Abstract
Human induced pluripotent stem cells (hiPSCs) have great therapeutic potential in repairing defective lung alveoli. However, genetic abnormalities caused by vector-integrations and low efficiency in generating hiPSCs, as well as difficulty in obtaining transplantable hiPSC-derived cell types, are still major obstacles. Here we report a novel strategy using a single non-viral site-specific-targeting vector with a combination of Tet-On inducible gene expression system, Cre/lox P switching gene expression system, and alveolar epithelial type II cell (ATIIC)-specific NeomycinR trangene expression system. With this strategy, a single copy of all of the required transgenes can be specifically knocked into a site immediately downstream of beta-2-microglobulin (B2M) gene locus at a high frequency, without causing B2M dysfunction. Thus, the expression of reprogramming factors, Oct4, Sox2, cMyc and Klf4, can be precisely regulated for efficient reprogramming of somatic cells into random-integration-free or genetic mutation-free hiPSCs. The exogenous reprogramming factor transgenes can be subsequently removed after reprogramming by transient expression of Cre recombinase, and the resulting random-integration-free and exogenous reprogramming-factor-free hiPSCs can be selectively differentiated into a homogenous population of ATIICs. In addition, we show that these hiPSC-derived ATIICs exhibit ultra-structural characteristics and biological functions of normal ATIICs. When transplanted into bleomycin-challenged mice lungs, hiPSC-derived ATIICs efficiently remain and re-epithelialize injured alveoli to restore pulmonary function, preventing lung fibrosis and increasing survival without tumorigenic side effect. This strategy allows for the first time efficient generation of patient-specific ATIICs for possible future clinical applications.
Keywords: Site-specific targeting strategy, Induced pluripotent stem cells, Differentiation and characterization, Lung alveolar epithelial type II cells
Introduction
Alveolar epithelial type II cells (ATIICs) are essential for maintaining homeostasis of alveoli. They synthesize and secrete pulmonary surfactant, playing a crucial role in reducing surface tension and preventing collapse of alveoli at end expiration. As progenitor cells for alveolar epithelial type I cells (ATICs), ATIICs are particularly crucial during re-epithelialization of the alveoli after lung injury. Loss of normal functions of ATIICs due to injuries or genetic deficiencies is thought to play an important role in the development of many life-threatening pulmonary diseases, including idiopathic pulmonary fibrosis (IPF) [1], fatal neonatal respiratory distress syndrome (RDS)[2, 3], and progressive familial and sporadic interstitial lung disease (ILD) [4]. Currently available treatments for these diseases at best alleviate symptoms within a limited time range, with poor long-term outcomes. Lung transplantation is the only therapeutic option for many patients. Therefore, there is an urgent need of novel therapeutic strategies. Recently, potential use of induced pluripotent stem cells (iPSCs) to regenerate injured lungs has attracted a lot of interest for disease modeling and repair of injured/diseased alveolar epithelium.
Previous techniques use retrovirus or lentivirus to integrate multiple copies of each reprogramming factor at random into the chromosomal DNA to reprogram somatic cells into iPSCs [5–7]. However, each random integration of viral DNA may cause unpredictable critical gene dysfunction, and the generated iPSCs may develop into tumors, or lose their self-renewal capacity or the potential to differentiate into the cell type needed for therapeutic transplantation. Recently, a technique using non-viral vector containing loxP targeting sequence in combination with inducible gene expression system has been developed for iPSC reprogramming [8]. However, random insertion of DNA or part of DNA sequences left behind after removal of factors is still a potential drawback. Several alternative techniques have been developed to improve safety in iPSC generation, including the use of adenovirus [9], sendai virus [10], minicircle vector [11], PiggyBac transposon [12], and episomal vectors [13]. Yet most of these techniques suffer from impractically low reprogramming efficiency, and the possibility of vector integrations remains. More recently, techniques to directly deliver proteins [14], RNAs [15] or mature microRNAs [16] for reprogramming have been developed, but require special treatment and multiple times of transduction/transfection with low reprogramming efficiency. Thus, efficient generation of iPSCs without causing genetic abnormalities continues to be a challenge. In addition, as iPSCs tend to spontaneously differentiate to various cell types in vitro, the spontaneously differentiated cultures of iPSCs are certainly not suitable for clinical application as they could cause deleterious side effects, even tumor formation. Therefore, lack of effective methodologies to derive iPSCs into transplantable cell lineages is also a major hurdle to their clinical application.
To overcome the above problems, here we report a novel beta-2-microglobulin (B2M) specific insertion targeting strategy using a single non-viral vector for reprogramming human somatic cells. By using tetracycline-controlled expression system (Tet-On) to conditionally express Oct4, Sox2, cMyc and Klf4 immediately downstream of B2M gene for efficient generation of random-integration-free human iPSCs (hiPSCs), this site-specific insertion does not cause B2M gene dysfunction. As the single targeting vector contains loxP targeting sequence and NeomycinR (NEOR) transgene controlled by ATIIC-specific surfactant protein C (SPC) promoter (SPCP-NEOR), the reprogramming factor transgenes can be subsequently removed and the hiPSCs can be selectively differentiated into a homogenous population of ATIICs for further exploration of their therapeutic potential.
Materials and Methods
Construction of 3′hprt.OSMK-LoxP.rtTA.SPCPNEOR.B2M targeting vector
One 4.2 kb AscI-BssHII DNA fragment homologous to 3′ region of B2M gene was cloned into AscI site of the 3′ hprt insertion targeting vector (a gift from Dr. Allan Bradley, The Wellcome Trust Sanger Institute, Cambridge, U.K.). The AscI and SalI digested TRE-PminCMV/Oct4/IRES/Sox2 fragment was isolated from pTRE-OIS vector (Supporting information Fig. S1A) and subcloned into the engineered MluI and SalI site downstream of PUROR. Similarly, XhoI and SalI digested TRE-PminCMV/Klf4/IRES/cMyc/LoxP fragment from pTRE-KIcML vector (Supporting information Fig. S1B) was subcloned into SalI site downstream of TRE-PminCMV/Oct4/IRES/Sox2 fragment with correct orientation. A 4.9 kb SPCP-NEOR transgene isolated from SPCP-NEOR vector (Supporting information Fig. S1C) was subsequently added into SalI and engineered NdeI site of the vector. In addition, EFaP-rtTA transgene was subcloned from EFαP-rtTA vector (Supporting information Fig. S1D) into AscI site of the insertion targeting vector. The resulting 3′hprt.OSMK-LoxP.rtTA.SPCPNEOR.B2M targeting vector is depicted in Figure 1. The EcoRI was used to linearize the vector and delete a 216 bp gap within Exon 4 of B2M fragment before transfection for gap repair targeting at the B2M gene locus [17].
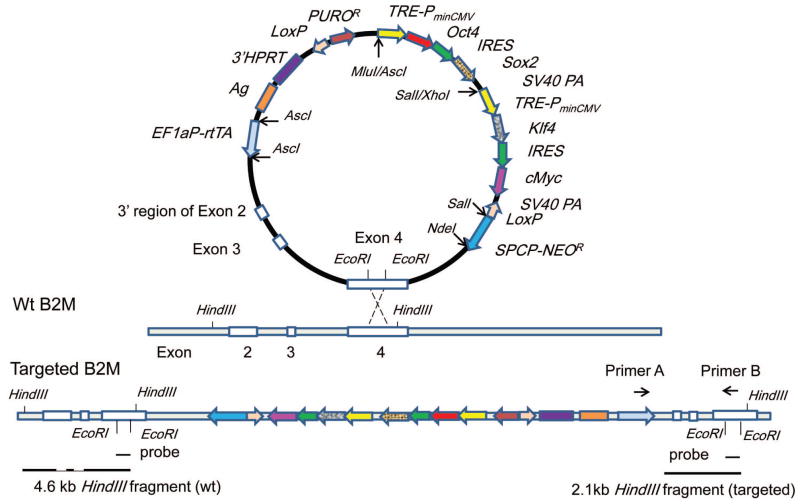
Figure 1. Schematic diagram of 3′hprt.OSMK-LoxP.rtTA.SPCPNEOR.B2M Targeting Vector.
The schematic diagram of the vector is to depict relevant positional information, hence it is not scaled proportionally based on sequence lengths. This vector harbors the reverse tetracycline controlled transactivator (rtTA) under the control of EF-1a promoter, and the four reprogramming factor (Oct4, Sox2, cMyc and Klf4) transgenes under the control of Tet-responsive promoter (TREPminCMV fragment). The two components of Tet-On inducible gene expression system, the regulator (rtTA) and the Tet-responsive promoter in the vector, allow induced expression of Oct4, Sox2, cMyc and Klf4 for hiPSC reprogramming more efficiently. In addition, the LoxP sites in the vector allow removal of puromycin and the four reprogramming factor transgenes by Cre-mediated recombination to generate exogenous reprogramming-factor-free hiPSCs. In order to selectively differentiate hiPSCs into ATIICs, the SPCP-NEOR trangene was added into the targeting vector. One 4.2 kb DNA fragment homologous to the 3′ region of B2M, including partial Exon2, Exon3, Exon4 and 3′ untranslated region, is included in the vector to specifically knock-in all of the transgenes into the 3′ region of B2M gene by homologous recombination. The homologous insertion strategy is expected to result in high targeting frequencies (10–30% on average) and a duplicate of the homologous region contained in the targeting vector [17]. As the insertion targeting vector harbors the 3′ region of B2M gene, the B2M gene should remain intact without causing gene dysfunction after site-specific targeting. To generate hiPSCs, EcoRI was used to linearize the vector and delete a 216 bp gap within Exon 4 of B2M fragment before transfection for gap repair targeting at the B2M gene locus. A 216 bp gap DNA fragment was used as probe, which can hybridize to 4.6 kb wt and 2.1 kb targeted B2M HindIII fragment. In addition, PCR can be performed by using primer A located within rtTA transgene and primer B in the gap region of B2M to identify B2M-targeted transgene.
