Abstract
Elderly humans show decreased humoral immunity to pathogens and vaccines, yet the effects of aging on B cells are not fully known. Chronic viral infection by cytomegalovirus (CMV) is implicated as a driver of clonal T cell proliferations in some aging humans, but whether CMV or Epstein-Barr virus (EBV) infection contributes to alterations in the B cell repertoire with age is unclear. We have used high-throughput DNA sequencing of immunoglobulin heavy chain (IGH) gene rearrangements to study the B cell receptor repertoires over two successive years in 27 individuals ranging in age from 20 to 89 years. Some features of the B cell repertoire remain stable with age, but elderly subjects show increased numbers of B cells with long CDR3 regions, a trend toward accumulation of more highly mutated IgM and IgG immunoglobulin genes, and persistent clonal B cell populations in the blood. Seropositivity for CMV or EBV infection alters B cell repertoires, regardless of the individual's age: EBV infection correlates with the presence of persistent clonal B cell expansions, while CMV infection correlates with the proportion of highly mutated antibody genes. These findings isolate effects of aging from those of chronic viral infection on B cell repertoires, and provide a baseline for understanding human B cell responses to vaccination or infectious stimuli.
Introduction
Many elderly individuals have a compromised immune system, leading to increased susceptibility to infectious diseases and decreased responses to vaccination (1). Aging has been reported to impair innate immunity, T cells and antibody-producing B cells (1-5). Humoral responses are critical for responding to pathogens such as Streptococcus pneumoniae and influenza viruses that cause increased morbidity and mortality in the elderly, but age-related changes in human B cells and immunoglobulin repertoires are only beginning to be understood (6-8).
Advanced age has been reported to lead to increased or decreased B cell counts in the peripheral blood, increased, decreased or unchanged proportions of naïve B cells, and increased CD5+ B cell populations (3, 5, 9-13). Changes in serum antibody production, including decreases in vaccine-specific antibodies, and isotype switching associated with lower expression of activation-induced cytidine deaminase (AID) in B cells have also been described (8, 10, 14). Understanding the effects of aging on B cell function is further complicated by the common chronic viral infections seen at higher rates in the aging population, such as cytomegalovirus (CMV) and Epstein-Barr virus (EBV). CMV infection is correlated with increased counts of LFA-1hi CD8+ memory T cells and reduced naïve CD8+ T cells, while total B cell counts in the blood are reportedly increased in CMV-seropositive individuals (15-17).
Following V(D)J rearrangement to generate functional immunoglobulin (Ig) genes in B cells, the Ig repertoire during a human's life span is further shaped by negative selection against self-antigens, clonal expansion of B cells stimulated by antigen, activation-induced mutation of immunoglobulin genes, and receptor editing, among other processes. Ineffective antibody responses in the elderly have been attributed to decreased diversity of antibody repertoires with accumulation of memory B cells and decrease of naïve B cell populations (18). Influenza vaccination responses in the elderly are associated with decreased numbers of vaccine-stimulated B cells (8), and a recent study that included 4 elderly subjects show decreased diversity of influenza vaccine-stimulated B cells (19). However, there is also evidence of relatively preserved Ig repertoire diversity in tonsillar tissue of aged humans, and increased proportions of naïve B cells in some elderly individuals (20). Mutation of IGHV in B cell populations reportedly changes with aging, with one study reporting modestly increased mutation in IgG but not memory IgM B cell populations in the blood, while data from tonsillar B cells indicate increased mutation in memory IgM B cells but not other subsets (20, 21). Most prior studies of IGH gene rearrangements in young versus elderly subjects have been limited to examination of tens to hundreds of sequences, from small numbers of individuals, and have not assessed the potentially confounding effects of chronic herpesvirus infections (20-23). Seropositivity for CMV, in particular, increases with age in human populations, and should be controlled for in studies of the effects of aging on the immune system (24).
Here, we characterize peripheral blood IGH repertoires measured with over 500,000 sequences from a cohort of healthy young (n=10) and older (n=17) people over two consecutive years, and analyze features that change with age, CMV or EBV infection. Some B cell repertoire features are stable with age, but we find that elderly individuals show increased numbers of B cells expressing long IGH CDR3 regions, and that the proportion of highly mutated B cells, particularly in IgM and IgG populations, shows a trend toward increasing with age, and is increased in subjects infected with CMV. Unusual large persistent clonal populations of B cells are common in the oldest individuals in our data set, and are absent from younger individuals; notably, the contribution of both large and small persistent B cell clones over the year-long time course is correlated with EBV seropositivity, regardless of age. Taken together, these findings isolate age-specific and CMV or EBV-associated alterations in the B cell repertoire, and provide a baseline for further study of impaired antigen-specific responses and increased autoreactivity in the elderly.
Materials and Methods
Specimen collection
Human peripheral blood was collected from a cohort of 27 healthy participants aged 20 to 89 years during each of 2 consecutive years: Y2 (2008) and Y3 (2009). Participant ID, age and other demographic information is described in Supplementary Table I. The participants were grouped into 3 age categories: 20-31 years (n=10), 61-69 years (n=7) and 72-89 years (n=10). Recruitment of patients, documentation of informed consent, collection of blood specimens, and experimental measurements were carried out with Institutional Review Board approval at Stanford University.
CMV and EBV serotyping
Serum was separated by centrifugation of clotted blood, and stored at -80°C before use. After all samples were collected, specimens were thawed at room temperature and assayed for the presence of CMV and EBV antibodies (both IgG and IgM) using the CMV or EBV ELISA Kit from Calbiotech (Spring Valley, CA), as recommended by the manufacturer. No specimens were positive for virus-specific IgM.
Genomic DNA (gDNA) and complementary DNA (cDNA) template preparation and PCR amplification
Isolation of peripheral blood mononuclear cells and isolation of gDNA and mRNA were performed as previously described (22). PCR of IGH variable region rearrangements from each sample was carried out with 6 independent 100ng gDNA aliquots to generate 6 independent barcoded libraries per sample. Multiplexed primer sets hybridizing to the FR1 or FR2 framework regions, and a J-primer were based on the BIOMED-2 design (25). For isotype-specific Ig libraries, mRNA was reverse-transcribed to cDNA using random hexamer primers and cDNA corresponding to 100ng mRNA was used for PCR using the same 5′- FR1 and FR2 multiplex primers and 3′- isotype specific primers located in the first exon of the constant region. Sample identity and replicate library identity were encoded by 10-nucleotide ‘barcode’ sequences in the primers. PCR was carried out with AmpliTaq Gold (Roche) following the manufacturer's instructions, and used a program of: 94°C 5 min, 35 cycles of (94°C 30 sec, 60°C 45 sec, 72°C 90 sec), and final extension at 72°C for 10 min. PCR products from each replicate library were quantitated, pooled in equimolar amounts, and then gel-purified and gel extracted (Qiagen). High-throughput sequencing was performed on the 454 (Roche) platform using Titanium chemistry.
