Abstract
The skeleton is originated from stem cells residing in the sclerotome and neural crest that undergo proliferation, migration and commitment. The development of the skeletal stem cells is influenced by many signaling pathways that govern cell fate determination, proliferation, differentiation and apoptosis. This review will focus on Notch signaling functions in regulating different cells types forming the skeletal system as well as the interplay between them to maintain homeostasis. Osteochondroprogenitors require Notch signaling to maintain the multipotency and to prevent from premature differentiation into osteoblast. Subsequently, over-activation of Notch signaling suppresses osteoblast maturation. Moreover, Notch signaling in osteochondroprogenitors is required for chondrocyte proliferation, hypertrophy and suppresses terminal differentiation. Translational studies demonstrated a crucial role of Notch signaling in osteosarcoma and osteoarthritis, where concepts derived from developmental pathways are often recapitulated. This brings hope of taking advantages of the molecular mechanisms learned from development to approach the pathological processes underlying abnormal bone/cartilage metabolism or tumorigenesis. Pharmacological agents that target Notch receptors or ligands in a tissue specific fashion would offer new opportunities for treating bone/cartilage diseases caused by dysregulation of Notch signaling.
Keywords: Notch signaling, osteoblast differentiation, chondrogenesis, osteoclastogenesis
Introduction
Notch signaling is an important evolutionally conserved pathway, and understanding of its pleiotropic functions involved in cell fate determination, proliferation, apoptosis and differentiation has been a continuous effort since its original description in the Notch Drosophila mutant by Morgan in 1917 [1]. Notch signaling contributes to tissue maintenance and/or renewal in the adult skin, hematopoietic system, and central nervous system and is implicated in various cancers. The Notch receptors and their ligands are transmembrane proteins that require cell-to-cell physical interaction between neighboring cells. Mammals have four Notch receptors (NOTCH 1-4) and twelve ligands grouped into four classes based on the structural motifs: 1) DSL (Delta/Serrate/LAG-2)/DOS (Delta and OSM-11-like proteins)-containing ligand: DLL1 (Delta-like 1), JAG1 and JAG2 [2], 2) DSL only ligand: DLL3 and DLL4, 3) DOS Co-ligands: DLK1, DLK2, 4) Non-canonical ligand: DNER, MAGP1 , MAGP2, F3/Contactin1, NB-3/Contactin6 [3]. Notch receptors undergo two sequential proteolytic cleavages upon binding to their cognate ligands presented on a neighboring cell surfaces [4]. The interaction between the ligand and Notch receptor results in a cleavage at the extracellural domain of the receptor by metalloproteinase tumor necrosis factor-α converting enzyme (TACE) and is followed by cleavage of the transmembrane domain by a γ-secretase complex consisting of Presenilin 1 and Presenilin 2 [5, 6]. Consequently, the Notch intracellular domain (Notch ICD, NICD) is released from the plasma membrane and translocates to the nucleus. In the nucleus, NICD interacts with RBPJ and Mastermind-Like (MAML), displacing the co-repressor complex bound by RBPJ to transform RBPJ into a transcription activator [7]. This transcriptionally active complex induces the expression of basic-helix-loop-helix (bHLH) family genes such as Hairy Enhancer of Split family genes: Hes1, 3, 5, and 7 and HES-related with a YRPF motif family genes Hey1, Hey2 and HeyL. Both the HES and HEY families function as transcriptional repressors regulating progenitor cell fate in many tissues such as muscle, neuron and hematopoietic cells [8, 9]. Less well understood is the process termed non-canonical Notch signaling, or Rbpj-independent Notch signaling. Here Rbpj deficiency does not recapitulate Notch deficiency as exemplified by Drosophila crystal cell differentiation [10], mammalian skin [11] or heart development [12].
Here, we will focus on the physiological role of Notch signaling in bone and cartilage development and in maintaining homeostasis, and extend consideration into its involvement in osteoarthritis and osteosarcoma. Finally, we will discuss the bone as a hematopoietic stem cell (HSC) niche whereby interaction with its microenvironment supports HSC homeostasis.
Notch signaling and human skeletal diseases
The role of Notch signaling during skeletogenesis was first identified in somitogenesis and patterning. Notch1 and Dll1 are highly expressed in the presomitic mesoderm of mouse embryos. Demonstratively, Notch1 null mouse embryos revealed significant delay and disorganization during somitogenesis [13]. Rbpj null embryos exhibited more severe defects in somitogenesis as a consequence of the complete loss of Notch signaling [14]. Presenilin1 null mice and Dll3 mutant mice (Pudgy mice) also have axial skeletal defects [15, 16]. Not surprisingly, human mutations in Notch signaling genes give rise to Spondylocostal dysostosis (SCDO), Alagille syndrome (AGS) and Adams-Oliver Syndrome (AOS) [17]. SCDO patients exhibit characteristic vertebral segmentation defects caused by disruption of Notch signaling due to homozygous mutations in DLL3 (Notch ligand), MESP2 (downstream target) or HES7 (downstream target), or LFNG (glycosylase). AGS, a multi-system disorder results from loss of function of JAG1 or NOTCH2, is a condition in which hemivertebrae and fractures are frequently seen and represent as diagnostic criteria [18]. Recently, heterozygous missense mutations in RBPJ were identified as causative mutations for autosomal dominant Adams-Oliver Syndrome of which distal limb defect is a prominent and consistent finding [17].
Apart from its role in embryonic axial skeleton patterning, recent studies have demonstrated that the Notch pathway also regulates developmental and homeostatic processes of cartilage and bone. Hajdu-cheney syndrome, an autosomal dominant disease, is characterized by craniofacial anomalies, acroosteolysis, Wormian bones, and osteoporosis [19], caused by heterozygous mutations in NOTCH2. These mutations result in a truncated protein lacking the proteolytic degradation domain of the NOTCH2 receptor. This leads to sustained NOTCH2 activation [20, 21]. Also, supporting Notch’s role in bone homeostasis is a genome wide association study demonstrating an association between bone mineral density and JAG1 polymorphisms in a Chinese population and in a population of mixed European and Chinese ancestry [22–24]. Hence, Notch signaling is essential for proper skeletal patterning during development and also for postnatal skeleton homeostasis.
Notch signaling during chondrocyte differentiation and osteoarthritis
The original shape and structure of the skeletal system are derived from cartilage tissue which plays roles in support and maintenance of the growth plate and articular cartilage [25]. Beginning with mesenchymal stem cell condensation, the chondrocyte is formed through an orderly differentiation process starting with resting cells to proliferating chondrocytes, transforming into pre-hypertrophic chondrocytes, then hypertrophic chondrocytes, and then finally terminally differentiating into hypertrophic chondrocytes [26] . Input from multiple signaling pathways, including transforming growth factor β (TGFβ), .broblast growth factor (FGF), Wnt, indian hedgehoge (IHH) and Notch signaling pathways, are integrated into a “morphogenetic rheostat” [27] to provide temporal and spatial cues for the proper differentiation [27–30]. Human mutations disrupting functions of these pathways underlie the mechanism of various skeletal disorders. A review from our lab [30] has provided a detailed description of these signaling pathways in skeletal dysplasias, here we will focus on the physiological and pathological functions of Notch signaling with implications for cancer therapy and regenerative medicine.
