Summary
Notch refers to a highly conserved cell-to-cell signaling pathway with essential roles in embryonic development and tissue maintenance. Dysfunctional signaling causes human disease, highlighting the importance of pathway regulation. Notch signaling ultimately results in the activation of target genes, which is regulated by the nuclear effector CSL. CSL dually functions as an activator and repressor of transcription through differential interactions with coactivator or corepressor proteins, respectively. While the structures of CSL-coactivator complexes have been determined, the structures of CSL-corepressor complexes are unknown. Here, using a combination of structural, biophysical, and cellular approaches, we characterize the structure and function of CSL in complex with the corepressor KyoT2. Collectively, our studies provide molecular insights into how KyoT2 binds CSL with high affinity and competes with coactivators, such as Notch, for binding CSL. These studies are important for understanding how CSL functions as both an activator and repressor of transcription of Notch target genes.
Introduction
The Notch pathway is an intercellular signaling mechanism conserved in metazoans (Kopan and Ilagan, 2009). Genetic ablation of Notch signaling results in embryonic lethality (Swiatek et al., 1994), while tissue specific ablation results in severe phenotypes and cellular dysfunction (Radtke et al., 1999), emphasizing the essential role Notch plays during prenatal development and the maintenance of adult tissues. Congruently, mutations in pathway components underlie the pathogenesis of many human diseases, including certain types of cancer and congenital defects (Gridley, 2003; Koch and Radtke, 2010). Given its prevalence with human disease, extensive efforts have been directed towards developing reagents that modulate Notch signaling for therapeutic purposes (Aster and Blacklow, 2012).
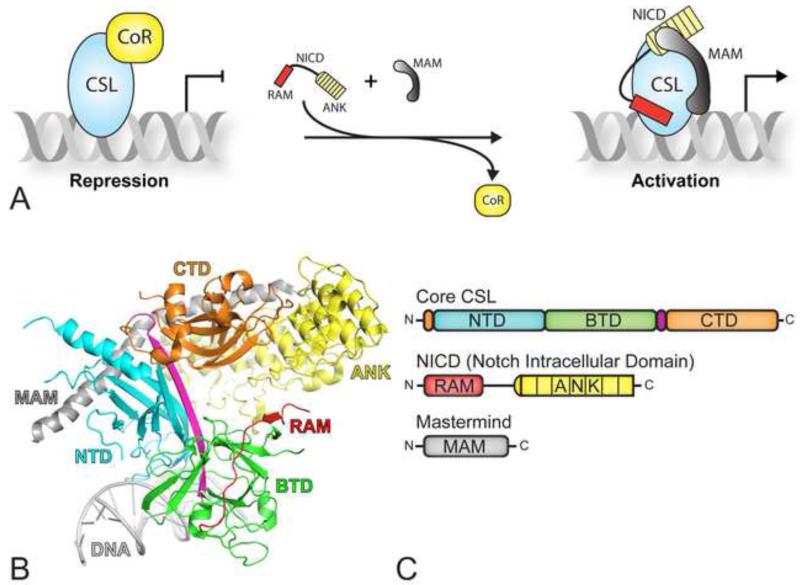
The central components consist of the receptor Notch, the ligand DSL (Delta, Serrate, Lag-2), and the DNA binding transcription factor CSL (CBF-1/RBP-J, Su(H), Lag-1) (Kovall and Blacklow, 2010). Notch-DSL complexation initiates signaling, which triggers proteolytic cleavage of Notch, thereby leading to release of its intracellular domain, termed NICD (Notch Intracellular Domain), from the cell membrane (Kopan and Ilagan, 2009). Subsequently, NICD translocates to the nucleus where it binds CSL and the transcriptional coactivator Mastermind (MAM). CSL, NICD, and MAM form a transcriptionally active ternary complex that binds at promoter and enhancer regions of Notch target genes, up-regulating transcription at these sites (Figure 1A).
Figure 1. CSL mediated transcriptional regulation.
(A) In the absence of a Notch signal, CSL keeps target genes in a repressed state through interactions with transcriptional corepressors (left). In the presence of a Notch signal, NICD (Notch intracellular domain) translocates to the nucleus, and interacts with CSL and the coactivator Mastermind (MAM), displacing corepressors from CSL and resulting in the activation of Notch target genes. (B) The X-ray structure of CSL in complex with NICD and MAM (PDB ID: 2FO1), referred to as the ternary complex (Wilson and Kovall, 2006). CSL is composed of the N-terminal domain (NTD-cyan), the β-trefoil domain (BTD-green), and the C-terminal domain (CTD-orange). The RAM (RBPJ-Associated Molecule, red) and ANK (ankyrin repeats, yellow) domains of NICD bind the BTD and CTD of CSL, respectively; MAM (grey) forms a long α-helix, which binds along a groove formed by ANK, CTD, and NTD. (C) Domain schematic of core CSL (top), NICD (middle) and MAM (bottom); domains are colored as in panel B.
CSL functions as a transcriptional repressor at some, but not all, Notch target genes by interacting with corepressor proteins, such as KyoT2, MINT/SHARP, and Hairless (Borggrefe and Oswald, 2009). Corepressors are components of large multi-protein, histone-modifying complexes, which link CSL to the transcriptional repression machinery in the nucleus. An early model in the field posited that CSL was constitutively bound to DNA, and upon pathway activation, NICD displaces corepressors from CSL (Figure 1A) (Hsieh and Hayward, 1995). However, more recent studies have shown that CSL binding to DNA is a much more dynamic process in vivo, whereby its occupancy at target genes is increased when Notch is active (Krejci and Bray, 2007). Whether NICD exclusively competes with corepressors for CSL binding or whether entire transcription complexes are exchanged and/or turned over at Notch target genes remains to be determined.
Our group and others have solved high-resolution X-ray structures of active Notch transcription complexes and assembly intermediates including CSL, CSL-RAM, and CSL-NICD-MAM ternary complexes bound to DNA (Kovall and Blacklow, 2010). As shown in Figure 1B and 1C, the structural core of CSL is composed of three domains: the NTD (N-terminal domain), the BTD (β-trefoil domain), and the CTD (C-terminal domain). DNA binding and specificity is mediated by the NTD and BTD. The RAM (Rbpj-Associated Molecule) and ANK (ankyrin repeats) domains of NICD interact with the BTD and CTD of CSL, respectively. MAM, which binds an interface formed by CTD-ANK and the NTD, locks the complex into an active conformation (Choi et al., 2012). Subsequent biochemical and biophysical studies have led to considerable insights into the assembly of the CSL-NICD-MAM ternary complex (Kovall and Blacklow, 2010); however, the structures of CSL-corepressor complexes are unknown and the molecular differences between corepressor and coactivator binding to CSL are poorly understood.