Sequence quality assessment and filtering
Sequences with matching barcodes were assigned to the appropriate replicates and samples and then were trimmed of bar codes and IGHV primer sequences. The V, D and J regions and V-D (N1), D-J (N2) junctions were identified by alignment with germline IGH sequences from IMGT and NCBI databases, using the alignment program iHMMune-align (26, 27). Amino acid sequences and location of CDR3 were defined by the conserved cysteine-104 and tryptophan-118 based on the IMGT numbering system (28). Sequences were filtered to remove non-IGH artifacts, sequences with V-gene insertion or deletions, and chimeric sequences. Samples with fewer than 100 sequences were excluded from further analysis (two samples from Y3). A total of 323,285 gDNA and 189,915 cDNA sequences were analyzed. The sequences have been deposited at the NCBI dbGAP online archive (http://www.ncbi.nlm.nih.gov/gap) with accession number phs000666.v1.p1.
V, D and J usage, junctional features and V-mutation analysis
Sequences from all replicates of each sample were pooled, and unique clones were defined by the same V and J gene usage and identical CDR3 amino acid sequence. To analyze V, D and J usage frequencies, CDR3 features, and mutations in V, reads belonging to each unique clone were collapsed to a single read. Hydrophobicity of CDR3 peptides was calculated using Kyte-Dolittle scale (29) and net charge was calculated using the Henderson–Hasselbalch equation (30). CDR3 segments from out-of-frame sequences were excluded from CDR3 feature analysis. Distribution of mutation along the V-region was obtained by alignment with gapped IMGT germline sequences and counting the mutation frequency per nucleotide position.
Clonal expansion analysis
To summarize the contribution of clonally-expanded B cells to the repertoire, while normalizing for sequencing depth obtained for each sample, we calculated a ‘clonality score’, adapted from a coincidence index used in cryptanalysis, sum(fi2), (where fi is the frequency of component i) (31). To compensate for the effect of PCR amplification on sequence counts, we calculated the clonality score as follows: sum of Nij*Nik (j!=k) over i,j,k, divided by the sum of Tj*Tk (j!=k) over j,k, where Nij and Nik are the copy numbers of clone i observed in independent replicate PCR libraries j and k generated from independent aliquots of template DNA; Tj and Tk are total read numbers in the corresponding replicate libraries. To quantitate the contribution of persistent B cell clonal populations detected in the two time points one year apart in the study, we calculated a ‘persistent clonality score’ exactly as above, except that only replicate libraries from different years were compared with each other. Large clones were defined as those observed in more than 4 of the 6 replicate libraries from a sample, and comprising greater than 1% of total reads from the sample.
Correlation and regression analysis for clonality
We tested for correlation between clonality score, persistent clonality score, IGHV segment mutation frequency, age, gender and seropositivity for EBV and CMV, using multiple linear regression, including all subjects in the analysis. Laplace smoothing was applied to avoid occurrence of zeros prior to calculating the logarithm of the clonality score.
Phylogenetic visualization of large clones
Immunitree is a dedicated algorithm and implementation for building immune receptor sequence trees from high-throughput DNA sequencing data (Laserson et al., PLOS Pathogens, accepted). The following briefly describes the conceptual basis for calculations in Immunitree, while additional details of the model-building and computation will be presented elsewhere. A variety of broadly-applied phylogenetic algorithms deal with sequences derived from species that are presumed to have evolved from common ancestors but where the intermediate shared ancestors are no longer present to be observed, and must therefore be inferred. In contrast, Immunitree takes into account the possibility that common ancestor B cells may still be present in the members of a clonal lineage that are observed.
In the Immunitree model of somatic hypermutation (SHM) of immunoglobulin gene rearrangements, potential nucleotides at each position on sequences representing parent and child subclones are modeled using a conditional multinomial distribution. Sequencing error is modeled similarly: the mutations from subclones to reads are also modeled using a conditional multinomial distribution. These distributions are subject to different priors: the SHM prior penalizes indels severely, while the sequencing error prior treats indels more permissively. This reflects the fact that true SHM is under selective pressure to produce in-frame antibodies, whereas 454 sequencing is known to generate homopolymer errors. In the Immunitree model, internal subclones may or may not generate reads; also, internal subclones may or may not have multiple subclones as children. In contrast, existing phylogenetic algorithms generally associate reads only to the leaves of a binary tree. Immunitree thus builds on and extends existing phylogenetic algorithms, implementing the aforementioned fine-tuning designed to account for the complex realities of immunoglobulin repertoire sequencing, and uses a single sampling and optimization approach to compute a lineage that optimizes the overall likelihood. Any specific lineage has a likelihood consisting of the following parameters: SHM rates from parent subclones (nodes) to children subclones, sequencing error of reads corresponding to a given subclone, the best V and J reference segments, the parent subclone of each subclone, and subclone birth and death rates. The parameters are sampled using a Metropolis-Hastings Markov Chain Monte Carlo approach. After running for 5000 iterations, each sampled lineage is examined and subjected to local optimization. The highest-likelihood lineage amongst the optimized sampled lineages is returned.
Results
V, D and J usage varies among individuals, but is comparable between young and older people
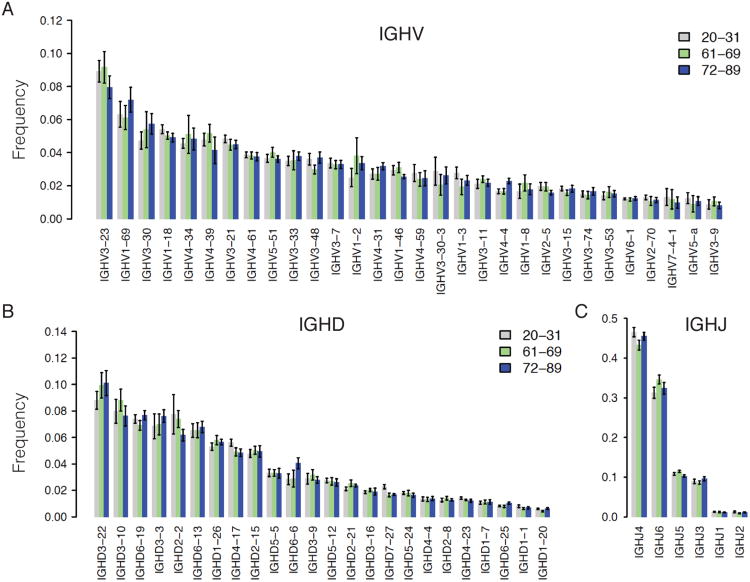
We first examined whether or not there were discernable age-related changes in the usage of V, D, or J gene in IGH rearrangements. Figure 1 shows that the proportion of different genes was comparable across three age groups of samples in 2008 (Y2), with gene frequencies consistent with literature reports (32). V, D and J gene usage from 2009 (Y3) follows a similar pattern and bears a high correlation with that from Y2 (data not shown). V gene segment usage in humans shows a different distribution in mutated versus nonmutated antibody gene rearrangements, so we separated these sequence subsets in our analysis (32). To be conservative in our mutation frequency estimates, given that 454 sequencing can demonstrate error rates of approximately 0.3-1% per base (22), we consider sequences with less than 1% V-mutation frequency as unmutated and those with greater than 1% as mutated. The V, D and J gene usage did not show a significant difference between younger and older groups within either mutated and unmutated categories. Variation between individuals in gene usage patterns was seen, similar to that previously reported (33), consistent with individual differences in the intrinsic mechanisms which generate VDJ rearrangements, including copy-number variation in IGH germline sequences, or RSS sequence features, or individual variation in the B cell selection process.