The in vivo function of Notch during the early steps of chondrogenesis was investigated by abolishing Notch signaling in limb bud mesenchymal stem cells by crossing Prx1-Cre with different conditional inactivation alleles of Notch signaling components. Presenilin 1/2 DKO (Presenilin1 flox/flox; Presenilin 2 −/−; Prx1-Cre , PPS mice), Notch 1/2 DKO (Notch1 flox/flox; Notch2 flox/flox; Prx1-Cre: PPN mice) [31], and Rbpj CKO (Rbpj flox/flox; Prx1-Cre) [32] exhibit a consistent phenotype: a delayed chondrocyte hypertrophy at E14.5-E15.5 and an elongated hypertrophic cartilage zone perinatally, suggesting Notch signaling is required to initiate chondrocyte hypertrophy. To explain the elongated hypertrophic zone, two possibilities are considered: 1) Accelerated hypertrophic chondrocyte differentiation; 2) Delayed chondrocyte apoptosis to remove terminally differentiated chondrocytes [33]. The length of the hypertrophic zone is reduced at the onset of hypertrophy (E14.5–E15.5) in all mutant mice identified above, negating the first explanation. Therefore, it is probable that a delay in terminal differentiation and programed cell death occurred as a consequence of Notch loss-of-function in chondrocyte progenitor cells.
Similarly, consequence of Notch signaling deficiency in chondrocyte progenitors was examined by using a different Cre line: Sox9 knock-in Cre, where endogenous Sox9 promoter drives Cre expression in the osteochondro-progenitors [34]. Inactivation of Rbpj with knock-in Sox9-Cre led to dwarfism, impaired matrix degradation and inhibited vasculature invasion. Molecularly, Mmp13 and Vegf expression were reduced, explaining the matrix and vasculature defects [41]. This corroborates with data from previous mutant mice using Prx1-Cre suggesting that Notch signaling is required for the initiation of chondrocyte maturation, i.e. hypertrophy and terminal differentiation.
A limitation of using Prx1-Cre and Sox9 knock-in Cre mouse lines is that the Cre is expressed in limb bud mesenchymal condensations, which contributes to osteoblastic and chondrogenic lineages. To further dissect the tissue-specific requirement of Notch in osteochondroprogenitors and chondrocytes, Col2a1-Cre and Col2a1-CreERT2 mouse lines were employed respectively. Deletion of Rbpj using Col2a1-Cre gives rise to a longer hypertrophic zone [35]. However, Col2-Cre; Rbpj flox/flox mice display milder degree of elongation than that of Prx1-Cre; Rbpj flox/flox. The Col2-Cre; Rbpj flox/flox mice display shorter hypertrophic zone marked by Col10a1 expression at E14.5 [32]. The reduction in length of the Mmp13 expression in chondrocytes strongly suggests a delay in chondrocyte terminal differentiation thereby reducing chondrocyte apoptosis and removal, which may account for the accumulation of hypertrophic chondrocytes at E18.5 and later time points.
To consider the pathological gain of Notch function in mesenchymal stem cells or osteochondroprogenitors, Prx1-Cre; RosaNotch1ICD and Col2a1-Cre; RosaNotch1ICD mutants were examined respectively. Both mutant mice shared similar phenotypes such as inhibition of cartilage formation [32, 35, 36], suppression of Sox9, Col2a1 and Aggrecan (Acan) expression, suggesting an inhibitory effect of Notch signaling on chondrocyte differentiation. Chondrocyte proliferation was inhibited in Col2a1-Cre; RosaNotch1ICD mice, while the mesenchymal stem cell proliferation was increased in the Prx1-Cre; RosaNotch1ICD mice [35, 36]. Taken together, Notch gain of function in chondrocytes inhibits chondrocyte differentiation and may have a differential effect on chondrocyte proliferation in context-dependent manner.
To assess the Notch gain of function (GOF) effects on hypertrophic chondrocytes, an in vitro assay was performed since Notch GOF mice have few mature chondrocytes in the limb. Enriched epiphyseal chondrocytes from RosaNotch1ICD mice were infected with adenovirus encoding Cre recombinase to induce NICD expression [37]. This leads to up-regulation of hypertrophic markers such as Col10a1 and Mmp13, and a notable down-regulation of Nfatc1 (nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1), a key regulator of osteoclast [38] and a target of Notch signaling in osteoblasts [39]. A rescue experiment shows that the expression of the constitutive form of Nfatc1 reversed the increased Col10a1 and Mmp13 in the Notch GOF chondrocytes, suggesting over-activated Notch signaling mis-regulates hypertrophic markers through suppressing Nfatc1 [37].
To understand the molecular mechanisms of Notch signaling during chondrocyte differentiation it is necessary to distinguish between the function of Rbpj-dependent (canonical) and Rbpj-independent (non-canonical) Notch signaling. Rbpj deletion in the context of Notch activation provides an insight into which aspects of Notch GOF phenotype are Rbpj-dependent. Dong et. al found that deletion of Rbpj in the Prx1-Cre; RosaNotch1ICD mice rescued the chondrodysplasia phenotype in the limbs. The restored cartilage in these mice was indistinguishable from the cartilage of Prx1-Cre; Rbpj flox/flox mice, supporting the concept that Rbpj is the sole mediator of Notch signaling function during limb cartilage development [36]. Kohn et. al removed Rbpj in the Col2a1-Cre ERT2;RosaNotch1ICD mice, which did not reverse the inhibited chondrocyte proliferation in the limbs but rather reversed the accelerated chondrocyte hypertrophy and terminal differentiation due to Notch activation [32]. Since cartilage development in the limbs is dependent upon canonical Notch signaling, a related question is whether this process depends on Rbpj during vertebral cartilage development. Our recent study showed the nearly complete loss of cartilage in the vertebral column from the Col2a1-Cre; RosaNotch1ICD mice [40]. Upon removal of Rbpj in the GOF background, cartilage formation was only partially restored, exhibiting a cleft between two hemivertebrae with nearly complete loss of ossification in the center of the vertebrae. This is in contrast to the vertebrae column of Col2a1-cre Rbpj flox/flox mice, which show no difference from the control mice, suggesting other factors than Rbpj may be recruited to mediate Notch GOF in the vertebrae. In terms of how Rbpj dependent Notch signaling regulated chondrogenesis in the vertebrae, Sox9 expression was reduced in the Notch GOF vertebrae, but was qualitatively restored in the GOF Rbpjflox/flox mice. It was also shown that Rbpj can associate with the Rbpj consensus binding motifs within the Sox9 promoter, leading to a direct regulation of Sox9 in trans. In summary, while Rbpj-dependent Sox9 suppression partially explains the chondrogenic phenotypes in the GOF mice, an Rbpj-independent Notch mechanism is postulated to govern chondrocyte differentiation and endochondral ossification.