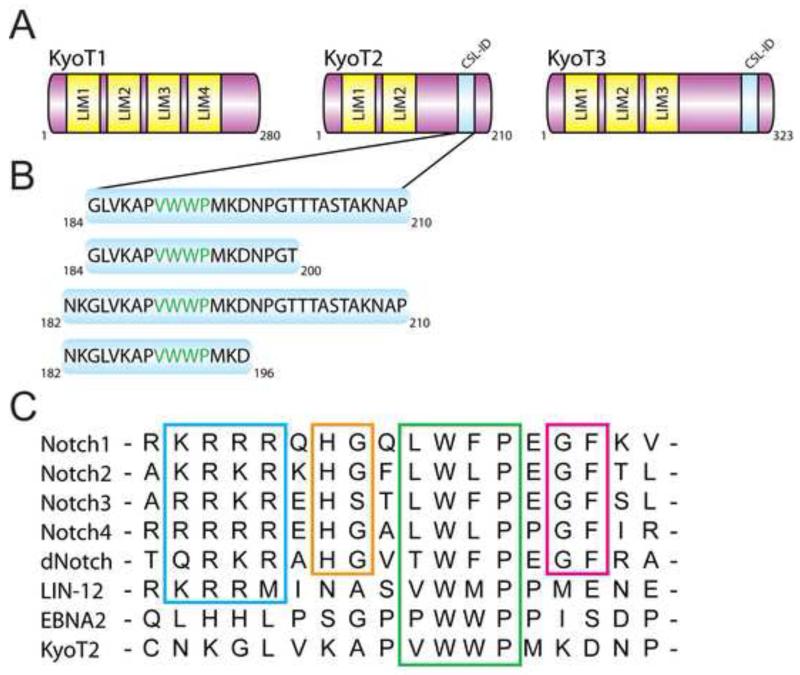
The corepressor KyoT2 was originally identified in a yeast two-hybrid screen for CSL binding partners (Taniguchi et al., 1998). KyoT2, along with KyoT1 and KyoT3, are splice variants of the KyoT gene (also known as Fhl1); KyoT1 are KyoT2 transcripts are found in a wide range of tissues, with particularly high levels in the skeletal muscle, heart, lung, and kidney (Taniguchi et al., 1998); KyoT3 is also expressed in a variety of tissues with higher levels found in the brain, kidney, and pancreas (Liang et al., 2008). All KyoT gene products encode proteins that have multiple N-terminal LIM domains (Figure 2A), and due to differential splicing, KyoT2 and KyoT3, but not KyoT1, contain a C-terminal region that binds CSL (CSL-ID; Figure 2A). Previous studies showed that KyoT2 and KyoT3 function as potent repressors of Notch mediated transcription in cells, and consistent with this role, the LIM domains have been shown to interact with Ring1 and HPC2 – members of the Polycomb group proteins involved in epigenetic silencing (Qin et al., 2005; Qin et al., 2004). As shown in Figure 2B and 2C, the CSL-ID of KyoT2 possesses a conserved hydrophobic tetrapeptide motif (φWφP; where φ is any hydrophobic residue) that is present in the RAM domain of Notch and the viral transactivator EBNA2 (Epstein-Barr virus nuclear antigen 2) (Ling and Hayward, 1995; Tamura et al., 1995). While the φWφP motif is essential for RAM and EBNA2 binding to the BTD of CSL, Johnson et al. identified additional conserved motifs in RAM that contribute to its high affinity interaction with BTD; these include an N-terminal basic region, and -HG- and -GF- dipeptide motifs (Figure 2C) (Johnson et al., 2010). Consistent with these motifs contributing to high affinity binding, EBNA2, which does not contain these additional motifs, binds CSL with ~50 fold less affinity (Johnson et al., 2010). Similar to EBNA2, KyoT2 also does not contain these additional motifs found in RAM (Figure 2C).
Figure 2. KyoT2 domain schematics and sequence comparison.
(A) The KyoT gene gives rise to three different isoforms: KyoT1, KyoT2, and KyoT3. KyoT1, KyoT2, and KyoT3 contain four, two, and three LIM domains, respectively. KyoT2 and KyoT3 contain a CSL-interaction domain (CSL-ID). (B) Figure shows KyoT2 constructs used in this study. (C) Sequence alignment of the RAM domains from murine Notch1-4, Drosophila Notch (dNotch), EBNA2 (Epstein-Barr virus nuclear antigen 2), the worm Notch receptor LIN-12, and the CSL-ID of KyoT2. The basic region, -HG- motif, φWφP motif, and -HG- motif are boxed and colored blue, orange, green, and magenta, respectively. See also Figure S1 for CD spectrum of KyoT2 (184-210).
The overall goals of this study are to understand at the structural level the complex formed between the corepressor KyoT2 and CSL, the molecular differences between KyoT2 and RAM binding to CSL, and how the CSL-KyoT2 complex functions to repress transcription of Notch target genes. Here, we used X-ray crystallography and isothermal titration calorimetry (ITC) to elucidate the structure and define the binding affinity, respectively, of the CSL-KyoT2 complex. We also analyzed mutants of both CSL and KyoT2, which revealed differences in the binding interactions between KyoT2 and RAM for CSL. Additionally, we used transcriptional reporter assays in cells to functionally validate our structural and binding studies. Taken together, our results provide the first high-resolution structure-function study of a CSL-corepressor complex, which is important for understanding how CSL functions as both a repressor and an activator of transcription from Notch target genes.
Results
Structure of the CSL-KyoT2-DNA Complex
In order to determine the high-resolution structure of the CSL-KyoT2 corepressor complex, we purified recombinant proteins from bacteria corresponding to mouse CSL (residues 53-474) and mouse KyoT2 (residues 184-210). As identified by Taniguchi et al., the 27 C-terminal residues of KyoT2 (184-210) correspond to the region of KyoT2 that is necessary to bind CSL (Taniguchi et al., 1998). CSL-KyoT2 complexes were mixed in equal molar ratios with an oligomeric 15mer DNA duplex, containing a single CSL binding site, and screened for crystals. Initial screens of the CSL-KyoT2 complex yielded no crystals; therefore, we designed and purified a surface entropy reduction mutant of CSL (R115T), which produced CSL-KyoT2-DNA crystals amenable to X-ray diffraction analysis. An orthorhombic crystal form (P21) of the CSL-KyoT2-DNA complex was obtained that diffracted to 2.85 Å. The structure was solved by molecular replacement, using previously determined CSL-DNA structures (Friedmann and Kovall, 2010; Friedmann et al., 2008), revealing that two CSL-KyoT2-DNA complexes were contained within the asymmetric unit. The final model, which consists of residues 53-472 of CSL, residues 184-196 of KyoT2, and the 15mer oligomeric DNA duplex, was refined to 2.85 Å with an Rfac and Rfree of 18.9% and 23.2%, respectively. The data collection, structure determination, and refinement statistics are summarized in Experimental Procedures and Table 1.
Table 1. Data collection and refinement statistics.
| Data Collection Statistics | |
| Beam Line | APS LS-CAT 21-ID-F |
| Resolution (Å) | 40.3 – 2.85 (2.95 – 2.85) |
| Space Group | P21 |
| Wavelength (Å) | 0.97872 |
| Unit Cell a, b, c (Å) | 62.16, 97.29, 144.10 |
| Unit Cell α, β, γ (°) | 90.00, 93.17, 90.00 |
| Rmerge | 0.053 (0.46) |
| I/σI | 28.1 (2.2) |
| Completeness (%) | 93.96 (66.6) |
| Redundancy | 5.6 (3.2) |
| Average mosaicity (°) | 0.19 – 0.28 |
| Refinement Statistics | |
| Rwork/Rfree (%) | 18.8 / 23.1 |
| Number of reflections | 37,773 |
| Number of atoms | 8,168 |
| Complexes/asymmetric unit | 2 |
| Wilson B/Mean B value (Å2) | 105.45 / 108.71 |
| RMSD Bond Lengths (Å) | 0.01 |
| RMSD Bond Angles (°) | 1.29 |
| Ramachandran (favored/outliers) | 94.67% / 0.23% |
Highest resolution shell shown in parentheses.
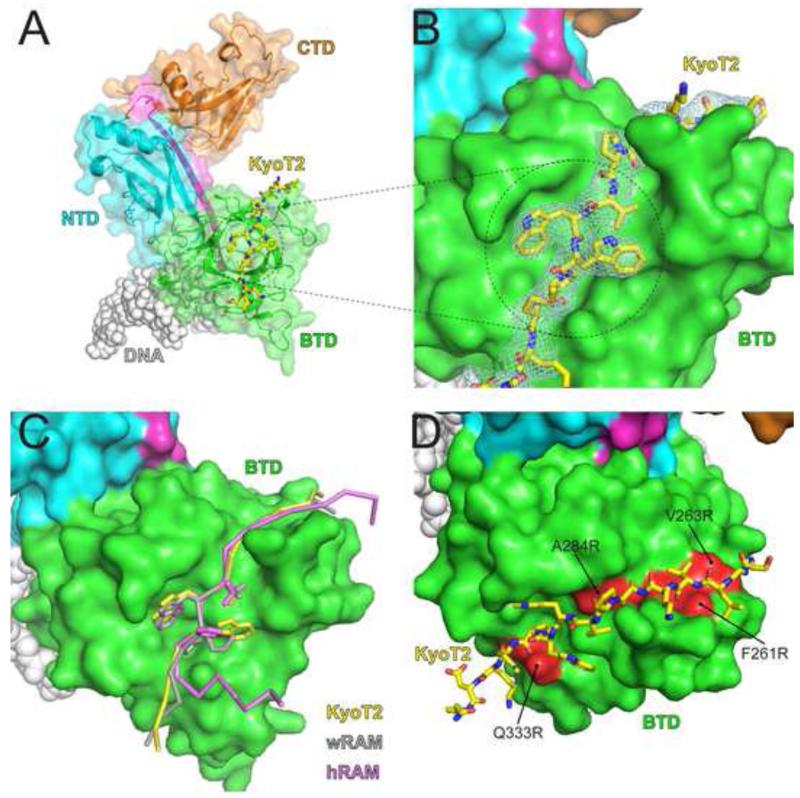
As shown in Figure 3, KyoT2 binds exclusively to the BTD of CSL in an extended conformation, wherein its N-terminal residues (184-187) form a β-strand with a large β-hairpin loop in the BTD of CSL and its φWφP motif (-VWWP-; residues 190-193) binds a prominent nonpolar pocket on the surface of the BTD. In isolation, residues 184-210 of KyoT2 have a circular dichroism spectrum that is consistent with this region being intrinsically disordered (Figure S1). Electron density for KyoT2 was only observed for residues 184-196, suggesting that residues 197-210 are not required for binding CSL, which was confirmed by our ITC binding studies of CSL-KyoT2 complexes detailed below.
