface representation of the amino acids that line the water-filled cavity. T338 and A403 are colored red, the other two positions mutated in this study are colored blue, and conserved catalytic residues are shown in gray. The two water molecules are shown as black spheres, the quinoline-3-carbonitrile moiety of bosutinib is shown as yellow sticks, and the hydrogen bond between the nitrile group of the drug and W1 is shown as a dashed yellow line. (b–d) IR spectra of bosutinib bound to WT Src (yellow) and p38 (b, green), EGFR (c, blue), PKA (d, red) and Src mutants bearing mutations that convert the cavity residues to those of the other kinases (dotted black lines). The IR spectrum of bosutinib bound to a triple mutant of PKA, in which the cavity residues are converted to those found in Src, is shown as a solid black line in d. Surface representations of the ATP-binding site, in which the positions of relevant substitutions are highlighted by thick black lines, are shown above the IR spectra.
The gatekeeper’s control over the hydrogen bond network is remarkably stringent, with only threonine and serine permitting bosutinib access, and even valine not being tolerated (in contrast, of five amino acids tested at the A403 position, only threonine blocked the hydrogen bond). Rather than merely occupying the cavity and perturbing W1, our structure of the Src T338M/M314L mutant shows that bulky gatekeeper substitutions can block access by inducing changes in bosutinib’s binding mode. The structure of bosutinib bound to WT Src suggests that a similar mechanism may apply even to the conservative threonine to valine substitution. In this structure the sidechain of T338 is tightly wedged between the nitrile group of the drug and the backbone carbonyl of the next residue (E339 in the kinase hinge region), with which it forms a hydrogen bond, and the T338V substitution would consequently create a steric clash with the carbonyl. Presumably a significant conformational change results, and is propagated to the neighboring hydrogen bond network (Supplementary Fig. 5).
Effects of sequence variation across divergent kinases
The mutational analysis above was limited to the residues lining the water-filled cavity, but when bosutinib is bound to divergent kinases more distant substitutions might also influence the hydrogen bond network. To test this, we measured the IR spectra of bosutinib bound to divergent kinases from three different families (p38α, EGFR and PKA), each of which differs from Src at one or more of the cavity-lining positions. We also measured IR spectra of the drug bound to chimeric Src proteins in which the amino acids that line the water-filled cavity were mutated to match those of the other kinases. Strikingly, in each case, the nitrile probe reports an almost identical environment in the chimera and in the kinase it is intended to mimic (Fig. 3b–d). Among the kinases tested, PKA exhibits the greatest difference in the probe frequency compared to Src: an 11 cm−1 red-shift, indicating that the probe’s hydrogen bond to W1 is lost in PKA. Although the proteins share only ~25% sequence identity, three substitutions in Src are sufficient to block hydrogen bonding and perfectly reproduce the chemical environment seen in PKA, as probed by the nitrile (Fig. 3d). Conversely, a PKA mutant bearing the Src cavity-lining residues restores the hydrogen bond and reproduces the chemical environment seen in Src (Fig. 3d). We conclude that the environment sensed by the probe is almost completely determined by the residues that line the water-filled cavity, and that these residues are therefore generally predictive of the status of the hydrogen bond network, even in highly divergent kinases.
Rationalizing bosutinib’s selectivity profile
The above results indicate that only kinases possessing a threonine or serine gatekeeper and lacking the A403T substitution are compatible with bosutinib’s participation in the hydrogen bond network. Strikingly, this observation accounts quite well for the selectivity of bosutinib across the human kinome, as most kinases that meet these criteria (referred to here as compatible kinases) bind bosutinib well, and most incompatible kinases do not (Fig. 4a). Note that the lack of bosutinib activity towards the TKL kinases is explained by the prevalence of the V323L substitution in this family, which, as noted above, interferes with bosutinib binding without compromising the hydrogen bond network.