Given the pivotal role of Notch in chondrocyte hypertrophy, an intriguing question is whether Notch signaling is associated with osteoarthritis (OA). OA, a degenerative disease of cartilage, presents with loss of articular cartilage markers, articular chondrocyte hypertrophy and subchondral bone remodeling. Loss of Notch signaling in adult cartilage, accomplished by Rbpj deletion with Col2a1-CreERT, a tamoxifen-inducible Cre line, ameliorates the progression of OA in a destabilization surgery model [41]. In relation to human articular cartilage, distribution of stem cell markers such as NOTCH1, STRO1 and vascular cell adhesion molecule 1 (VCAM1) were examined. NOTCH1 is expressed in 78% of the superficial zone of the articular cartilage cells significantly higher than in the middle or deep zone. In the fibrillated osteoarthritic cartilage, NOTCH1 positive cell population was significantly increased (up to 84%) in the superficial zone [42]. Similarly, another human study comparing the expression profiles of Notch signaling genes between OA cartilage and normal articular cartilage suggests that Notch pathway genes are expressed at a higher level in the OA cartilage which is consistent with the acceleration of chondrocyte hypertrophy observed in OA models [43–45]. This is supportive of the beneficial effect of reducing Notch signaling in the mouse model. It will be interesting to explore how the development of antibodies against Notch receptors and their ligands may influence OA treatment.
Notch Regulates Osteoblast commitment, proliferation and differentiation
Osteoblasts are bone-forming cells differentiated from mesenchymal precursors in the bone microenvironment. The role of Notch signaling in osteoblast differentiation has been examined by several in vitro studies and in vivo mouse models. In vitro studies suggested a potential role for Notch signaling in the regulation of osteoblast differentiation but with conflicting results. Tezuka et. al detected the expression of Notch1 in MC3T3-E1 osteoblastic cells at the early stages of differentiation. When Notch-ICD (NICD) was delivered by adenovirus vectors to MC3T3-E1 cells, a significant increase in calcified nodule formation was observed. Furthermore, Notch activation stimulated the multipotent mesenchymal cell line C3H10T1/2 to favor osteoblast differentiation rather than adipogenic differentiation [46]. Similarly, primary human bone marrow mesenchymal stem cells (hMSCs) also induced both spontaneous and stimulated osteoblastic cell differentiation by Notch activation. Consistently, the Notch target gene Hes-1 was shown to interact with Runx2 and increase the Runx2 transactivation effect on enhancers of osteoblast specific genes [47]. In addition, Maml (transcription activator of Notch signaling) was also shown to enhance transactivation of Runx2 in bone development. Overall, these observations suggest that osteoblast differentiation is stimulated by Notch signaling. On the other hand, others have shown a suppressive role of Notch signaling in osteoblast differentiation by inhibiting Wnt/β-catenin signaling after over-expressing Notch1 in ST-2 and MC3T3-E1 cells [48, 49].
The function of Notch in the osteoblast lineage and its differentiation was well defined by in vivo cell type specific Notch gain and loss of function mouse models. Transgenic mice expressing Notch1ICD under the control of the 2.3kb collagen type 1 (Col1a1) promoter drives the expression of the activated Notch receptor specifically in committed osteoblasts [50]. The transgenic mice showed a dramatic increase in osteoblast proliferation resulting in a severe osteosclerotic phenotype. However, the sclerotic bones were formed by disorganized woven bone, suggesting a defect in osteoblast maturation. This was confirmed by decreased expression of Osteocalcin (Ocn), a mature osteoblast marker from the calvarial tissue of mutant mice. Interestingly, Osterix was up-regulated along with Cyclin D and Cyclin E in this transgenic mouse, accounting for the dramatic expansion of the immature osteoblast population in these mutant mice. Mechanistically, Notch1ICD physically interacts with Runx2 and inhibits its transactivation on Ocn, demonstrating that Notch signaling suppresses Runx2 to maintain an immature osteoblast pool at the expense of mature osteoblast differentiation.
In contrast to these findings, expression of Notch1ICD under the control of the 3.6 kb Col1a1 promoter showed osteopenia due to inhibition of osteoblast progenitor differentiation [51]. This uncovers contrasting roles for Notch in two different cell populations. When Notch1ICD expression is restricted to committed osteoblasts in the case of the 2.3kb Col1a1 transgenic mice terminal differentiation of osteoblasts was inhibited allowing for expansion of immature progenitor cells leading to the increased formation of woven bone. However, when Notch1ICD expression is initiated in progenitor cells under the control of the 3.6 kb Col1a1 promoter, the early differentiation was compromised and led to a decreased number of mature osteoblasts and an osteopenic phenotype.
The loss of function of Notch signaling in committed osteoblasts was studied by the deletion of Presenilin1 and Presenilin2 using 2.3 kb Col1a1-Cre mice where the activation of all four Notch receptors was abolished in committed osteoblasts [50]. This double-knock out mouse did not show any immediately evident postnatal bone phenotype. However, the mice did show an age-related osteoporosis due to increased osteoclastogenesis caused by a decreased Opg level. To evaluate the function of Notch during early skeletal formation, Presenilin1 and Presenilin2 were deleted by using Prx1-Cre (PPS mice) in which Notch function was abolished in osteoblast progenitors [31]. Unlike Col1a1 2.3 kb - Cre mediated deletion of Presenilins, PPS mice exhibit a high bone mass phenotype at 2 months of age. Deletion of Notch1 and Notch2 via Prx1-Cre (PNN mice) mimicked the high bone mass phenotype of PPS mice. Interestingly, PPS and PNN mice progressed to a state of significant bone loss as they aged due to reduced osteoblast numbers and increased bone resorption. The bone marrow mesenchymal progenitors from PNN mice produced fewer CFU-f (colony forming unit-fibroblast) and CFU-ob (colony forming unit-osteoblast) and fewer differentiated adipocytes. The deletion of Rbpj in mesenchymal progenitors recapitulated the high-bone mass phenotype in PPS and PNN mice [39]. The progenitor specific Rbpj deficiency resulted in an increased osteoblast numbers, a diminished progenitor pool, and rapid age-related bone loss.