Figure 3. X-ray structure of CSL-KyoT2-DNA complex.
(A) Figure shows ribbon diagram of the CSL-KyoT2-DNA structure with transparent molecular surface. CSL is colored as in Figure 1. KyoT2, shown as a yellow stick representation (carbon, oxygen, and nitrogen atoms colored yellow, red, and blue, respectively) binds across the BTD of CSL. The DNA is represented as grey spheres. (B) Zoomed in view of the KyoT2 φWφP motif binding in the hydrophobic pocket of the BTD. KyoT2 electron density is shown and is derived from a simulated annealing composite omit map contoured at 1σ. (C) Structural overlay of KyoT2 with the RAM domains from the human Notch1 receptor (hRAM, pink) (Choi et al., 2012) and the C. elegans Notch receptor LIN-12 (wRAM, grey) (Friedmann et al., 2008). Corresponding cα atoms from KyoT2, wRAM, and hRAM were aligned and docked onto the BTD from the CSL-KyoT2-DNA complex structure. (D) Surface representation of the BTD showing where previously characterized mutations (colored red; F261R, V263R, A284R, Q333R) that affect RAM binding occur relative to KyoT2 binding (yellow, stick representation) (Yuan et al., 2012). See also Figure S2 for comparison of the two CSL-KyoT2-DNA complexes contained within the asymmetric unit.
Overall, the two CSL-KyoT2-DNA complexes contained within the asymmetric unit are very similar (Figure S2); the cα atoms of the two CSL molecules align with a 0.66 RMSD; and alignment of individual domains display greater correspondence with RMSDs of 0.27, 0.23, and 0.31 (cα atoms) when the NTD, BTD, and CTD domains, respectively, are overlayed. Despite very different crystal packing environments, the conformations of the two KyoT2 molecules are also very similar (0.24 RMSD for cα atoms; Figure S2E) and bury similar amounts of surface area at the BTD-KyoT2 interface (874 Å and 928 Å). The largest structural differences between the two complexes occur within the CTD of CSL, i.e. remote from where KyoT2 binds CSL. These include two β-hairpin loops in the CTD, which assume different conformations likely due to different crystal packing interactions (Figure S2, panels A and D). In addition, the two CTD domains assume different degrees of rotation relative to the NTD-BTD domains, resulting in differences of up to ~3 Å between corresponding cα atoms. Due to the lower overall temperature factors at the BTD-KyoT2 interface, chains C (CSL) and D (KyoT2) were used for all subsequent structural comparisons.
Given that both the RAM domain of Notch and KyoT2 exclusively bind the BTD, we next compared in detail the interactions of these ligands with CSL. Previously, four X-ray structures have been determined that contain the CSL-RAM interaction – one structure of the CSL-NICD-MAM ternary complex and two CSL-RAM structures for protein components from C. elegans, and one structure of the human CSL-NICD-MAM ternary complex that contains RAM (Choi et al., 2012; Friedmann et al., 2008; Kovall and Hendrickson, 2004; Wilson and Kovall, 2006). As shown in Figure 3C, when KyoT2 and the RAM domains are overlayed, there is an overall structural correspondence for how KyoT2 and RAM bind the BTD of CSL. In particular, residues proximal to the φWφP motif share the largest degree of structural similarity; however, the structural correspondence breaks down for N- and C-terminal residues distal from the φWφP motif. In particular, the basic region of RAM, as well as the -HG- and -GF- motifs (Figure 2), shares little to no structural similarity with KyoT2 (Figure 3C). It should also be mentioned that the C-terminal residues of the human Notch1 RAM structure adopt a strikingly different conformation than KyoT2 and the worm RAM structures. The authors of this study speculated that the tandem proline residues following the φWφP for worm RAM resulted in its elongated conformation, whereas human Notch1 RAM does not contain this motif, and therefore adopts a striking bend at this position (Choi et al., 2012). However, KyoT2 also does not contain this di-proline motif, yet adopts a conformation more similar to the worm RAM structures. The functional significance in these two different binding modes remains to be determined. Nonetheless, all CSL-RAM complex structures bury a similar amount of surface area as the CSL-KyoT2 binding interaction (Table S1).
Thermodynamic analysis of CSL-KyoT2 binding interactions
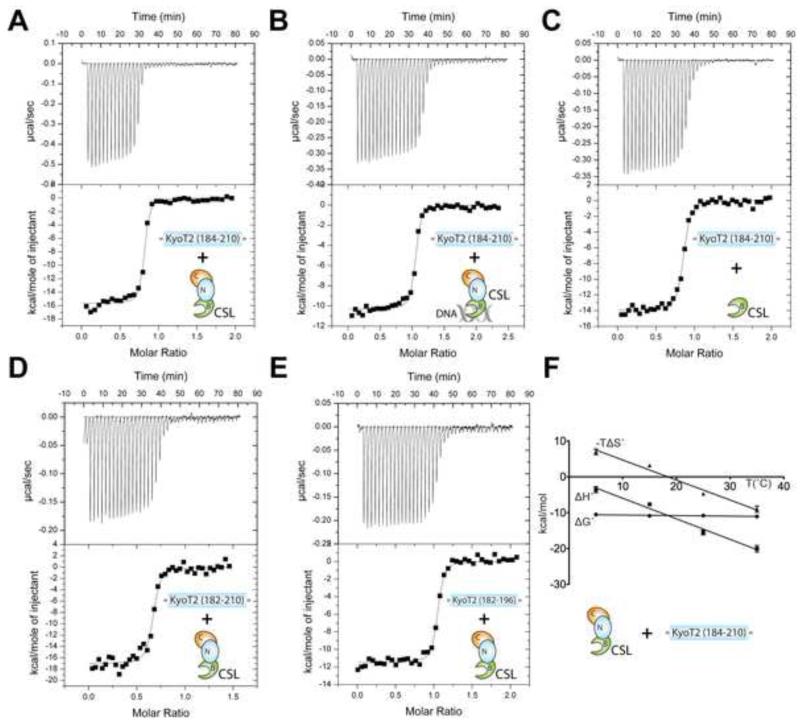
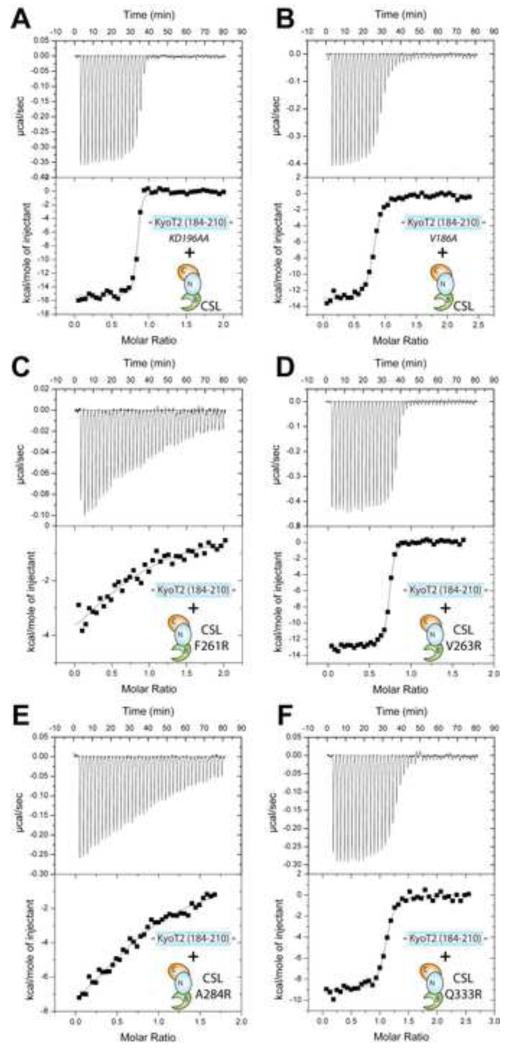
In order to characterize the binding interactions of CSL-KyoT2 complexes, we used ITC with our purified preparations of CSL and KyoT2. As shown in Figure 4A and Table 2, KyoT2 (184-210) forms a high affinity 1:1 interaction with CSL that is defined by a ~12nM dissociation constant (Kd), which is similar in magnitude to previous thermodynamic studies of CSL-RAM complexes (Friedmann et al., 2008; Johnson et al., 2010). Consistent with KyoT2 being intrinsically disordered prior to interactions with CSL, KyoT2 binding is enthalpically driven and entropically unfavorable. The overall affinity of CSL for KyoT2 is unchanged when CSL is bound to DNA (Figure 4B and Table 2); however, the enthalpic/entropic contributions to binding are affected by as much as 4 kcal/mol. Given that both KyoT2 and DNA bind the BTD of CSL, these data suggest that DNA binding to the BTD results in structural ordering of the BTD, which diminishes the entropic penalty for KyoT2-BTD complexation. Previously, we observed similar enthalpic/entropic compensation for RAM binding to CSL in the presence and absence of DNA (Friedmann et al., 2008). Consistent with our CSL-KyoT2 structure, the affinity of KyoT2 for the isolated BTD is comparable (i.e. within experimental error) to its binding interactions with the structural core of CSL (Figure 4C and Table 2).