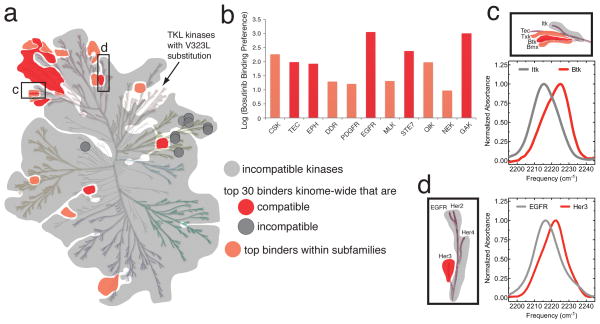
Figure 4. The impact of the water-mediated hydrogen bond network underlies the selectivity profile of bosutinib.
(a) Representation of the phylogenetic tree of the human kinome in which kinases with ATP-binding sites that are not compatible with bosutinib’s participation in the hydrogen bond network are shaded gray (referred to here as incompatible kinases, in contrast to kinases with compatible ATP-binding sites). Compatible kinases that are also in the top 30 bosutinib binders reported in reference 16 are shown in red, and compatible kinases that at least bind considerably better than their incompatible family members, are shown in light red. The dark gray circles represent incompatible kinases that nonetheless bind bosutinib tightly. (b) Bosutinib’s preference for compatible kinases within 11 kinase subfamilies is shown as the log of the ratio of the binding constants of compatible and incompatible family members (see Online Methods). (c) Enlarged view of the phylogenetic tree showing the TEC family, with IR spectra of bosutinib bound to Btk and Itk. The same coloring scheme is used as in a. (d) Enlarged view of the phylogenetic tree showing the EGFR family, with IR spectra of bosutinib bound to EGFR and Her3. The same coloring scheme is used as in a. The phylogenetic tree of the human kinome is reproduced with permission from Cell Signaling Technologies.
The impact of the hydrogen bond network is even more apparent when considering the selectivity of bosutinib within kinase subfamilies (Fig. 4a,b, Supplementary Fig. 6). Using the comprehensive bosutinib binding data from reference 16, we found that, almost without exception, across 11 different kinase subfamilies those family members possessing compatible ATP-binding sites bind better than their incompatible family members by factors ranging from 10–1000 (Fig. 4b).
To confirm that the hydrogen bond network is responsible for this subfamily selectivity we verified the hydrogen bond status in compatible and incompatible members of two kinase subfamilies, the TEC-family and the EGFR family. The TEC-family tyrosine kinases mostly possess threonine gatekeepers, and are targeted by bosutinib, with the notable exception of Itk, which has a phenylalanine gatekeeper. IR spectra of bosutinib bound to purified Btk and Itk confirm that the drug is engaged in the hydrogen bond network in Btk, but not in Itk (Fig. 4c). The EGFR family kinases all have threonine gatekeepers, but Her3 is one of the tightest bosutinib binders detected in ref 16, while the other EGFR family members bind bosutinib more weakly. This is accounted for by the fact that Her3 has an alanine residue at the position equivalent to A403 in Src, while the other three EGFR family kinases instead have a threonine at this position. IR spectra of bosutinib bound to purified Her3 and EGFR show that the nitrile is indeed hydrogen bonded in Her3 but not in EGFR (Fig. 4d), confirming that the A403 position also plays a role in bosutinib’s kinase selectivity (76 kinases possess a threonine at this position).
Interestingly, the disruption of the hydrogen bond network by the conservative Thr to Val gatekeeper substitution is also reflected in the selectivity profile, as bosutinib targets most Ephrin receptors (which have threonine gatekeepers), but not EphA6 and EphA7, which have valine and isoleucine gatekeepers, respectively (Supplementary Fig. 6). Bosutinib is also selective for the threonine-gatekeeper platelet derived growth factor receptor family (with the exception of Flt3, which has a phenylalanine gatekeeper) compared to the closely related FGFR and VEGFR families, which have valine gatekeepers (Supplementary Fig. 6).