Figure 1. V, D, J usage in young and older people is comparable.
The panels show the frequency of major V genes (higher than 1%) (A), D genes (B), and J genes (C). The color-coded bars represent the sample average within different age groups, 20-31 years (gray, n=10), 61-69 years (green, n=7), 72-89 years (blue, n=10) from Y2. Error bar is standard error. Pairwise comparison was performed with two-sided student t-test.
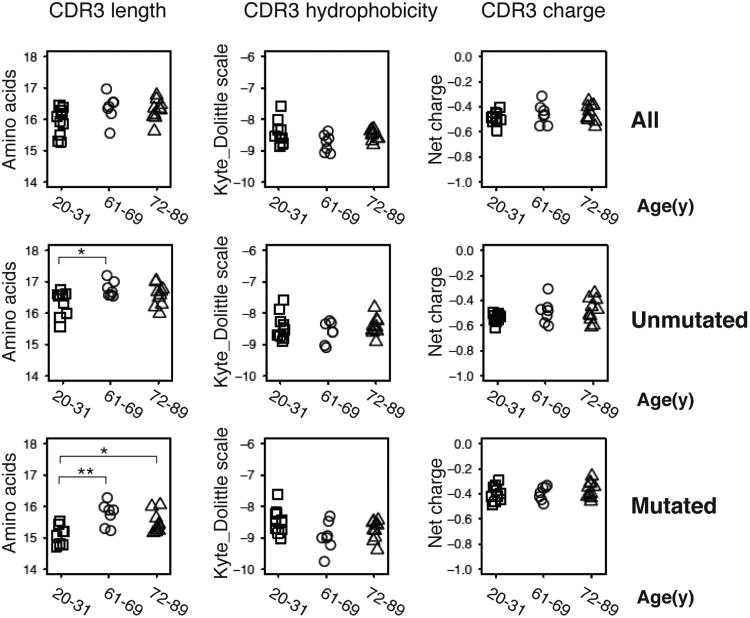
CDR3 length in both mutated and unmutated antibodies increases with age
We further analyzed biochemical features of the IGH CDR3 amino acid sequences, given the prominent role of this sequence region in determining antibody binding specificity, and the effects of B cell selection on the length of IGH CDR3 regions present in the B cell repertoire. It has been reported that B cell development in the bone marrow is accompanied by preferential removal of B cells expressing antibodies with long CDR3s, potentially due to the increased levels of autoreactivity demonstrated by such antibodies (34). Evidence of additional selection against B cells expressing antibodies with long CDR3s, or in favor of sequences with shorter CDR3s during antigen-driven peripheral responses, is seen in the decreased IGH CDR3s present in B cells expressing mutated Ig genes compared to unmutated ones (35). To look for age-associated alterations in these selection processes, we analyzed the CDR3 features of mutated and unmutated sequences separately in our dataset. Confirming prior observations, the unmutated sequences in our data set Figure 2 (middle panel) have longer CDR3s than mutated sequences Figure 2 (lower panel). Interestingly, the CDR3 regions of both unmutated and mutated sequences are significantly longer in older people compared with young individuals, especially within mutated sequences (Figure 2, middle and lower panel), while there is no obvious change in hydrophobicity or net charge with age. The presence of increased numbers of B cells with longer CDR3 segments in both unmutated and mutated antibody heavy chain rearrangements in elderly individuals suggests an overall decreased level of selection against antibodies with long CDR3s in the aging immune system, both in the generation of naïve B cells, as well as during antigen-driven responses.
Figure 2. Increased IGH CDR3 lengths in older individuals.
Average CDR3 length, hydrophobicity, and net charge of unmutated sequences (middle panel) and mutated sequences (lower panel) of each sample are grouped by age. Sequences are pooled from two time points Y2 and Y3. Pairwise comparison was performed with two-sided student t-test. * P < 0.05, ** P<0.01. For unmutated sequence CDR3 lengths (in amino acids), age 20-31 mean = 16.3, SE = 0.1; age 61-69 mean = 16.8, SE = 0.1; p-value = 0.013. For mutated sequence CDR3 lengths, age 20-31 mean = 15.1, SE = 0.1; age 61-69 mean = 15.8, SE = 0.1; age 72-89 mean = 15.5, SE = 0.1; p-value for age 20-31 vs. age 61-69 = 0.003; p-value for age 20-31 vs. age 72-89 = 0.022.
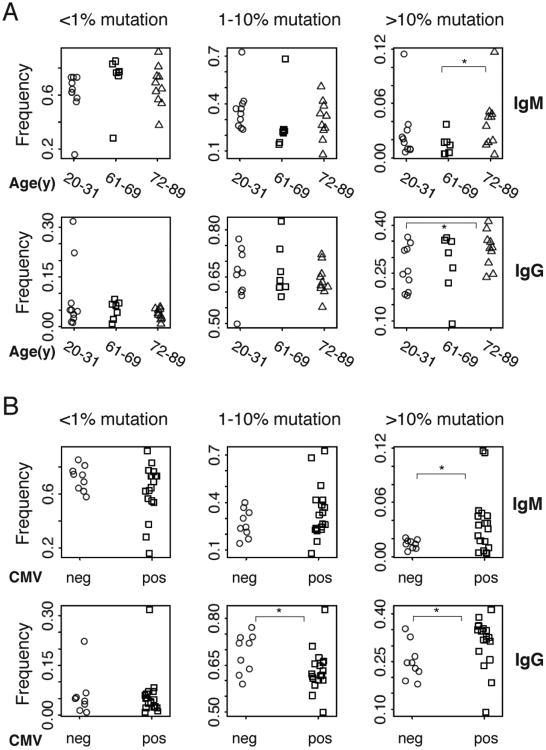
IgM and IgG show an age- and CMV- associated increase in mutation
We initially evaluated the levels of IGHV mutation regardless of antibody isotype in participants of various ages. We grouped sequences into three groups: less than 1% mutation frequency (unmutated), 1-10% mutation frequency (moderate), and greater than 10% mutation frequency (high). As shown in Figure 3A, the proportion of reads falling in these groups in each individual is highly correlated between two consecutive years, showing both the variation between individuals, and the stability of this feature of the B cell repertoire over time. Unmutated IGHV comprises, on average, 68%, 68%, 75% of the repertoire in individuals aged (20-31), (61-69), and (72-89) respectively (Figure 3B, upper panel).