Figure 4. Thermodynamic analysis of CSL-KyoT2 binding interactions.
This figure shows representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for KyoT2 binding to CSL. Forty titrations were performed per experiment, consisting of 7 μl injections that were spaced 120s apart. (A) Core CSL binding to KyoT2 residues 184-210. (B) Core CSL, pre-bound to a cognate DNA, binding to KyoT2 residues 184-210. (C) BTD binding to KyoT2 residues 184-210. (D) Core CSL binding to KyoT2 residues 182-210. (E) Core CSL binding to KyoT2 residues 182-196. (F) Thermodynamic profile of core CSL binding to KyoT2 residues 184-210. Thermodynamic parameters (ΔG°, ΔH°, and TΔS°) are plotted as a function of temperature (5, 15, 25, 35°C). The ΔCP of binding for the CSL-KyoT2 complex is −0.57 kcal/mol·K. See also Table S1 for buried surface at CSL-KyoT2 interface and Figure S3 for minimal KyoT2 peptide sequence that binds CSL.
TABLE 2. Calorimetric data for KyoT2 binding to CSL.
| Macromolecule | KyoT2 Ligand |
K (M−1) |
Kd
(μM) |
ΔG°
(kcal/mol) |
ΔH°
(kcal/mol) |
−TΔS°
(kcal/mol) |
|---|---|---|---|---|---|---|
| CSL | 184 - 210 | 8.5 ± 2.3 × 107 | 0.012 | −10.8 ± 0.2 | −15.5 ± 0.7 | 4.7 ± 0.5 |
| CSL + DNA | 184 - 210 | 1.1 ± 0.4 × 108 | 0.011 | −10.9 ± 0.3 | −11.0 ± 0.6 | 0.03 ± 0.3 |
| BTD | 184 - 210 | 4.0 ± 0.8 × 107 | 0.026 | −10.4 ± 0.1 | −13.6 ± 0.4 | 3.3 ± 0.5 |
| CSL | 184 - 200 | 3.1 ± 0.7 × 107 | 0.033 | −10.2 ± 0.1 | −13.1 ± 0.6 | 2.9 ± 0.5 |
| CSL | 182 - 210 | 7.1 ± 1.5 × 107 | 0.015 | −10.7 ± 0.1 | −16.0 ± 1.0 | 5.2 ± 0.9 |
| CSL | 182 - 196 | 1.0 ± 0.8 × 108 | 0.013 | −10.9 ± 0.1 | −10.8 ± 0.6 | −0.1 ± 0.6 |
All experiments were performed at 25°C. Values are the mean of at least three independent experiments and errors represent the standard deviation (S.D.) of multiple experiments. See also Table S2 for ITC data of minimal KyoT2 peptide sequence that binds CSL.
In order to glean additional thermodynamic insights into CSL-KyoT2 complexes, we determined the change in heat capacity (ΔCp) associated with KyoT2 binding to CSL. A negative ΔCp of binding is consistent with burial of nonpolar surface area upon complexation and/or folding coupled to binding (Sturtevant, 1977). As shown in Figure 4F and Table 3, we analyzed the thermodynamic parameters of binding (ΔH°, −TΔS°, and ΔG°) as a function of temperature. This analysis shows that the overall free energy of binding for CSL-KyoT2 complexes is independent of temperature and the ΔCp of binding is −0.57 kcal/mol·K. Previously, we found the ΔCp of binding for CSL-RAM complexes to be −0.62 kcal/mol·K (VanderWielen et al., 2011), which is very similar to the value determined here for CSL-KyoT2 interactions.
Table 3. Temperature dependence of KyoT2 binding to CSL.
| T (°C) | K (M−1) | Kd (μM) |
ΔG°
(kcal/mol) |
ΔH°
(kcal/mol) |
−TΔS°
(kcal/mol) |
|
|---|---|---|---|---|---|---|
| CSL + KyoT2 |
5 | 6.2 ± 1.4 × 107 | 0.017 | −9.9 ± 0.1 | −3.9 ± 0.4 | −6.0 ± 0.5 |
| 15 | 8.3 ± 2.3 × 107 | 0.013 | −10.4 ± 0.2 | −6.3 ± 0.3 | −4.2 ± 0.5 | |
| 25 | 9.4 ± 1.2 × 107 | 0.011 | −10.9 ± 0.1 | −14.6 ± 0.2 | 3.7 ± 0.2 | |
| 35 | 3.5 ± 0.6 × 107 | 0.029 | −10.6 ± 0.1 | −16.7 ± 0.7 | 6.1 ± 0.5 |
Values are the mean of at least three independent experiments and the errors represent the S.D. of multiple experiments.
Next, we sought to delineate the region in KyoT2 that maintains full affinity with CSL. Two C-terminal truncations in KyoT2 (184-200 & 182-196) resulted in overall similar binding to CSL as the KyoT2 construct 184-210 (Table 2 and Figure 4E). Similarly, N-terminally extended KyoT2 constructs (182-210 & 182-196) displayed no additional enhancements in binding affinity compared to the 184-210 KyoT2 construct (Figure 4D&E, Table 2). These data suggest that KyoT2 residues 184-196 are necessary and sufficient for binding CSL, which is entirely consistent with our CSL-KyoT2 structure. Subsequently, we pursued ITC binding studies to identify the shortest peptide sequence in KyoT2 that still retains measureable and specific binding to CSL. To address this question, we analyzed the binding of six additional KyoT2 peptides of decreasing lengths. Remarkably, the tetrapeptide -VWWP- of KyoT2 binds CSL with ~8uM affinity, which we confirmed was specific by showing that the interaction was abrogated with the BTD mutant F261R. It should also be mentioned that there is a significant enhancement in binding (~40fold) when the peptide -APVWWPMK- is lengthened by two residues to -KAPVWWPMKD- (Table S2 & Figure S3).