The phenocopy of Rbpj flox/flox;Prx1-cre with PNN or PPS mice suggests canonical Notch signaling is dominant in regulating osteoblast and osteoblast-osteoclast coupling. Furthermore, functions of canonical Notch targets such as Hes1, Hey1 and HeyL were examined in genetic mouse models or in vitro cell studies. Global Hey1 deletion leads to decreased BV/TV in adult mice, interestingly ubiquitous expression of Hey1 also leads to osteopenia. In both situations osteoblast differentiation and mineralization are inhibited and osteoclastogenesis is accelerated [52]. Osteoblast specific gain of function of Hes1 causes osteopenia by suppressing osteoblast differentiation. Loss of Hes1 in the background of Hes3 and 5 global null background manifest bone mass increase from 1 month up to 3 month, which is different from Rbpj deficient mice whose bone mass showed a significant decline by 4 month[53]. Moreover, both HeyL−/− and HeyL −/−, Hey1 +/−exhibit high bone mass at 2 month due to increase mature osteoblast number [39]. These studies support that canonical Notch signaling is a central regulator of osteoblast differentiation at early stages. A ChIP-seq (chromatin immunoprecipitation followed by high throughput sequencing) study demonstrated that Nfatc1 is a critical mediator of Notch function in osteoblasts. The recruitment of Hey1 to the promoter of Nfatc1 leads to transcription suppression of Nfatc1, and deletion of Rbpj relieved this repression in bone. Moreover, pharmacological inhibitor of Nfatc1 alleviated the abnormal high bone mass phenotype caused by deletion of Rbpj. Overall, Notch-Rbpj signaling functions in part via Hey1-mediated suppression of Nfatc1 to suppress osteoblastogenesis[39]. Considering another Nfat family member, Nfatc2, in vitro cell studies demonstrated that Notch1, Hey1 and Hey2 inhibit Nfatc2 transactivation by suppressing Gsk3β, which activates Nfatc2 by phosphorylation, in ST-2 stromal cells as well as in primary osteoblast cells from Notch GOF mice[37].
The consequences of activation of Notch signaling in osteocytes, terminally differentiated osteoblasts, was recently examined by using Dentin matrix protein1 (Dmp1) promoters-Cre [54]. Osteocytes are embedded in the mineralized matrix of the bone in distinct lacunae [55, 56]. Osteocytes are dendiritic cells that communicate through a canalicular network with each other and with osteoblasts and osteoclasts [57]. Osteocytes play a crucial role in mechanotransduction. When osteocytes were ablated, mice exhibited a severe bone loss and microstructural deterioration [58]. Dmp1-Cre;RosaNotch1ICD mice exhibited an increased bone volume due to initial inhibition of bone resorption at 1 month. This phenotype persisted at 3 months of age due to an increased osteoblast number, suggesting that Notch signaling also contributes to non-cell autonomous function of osteocytes in the regulation of osteoclasts and osteoblasts. Concordant with this, induction of Notch1 expression in an MLOY4 (osteocytic cell line) in response to a fluid flow shear stress demonstrated Notch may regulate mechanotrasduction in osteocytes [54].
Together, these studies show that during the early stages of osteogenesis, Notch maintains mesenchymal progenitor cells and differentiation potency. Upon loss of Notch signaling, the program of osteoblastic commitment is initiated in conjunction with loss of mesenchymal stem cell multi-potency. Once committed, Notch signaling is required for osteoblast turnover and osteoclast regulation during bone remodeling to maintain a sufficient pool of osteoblasts and an appropriate pace of bone remodeling. In ours and another study of Notch gain of function in committed osteoblasts causes abnormal proliferation of immature osteoblasts contributing to a high bone mass phenotype. Canonical Notch signaling is responsible for mediating the Notch functions during different stages of osteoblast differentiation as well as in the pathogenic GOF conditions [39, 59].
Notch regulates osteoclastogenesis
Osteoclastogenesis is modulated by the ratio of receptor activator of nuclear factor k-B ligand (RANKL), which induces osteoclast formation, and osteoprotegerin (OPG), a soluble protein that binds and inhibits RANKL [60]. In an In vitro study using immobilized Notch ligand Delta-1, it has suggested that osteoclastogenesis is inhibited when Notch is activated in the osteoclast lineage [61]. Additionally, the deletion of Notch1, Notch2 and Notch3 in murine myeloid cells was seen to enhance osteoclast precursor proliferation and differentiation compared to wild type in the response to the macrophage colony-stimulating factor, confirming the cell autonomous function of Notch signaling in osteoclast differentiation [62].
In vivo RANKL expression was up-regulated in PNN mice, and Opg levels were decreased [31]. Similarly, deletion of Presenillins1/2 in the committed osteoblast by Col1a1 2.3kb-Cre resulted in a decreased level of Opg and increased osteoclastogenesis thereby causing osteopenia [50]. These observations further indicate that Notch signaling regulates osteoclastogenesis in a non-cell autonomous manner through Opg expression by osteoblasts in vivo. The recent studies of osteocytes showed that these terminally differentiated cells from osteoblasts are actively involved in bone remodeling and bone resorption through the expression of RANKL. The suppression of bone resorption in Dmp1-Cre;RosaNotch1ICD mice is explained by an increased level of Opg consistent with previous studies [54].
In contrary, Notch signaling was shown to activate osteoclastogenesis in different contexts. Fukushima et. al demonstrated that Jagged1 could increase osteoclast differentiation in vitro by specifically activating Notch2 ICD which augmented RANKL-induced NFkB signaling [63]. Moreover, in a pathological context, tumor-expressed JAG1 acts as a potent mediator in promoting the vicious cycle of breast cancer metastasis to the bone by activating pre-osteoclast differentiation [64]. Consequently, γ-secretase inhibitor treatment reversed this pro-metastatic function of JAG1 by disrupting the Notch pathway. In a K/BxN serum-induced arthritic mouse model [65], the differential regulation of osteoclastogenesis by Notch2/Dll1 and Notch1/Jag1 axes was demonstrated. Blockade of Dll1 by anti-Dll1 monoclonal antibody ameliorated arthritis in this mouse model [66]. Collectively, these studies highlight that cell-autonomous Notch signaling plays a key role in osteoclast differentiation and is regulated through differential ligand-receptor interactions. The multiplicity of the cell types and ligand-receptor specificity employed by Notch signaling evokes attention to these contextual issues. Future identification and delineation of the mechanism underlying this complexity will help tease out the dominant ligand or cell type that takes part in a particular pathological context of bone-related diseases.
Osteoblasts regulate HSC via Notch signaling
In bone marrow, osteoblasts are active participants in regulating hematopoiesis by providing regulatory signals for hematopoietic cells. Homeostasis of the hematopoietic system is maintained by the capacity of hematopoietic stem cells (HSCs) to undergo differentiation forming mature blood cell types and self-renewal to replenish the HSC pool throughout life. Because of the juxtaposition of the endosteum and HSC, osteoblasts lining the endosteum are postulated to form the specific microenvironment (so called “niche”) to support HSC functions. The first functional study of osteoblast regulation of hematopoiesis in vivo was done by targeted expression of thymidine kinase in osteoblasts where gancyclovir administration induced cell death. The ablation of osteoblasts is correlated with a dramatic decrease in bone marrow cellularity and extramedullary hematopoiesis [67, 68]. Withdrawal of gancyclovir resulted in rapid recovery of hematopoiesis with increased bone formation.