Transfection of human skin fibroblasts for induction of pluripotency
Approximately 5×105 human adult fibroblasts (passages 4, provided by National Disease Research Interchange, NDRI) were re-suspended in 100 μl of supplemented Nucleofector Solution (VPD-1001, Lonza), mixed with 2 μg of the EcoRI-linearized targeting vector, and then transfected following manufacturers’ instruction. The transfected cells were cultured in DMEM containing 10% FBS (Gibco Invitrogene). Puromycin (0.5 μg/ml, Sigma) was added next day. After 3 day selection, the stably transfected cells that had survived puromycin-selection were replaced at 0.5×104/10-cm dish on mitomycin-treated MEF feeder cells in human embryonic stem cell (hESC) culture medium [18] in the presence of 2 μg/ml doxycycline (doxy, Sigma-Aldrich) [19].
Southern blot and PCR analysis of B2M-targeted hiPSC clones
Genomic DNA samples were digested with HindIII, SalI, KpnI, MluI and ClaI, respectively, for Southern blot analysis. A 216 bp PCR fragment homologous to the gap region within the Exon 4 of B2M (Fig. 1) or a 425 bp NEOR gene fragment was used as a probe. In addition, PCR was performed to confirm the site-specific targeting event by using primer A and B (Fig. 1).
Real-time quantitative reverse transcriptase-PCR (QRT-PCR) analysis
Total RNA samples were prepared for QRT-PCR analysis using TaqMan One-Step RT-PCR Master Mix Kit (AB Applied Biosystems) as previously reported [20] using 18S as endogenous control. All primers used in the study were listed in supporting information Table S1.
Differentiation and selection of hiPSC-derived ATIICs
To directly differentiate hiPSC-26B into ATIICs (hiPSC-ATIICs), cells were cultured on Matrigel-coated plate in differentiation medium (DM) with or without G418 (20 μg/ml, Gibco Invitrogen) for 14 days [18]. The DM contains 80% Knockout™ DMEM (Gibco Invitrogen), 20% FBS (HyClone), 1% nonessential amino acid, 1 mM L-glutamine, 100 μg/ml penicillin, and 100 μg/ml streptomycin.
Immunofluorescent staining
Differentiated and undifferentiated hiPSC-26B cells were washed twice with PBS and fixed in 4% paraformaldehyde for 15 min. Washed cells were permeabilized in 0.2% Triton X (Sigma) for 30 min before blocking in 10% goat or donkey serum for 2 hr. Differentiated cells were stained with 1:50 diluted mouse anti-human MHC-I (W6/32, Abcam), and visualized with Alexa Fluor 488 conjugated goat anti-mouse IgG (1:1000, Molecular Probes) with Draq5 (1:500, Biostatus) counterstaining. Undifferentiated cells were stained with either goat anti-human Nanog, goat anti-human Oct4, mouse anti-human SSEA4, mouse anti-SSEA1 (1:10 diluted, R&D System), or 1:500 diluted mouse anti TRA-1–60 (Abcam). Alexa Fluor 546 conjugated donkey anti-goat IgG (1:1000, Molecular Probes) was used to visualize expression of Nanog and Oct4, and Alexa Fluor 488 conjugated goat anti-mouse IgG to visualize expression of TRA-1–60, SSEA4, and SSEA1. To examine expression of surfactant protein A (SPA), B (SPB) and C, the G418-selected hiPSC-ATIICs on day 14 and human primary control ATIICs (hATIICs) were stained with the rabbit anti-human SPA, proSPB or proSPC (Chemicon) as previously reported [18]. The hATIICs were prepared as previously described[21]. The surfactant protein expression was visualized with Alexa Fluor 488 or 546 conjugated goat anti-rabbit IgG with Draq5 counterstaining. In addition, the rabbit anti T1α (Abgent), rabbit anti-human proSPC and mouse anti-human nuclei monoclonal antibody (Chemicon) were used for immunofluorescent staining of BLM-challenged lungs transplanted with hiPSC-ATIICs as previously reported [20].
Karyotype analysis and Teratoma formation
Chromosomal study using standard protocols for high-resolution G-banding was performed at the Clinical and Research Cytogenetic Laboratory, Texas Children’s Hospital. To examine in vivo pluripotency, hiPSC-26B cells were resuspended at 0.5×107 in hESC medium. The isoflurane anesthetized SCID mice were intramuscularly injected with 0.5×107 cells on left hind leg. Tumors were surgically dissected from mice 8 weeks after injection for histological analysis.
Electron microscopy
The G418-selected hiPSC-ATIICs on day 14 were prepared for ultra-structural examination using previously published protocol [18], which was performed by Electron Microscopy Laboratory, Department of Pathology, University of Medical School at Houston.
Regulated secretion of surfactant from cultured hiPSC-ATIICs
G418-selected hiPSC-ATIICs on day 14 were trypsinized and then seeded back onto fresh Matrigel-coated 10 cm culture plates with DM containing 3H-choline (1 mCi/plate, PerkinElmer) for 24 hr. Cells were then rinsed 3 times with PBS and cultured for 2 hr at 37°C with or without secretagogue (TPA, 50ng/ml; Sigma-Aldrich) [22]. The 3H-labeled phosphatidylcholine (PC) in the medium and cells were extracted and counted, respectively [23]. To examine surfactant protein secretion, the selected hiPSC-ATIICs were trypsinized on day 9 and then cultured by using Air-liquid-interface culture system in Small Airway Epithelial Cell Growth Medium (SAGM™, Chemicon Millipore) containing G418, with or without dibutyryl cAMP (Bt2cAMP, 1mM) + dexamethasone (Dex, 10−10M) for 5 days [21]. The proteins were harvested from culture medium and analyzed by Western blot using rabbit polyclonal antisera against SPB and SPC.
Transplantation of hiPSC-ATIICs into BLM-injured mice lungs
Pathogen-free, 8 to 10 week old, female SCID mice on a C57BL/6 genetic background (Jackson Laboratories, Bar Harbor, Maine) were exposed to bleomycin (BLM, 3.5 units/kg, Bristol-Myers Squibb Company) or sterile normal saline endotracheally via oropharynx intubation. On day 2 after BLM-challenge, mice received saline, human monocytes (hmonos) or G418-selected hiPSC-ATIICs harvested on day 14 (n=8). All cells were gently washed in sterile normal saline for 3 times and then re-suspended at 1.0×106 cells in 50 μl of sterile normal saline for transplantation via an endotracheal catheter [20]. The mice were weighed every another day for 10 days. The spontaneous lung tidal volumes and blood arterial oxygen saturation levels were measured at end time point as previously described [20]. Each group of mice was sacrificed on day 10 after euthanized and lungs were harvested for histological analysis. All procedures were approved by the Animal Welfare Committee, University of Texas Health Science Center at Houston.
Histological analysis and hydroxyproline assay
BLM-challenged lungs with and without transplantation of hiPSC-ATIICs or hmonos were harvested, lavaged, and fixed for embedding as previously described [20]. Tissue sections from three different levels of each lung were stained with hematoxylin-eosin for routine histology. BLM-induced lung damage was graded according to the injured lung area (0, < 10, 10 to 25, 25 to 50, 50 to 75 and > 75%) involved with cellular infiltration, interstitial thickening, and structure distortion [24]. Hydroxyproline assay was performed to evaluate content of collagen deposition in the BLM-injured lungs using previously described method [20].
Statistical analysis
The data were analyzed with the Student’s t-test or the Chi Square Frequency Test, and the resulting pairwise P values less than 0.05 were considered statistically significant.
Results
Efficient generation of random-integration-free and exogenous reprogramming-factor-free hiPSCs
This 3′HPRT.OSMK-LoxP.rtTA.SPCPNEOR.B2M targeting vector (Fig. 1) is designed to target the 3′ region of the B2M gene at a high targeting frequency, without disruption of the B2M function [17]. Of ~ 2.5×104 stably transfected fibroblasts seeded on MEF feeder cells, 96 hESC-like hiPSC colonies were picked up 3 weeks after transfection, representing a reprogramming frequency of 0.384%. Of these, 10 were found to contain a single copy of the correctly inserted transgenes immediately downstream of B2M, of which hiPSC clone 26 (hiPSC-26) was selected for further study. Southern blot hybridization demonstrated that hiPSC-26 contains one 4.6 kb wild-type (wt) and one 2.1 kb targeted B2M HindIII fragment hybridized to the gap probe (Fig. 2A upper panel), indicating that the targeting vector had been knocked into immediately downstream of B2M locus. To confirm this, PCR was performed with primer A and B. As primer A is located within rtTA transgene and primer B in the gap region of the B2M (Fig. 1), the PCR can only detect the B2M-targeted vector, but not those randomly inserted vectors. In support of Southern blot hybridization analysis, one 2.2 kb transgene fragment located in B2M locus was detected in the hiPSC-26 (Fig. 2A, lower panel). To analyze the number of integration sites, genomic DNA samples were prepared and digested by SalI, KpnI, MluI and ClaI, respectively, for Southern blot hybridization using a 425 bp DNA fragment homologous to neomycin transgene as probe. The hiPSC-26 was found to have a single copy of integrated vector in one site (Fig. 2B). Collectively, these results demonstrate that a single copy of B2M-targeted non-viral vector can be achieved for efficient reprogramming.