Although KyoT2 and the RAM have strikingly similar overall affinities for CSL, KyoT2 lacks the additional motifs identified in RAM that contribute to its high affinity binding (Johnson et al., 2010). This suggests that KyoT2 and RAM utilize different molecular interactions to achieve high affinity binding with CSL. In order to characterize these differences, we designed mutations in CSL, KyoT2, and RAM and analyzed their binding using ITC. Previously, we analyzed mutations in CSL that targeted the interfaces of the CSL-NICD-MAM coactivator complex (Yuan et al., 2012). Four mutations were targeted to the BTD of CSL (Figure 3D), all of which showed substantially reduced affinity for the RAM domains of Notch1 and Notch2 (Yuan et al., 2012). To determine whether these CSL mutants also affected KyoT2 binding, we measured the binding of KyoT2 with the BTD mutants F261R, V263R, A284R, and Q333R. As shown in Figure 5C&E and Table 4, the CSL mutants F261R and A284R have at least 500-fold reduced binding to KyoT2, which is similar to what was observed for these mutants interacting with RAM (Yuan et al., 2012). However, the BTD mutants V263R and Q333R had relatively modest effects on KyoT2 binding (Figure 5D&F, Table 4), which is significantly different than the ~20fold reduction in affinity observed for these mutants binding to RAM. Interestingly, the CSL mutants F261R and A284R lie at the interface with the φWφP motifs of KyoT2 and RAM (Figure 3D), whereas the CSL mutants V263R and Q333R target binding interactions outside of the φWφP motif, which is where KyoT2 and RAM binding interactions with CSL diverge.
Figure 5. Thermodynamic binding analysis of KyoT2 and CSL mutants.
This figure shows representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for KyoT2 mutants KD196AA (A) and V186A (B) binding to wild-type core CSL; and CSL mutants F261R (C), V263E (D), A284R (E), and Q333R (F) binding to wild-type KyoT2 (184-210). Forty titrations were performed per experiment, consisting of 7 μl injections that were spaced 120s apart.
TABLE 4. Calorimetric binding data for KyoT2 and CSL mutants.
| CSL | Ligand | K (M−1) |
Kd (μM) |
ΔG°
(kcal/mol) |
ΔH°
(kcal/mol) |
−TΔS°
(kcal/mol) |
ΔΔG°
(kcal/mol) |
|---|---|---|---|---|---|---|---|
| F261R | KyoT2 | 3.4 ± 3.9 × 105 | 6.3 | −6.8 ± 0.1 | −5.6 ± 0.8 | −1.2 ± 0.8 | 4.0 ± 0.2 |
| V263R | KyoT2 | 3.1 ± 0.7 × 107 | 0.019 | −10.5 ± 0.1 | −12.9 ± 0.1 | 2.4 ± 0.1 | 0.3 ± 0.2 |
| A284R | KyoT2 | 4.0 ± 0.8 × 105 | 7.2 | −7.0 ± 0.1 | −10.5 ± 0.4 | 3.4 ± 0.5 | 3.8 ± 0.2 |
| Q333R | KyoT2 | 2.0 ± 3.9 × 107 | 0.051 | −10.0 ± 0.1 | −11.7 ± 2.3 | 1.7 ± 2.2 | 0.8 ± 0.2 |
| WT | KyoT2 WW192AA |
NBD | --- | --- | --- | --- | --- |
| WT | KyoT2 KD196AA |
1.2 ± 0.7 × 108 | 0.012 | −10.9 ± 0.5 | −16.0 ± 1.9 | 5.1 ± 1.6 | −0.1 ± 0.5 |
| WT | KyoT2 V186A |
4.0 ± 0.8 × 105 | 0.069 | −9.8 ± 0.1 | −13.7 ± 1.7 | 3.9 ± 1.8 | 1.0 ± 0.2 |
| WT | RAM wild−type |
6.8 ± 0.8 × 107 | 0.015 | −10.7 ± 0.1 | −18.2 ± 1.7 | 7.5 ± 1.6 | --- |
| WT | RAM Q1763A |
2.0 ± 3.9 × 107 | 0.022 | −10.5 ± 0.2 | −7.50 ± 0.3 | 3.0 ± 0.2 | 0.2 ± 0.2 |
All experiments were performed at 25°C. Values are the mean of at least three independent experiments and errors represent the S.D. of multiple experiments. KyoT2 ligands are in the context of the construct 184-210.
Next, we analyzed mutations in KyoT2 that target motifs within and outside of the φWφP. As expected, mutation of the di-tryptophan motif within KyoT2 (WW192AA) completely abrogated binding (Table 4); this is consistent with the intimate interactions these residues make with the hydrophobic pocket on the BTD and previous work that demonstrated the importance of the φWφP motif in CSL-RAM binding (Johnson et al., 2010). We then tested the binding of the KyoT2 mutant construct KD196AA, which sequence-wise corresponds to the -GF- motif in the RAM domain of Notch (Figure 2C). Mutation of the -GF- motif in RAM results in an ~0.6 kcal/mol loss in binding energy (Johnson et al., 2010); conversely, the KyoT2 mutant KD196AA has no effect on CSL binding (Figure 5A and Table 4), which is consistent with our CSL-KyoT2 structure, as residues K196 and D197 are remote to the CSL-KyoT2 interface. The basic motif at the N-terminus of RAM (Figure 2C), which binds an electronegative patch on the BTD of CSL, contributes a substantial 2.5 kcal/mol to the overall binding energy of CSL-RAM complexation (Johnson et al., 2010). However, KyoT2 does not contain an equivalent set of basic residues, and the KyoT2 residues that correspond to the basic motif in RAM do not interact with the electronegative patch on the BTD. We surmised that the polar residue N182 and the basic residue K183 upstream of our KyoT2 construct 184-210 might be functioning in a similar manner to the basic motif in RAM. Nonetheless, extension of our KyoT2 construct to include residues N182/K183 had no effect on binding interactions with CSL (Table 1).
Given that the additional motifs identified in RAM by Johnson et al. are not present in KyoT2 and mutation of the corresponding residues have no affect on KyoT2 binding to CSL, we then carefully scrutinized our CSL-KyoT2 structure to identify residues in KyoT2 that likely provide greater contributions to binding than the corresponding residues in the RAM domain of Notch. We focused our analysis on V186 of KyoT2, which makes extensive hydrophobic contacts with residues F261 and V263 of CSL, whereas the corresponding residue in the human Notch1 RAM structure (Q1764) contributes far fewer interactions to the CSL-RAM interface. Because of these structural differences, we hypothesized that the KyoT2 mutant V186A would have a much greater effect on binding than the corresponding mutation in RAM (Q1764A). As shown in Figure 5B and Table 4, the KyoT2 mutant V186A resulted in at least a 5-fold reduction in binding (~1 kcal/mol); however, the RAM mutant Q940A bound with a comparable affinity to CSL as wild-type RAM.