Transgenic mice with expression of constitutively active parathyroid hormone/parathyroid hormone-related peptide receptor (PPR) under the control of the Col1a1 2.3 kb promoter demonstrated increased osteoblast numbers, an increased expression of Jag1, and a concomitant increase in the HSC population [69]. Moreover, the contact between the HSCs and Jag1-expressing osteoblasts were required for the HSC expansion, and the addition of a γ-secretase inhibitor to stromal cells abrogated the HSC expansion in vitro. These data demonstrated that the osteoblast is one of the regulatory components of HSCs and employs Notch signaling for such an action. In contrast, deletion of either Jag1 or Notch1 in the bone marrow cells with Mx1-Cre did not lead to HSC defects under basal conditions [70] When canonical Notch signaling was inhibited in adult HSC by either expressing a dominant-negative Mastermind-like1 fused to GFP (DNMML) or by conditionally deleting Rbpj, these HSCs failed to show any phenotype in a competitive repopulation assay [71]. Although not absolutely required, Notch2 enhances the rate of generation of long- and short-term repopulating HSCs and delays myeloid differentiation following bone marrow injury [72]. All together, these data suggest that Notch signaling may be dispensable for HSC development in the context of normal development, but may be partially required for HSC repopulation after bone marrow injury.
Dysregulation of Notch signaling and Osteosarcoma
Primary bone cancer consists of neoplastic cells in bone. The most common types of primary bone tumor are osteosarcoma (35%), chondrosarcoma (30%) and Ewing’s sarcoma (16%) [73]. As often seen in cancer, developmental pathways are recapitulated in crucial steps of cancer transformation, proliferation and metastasis. The association of Notch signaling to the keys steps of cancer progression has been illustrated by both human and animal studies. Expression of Notch pathway genes have been analyzed in osteosarcoma specimens, in which NOTCH2, JAG1, HEY1 and HEY2 are up-regulated while NOTCH1 and DLL1 are down-regulated, compared to those of normal human bone[74]. Similarly, we demonstrated NOTCH1, JAG1, HES1 and HEY1 expressions are significantly higher in human osteosarcoma compared to human osteoblasts [75]. Transcription profiling of osteosarcoma tumors from p53 heterozygous mice which develop osteosarcoma at a high frequency (24%) confirms the human findings, showing elevated Jag2, Hes1, Hey1 and Dll4 expressions [75–77]. A γ-secretase inhibitor, a potent agent to inhibit Notch signaling transduction, suppressed human osteosarcoma cell lines (SaOS2, HOS and 143B) proliferation. Importantly, xenograft of SaOS2 or 143B in nude mice showed that either a γ-secretase inhibitor or over-expression of dominant negative-Mastermind-like to abolish Notch transcriptional regulation can reduce primary tumor burden by 20–80% [74, 75]. The molecular mechanism is attributed to a down-regulation of CyclinD, CyclineE1, CyclineE2, SKP2 and c-Myc expressions when Notch signaling is inhibited [74].
Approximately 30% of patients with localized osteosarcoma and 80% of patients presenting metastatic disease experience relapse [73]. Analysis of Notch pathway gene expressions identified higher levels of Notch1, Notch2, Dll1 and Hes1 expression in metastatic osteosarcoma cell lines, compared to normal osteoblast or non-metastatic osteosarcoma [78]. A retrospective study on HES1 expression on human osteosarcoma specimen showed an inverse relationship between HES1 level and patients’ survival. The notion that Notch promotes metastasis is supported by the ability of the γ-secretase inhibitor or dominant negative-Mastermind-like to decrease invasiveness of osteosarcoma cell lines (OS187 and LM7) in a Matrigel transwell assay [79]. Notably, over-expression of DN-MML suppressed the metastasis potency of OS187 using an intra-tibial injection xenograft model, albeit not changing the primary tumor growth or primary osteolytic lesion formation [79]. Epigenetic regulation of Notch is also seen in osteosarcoma progression and invasion that role of miRNAs is relevant in these processes. Our recent study has demonstrated that miRNA-34c can directly regulate multiple components of Notch pathway (Notch1, Notch2 and Jag1) during osteoblast differentiation in in vivo mouse model [80]. Also, others showed the inverse correlation with an invasiveness of osteosarcoma tumors and expression of NOTCH1, NOTCH2 and DLL1 by miRNA-34 family [81]. The association of Deltex1 with activated Notch receptor restrains the invasiveness in osteosarcoma cell OS187, as Deltex1 over-expression leads to Notch degradation and reduced invasiveness [82].
In summary, pharmacological administration or genetic manipulation support that Notch signaling participates in the proliferation, invasion and metastasis of osteosarcoma progression.
Future Directions
Advances in understanding the role of Notch in various cells of the skeleton have provided important insights into the etiology of skeletal conditions like osteosclerosis, osteoporosis, chondrodysplasia, osteoarthritis or osteosarcoma. Attempts to modulate Notch signaling pathways have been made to treat diseases such as osteosarcoma, graft-versus-host disease, hematologic cancer in various mouse models. To translate these results into therapeutic reality, toxicity needs to be considered in terms of usage of γ-secretase inhibitors or pan-Notch receptor antibodies, which have demonstrated high intestinal toxicity in rodent [83, 84]. Antibodies that recognize Notch negative regulatory region and antagonize individual Notch receptor may reduce or limit this adverse effect [83–85]. Ligand specific antibodies against Dll1 and Dll4 further reduces the toxicity in treating a mouse model of graft-versus-host disease [85], suggesting rational design of ligand neutralizing antibodies specific for targeted tissues may shed promising light on its application. The skeletal defects exhibited from of Dll3 and Jag2 null mice and human skeletal diseases such as SCDO and AGS caused by mutations in DLL3 and JAG1 underscore the specific utilization of the Notch ligands in the context of skeletogenesis. Hence, defining functions and the molecular mechanisms of Notch ligands will allow us to delineate the signaling sending partners of Notch pathway and to develop ligand targeted therapy for skeletal disorders. Distinguishing canonical vs. non-canonical Notch signaling is second concern. Increasing evidence suggests Rbpj-independent Notch signaling governs differential response to Notch stimulation (at least under pathological conditions). Identification of the non-canonical components and downstream targets with screening tools like RNA-seq, ChIP-seq or Mass-spectrometry may serve as an informative guide to test biological functions of non-canonical Notch signaling genes in cell studies or animal models. This may also lead to lower toxicity or higher efficacy by targeting only a small subset of cell population where non-canonical signaling is active as demonstrated in heart development [12].
In conclusion, Notch signaling regulates multiple steps of osteoblast development including osteoblast commitment, proliferation and maturation as well as functional interaction with osteoclasts and HSCs in bone microenvironment (Figure 1). The requirement of Notch in cartilage during developmental processes and homeostasis provides new insights into skeletal disorders like SCDO and AGS beyond its well-understood function in patterning (Figure 2). Mis-regulation of Notch signaling and its association with cancer may lead to therapeutic innovations where manipulation of Notch signaling either canonically or non-canonically may be of benefit in reducing cancer risk, progression or metastasis.