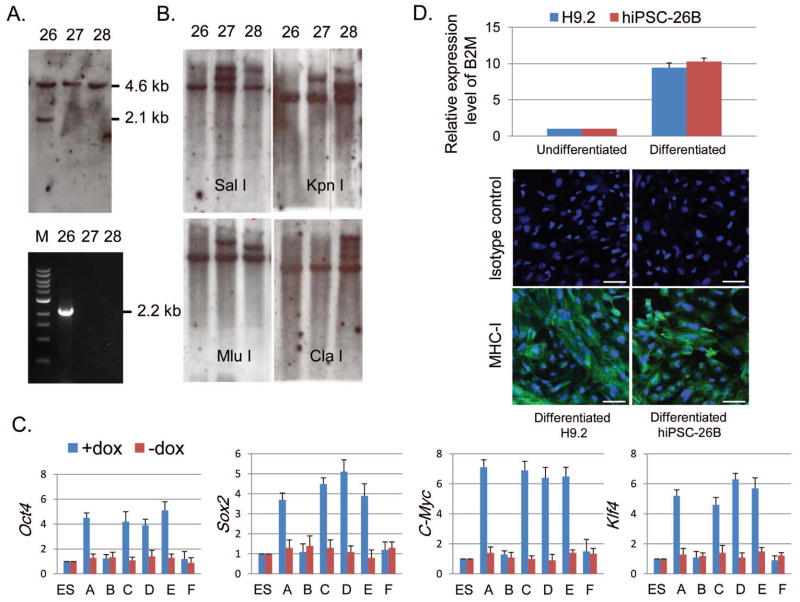
Figure 2. Identification of random-integration-free and reprogramming-factor-free hiPSCs.
(A) Southern blot and PCR analysis for targeting detection. One 4.6 kb wt B2M HindIII fragment was present in all hiPSC clones. One 2.1 kb targeted B2M HindIII fragment was detected by the gap probe in hiPSC-26, indicating a site-specific targeting event (upper panel). PCR analysis demonstrated a 2.2 kb B2M-targeted transgene fragment in hiPSC-26, but not in hiPSC clones 27 and 28 (lower panel). (B) Southern blot analysis of integration sites of targeting vector. A 425 bp NEOR DNA fragment was used as a hybridization probe to analyze SalI-, KpnI-, MluI-, and ClaI-digested genomic DNA of hiPSC clones, and demonstrated that hiPSC-26 contains a single copy of targeting vector in one site. (C) QRT-PCR analysis of endogenous and induced expression levels of Oct4, Sox2, Klf4 and cMyc to screen for exogenous reprogramming-factor-free hiPSC clones after transient expression of Cre recombinase. The expression levels were normalized to endogenous 18S rRNA control. Bar graphs depict relative RNA expression levels of hiPSC-26 clone A, B, C, D, E, and F with hESC control (n=5). (D) Expression of B2M and MHC-I. QRT-PCR was performed to analyze relative B2M RNA levels in differentiated and undifferentiated hiPSC-26B cells with hESCs as control. The expression levels of B2M were normalized to endogenous 18S rRNA control. (n=5) (upper panel). Immunofluorescent staining showed expression level of MHC-I on surface of differentiated hiPSC-26B and hESC control, with mouse IgG1+IgG3 as isotype control for the staining (lower panel, scale bar = 50μm).
To remove the 4 reprogramming factor transgenes including the cancer-promoting genes Klf4 and cMyc, the hiPSC-26 was transfected with Cre-IRES-PUROR vector (Addgene) by using Nucleofector II as previously described [18]. To reduce vector-integration, uncut vector was used. Six hiPSC-26 colonies (A, B, C, D, E, and F) were randomly selected on day 6 after transfecion to assess expression of exogenous reprogramming factors by QRT-PCR. Although significantly induced expression of each reprogramming factor can be demonstrated in the cultures of hiPSC-26 clones, A, C, D, and E in the presence of doxy, there was no evidence of induced expression of those reprogramming factors in the cultured hiPSC-26 clone B and F (Fig. 2C), representing exogenous reprogramming-factor-free hiPSC clones. PCR using Cre forward and reverse primer demonstrated no Cre vector-integration in the hiPSC-26 clone B (hiPSC-26B, Supporting information Fig. S2), indicating that transient expression of Cre recombinase had occurred for Cre mediated recombination in this clone. Thus, hiPSC-26B was used in the following study. To examine if this site-specific insertion has caused B2M gene dysfunction, QRT-PCR was performed using B2M forward and reverse primer to assess B2M expression. The iPSC-26B and hESC (H9.2) were cultured in DM and left to spontaneously differentiate for up to 14 days. In comparison to H9.2, the hiPSC-26B exhibited similar expression level of B2M RNA in the differentiated as well as undifferentiated culture (Fig. 2D). Since B2M is essential for cell surface expression of Major Histocompatibility Complex class I (MHC-I) molecules, aberrantly expressed B2M could lead to lack of MHC-I expression on cell surface. To further evaluate B2M function of the hiPSC-26B, immunocytochemistry was performed to examine MHC-I expression. In support of the B2M expression data, normal expression level of MHC-I was demonstrated on the surface of the differentiated hiPSC-26B cells (Fig. 2D). Taken together, these data demonstrate that hiPSC-26B is genetic mutation-free and exogenous reprogramming-factor-free clone.
The hiPSC-26B cells propagated robustly and maintained their self-renewal capacity, exhibiting typical hESC morphology for at least 68 passages in hESC supporting conditions in absence of doxy. As demonstrated by immunocytochemistry, the hiPSC-26B uniformly expressed the pluripotency markers Nanog, Oct4, SSEA4 and TRA-1-60, but not SSEA1, (Fig. 3A). The cytogenetic analysis using standard protocols for high-resolution G-banding showed the normal karyotype in all analyzed metaphase cells of hiPSC-26B (Fig. 3B). To examine in vivo pluripotency of hiPSC-26B, cells were transplanted intramuscularly on left hind leg of SCID mice to test for teratoma formation. Tumors were noted in 3 weeks, and harvested in 8 weeks after injection. The histological analysis revealed that the teratomas were comprised of a wide variety of cell types derived from all three embryonic germ layers (Fig. 3C). Collectively, these data demonstrate that genetic mutation-free and exogenous reprogramming-factor-free hiPSCs can be successfully generated with this novel, site-specific targeting strategy.
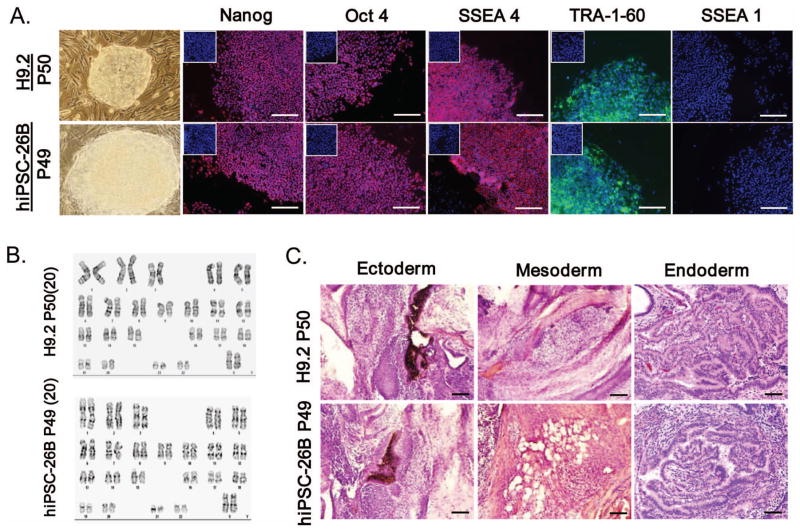
Figure 3. Characterization of hiPSC-26B cells.
(A) Immunofluorescent staining for a panel of pluripotent markers, Nanog, Oct4, SSEA4 and TRA-1–60, in hiPSC-26B and hESCs (scale bar = 100 μm), with isotype staining control shown in the small insert in each image. (B) Cytogenetic evaluation of harvested cells from hiPSC-26B and hESCs. Twenty metaphase cells were analyzed per sample. (C) Teratoma formation assay for in vivo pluripotency showed various tissues in hiPSC-26B and hESC teratoma (scale bar = 200 μm).
Differentiation and purification of hiPSC derived ATIICs
When cells were cultured on Matrigel-coated plate in DM, approximately 13.2% of cells in differentiated cultures of hiPSC-26B spontaneously differentiated into ATIICs on day 14 (Table 1), which was demonstrated by immunostaining for ATIIC-specific SPC expression. The percentage of SPC positive cells was comparable to that (14.0%) in differentiated H9.2 cell cultures. To enrich and purify hiPSC-ATIICs, hiPSC-26B cells were subjected to spontaneous differentiation in DM containing 20 μg/ml of G418 for 14 days. As mentioned above, hiPSC-26B cells already harbor the SPCP-NEOR transgene downstream of the B2M gene. Since SPC protein is specifically expressed by ATIICs, NEOR should be only expressed when hiPSC-26B cells are differentiated into ATIICs. Therefore only hiPSC-ATIICs can survive G418 selection. To determine the purity of G418-selected hiPSC-ATIICs in the differentiated cultures of hiPSC-26B, the differentiated cells were stained with anti-human proSPC. The number of SPC positive cells was counted per 1,000 cells based on DAPI staining on each plate. The results showed that the G418-selection leads to an enrichment of hiPSC-ATIICs to greater than 99% (Table 1). The few SPC negative cells were ATICs due to spontaneously differentiation of the ATIICs to ATICs after removal of G418.