These observations underscore the importance of the gatekeeper for bosutinib selectivity, with the A403 position playing a significant but secondary role. Although the importance of the gatekeeper has long been recognized, the steric mechanism usually invoked is unsatisfactory for the large number of inhibitors that, like bosutinib, leave a large cavity next to the gatekeeper. We have shown that the gatekeeper can nonetheless exert influence in such cases by controlling an inhibitor’s access to structured water molecules occupying the cavity. It is tempting to speculate that this mechanism applies generally to inhibitors that engage these conserved water molecules, but it is important to note that the precise constraints imposed on the ATP-binding site likely differ for each inhibitor. This is highlighted by contrasting results obtained using an isomer of bosutinib that differs only in the positions of the three substituents on the aniline ring. Authentic bosutinib and the bosutinib isomer have similar binding modes18, and their nitrile groups can both participate in the hydrogen bond network. However, we were surprised to find that the valine and isoleucine gatekeeper substitutions do not disrupt the hydrogen bond to W1 in the case of the bosutinib isomer (Supplementary Fig. 7a). This is supported by the x-ray structure of the isomer bound to the isoleucine gatekeeper kinase LOK (pdb code 4BC6), in which the hydrogen bond to W1 is observed (Supplementary Fig. 7b). The T338I substitution consequently has a much more severe effect on binding for authentic bosutinib than for the isomer (Supplementary Fig. 7c), underscoring that the water-mediated hydrogen bond network confers distinct selectivity properties on these similar molecules, and that the isomer may retain some activity against the bosutinib-resistant T315I gatekeeper mutation of BCR-Abl17.
DISCUSSION
In the light of our results with bosutinib it is interesting to consider the behavior of the chemically related 4-anilinoquinazoline inhibitors. While bosutinib does not target EGFR, which we attribute to the presence of the A403T substitution, the 4-anilinoquinazolines in fact include several high affinity inhibitors of EGFR. The carbonitrile group found in bosutinib is replaced by a heterocyclic nitrogen atom in these inhibitors (referred to as N3), and when they are bound to EGFR an ordered water molecule (which we refer to as W3 to distinguish it from W1 and W2) forms a hydrogen bond network that bridges the N3 atom of the inhibitor and the threonine gatekeeper38. Although W3 occupies a very similar position to the bosutinib nitrile, being less constrained it is nonetheless capable of forming a direct hydrogen bond to the threonine sidechain equivalent to A403T, thereby facilitating the targeting of EGFR by 4-anilinoquinazolines39,40 (Supplementary Fig. 8). The structure of the 4-anilinoquinazoline inhibitor saracatinib bound to Src shows that these compounds can also employ W3 as a bridge to W1, in cases where the latter is present41.
The structure of the Src A403T mutant indicated that the threonine sidechain directly interferes with the binding site for W1. Interestingly, our analysis of the protein databank uncovered several examples where an inhibitor forms a direct hydrogen bond to W1 even in the presence of a threonine at this position. In these cases, W1 shifts over slightly, simultaneously accommodating the threonine sidechain and forming a hydrogen bond to the inhibitor (Supplementary Fig. 9). Such a shift is not possible in the case of bosutinib as it would cause a clash with the aniline ring. We speculate that inhibitors that exploit this shifted positioning of W1 may be selective for kinases possessing the A403T substitution.
While structured water molecules are commonly observed participating in hydrogen bonding interactions in ligand/receptor interfaces42, the energetic significance of these interactions and their importance for selectivity have been difficult to establish. Our work highlights that the optimal engagement of such interactions depends in an unpredictable fashion on many interconnected factors, including the identity of nearby amino acids, and the chemical structure and binding mode of the ligand. The approach we have taken of exploiting intrinsic IR probes on the ligand is highly effective at dissecting these factors. In this context we note that the majority of type I kinase inhibitors that engage the conserved waters in the kinase active site use a carbonyl moiety to do so, and that our group has recently shown that with careful background subtraction specific carbonyl vibrations can be readily detected in proteins43. A key result of our work is the demonstration that these water-mediated interactions can make a critical contribution to an inhibitor’s ability to distinguish between receptor subtypes, suggesting that greater attention to structured water molecules in drug design may yield inhibitors with improved or altered selectivity properties.