Figure 3. V-mutation levels are increased in CMV positive individuals.
A, Frequencies of gDNA sequences with different V-mutation levels are correlated strongly between samples from two consecutive years of the same participants. V-mutation frequencies are categorized to 3 levels: <1% (unmutated), 1%-10% (moderate) and >=10% (high) mutation. B, Frequencies of sequences with different mutation levels are grouped by age, gender, CMV or EBV status. Sequences are pooled from two years for each participant. P values are calculated by two-sided student t-test, including all data points for each category. * P < 0.05.
We then asked whether gender or seropositivity for CMV or EBV correlate with the extent of IGHV mutation in each individual's total B cell repertoire. Figure 3B shows that highly mutated IGHV is increased in CMV-positive versus CMV-negative participants (P < 0.05). No significant differences in mutation rates were seen with regard to gender or EBV infection status. Multiple regression analysis of factors contributing to the proportion of highly mutated Ig showed that CMV has a stronger effect than that of age, gender or EBV infection, with P = 0.11 in the model.
To evaluate IGHV mutational frequencies in greater detail, we examined sequences from antibodies of particular isotypes to look for changes associated with aging or chronic viral infection. Figure 4A shows that highly mutated IgM and IgG sequences show a trend towards being increased in the oldest individuals (72-89y) compared with younger individuals, although one individual in the age 21-30 group (p65, described further below) had a proportion of >10% mutated IgM sequences that was unusually high for their age; this individual proved to be EBV and CMV seropositive. CMV infection is associated with an increased proportion of highly mutated sequences of IgM and IgG isotypes (Figure 4B). No significant age- or CMV-related increase in mutation frequencies was seen for other isotypes including IgA or IgD (Supplementary Figure 1). There were too few IgE sequences obtained for this analysis. The association between CMV seropositivity and elevated IgM and IgG mutation frequencies is stronger than the correlation between mutational frequencies and age, since the presence of variability of IgM and IgG mutation rates within each age group did not permit rejection of the ANOVA testing null hypothesis of there being no significant difference between age groups in the proportion of sequences with over 10% mutation (p=0.38 for IgM and p=0.19 for IgG). As with the overall B cell repertoire measured with gDNA, none of the isotypes show IGHV mutation frequencies significantly correlated with gender or EBV infection. As an incidental finding in the data set, we noticed occasional subjects showing IgM sequences that had accumulated unusually increased mutational loads. One of these participants, p65, also had an increase in their proportion of IgG sequences with low levels of mutation. These observations will require further investigation, but the subjects were healthy at the time of blood draws, without known infections or autoimmune disorders. The data indicate that phenotypic variation in IGHV mutational frequencies in human populations may be wider than has been previously reported.
Figure 4. Frequencies of highly mutated IgM and IgG sequences are increased in some elderly individuals, and correlate with CMV infection regardless of age.
A, Frequencies of IgM (upper panel) and IgG (lower panel) cDNA sequences with different levels of V-mutation grouped by age of participants. B, Frequencies of IgM (upper panel) and IgG (lower panel) cDNA sequences with different levels of V-mutation grouped by CMV status of participants. Sequences are pooled from two years. P values are calculated by two-sided student t-test, including all data points for each category. * P < 0.05.
IGHV gene usage differs between naïve and mutated IGH but is comparable in mutated IgM, IgG and IgA
Previous studies have reported evidence that memory IgM and isotype switched memory B cell populations utilize IGHV families with differing frequencies, with IgM memory cells having a lower frequency of IGHV1 gene usage and higher frequency of IGHV3 gene usage than IgG and IgA isotype-switched memory B cells (36). We considered IgM or IgD sequences with 1% or greater mutation frequency to be derived from IgM memory B cells, while those with <1% mutation were from naïve cells (37). Supplementary Figure 2 shows that naïve IgM and IgD sequences have significantly higher IGHV1 and lower IGHV3 usage than mutated IgM, IgD, IgG or IgA. However, we saw little difference in IGHV gene family usage between mutated IgM, IgD, IgG or IgA (Supplementary Figure 2). Similarly, age, CMV, or EBV infection had minimal impact on IGHV gene usage (Supplementary Figure 2).
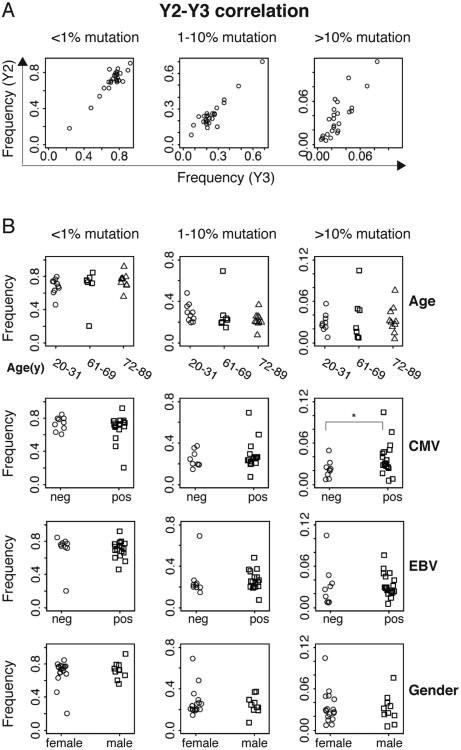
Age and chronic EBV infections are correlated with persistent B cell clonal expansion
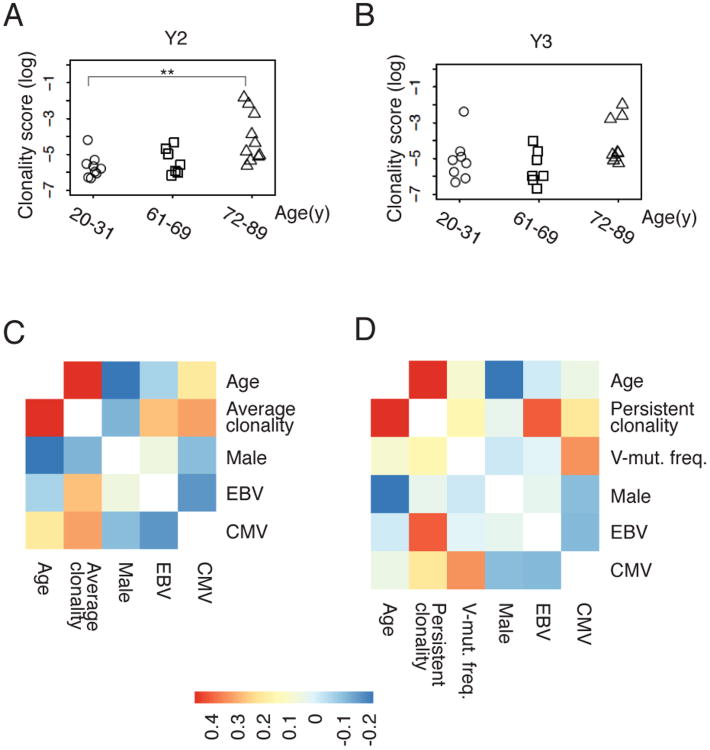
We quantified clonally-expanded B cells in each individual, using a conservative clone definition of sharing the same V and J genes and having identical CDR3 amino acid sequences. Six independent IGH libraries amplified from gDNA template were examined from each sample, to distinguish between PCR amplification bias and true clonal expansion of B cells, by requiring that the same rearrangement be detected in at least 2 independent libraries from the sample to be considered clonally expanded. We calculated a ‘clonality score’, as described in the Methods section, which can be thought of as the probability that two B cells selected at random from the patient's peripheral blood will belong to the same clonal lineage. Figure 5A and 5B show that most samples from healthy individuals sequenced at this depth show low levels of B cell clonality, indicating that almost all of the sequences observed in each replicate library from a given individual are unique. Significantly increased clonality scores are seen in the oldest group (72-89) compared with the youngest groups (20-31). Three older participants (ages 73, 85 and 88 years) showed markedly elevated clonal scores that persisted through Y2 and Y3 of the study, due to the presence of one or more large clonal B cell populations. In contrast, the only young individual (age 26) to show large B cell clones in the blood had them at a single time point (Y3), indicating that these clones may represent transient physiological B cell clonal expansions.