Cellular characterization of the CSL-KyoT2 corepressor complex
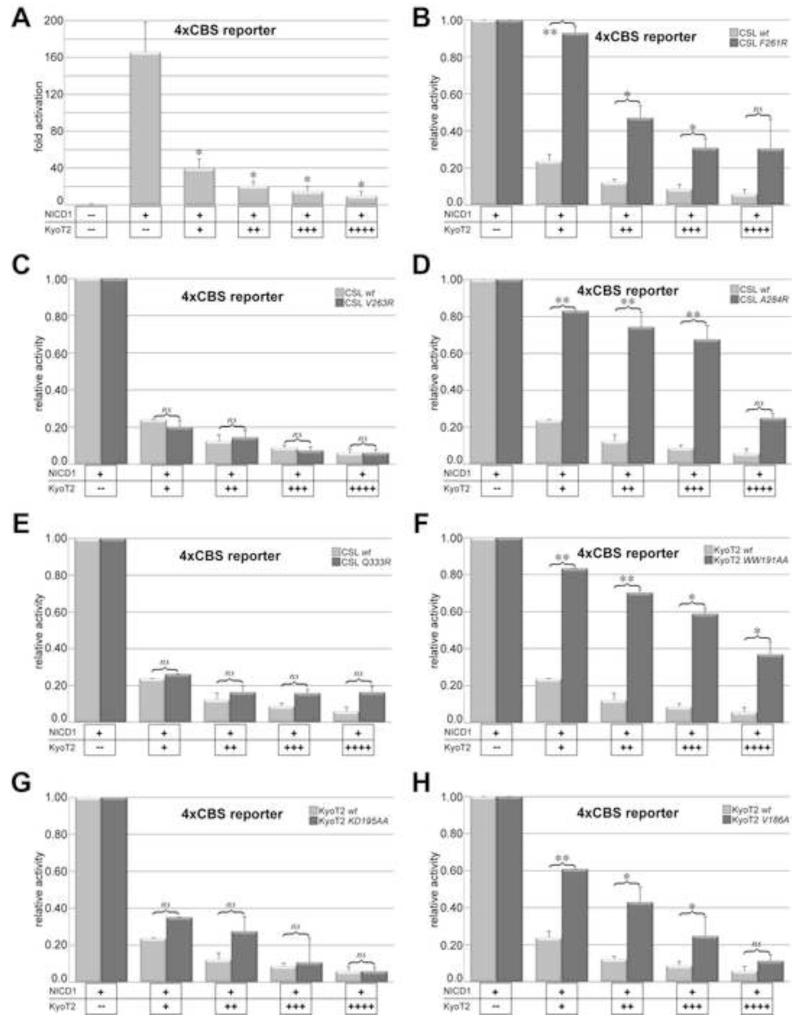
In order to validate the findings from our structural and binding studies of CSL-KyoT2 complexes, we characterized CSL and KyoT2 mutants in cells using transcriptional reporter assays. For these experiments, mouse embryonic fibroblasts (MEFs) derived from CSL-null embryos were cultured and transduced with a retrovirus that expresses either wild-type or mutant CSL molecules (Yuan et al., 2012). Notch signaling is activated in the MEFs by transfection of a construct that encodes the intracellular domain of the Notch1 receptor (NICD1), which is constitutively active in cellular assays (Jarriault et al., 1995). Notch activity is monitored by cotransfection of the luciferase reporter 4xCBS, which contains four iterative CSL binding sites upstream of the luciferase coding region (Ong et al., 2006). As shown in Figure 6A, transfection of NICD1 into MEFs expressing wild-type CSL results in robust ~160fold activation of the reporter compared to cells that were not transfected with NICD1. However, cotransfection of a plasmid that expresses wild-type KyoT2 severely blunts activation of the reporter, resulting in a ~16fold reduction in reporter activity (Figure 6A).
Figure 6. Analysis of KyoT2 and CSL mutants in cellular reporter assays.
Figure shows the effects KyoT2 and CSL mutations have on luciferase activity induced by NICD1 from the 4XCBS reporter in MEFs. (A) Addition of NICD1 results in robust activation from the reporter (~160fold); however, increasing concentrations of wild-type KyoT2 results in potent repression from the reporter. Data are normalized to cells without NICD1 and shown as fold activation. (B-E) Increasing concentrations of wild-type KyoT2 are titrated into MEFs expressing CSL mutants F261R (B), V263R (C), A284R (D), or Q333R (E). (F-H) Increasing concentrations of KyoT2 mutants [WW191AA (F), KD195AA (G), or V186AA (H)] are titrated into MEFs expressing wild-type CSL. For panels B-H, data are normalized to cells with NICD1, but without KyoT2, and shown as relative activity; mutant and wild-type data are shown in dark and light gray bars, respectively. The data shown are derived from three independent experiments performed in duplicate and represent the means ± S.E. Statistical significance was assessed using paired Student’s t tests with *, p ≤ 0.05; **, p ≤ 0.01; and ns, not significant.
Overall, we obtained excellent agreement between our in vitro binding studies of CSL and KyoT2 mutants, and the characterization of these mutants in cellular reporter assays. The CSL mutants F261R and A284R, which resulted in ~500fold reduction in binding by ITC, had significantly adverse effects on the ability of KyoT2 to repress Notch induced activation of the reporter (Figure 6B,D). Likewise, the CSL mutants V263R and Q333R, which only marginally affected KyoT2 binding to CSL in vitro, had no statistically significant effect on reporter activity compared to cells expressing wild-type CSL (Figure 6C,E). It should be noted that the four BTD mutants also affect to varying degrees the ability of NICD1 to activate the 4XCBS reporter, but do not completely abolish reporter activity (Yuan et al., 2012). Thus, Figure 6 (panels B-E) represents the ability of Kyot2 to repress the residual activity of the CSL mutants with NICD1. The KyoT2 mutant WW191AA, which displayed no binding by ITC, had the most deleterious effect on the ability of KyoT2 to repress NICD1 activation of the reporter (Figure 6F). As expected, the KyoT2 mutant KD195AA, which had no effect on CSL-KyoT2 interactions by ITC, also had no statistically significant effect on the ability of KyoT2 to repress transcription from the reporter (Figure 6G). Strikingly, the KyoT2 mutant V186A, which had a 5fold reduction of binding, had a commensurate decrease in the repressive effect on activity from the reporter compared to wild-type KyoT2 (Figure 6H).
Discussion
Canonical Notch signaling ultimately results in changes in gene expression, which is regulated by the transcription factor CSL (Kopan and Ilagan, 2009). CSL binds the regulatory elements of Notch target genes, and mediates transcriptional repression and activation at these sites by forming complexes with corepressors and coactivators, respectively (Borggrefe and Oswald, 2009). In contrast to CSL-coactivator complexes, e.g. the transcriptionally active CSL-NICD-MAM ternary complex, much less is known about the structure and function of CSL-corepressor complexes (Kovall and Blacklow, 2010). Moreover, the components, structure, and assembly mechanism of the activator complex are, by and large, well conserved between organisms; however, corepressor interactions with CSL are seemingly more diverse and species specific. For example, the mammalian corepressor KyoT2, which is not conserved in flies or worms, tethers tandem LIM domains to CSL via a C-terminal motif that resembles the RAM domain of Notch (Taniguchi et al., 1998). Nonetheless, characterizing the structure and function of both activation and repression complexes that regulate transcription at target genes is not only important for a detailed understanding of gene regulatory mechanisms in the Notch pathway, but will also inform drug discovery efforts that seek to modulate Notch signaling for therapeutic purposes (Aster and Blacklow, 2012). To this end, our studies may be particularly relevant to certain papillary renal cell carcinomas, in which it was shown that KyoT3 is significantly upregulated, thereby suppressing Notch signaling and contributing to tumorigenesis (Surendran et al., 2010).
Building upon our previous studies of Notch transcription complexes, here we pursued a comprehensive structure-function analysis of the CSL-KyoT2 corepressor complex, in which we determined the 2.85 Å X-ray structure of the CSL-KyoT2-DNA complex (Figure 3), defined the thermodynamic parameters of CSL-KyoT2 binding interactions (Figure 4), and validated our structural studies by characterizing CSL/KyoT2 mutants in ITC and cellular assays (Figures 5 and 6). Our data demonstrate that KyoT2 binds exclusively to the BTD of CSL with low nanomolar affinity (~10nM Kd; Table 2). The overall affinity of KyoT2 for CSL is unaffected by DNA, which due to the properties of linked equilibria (Wyman and Gill, 1990), implies that KyoT2 does not affect the affinity of CSL for DNA. Additionally, we had excellent agreement between our ITC binding studies of CSL and KyoT2 mutants, and characterization of these mutants in cellular reporter assays. Taken together, these data suggest that the repressive function of KyoT2 is largely dependent on its high affinity interaction with CSL, which localizes Polycomb repression complexes at Notch target genes. Moreover, the finding that both KyoT2 and NICD bind with high affinity to the BTD of CSL provides molecular insights into the putative competition of coregulators for binding to CSL.
At first glance, KyoT2 binds CSL in much the same manner as the RAM domain of NICD: (1) the main chain atoms of both KyoT2 and RAM have similar binding paths across the BTD, in which their φWφP motifs display the highest degree of structural correspondence (Figure 3C); (2) mutation of their respective φWφP motifs abrogates binding to CSL (Table 4) (Johnson et al., 2010); (3) both KyoT2 and RAM bury similar amounts of surface area upon complexation with CSL (Table S1); (4) both KyoT2 and RAM are intrinsically disordered prior to binding CSL (Figure S1) (Bertagna et al., 2008); and lastly, (5) both KyoT2 and RAM have similar high affinity for CSL, which is enthalpically driven and associated with a large and negative ΔCp of binding (approximately −0.6 kcal/mol·K) (Friedmann et al., 2008; Johnson et al., 2010). However, closer inspection of our CSL-KyoT2 structure and further characterization of CSL/KyoT2 mutants, reveal a number of molecular differences that distinguish between the binding modes of KyoT2 and RAM for CSL.