Online Methods
Expression and purification of Src WT and mutants
WT and mutant Src kinase domain constructs (residues 251-533, chicken c-Src numbering, with an N-terminal hexahistidine tag) were coexpressed in E.coli BL21 DE3 with YOP phosphatase at 18 °C overnight44. Cell pellets were resuspended in Ni buffer A (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 35 mM imidazole) lysed with a cell disrupter, and lysates subjected to centrifugation at 15000 rpm for 30 minutes. Cleared lysates were loaded on Ni NTA columns (GE healthcare), the columns were washed with Ni buffer A, and samples eluted with Ni buffer B (50 mM Tris-HCl pH 8.0, 500 mM imidazole). For protein intended for crystallization the samples were digested overnight with TurboTev protease (Eton Bioscience), in order to remove the N-terminal hexahistidine tags, whereas the samples for IR and fluorescence proceeded directly to the anion exchange step. Samples were desalted into Q buffer A (50 mM Tris-HCl pH 8.0, 10% glycerol, 1mM DTT), loaded on HiTrap Q anion exchange columns (GE Healthcare) and eluted with a 20 column volume gradient from 0% to 100% Q buffer B (50 mM Tris-HCl pH 8.0, 1M NaCl, 10% glycerol, 1mM DTT). The identity of each mutant was confirmed by DNA sequencing combined with mass spectrometry of the purified proteins, and the catalytic activity of most mutants was confirmed with kinase assays (Supplementary Fig. 10).
In addition to the 30 single ATP-binding site mutants, the following compound mutants were prepared. The T338M gatekeeper mutation was combined with M314L (tandem substitutions found in 101 human kinases), V323T (18 kinases), A403C/S/T (16/27/34 kinases), A403T+V323T (4 kinases), and M314L+A403T (20 kinases). The T338M M314L A403T triple mutant served as a mimic of PKA, and the V323C A403T double mutant was made as a mimic of EGFR. The single mutations M314L and T338F served as the mimics of p38 and Itk, respectively.
Preparation of other kinases
Murine Btk and Itk (residues 394-657 and 356-619, respectively, fused to a C-terminal hexahistidine tag) were expressed in ArcticExpress cells at 12 °C O/N and purified as described45. Murine PKA WT and mutants (residues 1-351 fused to an N-terminal hexahistidine tag) were expressed in BL21 (DE3) at 18 °C O/N and purified by Ni-affinity chromatography and cation exchange chromatography as described46. Purified p38 was obtained from the QB3 MacroLab at UC Berkeley. Purified Her3 kinase domain was a gift from Natalia Jura and Peter Littlefield and purified EGFR was a gift from Kate Engel and John Kuriyan.
Kinase assays
The kinase activity of selected Src mutants was measured to verify that the proteins are active, and to assess the effects of mutations on inhibition by bosutinib (Supplementary Fig. 10). Measurements were performed using a coupled kinase assay as described18, with 100 mM Tris-HCl pH 8.0, 1 mM phosphoenolpyruvate (Sigma), 0.6 mg/ml reduced NADH (Sigma), 112.5/157.5 units Pyruvate Kinase/Lactate Dehydrogenase (Sigma), 500 μM ATP, 10 mM MgCl2, 1 mM DTT, 0.6 mg/ml polyGlu,Tyr substrate peptide, 100 or 200 nM kinase and inhibitor in DMSO to a final DMSO concentration of 5%. The absorbance of NADH at 340 nm was monitored as a function of time using an absorbance plate reader. Under these conditions the rates were approximately constant for the first five minutes of the reactions, and were determined from a linear fit of the time-dependent absorbance.