Figure 5. Age and chronic EBV infections are correlated with persistent B cell clonal expansion.
A, B, Clonality scores of the Ig repertoire of each sample are grouped by age for time point Y2 (A, n=27) and Y3 (B, n=25). The pairwise comparison between different age groups is performed with two-sided Wilcoxon tests. ** P < 0.01. C, Correlation heat map of Y2-Y3 average clonality score (log) for each participant's B cell repertoire, according to age, gender, CMV seropositivity and EBV seropositivity in 27 individuals. D, Correlation heat map of persistent clonality score (log) for clones detected in Y2 and Y3 of the study, according to age, gender, CMV, EBV and frequency of highly mutated sequences (V-mut. freq.), in 25 individuals. The colored scale bar represents the strength of correlation for each variable pair. Details of the clonality score calculations, which yield a measure of the proportion of the B cell repertoire contributed by expanded clonal populations, normalized for the depth of sequencing carried out in each sample, are presented in the Methods section.
We evaluated the correlation between B cell repertoire clonality and an individual's age, gender and serological status for EBV and CMV infection. To combine sequences from two years, we took the average of the clonality score from the two years. The heat map of Figure 5C shows that age is correlated with B cell clonal expansions detected within each year of the study, with a correlation of 0.46, and two-sided P value of 0.016. We used multiple linear regression to address potential confounding variables, using gender, CMV status, and EBV status as covariates, correcting for multiple hypotheses. Age remained a significant factor, with a Benjamini-Hochberg corrected false discovery rate of 0.061, and EBV infection status showed significance with a false discovery rate of 0.083. Persistent clonality scores normalized for sequencing depth were calculated as described in the Methods section, to quantitate B cell clones persisting in an individual through the year-long time course of the study. Correlation coefficients (Figure 5D) showed that the persistent clonality score (i.e., the levels of B cell clones that were found to persist in a particular individual and were detected in both year Y2 and year Y3) was correlated with age and EBV status. Multiple linear regression analysis demonstrated that the effects of age and EBV infection on the persistent clonality score are significant after multiple hypothesis adjustment (P=0.001 for age, P= 0.016 for EBV infection). There was no significant association between the persistent clonality score and CMV status, gender, or the proportion of highly mutated sequences in the individual's B cell repertoire.
Persistent B cell clones in the elderly form discrete lineage tree categories
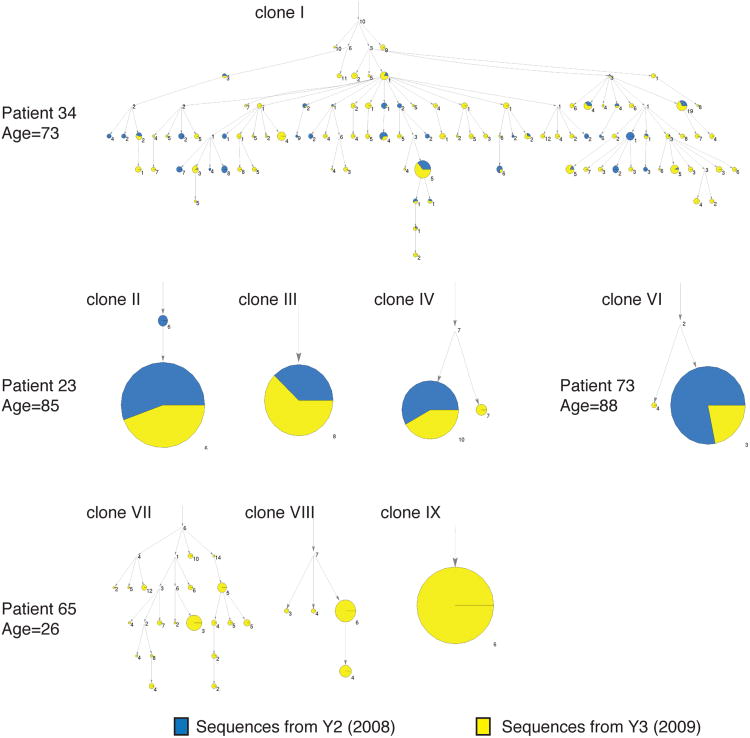
B cell clonal expansion has been described in aged population, but the antibody gene sequences of expanded clones have not been characterized (38). The sequencing depth in our study enabled us to examine the IGH sequences of the expanded clones, and infer the lineage relationships of clone members in detail. We searched for the biggest clones in the sequence data from each individual, focusing on clones that appeared in at least four out of six replicate sub-libraries and comprised more than 1% of total sequence reads. One elderly individual (p23) showed three distinct larger persistent clones, while the other two elderly individuals showed a single clonal sequence and an out-of-frame sequence, or several closely related CDR3 sequences differing only by apparent mutation changes (Supplementary Table II). The large clones showed mutated IGHV regions, with mutation frequencies of 2-9%, consistent with prior or ongoing affinity maturation. All big clones in the elderly were observed in both years while those in the young individual (p26) were only found in a single year (Y3).