Table 1.
Relative content of hiPSC-ATIICs in G418 selected and nonselected cultures of differentiated hiPSC-26B
| G418 selection | SPC+ cells | SPC− cells | SPC+ cells %* | |
|---|---|---|---|---|
| H9.2 | − | 140 ± 8.03 | 860 ± 8.03 | 14.0 ± 0.8** |
| hiPSC-26B | − | 132 ± 8.68 | 868 ± 8.68 | 13.2 ± 0.86 |
| 4hiPSC-26B | + | 992 ± 2.55 | 8 ± 2.55 | 99.2 ± 2.55 |
The percentage of SPC positive cells was determined by visually counting 1,000 cells based on DAPI staining in the differentiated cultures on day 14.
P=0.991 versus nonselected hiPSC-26B group
Ultra-structure and biological functions of hiPSC-ATIICs
Ultra-structural examination by transmission electron microscopy demonstrated that hiPSC-ATIICs are morphologically normal and exhibit typical lamellar bodies with normal concentric, tightly packed lamellae (Fig. 4A), which is a characteristic hallmark of hATIICs. Papanicolaous staining demonstrated that all G418-selected hiPSC-ATIICs contain lamellar bodies (Fig. S3B). As hATIIC control, G418-selected hiPSC-ATIICs expressed SPA, SPB, SPC and thyroid transcription factor 1 (TTF1) (Fig. 4A). Recently, SPC+CCSP+ bronchioalveolar stem cells (BASCs) have been identified at bronchioalveolar duct junction [25]. These SPC+CCSP+ BASCs may serve as progenitor for both ATIICs and Clara cells. To examine the possibility if the hiPSC-26B derived BASCs were also present in the G418-selected cultures, the cells were stained with CCSP. No CCSP positive cells were observed in the G418-selected cultures on day 14 (Fig. 4A), suggesting that production of the more primitive lung precursor cells derived from hiPSCs/hESCs may be difficult. It has been reported that hATIICs in primary culture system retain regulated expression and secretion of surfactant in a manner that recapitulates that found in lungs [21]. To examine if the hiPSC-ATIICs possess the same ability, the cultured cells were labeled with 3H-choline and examined for stimulated surfactant lipid secretion in response to secretagogue. We found that secretion of the 3H-labeled PC (as percent of total 3H-labeled PC) by hiPSC-ATIICs was in a regulated fashion as that seen in the control hATIICs (Fig. 4B), which demonstrated the active surfactant metabolism pathway in the hiPSC-ATIICs. To further examine if the hiPSC-ATIICs can be induced to express and secrete surfactant proteins, the selected hiPSC-ATIICs were cultured by using Air-liquid-interface culture system in SAGM™ in the absence or presence of Bt2cAMP + Dex for 5 days. We found that hATIICs and hiPSC-ATIICs synthesize and secrete SPC and SPB at very similar levels, which can be significantly induced in response to the treatment of Bt2cAMP + Dex (Fig. 4C).
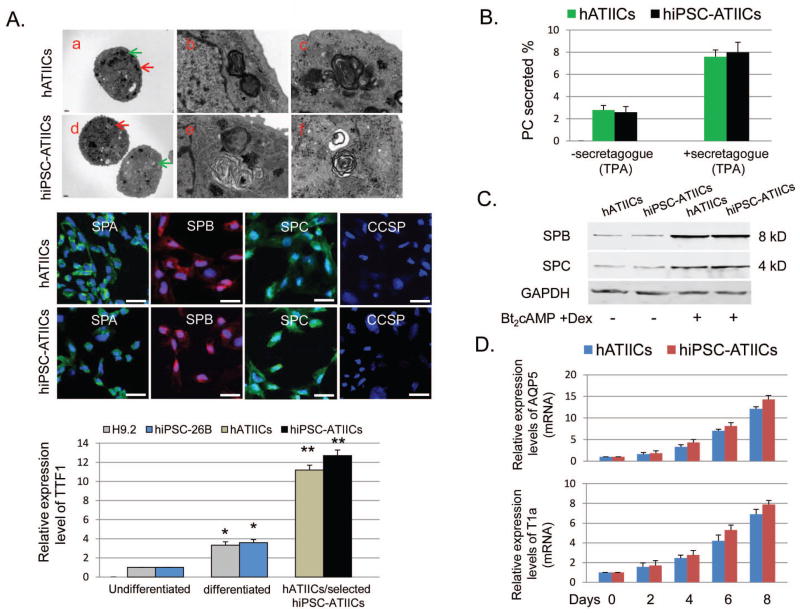
Figure 4. Characterization of hiPSC-ATIICs.
(A, top panel) Electron micrographs showed well-developed lamellar bodies and other morphological characteristics in the selected hiPSC-ATIICs on day 14 (d, scale bar = 1μm) as observed in hATIICs (a, scale bar = 1 μm). (b and c) Magnified views of regions indicated by red and green arrow in a, respectively, showing clear structures of lamellar bodies (Scale bar = 0.1 μm). (e and f) Magnified views of regions indicated by red and green arrow in d, respectively, clearly showing the lamellar structures (Scale bar = 0.1 μm). (A, middle panel) Immunofluorescent staining demonstrated that G418-selected hiPSC-ATIICs express SPA, SPB and SPC as control hATIICs, but do not express CCSP (Scale bar = 25 μm). (A, bottom panel) QRT-PCR was performed to determine expression levels of human TTF1 in the G418-selected and nonselected cultures of differentiated hiPSC-26B on day 14, using 18S rRNA as endogenous control. The TTF1 primers (Table S1) were designed to detect human TTF1 mRNA. The expression levels of TTF1 were found to significantly increase in the spontaneously differentiated cultures of H9.2 and hiPSC-26B on day 14 (n=5), and compared to these increases, the increases in the expression levels of TTF1 in hATIIC and G418-selected hiPSC-ATIIC cultures were much higher (* P<0.01 versus undifferentiated H9.2 or hiPSC-26B; ** P<0.001 versus differentiated H9.2 and hiPSC-26B). (B) Stimulated surfactant secretion from cultured hiPSC-ATIICs in response to secretagogue (TPA, 50ng/ml). Secretion of 3H-labeled PC was shown as percent of total 3H-labeled PC in the absence or presence of secretagogue (n=5). (C) Secretion of SPB and SPC from cultured hiPSC-ATIICs. Media collected from cultured hiPSC-ATIICs and hATIICs in the absence or presence of Bt2cAMP+Dex were analyzed by immunoblotting for SPB and SPC. The GAPDH expression in each sample was shown in the bottom plot as a control. (D) QRT-PCR was performed to analyze expression levels of ATIC markers, AQP5 and T1 α in the differentiating cultures of hiPSC-ATIICs on days 0, 2, 4, 6, and 8, using 18S rRNA as endogenous control (n=5).
One of the key functions of ATIICs is to serve as progenitor cells for maintaining alveolar homeostasis. It is critical that the hiPSC-ATIICs also possess the ability to proliferate and differentiate to ATICs for developing cell-based therapies. To demonstrate this ability, the G418-selected hiPSC-ATIICs were cultured in six-well culture dishes with DMEM containing 10% FBS and left to spontaneously differentiate for up to 8 days [20]. Total RNA was isolated from the cultures on days 0, 2, 4, 6, and 8. Differentiation to ATICs was assessed by QRT-PCR to detect gene expression of Aquaporin-5 (AQP5) and Podoplanin (T1α), which are frequently used as markers for ATICs [26–30]. As hATIICs, differentiated hiPSC-ATIICs exhibited increased expression of AQP5 and T1α at a similar level, indicating differentiation over time into ATICs (Fig. 4D). To examine their proliferation capacity, the selected hiPSC-ATIICs were cultured on Matrigel-coated 6-well plates at 1.0×104 cells/well in MEF conditioned DMEM for up to 7 days to test the colony formation ability [20]. A few small colonies were observed on day 4, but more colonies (~16 colonies/well) composed of 15–21 cells were present on day 7’s cultures, indicating that hiPSC-ATIICs proliferated in vitro. Proliferation capacity of hiPSC-ATIICs can be promoted by addition of recombinant keratinocyte growth factor (rhKGF, 30ng/ml; R&D Systems) in the culture medium, leading to significantly increased number and size of colonies (average 45 colonies/well and 27–40 cells/colony, Supporting information Fig. S3). Taken together, these data have demonstrated that hiPSC-26B cells can be derived into an enriched population of ATIICs that possess normal important biological functions and ultra-structure characteristics of hATIICs. Thus, the hiPSCs generated by our novel strategy will provide reliable source of cells to explore their therapeutic potential in lung tissue regeneration.