Crystallography
For Src kinases, a molar excess of bosutinib (Tocris Bioscience) made up in DMSO was added to the protein samples to a final concentration of ~100 μM and the protein samples were concentrated to 1 mM, diluted 10 fold with crystallization buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol, 2 mM DTT) and reconcentrated to 10–15 mg/ml.
Crystals were obtained in hanging drops using 0–4% PEG 3350, 0.2M Ammonium Acetate, 0.1 M Hepes pH 7.5 in the mother liquor. Crystals were cryoprotected in mother liquor plus 35% glycerol and flash frozen in liquid nitrogen. X-ray diffraction data were collected at beamline 11-1 at the Stanford Synchrotron Radiation Lightsource.
Data processing was performed with mosflm47 and scalepack48, and refinement with Phenix49. In the case of WT Src and the T338M/M314L mutant, all datasets collected on single crystals were incomplete, and data collected from many different crystals were studied to identify pairs that could be merged to obtain complete sets of structure factor amplitudes. In the case of the T338M/M314L mutant this led to somewhat higher Rmerge values in the lowest resolution shell. Nonetheless, the electron density map was of excellent quality and the rotation of the bosutinib aniline ring was readily apparent in both molecules in the asymmetric unit prior to performing refinement.
The structure of WT Src was solved by molecular replacement in Phenix using a structure of Src bound to a type II inhibitor50 (pdb code 3G6G), from which the ligand was removed, as a search model. The activation loop was subsequently rebuilt in the active conformation in Coot51 prior to refinement. The final model of WT Src bound to bosutinib was used as the starting point for refinement of the A403T and T338M M314L mutants, with the same test sets used for all three structures.
The nitrile-W1 hydrogen bond length in the molecular model refined with default geometry restraints is 3.1 Å. This is slightly longer than typical nitrile-water hydrogen bonds (the average length is 2.94 Å for hydrogen bonds between a nitrile acceptor and a water OH donor observed in high resolution small-molecule crystal structures in the Cambridge Structural Database52). However, the electron density map showed evidence of a significant puckering of the quinoline-3-carbonitrile moiety of bosutinib, with the nitrile group tilted out of plane from the quinoline group towards W1 (Supplementary Fig. 11). We attempted to account for this by relaxing the geometry restraints for the quinoline and nitrile groups to varying extents prior to performing refinement. The best fit to the electron density was obtained by restraining the quinoline-3-carbonitrile moiety into three separate planes, comprising the benzene ring, the pyridine ring, and the nitrile group. This results in kinks between each group of between 5–10°. Such small out of plane distortions of aromatic systems are frequently observed and are thought to incur minimal energetic penalties53. In the model refined with these relaxed restraints the distance between the nitrile nitrogen atom and the W1 oxygen atom shortens to 2.9–3.0 Å (Supplementary Fig. 11), demonstrating that the bosutinib-water hydrogen bond length is, in fact, typical of this chemical class of hydrogen bond.
IR spectroscopy
Bosutinib (Tocris Bioscience) in DMSO was added to ~50 μM protein samples in IR buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 2mM DTT) to a final concentration of 100 μM bosutinib and 5% DMSO, and the samples concentrated to ~1 mM. Samples were loaded into a demountable IR cell containing sapphire windows separated by offset 75 and 100 μm Teflon spacers to avoid interference fringes in the spectra. IR Spectra were recorded on a Vertex 70 FTIR spectrometer (Bruker) equipped with a liquid nitrogen cooled indium antimonide detector and a 2000–2500 cm−1 bandpass filter (Spectrogon), using 2 cm−1 resolution, and averaged across 256 scans. Spectra were background-subtracted using the buffer flow-through from the final sample concentration step and baselined using the polynomial method in the Opus software (Bruker). For a 1 mM sample, the bosutinib nitrile stretching band had an absorbance value of ~1.5 mAU, corresponding to an extinction coefficient of ~180 M−1/cm−1, whereas the noise in the baseline was typically ~0.01 mAU. For each sample 3 spectra were recorded and baselined separately, and the peak positions calculated in Opus. The standard deviation of the peak positions for these replicates was never greater than 0.2 cm−1.