Large persistent B cell clones in elderly individuals could represent abnormal neoplastic monoclonal B cell proliferations (39-41). Alternatively, such clones could result from antigenic stimulation by endogenous viruses or other antigens. We examined the extent and diversity of mutation in the large persistent clones, using a Bayesian probabilistic phylogenetic model implemented in the Immunitree program (see Methods). The lineage trees in Figure 6 show that the large persistent clones detected in elderly individuals include examples of clones with very little mutational diversification, in which almost all sequences are mutated at identical positions (clones II, III, IV and VI), as well as examples where there is an extensive tree of mutation variants derived from a common precursor (clone I). The three large clonal lineages detected during study year Y3 in the 26-year-old participant p65 also include an example with limited intraclonal mutational variation (clone IX), as well as two clones with more extensively varying mutation (clones VII and VIII). All of the clonal lineages detected in the study participants showed some level of mutation, with no fully unmutated lineages seen.
Figure 6. Phylogenetic visualization of large clones.
The large clones are visualized via immunitree. Each arrow points from an ancestral clone to a descendent clone. Sizes of the nodes correspond to the number of reads observed; the blue portion of the pie chart represents Y2, while the yellow portion represents Y3. The clones are labeled with the number of mutations from their immediate parent subclone. The root node's numerical label counts the number of mutations from the best matching germline reference V segment.
Discussion
We have analyzed over 500,000 rearranged immunoglobulin heavy chain sequences derived from the peripheral blood of young adults and elderly individuals, in serial samples taken from each subject at two time points separated by approximately one year. Our data indicate that overall V, D, J gene usage are similar in young and older individuals, but that the aging B cell repertoire shows prominent changes in the lengths of IGH CDR3 regions that are present in B cells, and in the levels of somatic mutation in antibody sequences. Importantly, the individuals studied here were of known seropositivity status for CMV and EBV chronic infection, and there were sufficient numbers of seropositive and seronegative individuals to allow analysis of the impact of these viral infections on the B cell repertoire, independent of patient age. Our findings highlight an association between CMV seropositivity and levels of antibody gene somatic mutation, while EBV seropositivity is correlated with the contribution of persistent clonal populations to the B cell repertoire.
Increased levels of B cells expressing long IGH CDR3 regions in the elderly are seen in our data both in unmutated sequences, and in mutated sequences. Selection against B cells expressing antibodies with long CDR3s during the process of generating the naïve B cell repertoire has previously been correlated with the higher frequency of autoreactivity in antibodies with long CDR3 regions (34). The effects of this selection process can be detected in the difference between the longer CDR3 regions present in unproductively rearranged IGH genes compared to productive expressed IGH sequences (42). The presence of longer CDR3 regions in mutated IGH sequences in the elderly in our data may simply reflect the fact that mutated sequences derive from the initially unmutated pool, or could reflect additional age-related impairment in positive selection for B cells expressing antibodies with shorter CDR3s in antigen-driven responses. Prior literature has documented increased levels of autoantibodies in the sera of elderly individuals compared to younger individuals (43-45). The results presented here may indicate that increased autoimmunity in the elderly stems from two different defects: impaired selection against antibodies with long CDR3 regions in generation of the naïve B cell repertoire, and decreased selection against long CDR3 antibodies in B cell responses associated with antibody mutation and isotype switching. Other CDR3 features that have been associated with autoreactivity, such as hydrophobicity, did not show age-related changes in our data.
Both IgM and IgG mutated memory B cell pools in our data set show a trend toward age-associated accumulation of highly mutated sequences, although there is considerable variability between individuals in all age groups examined. These findings could suggest that progressive antigenic exposure stimulating B cell proliferation and targeted immunoglobulin gene mutation can lead to the accumulation of increasingly highly mutated IGHV genes over the course of a human lifespan. Overall, there is considerable overlap in the levels of IGHV mutation seen in the B cell repertoires of young and elderly individuals. Implications of the latter result are that most long-lived memory B cells may be formed primarily from the relatively less-mutated members of a clonal proliferation, or that memory B cells formed from more highly-mutated clone members may have a shorter lifetime in the host. Indeed, very long-lasting memory B cells, such as those specific for the 1918 pandemic influenza strain in nonagenarian or centenarian survivors of the pandemic have shown IGHV mutation levels of approximately 10%, within the range of mutation frequencies that can be seen in memory B cells in younger individuals (46). The origin of IgM memory B cells is a subject of debate, with some reports proposing novel developmental pathways that are independent of germinal center stimulation, or independent of antigen stimulation entirely, while other interpretations describe these cells as the output of immune responses associated with decreased germinal center function, or simply a subset of memory B cells typically produced in T cell-dependent immune responses (37, 47). If the generation of IgM memory cells is a consequence of impaired germinal center function, then age-associated increases in IgM mutation frequencies could be a consequence of decreased germinal center activity in the elderly, leading to greater reliance on B cell clonal lineages that show some Ig mutation, but have not been able to isotype switch.
We identified an independent correlation between CMV infection and the mutation levels in both IgM and IgG-expressing B cells in individuals regardless of age. This could be a direct effect of some B cells being specific for CMV, or alternately, could reflect the reported superantigen-like effects reported for CMV phosphoprotein pUL32 or other CMV proteins (48). Arguing against the latter possibility, the described superantigen-like activity of pUL32 was for B cells expressing IGHV1-69 or IGHV3-21 gene segments, and we did not see such segments overrepresented among highly mutated sequences in our data.
We identified large B cell clones that persisted over the year-long period of the study only in individuals over 70 years old (3 of 10), although one younger subject showed comparably large clones at a single time point. The mechanisms leading to persistent B cell clones are of great interest, and tie our findings to reports in the hematology-oncology and infectious disease literatures. Current classification of B cell lymphomas and leukemias defines numerous subtypes of overt malignancy, but also includes the category “monoclonal B cell lymphocytosis” (MBL) an age-associated condition in which abnormal persistent clones of B cells with the immunophenotype of chronic lymphocytic leukemia (CLL) are identified, but at lower cell counts than the minimum required for a CLL diagnosis (5×103 cells/μL) (39-41). Although flow cytometry data to test for such abnormal B cell populations are not available for the subjects in this study, it is likely that some of the persistent large clones identified by IGH sequencing correspond to MBL populations. Indeed, the clonal lineage tree analysis showed that several of the large persistent clones were mutated, but were fixed with all members showing essentially identical mutation patterns, a characteristic feature of mutated CLL clones (41).