Functional repair of bleomycin-induced acute alveolar injury in mice by transplanted hiPSC-ATIICs
We used BLM-induced acute lung injury model, which is well characterized in mice and used routinely as a general model of pulmonary fibrosis [31, 32], to evaluate the capacity of hiPSC-ATIICs to engraft and repair damaged alveoli. To avoid graft rejection of hiPSC-ATIICs, immune deficient SCID mice were used in the study. As described in Experimental procedures, mice were transplanted with hiPSC-ATIICs (1.0×106 in 50 μl of saline) endotracheally on day 2 after BLM exposure. In addition, BLM-challenged mice transplanted with same number of hmonos were used to verify the specificity of hiPSC-ATIICs in engrafting and repairing the injured alveoli. The engraftment efficiency as well as the capacity of transplanted hiPSC-ATIICs to differentiate into ATICs in lungs was determined by immunohistochemical staining (IHC) of end point lung sections using anti-human nuclei, anti-proSPC and anti-T1α antibody [20]. There was no human nuclei positive cell observed in control mice and BLM-treated mice receiving either saline or hmonos (Fig. 5A and Table 2). In contrast, numerous human nuclei positive cells were identified in the BLM-challenged lungs transplanted with hiPSC-ATIICs, most of which were co-stained with proSPC (indicated by arrows), representing the retention of transplanted hiPSC-ATIICs. QRT-PCR analysis of expression of human TTF1 in the BLM-challenged lungs with and without transplantation of hiPSC-ATIICs supported this observation (Fig. S3A). In addition, co-staining of human nuclei positive cells with T1α (Fig. 5B) in the BLM-challenged lungs transplanted with hiPSC-ATIICs demonstrated that some of transplanted hiPSC-ATIICs had differentiated into ATICs. To determine percentage of remained hiPSC-ATIICs that had been differentiated into ATICs, 500 SPC positive cells were counted in at least 3 slides per mouse lung receiving hiPSC-ATIICs; the number of human nuclei positive cells was determined in the same counted area of 500 SPC positive cells. Of these human nuclei positive cells, ~59.1% of cells were co-stained with proSPC and 40.9% of cells proSPC negative (Table 2), indicating numerous remained hiPSC-ATIICs had differentiated into ATICs. The percentage of hiPSC-ATIICs differentiated into ATICs was confirmed by counting human nuclei positive cells co-stained with T1α in BLM-challenged lungs. Collectively, these data demonstrate the capacity of transplanted hiPSC-ATIICs to remain and differentiate into ATICs in injured alveoli.
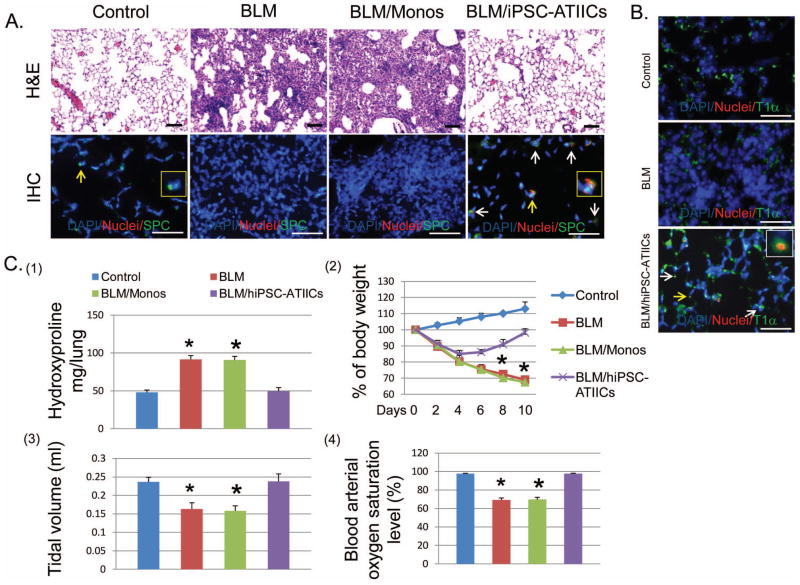
Figure 5. Functional repair of BLM-injured lungs by transplanted hiPSC-ATIICs in mice.
(A) Hematoxylin–eosin (H&E) staining of representative BLM-injured lung sections (upper panel, scale bar = 200 μm) showed that BLM-challenge caused severe alveolar architecture damage in 73.1 ± 5.3% of lung area with extensive inflammatory infiltrates and edema (the second image) compared with saline control (the left image). Functional transplantation of hiPSC-ATIICs was noted by significantly reduced alveolar structure damage in only 6.2 ± 2.3% of lung tissue (the right image). In contrast, the severity of alveolar structure damage in the BLM-challenged lung transplanted with hmonos (71.8 ± 5.9%) was similar to BLM-challenged lungs without hiPSC-ATIIC transplantation (the third image). Immunofluorescent staining of representative lung sections from BLM-challenged mice with and without transplanted hiPSC-ATIICs or hmonos was performed by using a mouse anti-human nuclei monoclonal antibody and rabbit anti-human proSPC antibody with DAPI counterstaining (lower panel, scale bar = 100 μm). The human nuclei monoclonal antibody recognizes cells of human origin in mouse lungs (red); the rabbit anti-human proSPC stains both mouse and human ATIICs that express SPC (green). The normal distribution of mouse ATIICs was observed in control lungs (the first image). A magnified view of a SPC positive cell indicated by the yellow arrow is inserted. In compared to control lungs, no ATIICs existed in BLM-injured lung area of mice received either saline or hmonos (the second and the third image). Human nuclei positive cells (red) were identified only in the BLM-challenged lungs transplanted with hiPSC-ATIICs. Some of these human nuclei positive cells (red) were co-stained with SPC (green), representing remained hiPSC-ATIICs (the right image, indicated by arrows) in the injured lungs. The magnified view of a human nuclei positive and SPC positive cell indicated by the yellow arrow is inserted. Those human nuclei positive cells that were SPC negative in the BLM-injured lungs suggested that some of transplanted hiPSC-ATIICs had differentiated into ATICs in the lungs. (B) Immunofluorescent staining of representative lung sections from BLM-challenged mice with and without transplanted hiPSC-ATIICs was performed by using a mouse anti-human nuclei monoclonal antibody and 1:50 diluted rabbit anti-human T1α antibody (Abgent, San Diego, CA), with DAPI counterstaining, for identifying human derived ATICs. The rabbit anti-human T1α antibody stains both mouse and human ATICs that express T1α (green). The T1α positive cells were observed in control lungs (control panel, scale bar = 100 μm) and BLM-challenged lings (BLM panel). Human nuclei positive cells (red) were identified only in the BLM-challenged lungs transplanted with hiPSC-ATIICs (BLM/hiPSC-ATIICs panel). Some of these human nuclei positive cells were co-stained with T1α (indicated by the arrows), indicating the cells with ATIC phenotype derived from the transplanted hiPSC-ATIICs in the injured lungs. A magnified view of a human derived cell with ATIC phenotype indicated by yellow arrow was inserted in the image. (C) (1). Hydroxyproline levels in BLM-challenged lungs (91.56 ± 4.93 μg/lung) were significantly increased compared with that in saline control lungs (47.90 ± 3.30 μg/lung) at end of time point. Transplantation of hiPSC-ATIICs significantly reduced the content of hydroxyproline (49.71 ± 4.46μg/lung) in BLM-challenged lungs to the control levels. Consistent with histological analysis, transplanted hmonos did not reduce collagen deposition (90.7 ± 4.87 μg/lung) in the BLM-challenged lungs (n=8). (2). During the time course of BLM-challenge, body weight of BLM-challenged mice received either saline or hmonos significantly decreased over time and was down to ~70% of their initial body weight at end of time point. Although BLM-challenged mice that received hiPSC-ATIICs experienced an initial body weight loss, they started their body weight recovery on day 6 after lung injury and had recovered ~98% of their initial body weight on day 10 (n=8). (3). The lung tidal volumes were measured in BLM-challenged mice with and without transplantation of hiPSC-ATIICs or hmonos at end of time point. BLM-challenge caused a significant decrease in tidal volume (0.163 ± 0.017 ml) compared to saline control (0.236 ± 0.013 ml). In comparison, transplanted hiPSC-ATIICs (0.238 ± 0.020 ml), but not transplanted hmonos (0.158± 0.013 ml), restored tidal volume in BLM-challenged mice to the control levels. (n=8) (4). The blood arterial oxygen saturation levels were recorded in BLM-challenged mice with and without transplantation of hiPSC-ATIICs or hmonos at end of time point. The blood arterial oxygen saturation levels significantly decreased in BLM-challenged mice (69.25 ± 2.11%) and BLM-challenged mice that received hmonos (69.66 ± 2.44%) compared to that in saline control mice (97.73 ± 0.33%). In contrast, BLM-challenged mice transplanted with hiPSC-ATIICs had completely recovered normal arterial oxygen saturation levels (97.66 ± 0.49%) (n=8). * P<0.001 versus BLM/hiPSC-ATIICs and control group.
Table 2.