Fluorescence binding assay
Binding measurements were performed as reported earlier, with minor modifications18. A dilution series of bosutinib was prepared in DMSO and added stepwise to 5 nM protein samples in fluorescence buffer (20 mM Tris pH 8.0, 2 mM DTT), to a maximum final DMSO concentration of 6%. Fluorescence emission spectra were recorded on a Perkin Elmer LS 55 spectrometer with excitation at 280 nm. Binding curves were prepared by plotting either the decrease in fluorescence emission at 340 nm or the increase in emission at 480 nm, as a function of the bosutinib concentration, with both methods giving very similar results. The 340 nm emission data were corrected for the effect of DMSO on the protein fluorescence, whereas the 480 nm data were corrected for the background fluorescence of free bosutinib. Dissociation constants were determined by fitting of the binding curves to the analytical solution to the one-to-one binding equation in Mathematica (Wolfram Research), which accounts for the effects of ligand depletion, allowing accurate determination of binding constants with values as small as 0.5 nM. When necessary to facilitate comparison the fluorescence binding curves were normalized to a maximum fluorescence of 1.
Analysis of type I kinase inhibitor structures in the Protein Databank
The full protein databank was searched for structures containing the “XDFGX” motif, where the backbone torsion angles φ/ψ of those five amino acids where constrained to −150/−180, 60/70, −110/0, −60/−60, and −110/0 degrees (these values are derived from our structure of WT Src bound to bosutinib). From multiple searches, a +/− 30 degree tolerance was found to be optimal, in that it gave the largest number of hits without including a significant number of non-kinase structures. Nucleotide complexes were excluded by adding an adenine ring to the search criteria, and 146 apo structures were manually excluded. Hydrogen bond lengths were determined using PyMOL. The full set of structures possessing a ligand/W1 hydrogen bond is given in a Supplementary Data Set.
Analysis of bosutinib selectivity within kinase subfamilies
The data plotted in Fig. 4b was obtained in the following manner. For each member of a given kinase subfamily with a “compatible” ATP-binding site we determined the ratio of its binding constant and the average binding constant for all incompatible members of the subfamily (i.e. kinases on the same branch of the kinome) using data from reference 16. For subfamilies possessing more than one compatible kinase the average value of these ratios is plotted. Specifically, the kinases included were: 1) CSK versus CTK, 2) BTK, TEC, TXK and BMX versus ITK, 3) EPHA2-5, EPHA8, EPHB1-4 versus EPHA6 (while EPHA7 is also incompatible its binding was too weak to be measurable), 4) DDR1-2 versus TRKA-C, 5) PDGFRa-b, CSF1R, KIT versus FLT3, TIE1-2, 6) HER3 versus EGFR, HER2, HER4, 7) ZAK versus LZK, DLK, MLK1,MLK3, 8) MEK5 versus MEK1-6, MKK7, 9) SIK2, SIK and QSK versus SNARK, ARK5, MARK1-4, AMPK-alpha1, 10) NEK11 versus NEK1-9, 11) GAK versus BMP2K, AAK1.
Supplementary Material
Acknowledgments
We would like to thank Natalia Jura and Peter Littlefield for providing purified Her3, and John Kuriyan and Kate Engel for providing purified EGFR. We also thank Natalia Jura and Kevan Shokat for critical reading of the manuscript, and Aina Cohen for help with x-ray crystallography. This work is supported by a K99/R00 Pathway to Independence Award (1K99GM102288-01) to NML, and a long-standing grant from the NIH (GM27738) to SGB.