Epidemiological studies report correlations between infectious disease exposure and the development of MBL or CLL, and stereotypic IGHV rearrangements in CLL have been proposed as evidence of antigen drive contributing to oncogenesis (49-51). The CDR3 sequences of the clones in our study differed from known stereotyped CLL CDR3 sequences, but we find a correlation between the levels of persistent B cell clones and EBV seropositivity, suggesting that EBV infection may contribute to age-associated B cell clonal expansions. Large clonal proliferations of CD8+ T cells associated with CMV infection are well-described, but effects of CMV or EBV on B cell populations have been less clear (2, 15). Although the elderly participants who have the largest clonal expansions in our study are all positive for both CMV and EBV infection, our multiple regression analysis found that age and EBV status were the most significant contributors to the overall levels of clonally expanded B cells when all participants were considered.
These measurements of peripheral blood IGH sequences from young and elderly individuals highlight effects of aging and chronic CMV or EBV infection on the B cell repertoire. Increased age is associated with the development of large persistent clonal B cell populations, and a trend toward increased IGHV mutation levels in B cells expressing IgM or IgG, while features of the repertoire such as V, D and J segment usage are relatively age-invariant. B cell repertoires in the elderly show decreased selection against antibodies with long CDR3 regions, both in unmutated antibodies likely associated with naïve B cells, as well as in presumably antigen-experienced B cells expressing mutated antibodies. CMV infection is correlated with high mutational levels in IgM and IgG-expressing B cells, while EBV infection correlates with the proportion of persistent B cell clones in the blood over a year's time, revealing distinct and important shaping of the B cell repertoire by these common chronic viral infections. Studies of antigen-specific responses stimulated by vaccination or infection in the elderly hold the promise of revealing further age- or chronic viral infection-associated deficiencies.
Supplementary Material
Acknowledgments
The authors wish to thank Sally Mackey for project, regulatory and data management; Research Nurses Sue Swope and Cynthia Walsh; Phlebotomist Michele Ugur and Research Assistant Kyrsten Spann for scheduling and conducting the study visits.
Grant Support: The study was supported in part by Stanford Center for Clinical and Translational Education and Research (SCCTER) NIH grant 1UL1 RR025744 from the National Center for Research Resources, NIH grant U19 AI090019, and grants from the Ellison Medical Foundation to Mark Davis and Scott Boyd.
References
- 1.Dorshkind K, Montecino-Rodriguez E, Signer RA. The ageing immune system: is it ever too old to become young again? Nature reviews Immunology. 2009;9:57–62. doi: 10.1038/nri2471. [DOI] [PubMed] [Google Scholar]
- 2.Pawelec G, Akbar A, Caruso C, Solana R, Grubeck-Loebenstein B, Wikby A. Human immunosenescence: is it infectious? Immunol Rev. 2005;205:257–268. doi: 10.1111/j.0105-2896.2005.00271.x. [DOI] [PubMed] [Google Scholar]
- 3.Dunn-Walters DK, Ademokun AA. B cell repertoire and ageing. Curr Opin Immunol. 2010;22:514–520. doi: 10.1016/j.coi.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Frasca D, Blomberg BB. Effects of aging on B cell function. Curr Opin Immunol. 2009;21:425–430. doi: 10.1016/j.coi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiskopf D, Weinberger B, Grubeck-Loebenstein B. The aging of the immune system. Transpl Int. 2009;22:1041–1050. doi: 10.1111/j.1432-2277.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E, Kaye D, Abrutyn E, Gross P, Dorfman M, Murasko DM. Immune response to influenza vaccination in a large healthy elderly population. Vaccine. 1999;17:82–94. doi: 10.1016/s0264-410x(98)00117-0. [DOI] [PubMed] [Google Scholar]
- 7.Sankilampi U, Isoaho R, Bloigu A, Kivela SL, Leinonen M. Effect of age, sex and smoking habits on pneumococcal antibodies in an elderly population. Int J Epidemiol. 1997;26:420–427. doi: 10.1093/ije/26.2.420. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki S, Sullivan M, Narvaez CF, Holmes TH, Furman D, Zheng NY, Nishtala M, Wrammert J, Smith K, James JA, Dekker CL, Davis MM, Wilson PC, Greenberg HB, He XS. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. The Journal of clinical investigation. 2011;121:3109–3119. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ademokun A, Wu YC, Dunn-Walters D. The ageing B cell population: composition and function. Biogerontology. 2010;11:125–137. doi: 10.1007/s10522-009-9256-9. [DOI] [PubMed] [Google Scholar]
- 10.Frasca D, Landin AM, Lechner SC, Ryan JG, Schwartz R, Riley RL, Blomberg BB. Aging down-regulates the transcription factor E2A, activation-induced cytidine deaminase, and Ig class switch in human B cells. J Immunol. 2008;180:5283–5290. doi: 10.4049/jimmunol.180.8.5283. [DOI] [PubMed] [Google Scholar]
- 11.Kogut I, Scholz JL, Cancro MP, Cambier JC. B cell maintenance and function in aging. Seminars in immunology. 2012;24:342–349. doi: 10.1016/j.smim.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Yamazaki T, Okubo Y, Uehara Y, Sugane K, Agematsu K. Regulation of aged humoral immune defense against pneumococcal bacteria by IgM memory B cell. J Immunol. 2005;175:3262–3267. doi: 10.4049/jimmunol.175.5.3262. [DOI] [PubMed] [Google Scholar]
- 13.Weksler ME, Goodhardt M, Szabo P. The effect of age on B cell development and humoral immunity. Springer seminars in immunopathology. 2002;24:35–52. doi: 10.1007/s00281-001-0094-3. [DOI] [PubMed] [Google Scholar]
- 14.Frasca D, Diaz A, Romero M, Landin AM, Phillips M, Lechner SC, Ryan JG, Blomberg BB. Intrinsic defects in B cell response to seasonal influenza vaccination in elderly humans. Vaccine. 2010;28:8077–8084. doi: 10.1016/j.vaccine.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chidrawar S, Khan N, Wei W, McLarnon A, Smith N, Nayak L, Moss P. Cytomegalovirus-seropositivity has a profound influence on the magnitude of major lymphoid subsets within healthy individuals. Clin Exp Immunol. 2009;155:423–432. doi: 10.1111/j.1365-2249.2008.03785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Looney RJ, Falsey A, Campbell D, Torres A, Kolassa J, Brower C, McCann R, Menegus M, McCormick K, Frampton M, Hall W, Abraham GN. Role of cytomegalovirus in the T cell changes seen in elderly individuals. Clin Immunol. 1999;90:213–219. doi: 10.1006/clim.1998.4638. [DOI] [PubMed] [Google Scholar]
- 17.Ouyang Q, Wagner WM, Wikby A, Walter S, Aubert G, Dodi AI, Travers P, Pawelec G. Large numbers of dysfunctional CD8+ T lymphocytes bearing receptors for a single dominant CMV epitope in the very old. Journal of clinical immunology. 2003;23:247–257. doi: 10.1023/a:1024580531705. [DOI] [PubMed] [Google Scholar]