While there is a high degree of structural correspondence between the φWφP motifs and neighboring residues for KyoT2 and RAM when in complex with CSL (Figure 3C), the structural similarity between KyoT2 and RAM is noticeably reduced for N- and C-terminal regions that are remote from the φWφP motif. This lack of structural correspondence is reflected in the primary sequences of KyoT2 and RAM, which diverge in regions outside of the φWφP motif (Figure 2C). Importantly, Johnson et al. showed that RAM has additional motifs (basic, -HG-, and -GF-) outside of the φWφP that make significant contributions to the high affinity CSL-RAM interaction (Figure 2C) (Johnson et al., 2010). However, KyoT2 lacks these additional motifs and mutation of the corresponding residues in KyoT2 has little to no effect on complex formation (Table 4), yet KyoT2 binds CSL as strongly as RAM. This raises the question of how KyoT2 achieves nanomolar affinity for CSL and what residues outside of its φWφP motif contribute to binding. While a comprehensive binding study of CSL-KyoT2 mutants is needed to completely address this question, which is outside the scope of our current study, a keener examination of our structure revealed one residue in KyoT2 that plays a more important role in binding than the corresponding residue in RAM. Namely, the branched hydrophobic side chain of V186, which is a glutamine in Notch1 RAM (Figure 2C), contributes ~1 kcal/mol of binding energy to CSL-KyoT2 complex formation, whereas the glutamine side chain in Notch1 RAM contributes a modest 0.2 kcal/mol to binding CSL. This is a remarkable result, given that both KyoT2 and RAM bury similar amounts of nonpolar surface area when in complex with CSL (Table S1), and have similar ΔCp of binding values, which is correlated to the burial of nonpolar surface area. Taken together, these data suggest that the φWφP motifs of KyoT2 and RAM contribute similarly to CSL binding; however, the KyoT2 and RAM residues outside of the φWφP motif make significantly different energetic contributions to complexes formed with CSL.
Our analysis of mutations in the BTD of CSL revealed additional differences in the binding modes of KyoT2 and RAM. Previously, we designed four disruptive mutations in the BTD of CSL (F261R, V263R, A284R, Q333R) that targeted its interaction with the RAM domain of Notch and were based on our structure of the CSL-NICD-MAM activation complex bound to DNA (Yuan et al., 2012). As expected, all of these CSL mutants had severely reduced affinity for the RAM domains of Notch1 and Notch2 (from ~6-600fold), with F261R having the most detrimental effect on the binding of both Notch1 and Notch2 RAM (Table S2) (Yuan et al., 2012). However, a subset of these CSL mutants behaved very differently in binding experiments with KyoT2. While the mutants F261R and A284R had drastically reduced affinity for KyoT2 (>500fold), the BTD mutant V263R had no significant effect on binding (~1.6fold) and the Q333R mutant only modestly affected binding (~4.3fold) to CSL (Table S2). Moreover, these results were consistent with our cellular studies of KyoT2 with these CSL mutants. This is in stark contrast to the effect V263R and Q333R have on RAM binding (Table S2). Consistent with the observed changes in affinity, the mutants F261R and A284R map to regions on the BTD where there is a high degree of structural similarity between KyoT2 and RAM, whereas V263R and Q333R map to regions where the binding paths of KyoT2 and RAM bound diverge (Figure 3C and 3D). Interestingly, our binding analysis of CSL mutants largely mirrors previous studies, which identified mutations in the BTD of CSL that discriminated between RAM and EBNA2 binding (Fuchs et al., 2001). Through random mutagenesis and yeast two-hybrid approaches, Fuchs et al. identified mutations at residues F261, K275, A284, and Q333 in the BTD of CSL that retained binding of either RAM or EBNA2, but not both (Fuchs et al., 2001); three of these mutants were later confirmed in ITC binding studies by Johnson et al. (Johnson et al., 2010). Taken together, the data presented here, as well as previous studies, strongly suggest that while the structures of KyoT2 or RAM bound to CSL are generally similar, the peripheral regions of the KyoT2/RAM binding sites on the BTD display different selectivity towards different ligands, which leads to differential binding modes by KyoT2 and RAM, as well as EBNA2, for CSL. While the in vivo significance of these findings remains to be determined, it should be mentioned that a number of CSL-interacting coregulators have been identified, both in metazoans and viruses, that have motifs loosely resembling the φWφP motif and are thought to interact with the BTD of CSL; for example, the Epstein-Barr virus corepressor EBNA3C (Calderwood et al., 2011), the Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4/K10) (Heinzelmann et al., 2010), and the mammalian corepressor RITA (Wacker et al., 2011). Certainly, it will be of interest in future studies to characterize the structure and binding of these coregulators in complex with CSL, and compare these findings with our current and previous studies of CSL-KyoT2 and CSL-RAM complexes.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression and Purification
The core domain of mouse CSL (53-474) or the BTD of CSL (203-393) was cloned into the pGEX6P1 vector, expressed and purified as previously described (Friedmann and Kovall, 2010). Briefly, GST-CSL fusion constructs were overexpressed in bacteria and further isolated via glutathione-Sepharose affinity chromatography. The fusion protein was then cleaved with PreScission Protease, and the resulting CSL construct was purified to homogeneity using ion exchange and size exclusion chromatography. Mutants of CSL were made using site-directed mutagenesis and purified in the same manner as wild-type CSL.
Residues 184-210 and 184-200 of KyoT2 were cloned into the pSMT3 vector, which has the SMT3 (suppressor of Mif2 temperature-sensitive mutant 3) gene inserted into the pET28b(+) backbone (Mossessova and Lima, 2000). Expression from this plasmid results in a His-SMT3-KyoT2 fusion protein. KyoT2 constructs were transformed into bacteria BL21(DE3) Tuner cells, and were grown and overexpressed using autoinduction medium and methods (Studier, 2005). Bacteria were harvested using centrifugation and lysed using a high-pressure homogenizer. Bacterial lysates were cleared by centrifugation and incubated with Ni-NTA resin (Qiagen). The bead slurry was poured into a column, washed with buffer, and eluted with buffer containing imidazole, following the manufacturer’s protocol. Ulp-1 protease was used to cleave the His-SMT3-KyoT2 fusion protein, which cleaves after the SMT3 moiety, leaving an additional serine residue attached to the N-terminus of KyoT2. A His hi-trap column (GE Healthcare Life Sciences) was used to separate His-SMT3 from KyoT2, and KyoT2 was purified to homogeneity using size exclusion chromatography. KyoT2 mutants and variants were made using site-directed mutagenesis and purified in the same manner as KyoT2 wild-type proteins. A peptide corresponding to KyoT2 residues 182-196 was chemically synthesized and purified using reverse phase HPLC. Peptides corresponding to KyoT2 residues 184-197, 186-197, 187-196, 188-195, 189-194, 190-193 were chemically synthesized and ordered 95% pure from Peptide2.0. All KyoT2 constructs and mutants were verified through DNA sequencing and MALDI-TOF mass spectrometry analysis.
Crystallization and data collection
CSL-KyoT2-DNA complexes, which consist of residues 53-474 of the CSL surface entropy reduction mutation R115T, KyoT2 (184-210), and an oligomeric DNA duplex containing a single CSL binding site with single-stranded TT/AA overhangs (TTACTGTGGGAAAGA, AATCTTTCCCACAGT) were formed by mixing the components in a 1:1.1:1.1 ratio. Initial crystallization conditions were identified using the Hampton Research Index screen and an Art Robbins Phenix Crystallization Robot. Crystals were further optimized using microbatch under oil methods and the final optimized conditions were 0.1M HEPES pH 7.2, 21% PEG 3350, 0.2M Ammonium Acetate. Crystals were cryoprotected with ethylene glycol and flash frozen in liquid nitrogen. Data were collected at the Advanced Photon Source, beamline LS-CAT. Crystals nominally diffracted to 2.85 Å and belong to the space group P21 with unit cell dimensions a = 62.16 Å, b = 97.29 Å, c = 144.10 Å (see Table 1). All data were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997).