Relative content of human derived ATIICs and ATICs in BLM-mouse lung tissue
| SPC+ | Nuclei+ (#) | SPC+/Nuclei+ | SPC+/Nuclei− | SPC−/Nuclei+ | SPC+ Nuclei+ (%*) | SPC−Nuclei+ (%**) | |
|---|---|---|---|---|---|---|---|
| Saline-SCID | 500.0 | - | - | 500.0 | - | - | - |
| BLM-SCID | 100.0 | - | - | 100.0 | - | - | - |
| BLM-SCID/Monos | 100.0 | - | - | 100.0 | - | - | - |
| BLM-SCID/ATIICs | 500.0 | 232.0 ± 4.8 | 137.0 ± 4.0 (59.1%)a | 363.0 ± 4 | 95 ± 8.6 (40.9%)b | 37.74 ± 1.5 | 69.5 ± 8.2 |
SPC+: ATIICs (mouse and human ATIICs)
Nuclei+: human derived cells
SPC+/Nuclei+: hiPSC-ATIICs
SPC+/Nuclei−: mouse ATIICs
number of nuclei+ cells in the counted area of 500 SPC+ cells
ratio of hiPSC-ATIICs to mouse ATIICs, %**: ratio of SPC− human cells to hiPSC-ATIICs
ratio of hiPSC-ATIICs to human derived cells,
ratio of SPC− human cells to human derived cells
Exposure to this relatively high dose of BLM severely impaired the endogenous repair capacity, thus damaged alveolar epithelium could not be effectively repaired, resulting in severe alveolar structure damage in 73.1 ± 5.3% of area showing extensive accumulation of inflammatory cells and interstitial thickening on day 10 (Fig. 5A). To evaluate BLM mediated collagen deposition in the lungs, level of hydroxyproline, a modified amino acid specifically derived from collagen, was assessed. BLM-exposed lungs showed a significantly increased hydroxyproline content when compared with control lungs (Fig. 5C1). Transplantation of hmonos after BLM-challenge did not significantly reduce BLM-induced lung damage (71.8 ± 5.9%) and collagen deposition. In contrast, the extent of parenchymal damage within BLM-challenged lungs transplanted with hiPSC-ATIICs was greatly reduced to only 6.2 ± 2.3% of the lung area as evidence by a few isolated, small destructed regions, whereas much larger areas showed tissue with normal alveolar organization (Fig. 5A). In addition, the level of hydroxyproline in the BLM-injured lungs that received hiPSC-ATIICs was significantly reduced to control level, suggesting that transplanted hiPSC-ATIICs reversed or prevented development of BLM-induced fibrosis.
BLM-induced destruction of alveolar architecture could result in severely impaired pulmonary function associated with significant loss of body weight. As illustrated in Figure 5A and 5C1, BLM-induced alveolar damage and collagen deposition were significantly repaired by transplanted hiPSC-ATIICs; therefore, it was expected that impaired lung function of BLM-exposed mice can be restored by transplanted hiPSC-ATIICs. To test this hypothesis, body weight and lung function were examined. The body weight of BLM-challenged mice receiving either saline or hmonos significantly decreased on day 2 and continued to decrease over time to ~70% of their starting body weight on day 10 (Fig. 5C2). Although BLM-challenged mice transplanted with hiPSC-ATIICs experienced an initial decrease in body weight, their body weight was significantly increased on day 6 and almost completely recovered on day 10. The lung tidal volumes of BLM-challenged mice without transplantation of hiPSC-ATIICs significantly decreased. Likewise, blood oxygen levels of these mice declined drastically (Fig. 5C3 and 4), indicating that the mice experienced respiratory failure. As expected, the BLM-challenged mice transplanted with hiPSC-ATIICs showed normal lung tidal volumes and blood arterial oxygen saturation levels, demonstrating functional repair of injured alveoli by the transplanted hiPSC-ATIICs.
Any pluripotent cells or incompletely differentiated hiPSCs remained in G418 selected hiPSC-ATIIC cultures may cause either teratomas or lung epithelial tumors after transplantation into recipients. To assess the possibility, 24 female SCID mice were subjected to BLM induced acute lung injury, and treated with saline, hmonos or hiPSC-ATIICs (8 mice/group) as above. All the BLM-challenged mice without the treatment of hiPSC-ATIICs died of severe respiratory failure before day 12 after BLM-challenge (Supporting information Fig. S4A). Impressively, all the BLM-challenged mice transplanted with hiPSC-ATIICs remained healthy with normal pulmonary function and no evidence of teratoma formation during the study period of 11 months after transplantation (Supporting information Fig. S4), indicating that the selection procedure produces pluripotent cell-free hiPSC-ATIICs. Immunofluorescent staining demonstrated that ~ 5% of the hiPSC-ATIICs and ~8.4% of the hiPSC-ATIIC derived ATICs remained in the BLM-challenged lungs 330 days after transplantation (Supporting information Fig. S4C, Table S2). The percentage of remaining hiPSC-ATIICs as well as their derived ATICs reflects the turnover of transplanted hiPSC-ATIICs in the BLM-injured lungs, which is comparable to what we found in our previous study [20]. Collectively, these data demonstrate the capacity of transplanted hiPSC-ATIICs to functionally repair injured alveolar epithelium with a long-term benefit, without side effect.
Discussion
With our novel strategy, a single copy of all of the required transgenes can be specifically knocked into a site immediately downstream of B2M gene locus at a high frequency to generate random-integration-free and exogenous reprogramming-factor-free hiPSCs, without causing B2M dysfunction. Thus the risk of genetic abnormalities caused by vector integrations, a major concern for clinical use of hiPSCs, is completely eliminated. Previously it was reported that a significant number of iPSCs generated with vector-based methodologies was not fully reprogrammed to pluripotency [33, 34], possibly because of stochastic expression variation of reprogramming factors and/or disrupted reprogramming pathways due to insertion mutations. Non-genetic techniques have been developed to generate vector-integration-free iPSCs, yet they require repeated delivery of reprogramming factors, which is still stochastic. A major advantage of our new strategy is that a single copy of 4 reprogramming factor transgenes can be inserted downstream of the B2M gene locus, thereby facilitating the efficient reprogramming somatic cells into genetic mutation-free hiPSCs. As expected, the derived hiPSCs express a panel of stem cell markers, including TRA-1-60 previously reported not to be expressed in partially reprogrammed clones [33], and strongly resemble characteristics of hESCs with a long-term self-renewal capacity.
Development of safe and efficient methodologies to derive pluripotent stem cells into transplantable tissue cell types is critical to explore the therapeutic potential of hiPSCs for tissue regeneration. Efficiency of differentiation of embryonic stem cells (ESCs) into ATIICs is very low [35]. Use of factors to manipulate signaling pathways for efficient differentiation of ESCs into ATIIC cells has been reported [36], but not yet successful for generating transplantable ATIICs. In addition, it remains unclear whether or not those ESC derived ATIIC cells possess normal biological functions of hATIICs. Recently, a pure population of ATIICs derived from genetic modified hESCs has shown their therapeutic potential for functional repair of injured alveolar epithelium [20]. However, immune rejection toward hESC derived ATIICs and random genetic modification limit their therapeutic application. As our novel technique combines the ATIIC-specific NEOR expression strategy, the derived random-integration-free and exogenous reprogramming-factor-free hiSPCs can be selectively differentiated into a homogenous population of ATIICs without the need of additional genetic modification. Our data reported for first time that the hiPSC-ATIICs are fully functional with characteristics of hATIICs. A relatively low dose of G418 was used during the entire time course of differentiation for ATIIC selection, which allows more efficient in vitro production of hiPSC-ATIICs compared with a high dose of G418 used to eliminate all other cell types in the last two days (Supporting information Table S3). Treatment with low dose of G418 from the beginning may promote activation of the signaling pathways required for derivation of ATIICs. Although the underlying mechanism still awaits deciphering, our results demonstrate that this novel strategy is reliable for producing sufficient number of functional hiPSC-ATIICs for regeneration of injured alveolar epithelium.
To explore the therapeutic potential of hiPSC-ATIICs, a relative high dose of BLM was used to eliminate the endogenous repair capacity of the injured alveoli. Therefore, no ATIICs survived in the damaged alveolar areas of BLM-challenged mice treated with saline or hmonos, causing extensive alveolar structure damage. All of these mice died 12 days after BLM-challenge. Our data have demonstrated the retention and differentiation of hiPSC-ATIICs in the BLM-injured lungs that received hiPSC-ATIICs on day 10, suggesting that functional transplantation of hiPSC-ATIICs had prevented or reversed development of pulmonary fibrosis. It was observed that numerous endogenous mouse ATIICs survived in the BLM-injured lungs of hiPSC-ATIIC transplanted mice, suggesting that structural engraftment and differentiation of hiPSC-ATIICs is not the only beneficial event provided by transplanted hiPSC-ATIICs. In vitro studies have suggested that reciprocal interaction between ATIICs and messenchymal cells may play an essential role in maintaining the homeostasis of vascular and alveolar epithelial compartment in lung [37, 38]. Therefore the remained hiPSC-ATIICs may provide an appropriate signal microenvironment via their paracrine effect to promote endogenous repair capacity. Thus, structural engraftment and differentiation of hiPSC-ATIICs coupled with activated endogenous ATIICs accounted for robust re-epithelialization of injured alveoli. Further investigations are needed to characterize signaling communication between ATIICs for maintaining alveolar homeostasis. In addition, unlike BLM-challenged mice treated with saline or hmonos, all BLM-injured mice transplanted with hiPSC-ATIICs maintained normal pulmonary function well with no evidence of tumorigenesis for at least up to 11 months, suggesting that the strategy is safe and efficient to generate transplantable hiPSC-ATIICs.