Footnotes
Author contributions
N.M.L. designed and performed experiments, and wrote the manuscript. S.G.B. designed the experiments and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Accession codes
PDB: The coordinates and structure factors for the structures of bosutinib bound to WT Src, Src A403T and Src T338M/M314L have been deposited in the protein databank under accession codes 4MXO, 4MXX and 4MXZ, respectively.
References
- 1.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762298/5600/1912. [pii] [DOI] [PubMed] [Google Scholar]
- 4.Wong S, et al. Sole BCR-ABL inhibition is insufficient to eliminate all myeloproliferative disorder cell populations. Proc Natl Acad Sci U S A. 2004;101:17456–17461. doi: 10.1073/pnas.0407061101. 0407061101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461347/7/472. [pii] [DOI] [PubMed] [Google Scholar]
- 6.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. S1074-5521(05)00123-7 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. nbt1068 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. nchembio799 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. 8823 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Cortes JE, et al. Safety and efficacy of bosutinib (SKI-606) in patients (pts) with chronic phase (CP) chronic myeloid leukemia (CML) following resistance or intolerance to imatinib (IM) J Clin Oncol. 2010;28:487. doi: 10.1182/blood-2011-05-355594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campone M, et al. Phase II study of single-agent bosutinib, a Src/Abl tyrosine kinase inhibitor, in patients with locally advanced or metastatic breast cancer pretreated with chemotherapy. Ann Oncol. 2012;23:610–617. doi: 10.1093/annonc/mdr261. mdr261 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Blencke S, et al. Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem Biol. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. S1074552104001164 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, et al. Structural basis for selective inhibition of Src family kinases by PP1. Chem Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. cm6911 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. 1062538 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 16.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. nbt.1990 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Redaelli S, et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J Clin Oncol. 2009;27:469–471. doi: 10.1200/JCO.2008.19.8853. JCO.2008.19.8853 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Levinson NM, Boxer SG. Structural and spectroscopic analysis of the kinase inhibitor bosutinib and an isomer of bosutinib binding to the Abl tyrosine kinase domain. PLoS One. 2012;7:e29828. doi: 10.1371/journal.pone.0029828. PONE-D-11-17300 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan Y, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci U S A. 2009;106:139–144. doi: 10.1073/pnas.0811223106. 0811223106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 22.Lowe ED, et al. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 1997;16:6646–6658. doi: 10.1093/emboj/16.22.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madhusudan Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002;9:273–277. doi: 10.1038/nsb780. nsb780 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. S0092867402007419 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Golovin A, Henrick K. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics. 2008;9:312. doi: 10.1186/1471-2105-9-312. 1471-2105-9-312 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijer L, et al. Inhibition of cyclin-dependent kinases, GSK-3beta and CK1 by hymenialdisine, a marine sponge constituent. Chem Biol. 2000;7:51–63. doi: 10.1016/s1074-5521(00)00063-6. S1074-5521(00)00063-6 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Kothe M, et al. Selectivity-determining residues in Plk1. Chem Biol Drug Des. 2007;70:540–546. doi: 10.1111/j.1747-0285.2007.00594.x. JPP594 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Hughes S, et al. Crystal structure of human CDC7 kinase in complex with its activator DBF4. Nat Struct Mol Biol. 2012;19:1101–1107. doi: 10.1038/nsmb.2404. nsmb.2404 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Davies TG, et al. Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor. Nat Struct Biol. 2002;9:745–749. doi: 10.1038/nsb842. nsb842 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Tahtouh T, et al. Selectivity, cocrystal structures, and neuroprotective properties of leucettines, a family of protein kinase inhibitors derived from the marine sponge alkaloid leucettamine B. J Med Chem. 2012;55:9312–9330. doi: 10.1021/jm301034u. [DOI] [PubMed] [Google Scholar]