- 18.Gibson KL, Wu YC, Barnett Y, Duggan O, Vaughan R, Kondeatis E, Nilsson BO, Wikby A, Kipling D, Dunn-Walters DK. B-cell diversity decreases in old age and is correlated with poor health status. Aging cell. 2009;8:18–25. doi: 10.1111/j.1474-9726.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, Wilson PC, Greenberg HB, Davis MM, Fisher DS, Quake SR. Lineage structure of the human antibody repertoire in response to influenza vaccination. Science translational medicine. 2013;5:171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolar GR, Mehta D, Wilson PC, Capra JD. Diversity of the Ig repertoire is maintained with age in spite of reduced germinal centre cells in human tonsil lymphoid tissue. Scandinavian journal of immunology. 2006;64:314–324. doi: 10.1111/j.1365-3083.2006.01817.x. [DOI] [PubMed] [Google Scholar]
- 21.Chong Y, Ikematsu H, Yamaji K, Nishimura M, Kashiwagi S, Hayashi J. Age-related accumulation of Ig V(H) gene somatic mutations in peripheral B cells from aged humans. Clin Exp Immunol. 2003;133:59–66. doi: 10.1046/j.1365-2249.2003.02185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Fire AZ. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Science translational medicine. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YC, Kipling D, Dunn-Walters DK. Age-Related Changes in Human Peripheral Blood IGH Repertoire Following Vaccination. Frontiers in immunology. 2012;3:193. doi: 10.3389/fimmu.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths PD, Baboonian C, Rutter D, Peckham C. Congenital and maternal cytomegalovirus infections in a London population. British journal of obstetrics and gynaecology. 1991;98:135–140. doi: 10.1111/j.1471-0528.1991.tb13358.x. [DOI] [PubMed] [Google Scholar]
- 25.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 26.Gaeta BA, Malming HR, Jackson KJ, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 27.Jackson KJ, Boyd S, Gaeta BA, Collins AM. Benchmarking the performance of human antibody gene alignment utilities using a 454 sequence dataset. Bioinformatics. 2010;26:3129–3130. doi: 10.1093/bioinformatics/btq604. [DOI] [PubMed] [Google Scholar]
- 28.Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Developmental and comparative immunology. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of molecular biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Winzor DJ. Protein charge determination. In: Coligan John E, et al., editors. Current protocols in protein science. Unit 2. Chapter 2. 2005. p. 10. [DOI] [PubMed] [Google Scholar]
- 31.Friedman WF. The index of coincidence and its applications in cryptanalysis. Aegean Park Press; 1987. [Google Scholar]
- 32.Glanville J, Kuo TC, von Budingen HC, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, Cox DR, Rajpal A, Pons J. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 35.Rosner K, Winter DB, Tarone RE, Skovgaard GL, Bohr VA, Gearhart PJ. Third complementarity-determining region of mutated VH immunoglobulin genes contains shorter V, D, J, P, and N components than non-mutated genes. Immunology. 2001;103:179–187. doi: 10.1046/j.1365-2567.2001.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tangye SG, Good KL. Human IgM+CD27+ B cells: memory B cells or “memory” B cells? J Immunol. 2007;179:13–19. doi: 10.4049/jimmunol.179.1.13. [DOI] [PubMed] [Google Scholar]
- 38.Ademokun A, Wu YC, Martin V, Mitra R, Sack U, Baxendale H, Kipling D, Dunn-Walters DK. Vaccination-induced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging cell. 2011;10:922–930. doi: 10.1111/j.1474-9726.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawstron AC, Green MJ, Kuzmicki A, Kennedy B, Fenton JA, Evans PA, O'Connor SJ, Richards SJ, Morgan GJ, Jack AS, Hillmen P. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002;100:635–639. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 40.Rawstron AC, Yuille MR, Fuller J, Cullen M, Kennedy B, Richards SJ, Jack AS, Matutes E, Catovsky D, Hillmen P, Houlston RS. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood. 2002;100:2289–2290. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 41.Swerdlow AJ, Campo E, Harris NL, Jaffe ES. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2008. [Google Scholar]
- 42.Larimore K, McCormick MW, Robins HS, Greenberg PD. Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol. 2012;189:3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 43.Mariotti S, Chiovato L, Franceschi C, Pinchera A. Thyroid autoimmunity and aging. Experimental gerontology. 1998;33:535–541. doi: 10.1016/s0531-5565(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 44.Tomer Y, Shoenfeld Y. Ageing and autoantibodies. Autoimmunity. 1988;1:141–149. doi: 10.3109/08916938809001927. [DOI] [PubMed] [Google Scholar]
- 45.Hijmans W, Radl J, Bottazzo GF, Doniach D. Autoantibodies in highly aged humans. Mechanisms of ageing and development. 1984;26:83–89. doi: 10.1016/0047-6374(84)90167-2. [DOI] [PubMed] [Google Scholar]
- 46.Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, Stevens J, Wilson IA, Aguilar PV, Altschuler EL, Basler CF, Crowe JE., Jr Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weller S, Braun MC, Tan BK, Rosenwald A, Cordier C, Conley ME, Plebani A, Kumararatne DS, Bonnet D, Tournilhac O, Tchernia G, Steiniger B, Staudt LM, Casanova JL, Reynaud CA, Weill JC. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steininger C, Widhopf GF, 2nd, Ghia EM, Morello CS, Vanura K, Sanders R, Spector D, Guiney D, Jager U, Kipps TJ. Recombinant antibodies encoded by IGHV1-69 react with pUL32, a phosphoprotein of cytomegalovirus and B-cell superantigen. Blood. 2012;119:2293–2301. doi: 10.1182/blood-2011-08-374058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casabonne D, Almeida J, Nieto WG, Romero A, Fernandez-Navarro P, Rodriguez-Caballero A, Munoz-Criado S, Diaz MG, Benavente Y, de Sanjose S, Orfao A. Common Infectious Agents and Monoclonal B-Cell Lymphocytosis: A Cross-Sectional Epidemiological Study among Healthy Adults. PLoS One. 2012;7:e52808. doi: 10.1371/journal.pone.0052808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghia P, Chiorazzi N, Stamatopoulos K. Microenvironmental influences in chronic lymphocytic leukaemia: the role of antigen stimulation. J Intern Med. 2008;264:549–562. doi: 10.1111/j.1365-2796.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 51.Landgren O, Rapkin JS, Caporaso NE, Mellemkjaer L, Gridley G, Goldin LR, Engels EA. Respiratory tract infections and subsequent risk of chronic lymphocytic leukemia. Blood. 2007;109:2198–2201. doi: 10.1182/blood-2006-08-044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.