Structure determination, model building, and refinement
Phaser (Storoni et al., 2004), within the Phenix GUI (Adams et al., 2010), was used to solve the CSL-KyoT2-DNA structure, using our previous mouse CSL-DNA structures (3BRG & 3IAG) (Friedmann and Kovall, 2010; Friedmann et al., 2008). The Phaser solution revealed that there are two complexes within the asymmetric unit. Coot (Emsley and Cowtan, 2004) was used to manually build the KyoT2 residues into density, and the rebuilding of the model following refinement. Refinement of the CSL-KyoT2 structure was initially performed using Phenix, and Buster (Smart et al., 2012) was used for later rounds of refinement. Both NCS and TLS were used during refinement. The structure was validated with Molprobity (Chen et al., 2010) and deposited into the PDB under the accession code 4J2X. Pymol was used to create all structure figures, as well as perform all structural comparisons (Schrodinger, 2010). This PISA server was used for buried surface area calculations (Krissinel and Henrick, 2007).
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were carried out using a MicroCal VP-ITC microcalorimeter. All experiments were performed at 25°C in a buffer composed of 50 mM sodium phosphate pH 6.5 and 150 mM NaCl. CSL and KyoT2 proteins were degassed and buffer-matched using dialysis and size exclusion chromatography, respectively. A typical experiment was carried out with 10 uM CSL in the cell and 100 uM KyoT2 in the syringe. The data were analyzed using ORIGIN software and fit to a one-site binding model.
Cellular assays
As described previously, mouse embryonic fibroblasts (MEFs) derived from CSL knockout embryos (OT11) were transduced with retroviruses that express either wild-type or mutant CSL proteins (Yuan et al., 2012). Briefly, retrovirally transduced MEFs were grown to ~80% confluence in six-well plates and transiently transfected with an NICD1 construct (murine Notch1, residues 1744-2531, which is constitutively active), the luciferase reporter 4xCBS that contains four iterative CSL-binding sites, and the construct phRL, which expresses Renilla luciferase to normalize for transfection efficiency. In addition, full length wild-type or mutant (V186A, KD195AA, WW192AA) KyoT2 was cotransfected in increasing concentrations to measure the repressive effects of KyoT2 upon Notch mediated activation of the 4xCBS reporter. The SatisFection reagent (Agilent Technologies, Santa Clara, CA) was used for all transfections, following the manufacturer’s protocol. The amount of transfected DNA was normalized using pBluescript (Stratagene). Cells were harvested 48 hours post-transfection and luciferase activity was measured using the Dual Luciferase Kit (Promega). Firefly luciferase activity from the 4xCBS reporter was first normalized to Renilla luciferase activity and either reported as fold activation or relative activity. Average values, errors, and standard deviations were determined from three independent experiments performed in duplicate.
Circular dichroism
CD measurements were taken in triplicate using an Aviv Circular Dichroism Spectrometer Model 215 at 25 °C in a 0.02-cm cuvette. Wavelength scans were performed between 190 and 290 using 1.0-nm increments. KyoT2 (184-210) was characterized in a buffer containing 50 mM sodium phosphate pH 6.5 and 75 mM NaCl at a concentration of 0.35 mg/ml (120 uM). CD data were analyzed on DiChroweb using CDSSTR analysis program with reference set 7 (Whitmore and Wallace, 2004).
Supplementary Material
Highlights.
-
➢
We determined the 2.85 Å X-ray structure of the CSL-KyoT2 corepressor complex.
-
➢
Using ITC we defined the thermodynamic binding parameters of CSL-KyoT2 interactions.
-
➢
We showed that CSL-KyoT2 interactions correlate with repression potency in cells.
-
➢
Notch and KyoT2 bind CSL similarly, but utilize different molecular interaction modes.
Acknowledgements
We thank the members of the Kovall laboratory for their technical support and insightful discussion. We also thank Drs. Franz Oswald, Tilman Borgrrefe, and Rafi Kopan for constructs used in this study. This work was supported, in whole or in part, by National Institutes of Health Grant CA178974, a Leukemia and Lymphoma Society Scholar Award to R. A. K., and a predoctoral fellowship from the American Heart Association to K. J. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Blacklow SC. Targeting the Notch pathway: twists and turns on the road to rational therapeutics. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:2418–2420. doi: 10.1200/JCO.2012.42.0992. [DOI] [PubMed] [Google Scholar]
- Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochemical Society transactions. 2008;36:157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MA, Lee S, Holthaus AM, Blacklow SC, Kieff E, Johannsen E. Epstein-Barr virus nuclear protein 3C binds to the N-terminal (NTD) and beta trefoil domains (BTD) of RBP/CSL; only the NTD interaction is essential for lymphoblastoid cell growth. Virology. 2011;414:19–25. doi: 10.1016/j.virol.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Wales TE, Nam Y, O’Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational locking upon cooperative assembly of notch transcription complexes. Structure. 2012;20:340–349. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friedmann DR, Kovall RA. Thermodynamic and structural insights into CSL-DNA complexes. Protein Sci. 2010;19:34–46. doi: 10.1002/pro.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs KP, Bommer G, Dumont E, Christoph B, Vidal M, Kremmer E, Kempkes B. Mutational analysis of the J recombination signal sequence binding protein (RBP-J)/Epstein-Barr virus nuclear antigen 2 (EBNA2) and RBP-J/Notch interaction. European journal of biochemistry / FEBS. 2001;268:4639–4646. doi: 10.1046/j.1432-1327.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Human molecular genetics. 2003;12(Spec No 1):R9–13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Heinzelmann K, Scholz BA, Nowak A, Fossum E, Kremmer E, Haas J, Frank R, Kempkes B. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4/K10) is a novel interaction partner of CSL/CBF1, the major downstream effector of Notch signaling. Journal of virology. 2010;84:12255–12264. doi: 10.1128/JVI.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Johnson SE, Ilagan MX, Kopan R, Barrick D. Thermodynamic analysis of the CSL × Notch interaction: distribution of binding energy of the Notch RAM region to the CSL beta-trefoil domain and the mode of competition with the viral transactivator EBNA2. J Biol Chem. 2010;285:6681–6692. doi: 10.1074/jbc.M109.019968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411–455. doi: 10.1016/S0070-2153(10)92013-9. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. The EMBO journal. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes & development. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Liang L, Zhang HW, Liang J, Niu XL, Zhang SZ, Feng L, Liang YM, Han H. KyoT3, an isoform of murine FHL1, associates with the transcription factor RBP-J and represses the RBP-J-mediated transactivation. Biochim Biophys Acta. 2008;1779:805–810. doi: 10.1016/j.bbagrm.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Ling PD, Hayward SD. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. Journal of virology. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Qin H, Du D, Zhu Y, Li J, Feng L, Liang Y, Han H. The PcG protein HPC2 inhibits RBP-J-mediated transcription by interacting with LIM protein KyoT2. FEBS Lett. 2005;579:1220–1226. doi: 10.1016/j.febslet.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Qin H, Wang J, Liang Y, Taniguchi Y, Tanigaki K, Han H. RING1 inhibits transactivation of RBP-J by Notch through interaction with LIM protein KyoT2. Nucleic acids research. 2004;32:1492–1501. doi: 10.1093/nar/gkh295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci U S A. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran K, Selassie M, Liapis H, Krigman H, Kopan R. Reduced Notch signaling leads to renal cysts and papillary microadenomas. Journal of the American Society of Nephrology: JASN. 2010;21:819–832. doi: 10.1681/ASN.2009090925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes & development. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Current biology: CB. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWielen BD, Yuan Z, Friedmann DR, Kovall RA. Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J Biol Chem. 2011;286:14892–14902. doi: 10.1074/jbc.M110.181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker SA, Alvarado C, von Wichert G, Knippschild U, Wiedenmann J, Clauss K, Nienhaus GU, Hameister H, Baumann B, Borggrefe T, et al. RITA, a novel modulator of Notch signalling, acts via nuclear export of RBP-J. The EMBO journal. 2011;30:43–56. doi: 10.1038/emboj.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic acids research. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Wyman J, Gill SJ. Binding and linkage: functional chemistry of biological macromolecules. University Science Books; Mill Valley, Ca: 1990. [Google Scholar]
- Yuan Z, Friedmann DR, VanderWielen BD, Collins KJ, Kovall RA. Characterization of CSL (CBF-1, Su(H), Lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. J Biol Chem. 2012;287:34904–34916. doi: 10.1074/jbc.M112.403287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.