- 31.Liddle J, et al. Discovery of GSK143, a highly potent, selective and orally efficacious spleen tyrosine kinase inhibitor. Bioorg Med Chem Lett. 2011;21:6188–6194. doi: 10.1016/j.bmcl.2011.07.082. S0960-894X(11)01028-6 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka D, Nakasako M. Probability distributions of hydration water molecules around polar protein atoms obtained by a database analysis. J Phys Chem B. 2009;113:11274–11292. doi: 10.1021/jp902459n. [DOI] [PubMed] [Google Scholar]
- 33.Atwell S, et al. A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase. J Biol Chem. 2004;279:55827–55832. doi: 10.1074/jbc.M409792200. M409792200 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Ladbury JE. Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem Biol. 1996;3:973–980. doi: 10.1016/s1074-5521(96)90164-7. [DOI] [PubMed] [Google Scholar]
- 35.Reimers JR, Hall LE. The solvation of acetonitrile. Journal of the American Chemical Society. 1999;121:3730–3744. [Google Scholar]
- 36.Choi JH, Oh KI, Lee H, Lee C, Cho M. Nitrile and thiocyanate IR probes: Quantum chemistry calculation studies and multivariate least-square fitting analysis. J Chem Phys. 2008;128:Artn 134506. doi: 10.1063/1.2844787. [DOI] [PubMed] [Google Scholar]
- 37.Weiss EL, Bishop AC, Shokat KM, Drubin DG. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- 38.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. M207135200 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Gajiwala KS, et al. Insights into the aberrant activity of mutant EGFR kinase domain and drug recognition. Structure. 2013;21:209–219. doi: 10.1016/j.str.2012.11.014. S0969-2126(12)00429-7 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Park JH, Liu Y, Lemmon MA, Radhakrishnan R. Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem J. 2012;448:417–423. doi: 10.1042/BJ20121513. BJ20121513 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hennequin LF, et al. N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J Med Chem. 2006;49:6465–6488. doi: 10.1021/jm060434q. [DOI] [PubMed] [Google Scholar]
- 42.Poornima CS, Dean PM. Hydration in drug design. 1. Multiple hydrogen-bonding features of water molecules in mediating protein-ligand interactions. J Comput Aided Mol Des. 1995;9:500–512. doi: 10.1007/BF00124321. [DOI] [PubMed] [Google Scholar]
- 43.Fried SD, Bagchi S, Boxer SG. Measuring electrostatic fields in both hydrogen-bonding and non-hydrogen-bonding environments using carbonyl vibrational probes. J Am Chem Soc. 2013;135:11181–11192. doi: 10.1021/ja403917z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seeliger MA, et al. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. ps.051750905 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph RE, Andreotti AH. Bacterial expression and purification of interleukin-2 tyrosine kinase: single step separation of the chaperonin impurity. Protein Expr Purif. 2008;60:194–197. doi: 10.1016/j.pep.2008.04.001. S1046-5928(08)00099-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steichen JM, et al. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J Biol Chem. 2012;287:14672–14680. doi: 10.1074/jbc.M111.335091. M111.335091 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 48.CCP4. The CCP4 Suite: programs for protein crystallography. Acta CrystD. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. S0907444909052925 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeliger MA, et al. Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. 0008-5472.CAN-08-3953 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. S0907444910007493 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steiner T. The hydrogen bond in the solid state. Angew Chem Int Ed Engl. 2002;41:49–76. doi: 10.1002/1521-3773(20020104)41:1<48::AID-ANIE48>3.0.CO;2-U. [pii] [DOI] [PubMed] [Google Scholar]
- 53.Bachrach SM. DFT study of [2.2]-, [3.3]-, and [4.4]paracyclophanes: strain energy, conformations, and rotational barriers. J Phys Chem A. 2011;115:2396–2401. doi: 10.1021/jp111523u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.