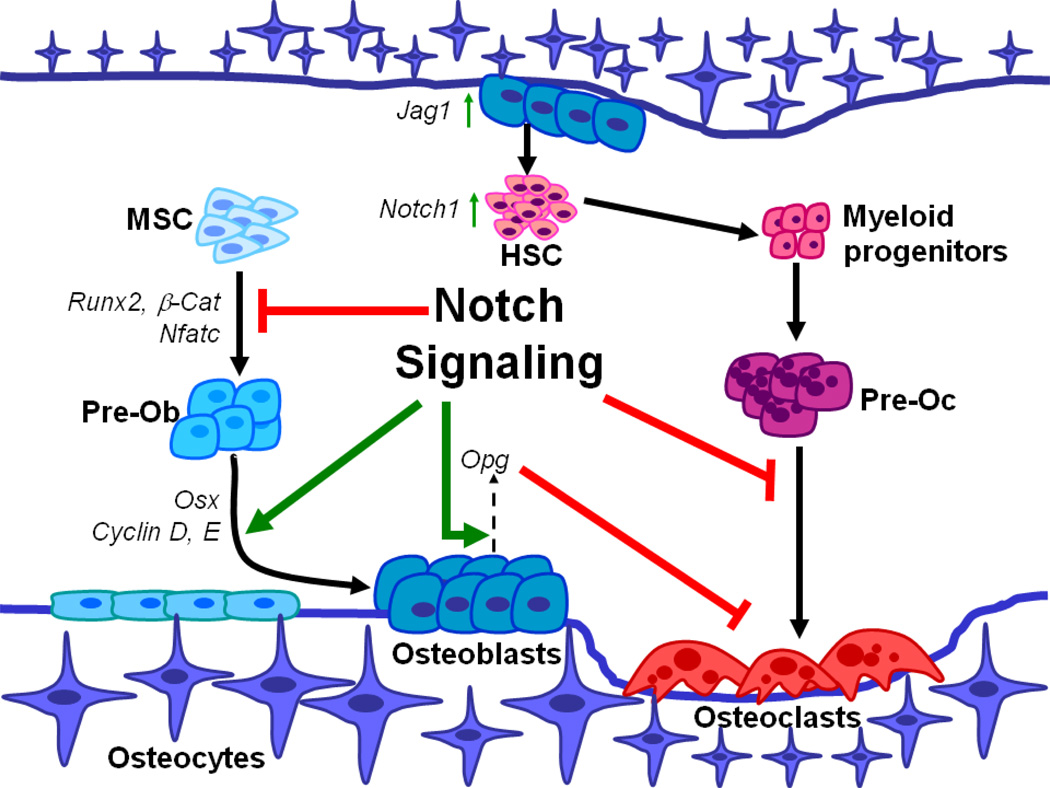
Figure 1.
Notch signaling in bone homeostasis and bone microenvironment. Notch maintains mesenchymal stem cells (MSCs) in an undifferentiated stage by repressing Runx2. The role of Notch signaling in inhibiting osteoblast differentiation is mediated by Nfatc1 and by inhibition of Wnt/β-catenin. Notch gain of function increases immature pre-osteoblasts (Pre-Ob) pool by up-regulating transcription of Osx, Cyclin D, and Cyclin E. Notch signaling regulates osteoclastogenesis in a non-cell autonomous manner through Opg expression by osteoblasts in vivo. Additionally, osteoclastogenesis is repressed by cell autonomous function of Notch signaling in Pre-osteoclast (Pre-Oc). At the interface of osteoblast and HSC within the bone microenvironment, osteoblasts act as signal sending cells by expressing Jag1 and activate Notch1 signaling in hematopoietic stem cells (HSCs) resulting in an expansion of this cell population.
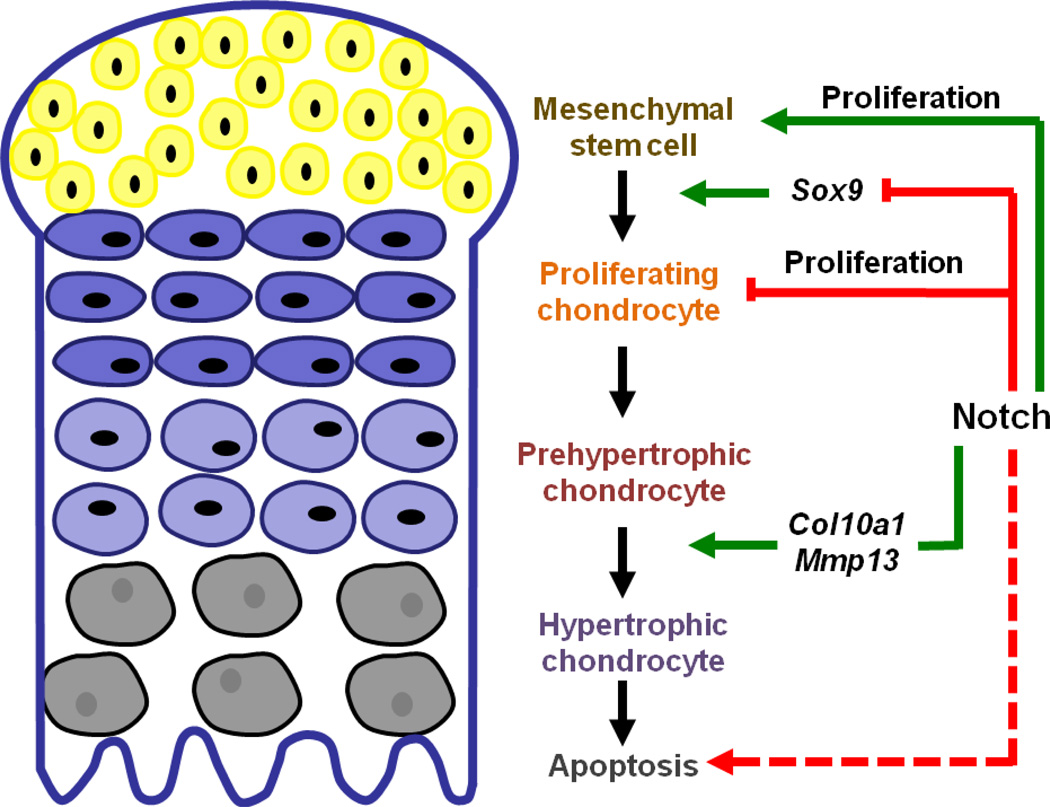
Figure 2.
Notch signaling in cartilage development. Notch signaling is necessary and sufficient to suppress Sox9 expression during early mesenchymal stem cell commitment. Notch signaling promotes mesenchymal stem cell proliferation but inhibits chondrocyte proliferation. Notch signaling is also requried for the onset ofchondrocyte hypertrophy via regulating Col10a1 and Mmp13 expressions. It is postulated that Notch signaling promotes terminal differentiation of hypertrophic chondrocytes as evidenced by the accumulation of hypertrophic chondrocytes in the growth plate of Notch deficient mice.
Acknowledgments
The authors thank Terry Bertin for critical reading and editing the manuscript. Some of the works are supported by National Institute of Health grants DE016990 (BL) and HD022657 (BL), and Cancer Prevention Institute of Texas grant RP 101017 (BL).
Footnotes
The authors have stated that they have no conflict of interest.