Summary
In conclusion, we have successfully established a novel site-specific insertion targeting strategy for efficiently generating genetic mutation-free and reprogramming-factor-free hiPSCs. Importantly, the derived hiPSCs can be subsequently differentiated into a homogenous population of functional ATIICs. This novel strategy makes it plausible for the first time to explore the clinical application of patient-specific iPSC-derived ATIICs. Furthermore, this technology can be applied generally for generation of hiPSC for transplantation in different organs. For example, the ATIIC-specific NEOR expression transgene in the vector can be easily replaced with any cell type-specific NEOR expression transgene to develop a platform for generating patient-specific neuron progenitor, heart cells, pancreatic beta-cells and other for tissue regeneration or disease modeling.
Supplementary Material
Figure S1. Structures of pTRE-OIS vector, pTRE-KIcML vector, SPCP-NEOR vector, and EFαP-rtTA vector, related to Figure 1
The schematic diagram of each vector is to depict relevant positional information, hence not scaled proportionally based on sequence lengths. (A) pTRE-OIS vector contains Oct4 and Sox2 transgene. The internal ribosome entry site (IRES) of the encephalomyocarditis virus located between the two trangenes allows co-translating Oct4 and Sox2 from a single RNA transcript under the control of TRE-PminCMV promoter. (B) pTRE-KIcML vector harbors Klf4 and cMyc transgene. The IRES is included in this vector for co-translation of the two transgenes under the control of TRE-PminCMV promoter. This vector also contains LoxP targeting sequence downstream of cMyc cDNA. (C) SPCP-NEOR vector contains NEOR transgene under the control of ATIIC-specific SPC promoter (SPCP). (D) EFαP-rtTA vector contains rtTA cDNA downstream of EF-1a promoter (EFαP).
Figure S2. PCR analysis of vector-integration of Cre-IRES-PUROR, related to Figure 2
PCR was performed to examine Cre-IRES-PUROR vector-integration in chromosomal DNA of hiPSC-26 clones A, B, C, D, E, and F after transfection. PCR using Cre forward and Cre reverse primer generated one 745 bp Cre transgene DNA fragment in hiPSC-26 clone F, but not in clones A, B, C, D, and E, indicating that vector-integration event had occurred in hiPSC-26 clone F.
Figure S3. Characterization of hiPSC-ATIICs, related to Figure 4
(A) To examine if human thyroid transcription factor 1 (TTF1) can be detected in the BLM-challenged lungs transplanted with hiPSC-ATIICs, QRT-PCR was performed using total RNA isolated from the BLM-challenged lungs with and without transplantation of hiPSC-ATIICs on day 10 (n=5). The TTF1 primers (Table S1) were designed to detect human TTF1 mRNA in mouse lungs using 18S rRNA as endogenous control. TTF1 RNA was detected in the BLM-challenged lungs transplanted with hiPSC-ATIICs, but not in the control lungs and BLM-challenged lungs without hiPSC-ATIIC transplantation. (B) Papanicolaous staining of lamellar bodies. To show a group of hiPSC-ATIICs containing lamellar bodies, the modified Papanicolaous staining, which is another procedure routinely used for identifying lamellar bodies, was performed. As control hATIICs (the upper image, x630), all of the G418 selected hiPSC-ATIICs contained abundant lamellar bodies on day 14 (the lower image, x630). (C) Proliferation capacity of hiPSC-ATIICs in vitro. The hiPSC-ATIIC colonies (~16 colonies/well) were observed when cells were cultured on Matrigel-coated plates in mouse MEF-conditioned DMEM containing 10% FBS for 7 days, and significantly larger number of hiPSC-ATIIC colony (~45 colonies/well) were present in the rhKGF (30ng/ml) treated cultures compared to control (upper panel). The rhKGF-promoted proliferation was also demonstrated by the evidence of larger hiPSC-ATIIC colonies containing 27–40 cells in the rhKGF-treated cultures compared with those containing 15–21 cells in the untreated cultures.
Figure S4. Percent survival and pulmonary function of BLM-challenged mice with and without transplantation of hiPSC-ATIICs, related to Figure 5
To assess whether the therapeutic benefits provided by hiPSC-ATIICs is long-term without teratoma formation, 24 female SCID mice were subjected to BLM induced acute lung injury, and received transplantation using either saline, hmonos or hiPSC-ATIICs (8 mice/group) on day 2 after BLM challenge. (A) On day 7 after BLM-challenge, 1 of 8 BLM-challenged mice that received saline and 2 of 8 BLM-challenged mice transplanted with hmonos had died. Between 7 and 10 days, another 2 BLM-challenged mice that received saline and 1 BLM-challenged mouse transplanted with hmonos had died. All of the remaining BLM-challenged mice received either saline or hmonos had died by 12 days after BLM treatment. In contrast, all of the BLM-challenged mice treated with hiPSC-ATIICs survived at least up to 11 months. (B) The BLM-challenged mice with transplantation of hiPSC-ATIICs maintained normal blood arterial oxygen saturation levels (97.125 ± 0.59, upper panel) at end of time point (11 months) compared with saline control mice (97.26 ± 0.67). Similarly, the lung tidal volumes (0.241 ± 0.006, lower panel) measured in these mice were comparable to that (0.243 ± 0.007) in saline control mice. (C) Immunofluorescent staining of representative lung sections from BLM-challenged mice transplanted with hiPSC-ATIICs at end of time point (11 months) using mouse anti human nuclei monoclonal antibody and rabbit anti human pro-SPC antibody. The mouse anti human nuclei monoclonal antibody recognizes cells of human origin in mouse lungs (red); and the rabbit anti human pro-SPC stains both mouse and human ATIICs that express SPC (green). The cells expressing both human nuclei and SPC were present in BLM-challenged lungs transplanted with hiPSC-ATIICs after 11 months (right image, indicated by arrow; scale bar = 100 μm), but not in saline control lungs (left image). The magnified view of human derived ATIIC indicated by yellow arrow was inserted in the right image. (D). Immunofluorescent staining of representative lung sections from BLM-challenged mice transplanted with hiPSC-ATIICs at end of time point (11 months) using mouse anti human TTF1 (PM 087 AA, Biocare Medical). The mouse anti human TTF1 identifies human alveolar epithelial cells, but not mouse alveolar epithelial cells, in mice lungs. The human TTF1 positive cells were observed in BLM-challenged lungs transplanted with hiPSC-ATIICs after 11 months (red, the right image, scale bar = 200 μm) with normal lung histology, but not in the control lungs (the left image). The magnified view of human TTF1 positive cell indicated by yellow arrow was inserted in the right image. These TTF1 positive cells distributed individually in the lungs, and did not expand to form any abnormal structure or cell cluster in the lung, suggesting the remaining hiPSC-ATIICs had not developed into a cancer tissue in the lungs after 11 months.
Acknowledgments
This work was supported by The Brown Foundation Institute of Molecular Medicine, University of Texas Medical School at Houston, U.S. Public Health Service National Institutes Health Grants R21 HL102833-01(D.W) and R01 HL168116 (J.L.A), and the Nancy and Clive Runnells Embryonic Stem Cell Research Fund (R.A.W).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have no financial conflict of interests to declare.
Author Contributions: Q.Y., Y.Q., and D.W.: conception and design, collection and assembly data, data analysis and interpretation, and manuscript writing; H.S., X.P., Z.Z. and J.L.A.: collection of data, data analysis and interpretation; R.A.W.: administrative support.