DISCLOSURES: NONE
References
- 1.Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- 2.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 3.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 5.De Strooper B, Woodgett J. Alzheimer's disease: Mental plaque removal. Nature. 2003;423:392–393. doi: 10.1038/423392a. [DOI] [PubMed] [Google Scholar]
- 6.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 7.Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- 8.Zanotti S, Canalis E. Notch regulation of bone development and remodeling and related skeletal disorders. Calcif Tissue Int. 2012;90:69–75. doi: 10.1007/s00223-011-9541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engin F, Lee B. NOTCHing the bone: insights into multi-functionality. Bone. 2010;46:274–280. doi: 10.1016/j.bone.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, Grigsby PW, Miner JH, Farr AG, Kopan R. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 14.Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 15.Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 16.Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- 17.Hassed SJ, Wiley GB, Wang S, Lee JY, Li S, Xu W, Zhao ZJ, Mulvihill JJ, Robertson J, Warner J, Gaffney PM. RBPJ mutations identified in two families affected by Adams-Oliver syndrome. Am J Hum Genet. 2012;91:391–395. doi: 10.1016/j.ajhg.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narumi Y, Min BJ, Shimizu K, Kazukawa I, Sameshima K, Nakamura K, Kosho T, Rhee Y, Chung YS, Kim OH, Fukushima Y, Park WY, Nishimura G. Clinical consequences in truncating mutations in exon 34 of NOTCH2: Report of six patients with Hajdu-Cheney syndrome and a patient with serpentine fibula polycystic kidney syndrome. Am J Med Genet A. 2013;161:518–526. doi: 10.1002/ajmg.a.35772. [DOI] [PubMed] [Google Scholar]
- 20.Isidor B, Lindenbaum P, Pichon O, Bezieau S, Dina C, Jacquemont S, Martin-Coignard D, Thauvin-Robinet C, Le Merrer M, Mandel JL, David A, Faivre L, Cormier-Daire V, Redon R, Le Caignec C. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet. 2011;43:306–308. doi: 10.1038/ng.778. [DOI] [PubMed] [Google Scholar]
- 21.Majewski J, Schwartzentruber JA, Caqueret A, Patry L, Marcadier J, Fryns JP, Boycott KM, Ste-Marie LG, McKiernan FE, Marik I, Van Esch H, Michaud JL, Samuels ME. Mutations in NOTCH2 in families with Hajdu-Cheney syndrome. Hum Mutat. 2011;32:1114–1117. doi: 10.1002/humu.21546. [DOI] [PubMed] [Google Scholar]
- 22.Hsu YH, Kiel DP. Clinical review: Genome-wide association studies of skeletal phenotypes: what we have learned and where we are headed. J Clin Endocrinol Metab. 2012;97:E1958–E1977. doi: 10.1210/jc.2012-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kung AW, Xiao SM, Cherny S, Li GH, Gao Y, Tso G, Lau KS, Luk KD, Liu JM, Cui B, Zhang MJ, Zhang ZL, He JW, Yue H, Xia WB, Luo LM, He SL, Kiel DP, Karasik D, Hsu YH, Cupples LA, Demissie S, Styrkarsdottir U, Halldorsson BV, Sigurdsson G, Thorsteinsdottir U, Stefansson K, Richards JB, Zhai G, Soranzo N, Valdes A, Spector TD, Sham PC. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet. 2010;86:229–239. doi: 10.1016/j.ajhg.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao SM, Kung AW, Gao Y, Lau KS, Ma A, Zhang ZL, Liu JM, Xia W, He JW, Zhao L, Nie M, Fu WZ, Zhang MJ, Sun J, Kwan JS, Tso GH, Dai ZJ, Cheung CL, Bow CH, Leung AY, Tan KC, Sham PC. Post-genome wide association studies and functional analyses identify association of MPP7 gene variants with site-specific bone mineral density. Hum Mol Genet. 2012;21:1648–1657. doi: 10.1093/hmg/ddr586. [DOI] [PubMed] [Google Scholar]
- 25.Cole AG. A review of diversity in the evolution and development of cartilage: the search for the origin of the chondrocyte. Eur Cell Mater. 2011;21:122–129. doi: 10.22203/ecm.v021a10. [DOI] [PubMed] [Google Scholar]
- 26.van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage. 2012;20:223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Wuelling M, Vortkamp A. Transcriptional networks controlling chondrocyte proliferation and differentiation during endochondral ossification. Pediatr Nephrol. 2010;25:625–631. doi: 10.1007/s00467-009-1368-6. [DOI] [PubMed] [Google Scholar]
- 28.Miraoui H, Marie PJ. Fibroblast growth factor receptor signaling crosstalk in skeletogenesis. Sci Signal. 2010;3:re9. doi: 10.1126/scisignal.3146re9. [DOI] [PubMed] [Google Scholar]
- 29.Plaas A, Velasco J, Gorski DJ, Li J, Cole A, Christopherson K, Sandy JD. The relationship between fibrogenic TGFbeta1 signaling in the joint and cartilage degradation in post-injury osteoarthritis. Osteoarthritis Cartilage. 2011;19:1081–1090. doi: 10.1016/j.joca.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Baldridge D, Shchelochkov O, Kelley B, Lee B. Signaling pathways in human skeletal dysplasias. Annu Rev Genomics Hum Genet. 2010;11:189–217. doi: 10.1146/annurev-genom-082908-150158. [DOI] [PubMed] [Google Scholar]
- 31.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, O'Keefe RJ, Hilton MJ. Cartilage-specific RBPjkappa-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139:1198–1212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro IM, Adams CS, Freeman T, Srinivas V. Fate of the hypertrophic chondrocyte: microenvironmental perspectives on apoptosis and survival in the epiphyseal growth plate. Birth Defects Res C Embryo Today. 2005;75:330–339. doi: 10.1002/bdrc.20057. [DOI] [PubMed] [Google Scholar]
- 34.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci U S A. 2009;106:14420–14425. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, O'Keefe RJ, Hilton MJ. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanotti S, Canalis E. Notch suppresses nuclear factor of activated T cells (NFAT) transactivation and Nfatc1 expression in chondrocytes. Endocrinology. 2013;154:762–772. doi: 10.1210/en.2012-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song I, Kim JH, Kim K, Jin HM, Youn BU, Kim N. Regulatory mechanism of NFATc1 in RANKL-induced osteoclast activation. FEBS Lett. 2009;583:2435–2440. doi: 10.1016/j.febslet.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 39.Tu X, Chen J, Lim J, Karner CM, Lee SY, Heisig J, Wiese C, Surendran K, Kopan R, Gessler M, Long F. Physiological notch signaling maintains bone homeostasis via RBPjk and Hey upstream of NFATc1. PLoS Genet. 2012;8:e1002577. doi: 10.1371/journal.pgen.1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S, Tao J, Bae Y, Jiang MM, Bertin T, Chen Y, Yang T, Lee B. Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. J Bone Miner Res. 2013;28:649–659. doi: 10.1002/jbmr.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosaka Y, Saito T, Sugita S, Hikata T, Kobayashi H, Fukai A, Taniguchi Y, Hirata M, Akiyama H, Chung UI, Kawaguchi H. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proc Natl Acad Sci U S A. 2013;110:1875–1880. doi: 10.1073/pnas.1207458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grogan SP, Miyaki S, Asahara H, D'Lima DD, Lotz MK. Mesenchymal progenitor cell markers in human articular cartilage: normal distribution and changes in osteoarthritis. Arthritis Res Ther. 2009;11:R85. doi: 10.1186/ar2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahjoub M, Sassi N, Driss M, Laadhar L, Allouche M, Hamdoun M, Romdhane KB, Sellami S, Makni S. Expression patterns of Notch receptors and their ligands in human osteoarthritic and healthy articular cartilage. Tissue Cell. 2012;44:182–194. doi: 10.1016/j.tice.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Sassi N, Laadhar L, Driss M, Kallel-Sellami M, Sellami S, Makni S. The role of the Notch pathway in healthy and osteoarthritic articular cartilage: from experimental models to ex vivo studies. Arthritis Res Ther. 2011;13:208. doi: 10.1186/ar3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsson C, Brantsing C, Egell S, Lindahl A. Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs. 2008;188:287–298. doi: 10.1159/000121610. [DOI] [PubMed] [Google Scholar]