References
- 1.Selman M, et al. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med. 2001;134(2):136–51. doi: 10.7326/0003-4819-134-2-200101160-00015. [DOI] [PubMed] [Google Scholar]
- 2.Nogee LM, et al. A mutation in the surfactant protein B gene responsible for fatal neonatal respiratory disease in multiple kindreds. J Clin Invest. 1994;93(4):1860–3. doi: 10.1172/JCI117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nogee LM, et al. Brief report: deficiency of pulmonary surfactant protein B in congenital alveolar proteinosis. N Engl J Med. 1993;328(6):406–10. doi: 10.1056/NEJM199302113280606. [DOI] [PubMed] [Google Scholar]
- 4.Hamvas A, et al. Progressive lung disease and surfactant dysfunction with a deletion in surfactant protein C gene. Am J Respir Cell Mol Biol. 2004;30(6):771–6. doi: 10.1165/rcmb.2003-0323OC. [DOI] [PubMed] [Google Scholar]
- 5.Yamanaka S, Takahashi K. Induction of pluripotent stem cells from mouse fibroblast cultures. Tanpakushitsu Kakusan Koso. 2006;51(15):2346–51. [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771–5. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadtfeld M, et al. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–9. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusaki N, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia F, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7(3):197–9. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–70. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–6. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8(6):633–8. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Zheng B, Mills AA, Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 1999;27(11):2354–60. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, et al. A pure population of lung alveolar epithelial type II cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(11):4449–54. doi: 10.1073/pnas.0700052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26(8):916–24. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, et al. Transplantation of human embryonic stem cell-derived alveolar epithelial type II cells abrogates acute lung injury in mice. Mol Ther. 2010;18(3):625–34. doi: 10.1038/mt.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alcorn JL, et al. Primary cell culture of human type II pneumonocytes: maintenance of a differentiated phenotype and transfection with recombinant adenoviruses. Am J Respir Cell Mol Biol. 1997;17(6):672–82. doi: 10.1165/ajrcmb.17.6.2858. [DOI] [PubMed] [Google Scholar]
- 22.Sano K, Voelker DR, Mason RJ. Effect of secretagogues on cytoplasmic free calcium in alveolar type II epithelial cells. Am J Physiol. 1987;253(5 Pt 1):C679–86. doi: 10.1152/ajpcell.1987.253.5.C679. [DOI] [PubMed] [Google Scholar]
- 23.Romero EJ, et al. Interaction of an artificial surfactant in human pulmonary epithelial cells. Pediatr Pulmonol. 2005;39(2):167–77. doi: 10.1002/ppul.20166. [DOI] [PubMed] [Google Scholar]
- 24.Chen ES, et al. Attenuation of lung inflammation and fibrosis in interferon-gamma-deficient mice after intratracheal bleomycin. Am J Respir Cell Mol Biol. 2001;24(5):545–55. doi: 10.1165/ajrcmb.24.5.4064. [DOI] [PubMed] [Google Scholar]
- 25.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Kreda SM, et al. Expression and localization of epithelial aquaporins in the adult human lung. Am J Respir Cell Mol Biol. 2001;24(3):224–34. doi: 10.1165/ajrcmb.24.3.4367. [DOI] [PubMed] [Google Scholar]
- 27.Ramirez MI, et al. T1alpha, a lung type I cell differentiation gene, is required for normal lung cell proliferation and alveolus formation at birth. Dev Biol. 2003;256(1):61–72. doi: 10.1016/s0012-1606(02)00098-2. [DOI] [PubMed] [Google Scholar]
- 28.Vanderbilt JN, et al. Directed expression of transgenes to alveolar type I cells in the mouse. Am J Respir Cell Mol Biol. 2008;39(3):253–62. doi: 10.1165/rcmb.2008-0049OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, et al. Isolation of highly pure alveolar epithelial type I and type II cells from rat lungs. Lab Invest. 2004;84(6):727–35. doi: 10.1038/labinvest.3700095. [DOI] [PubMed] [Google Scholar]
- 30.Millien G, et al. Alterations in gene expression in T1 alpha null lung: a model of deficient alveolar sac development. BMC Dev Biol. 2006;6:35. doi: 10.1186/1471-213X-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore BB, Hogaboam CM. Murine models of pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;294(2):L152–60. doi: 10.1152/ajplung.00313.2007. [DOI] [PubMed] [Google Scholar]
- 32.Chua F, Gauldie J, Laurent GJ. Pulmonary fibrosis: searching for model answers. Am J Respir Cell Mol Biol. 2005;33(1):9–13. doi: 10.1165/rcmb.2005-0062TR. [DOI] [PubMed] [Google Scholar]
- 33.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27(11):1033–7. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 34.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136(2):364–77. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rippon HJ, et al. Derivation of distal lung epithelial progenitors from murine embryonic stem cells using a novel three-step differentiation protocol. Stem Cells. 2006;24(5):1389–98. doi: 10.1634/stemcells.2005-0465. [DOI] [PubMed] [Google Scholar]
- 36.Roszell B, et al. Efficient derivation of alveolar type II cells from embryonic stem cells for in vivo application. Tissue Eng Part A. 2009;15(11):3351–65. doi: 10.1089/ten.tea.2008.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefanie Zuegel PBaHM. Co-culture of alveolar epithelial and endothelial cells blunts failure of alveolar barrier function in hypoxia. The FASEB. 2008;22:932.8–932. [Google Scholar]
- 38.Wendt CH, et al. Alveolar epithelial cells regulate the induction of endothelial cell apoptosis. Am J Physiol. 1994;267(4 Pt 1):C893–900. doi: 10.1152/ajpcell.1994.267.4.C893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Structures of pTRE-OIS vector, pTRE-KIcML vector, SPCP-NEOR vector, and EFαP-rtTA vector, related to Figure 1
The schematic diagram of each vector is to depict relevant positional information, hence not scaled proportionally based on sequence lengths. (A) pTRE-OIS vector contains Oct4 and Sox2 transgene. The internal ribosome entry site (IRES) of the encephalomyocarditis virus located between the two trangenes allows co-translating Oct4 and Sox2 from a single RNA transcript under the control of TRE-PminCMV promoter. (B) pTRE-KIcML vector harbors Klf4 and cMyc transgene. The IRES is included in this vector for co-translation of the two transgenes under the control of TRE-PminCMV promoter. This vector also contains LoxP targeting sequence downstream of cMyc cDNA. (C) SPCP-NEOR vector contains NEOR transgene under the control of ATIIC-specific SPC promoter (SPCP). (D) EFαP-rtTA vector contains rtTA cDNA downstream of EF-1a promoter (EFαP).
Figure S2. PCR analysis of vector-integration of Cre-IRES-PUROR, related to Figure 2
PCR was performed to examine Cre-IRES-PUROR vector-integration in chromosomal DNA of hiPSC-26 clones A, B, C, D, E, and F after transfection. PCR using Cre forward and Cre reverse primer generated one 745 bp Cre transgene DNA fragment in hiPSC-26 clone F, but not in clones A, B, C, D, and E, indicating that vector-integration event had occurred in hiPSC-26 clone F.
Figure S3. Characterization of hiPSC-ATIICs, related to Figure 4
(A) To examine if human thyroid transcription factor 1 (TTF1) can be detected in the BLM-challenged lungs transplanted with hiPSC-ATIICs, QRT-PCR was performed using total RNA isolated from the BLM-challenged lungs with and without transplantation of hiPSC-ATIICs on day 10 (n=5). The TTF1 primers (Table S1) were designed to detect human TTF1 mRNA in mouse lungs using 18S rRNA as endogenous control. TTF1 RNA was detected in the BLM-challenged lungs transplanted with hiPSC-ATIICs, but not in the control lungs and BLM-challenged lungs without hiPSC-ATIIC transplantation. (B) Papanicolaous staining of lamellar bodies. To show a group of hiPSC-ATIICs containing lamellar bodies, the modified Papanicolaous staining, which is another procedure routinely used for identifying lamellar bodies, was performed. As control hATIICs (the upper image, x630), all of the G418 selected hiPSC-ATIICs contained abundant lamellar bodies on day 14 (the lower image, x630). (C) Proliferation capacity of hiPSC-ATIICs in vitro. The hiPSC-ATIIC colonies (~16 colonies/well) were observed when cells were cultured on Matrigel-coated plates in mouse MEF-conditioned DMEM containing 10% FBS for 7 days, and significantly larger number of hiPSC-ATIIC colony (~45 colonies/well) were present in the rhKGF (30ng/ml) treated cultures compared to control (upper panel). The rhKGF-promoted proliferation was also demonstrated by the evidence of larger hiPSC-ATIIC colonies containing 27–40 cells in the rhKGF-treated cultures compared with those containing 15–21 cells in the untreated cultures.
Figure S4. Percent survival and pulmonary function of BLM-challenged mice with and without transplantation of hiPSC-ATIICs, related to Figure 5
To assess whether the therapeutic benefits provided by hiPSC-ATIICs is long-term without teratoma formation, 24 female SCID mice were subjected to BLM induced acute lung injury, and received transplantation using either saline, hmonos or hiPSC-ATIICs (8 mice/group) on day 2 after BLM challenge. (A) On day 7 after BLM-challenge, 1 of 8 BLM-challenged mice that received saline and 2 of 8 BLM-challenged mice transplanted with hmonos had died. Between 7 and 10 days, another 2 BLM-challenged mice that received saline and 1 BLM-challenged mouse transplanted with hmonos had died. All of the remaining BLM-challenged mice received either saline or hmonos had died by 12 days after BLM treatment. In contrast, all of the BLM-challenged mice treated with hiPSC-ATIICs survived at least up to 11 months. (B) The BLM-challenged mice with transplantation of hiPSC-ATIICs maintained normal blood arterial oxygen saturation levels (97.125 ± 0.59, upper panel) at end of time point (11 months) compared with saline control mice (97.26 ± 0.67). Similarly, the lung tidal volumes (0.241 ± 0.006, lower panel) measured in these mice were comparable to that (0.243 ± 0.007) in saline control mice. (C) Immunofluorescent staining of representative lung sections from BLM-challenged mice transplanted with hiPSC-ATIICs at end of time point (11 months) using mouse anti human nuclei monoclonal antibody and rabbit anti human pro-SPC antibody. The mouse anti human nuclei monoclonal antibody recognizes cells of human origin in mouse lungs (red); and the rabbit anti human pro-SPC stains both mouse and human ATIICs that express SPC (green). The cells expressing both human nuclei and SPC were present in BLM-challenged lungs transplanted with hiPSC-ATIICs after 11 months (right image, indicated by arrow; scale bar = 100 μm), but not in saline control lungs (left image). The magnified view of human derived ATIIC indicated by yellow arrow was inserted in the right image. (D). Immunofluorescent staining of representative lung sections from BLM-challenged mice transplanted with hiPSC-ATIICs at end of time point (11 months) using mouse anti human TTF1 (PM 087 AA, Biocare Medical). The mouse anti human TTF1 identifies human alveolar epithelial cells, but not mouse alveolar epithelial cells, in mice lungs. The human TTF1 positive cells were observed in BLM-challenged lungs transplanted with hiPSC-ATIICs after 11 months (red, the right image, scale bar = 200 μm) with normal lung histology, but not in the control lungs (the left image). The magnified view of human TTF1 positive cell indicated by yellow arrow was inserted in the right image. These TTF1 positive cells distributed individually in the lungs, and did not expand to form any abnormal structure or cell cluster in the lung, suggesting the remaining hiPSC-ATIICs had not developed into a cancer tissue in the lungs after 11 months.