- 46.Tezuka K, Yasuda M, Watanabe N, Morimura N, Kuroda K, Miyatani S, Hozumi N. Stimulation of osteoblastic cell differentiation by Notch. J Bone Miner Res. 2002;17:231–239. doi: 10.1359/jbmr.2002.17.2.231. [DOI] [PubMed] [Google Scholar]
- 47.McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 48.Sciaudone M, Gazzerro E, Priest L, Delany AM, Canalis E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology. 2003;144:5631–5639. doi: 10.1210/en.2003-0463. [DOI] [PubMed] [Google Scholar]
- 49.Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J Biol Chem. 2006;281:6203–6210. doi: 10.1074/jbc.M508370200. [DOI] [PubMed] [Google Scholar]
- 50.Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149:3890–3899. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salie R, Kneissel M, Vukevic M, Zamurovic N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B, Susa M. Ubiquitous overexpression of Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. Bone. 2010;46:680–694. doi: 10.1016/j.bone.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 53.Zanotti S, Smerdel-Ramoya A, Canalis E. HES1 (hairy and enhancer of split 1) is a determinant of bone mass. J Biol Chem. 2011;286:2648–2657. doi: 10.1074/jbc.M110.183038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canalis E, Parker K, Feng JQ, Zanotti S. Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology. 2012;154:623–634. doi: 10.1210/en.2012-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aarden EM, Burger EH, Nijweide PJ. Function of osteocytes in bone. J Cell Biochem. 1994;55:287–299. doi: 10.1002/jcb.240550304. [DOI] [PubMed] [Google Scholar]
- 56.Bonewald LF. The amazing osteocyte. J Bone Miner Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong J, O'Brien CA. Osteocyte RANKL: new insights into the control of bone remodeling. J Bone Miner Res. 2012;27:499–505. doi: 10.1002/jbmr.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatsumi S, Ishii K, Amizuka N, Li M, Kobayashi T, Kohno K, Ito M, Takeshita S, Ikeda K. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 2007;5:464–475. doi: 10.1016/j.cmet.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Tao J, Chen S, Yang T, Dawson B, Munivez E, Bertin T, Lee B. Osteosclerosis owing to Notch gain of function is solely Rbpj-dependent. J Bone Miner Res. 2010;25:2175–2183. doi: 10.1002/jbmr.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teitelbaum SL. Osteoclasts: what do they do and how do they do it? Am J Pathol. 2007;170:427–435. doi: 10.2353/ajpath.2007.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101:2227–2234. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- 62.Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem. 2008;283:6509–6518. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- 63.Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, Okabe K. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol Cell Biol. 2008;28:6402–6412. doi: 10.1128/MCB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 66.Sekine C, Koyanagi A, Koyama N, Hozumi K, Chiba S, Yagita H. Differential regulation of osteoclastogenesis by Notch2/Delta-like 1 and Notch1/Jagged1 axes. Arthritis Res Ther. 2012;14:R45. doi: 10.1186/ar3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visnjic D, Kalajzic I, Gronowicz G, Aguila HL, Clark SH, Lichtler AC, Rowe DW. Conditional ablation of the osteoblast lineage in Col2.3deltatk transgenic mice. J Bone Miner Res. 2001;16:2222–2231. doi: 10.1359/jbmr.2001.16.12.2222. [DOI] [PubMed] [Google Scholar]
- 68.Visnjic D, Kalajzic Z, Rowe DW, Katavic V, Lorenzo J, Aguila HL. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- 69.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 70.Mancini SJ, Mantei N, Dumortier A, Suter U, MacDonald HR, Radtke F. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 2005;105:2340–2342. doi: 10.1182/blood-2004-08-3207. [DOI] [PubMed] [Google Scholar]
- 71.Maillard I, Koch U, Dumortier A, Shestova O, Xu L, Sai H, Pross SE, Aster JC, Bhandoola A, Radtke F, Pear WS. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008;2:356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varnum-Finney B, Halasz LM, Sun M, Gridley T, Radtke F, Bernstein ID. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J Clin Invest. 2011;121:1207–1216. doi: 10.1172/JCI43868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gralow JR, Biermann JS, Farooki A, Fornier MN, Gagel RF, Kumar RN, Shapiro CL, Shields A, Smith MR, Srinivas S, Van Poznak CH. NCCN Task Force Report: Bone Health in Cancer Care. J Natl Compr Canc Netw 7 Suppl. 2009;3:S1–S32. doi: 10.6004/jnccn.2009.0076. quiz S33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka M, Setoguchi T, Hirotsu M, Gao H, Sasaki H, Matsunoshita Y, Komiya S. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100:1957–1965. doi: 10.1038/sj.bjc.6605060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet. 2009;18:1464–1470. doi: 10.1093/hmg/ddp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, El-Naggar AK, Lozano G. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Hughes DP. How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. Cancer Treat Res. 2009;152:479–496. doi: 10.1007/978-1-4419-0284-9_28. [DOI] [PubMed] [Google Scholar]
- 79.Zhang P, Yang Y, Zweidler-McKay PA, Hughes DP. Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin Cancer Res. 2008;14:2962–2969. doi: 10.1158/1078-0432.CCR-07-1992. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun. 2009;388:35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 82.Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DP. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene. 2010;29:2916–2926. doi: 10.1038/onc.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 84.Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, Huang L, Vitelli S, Vo KT, Haytko P, Zhao JZ, Baleydier F, L'Heureux S, Wang H, Gordon WR, Thoryk E, Andrawes MB, Tiyanont K, Stegmaier K, Roti G, Ross KN, Franlin LL, Wang F, Chastain M, Bett AJ, Audoly LP, Aster JC, Blacklow SC, Huber HE. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tran IT, Sandy AR, Carulli AJ, Ebens C, Chung J, Shan GT, Radojcic V, Friedman A, Gridley T, Shelton A, Reddy P, Samuelson LC, Yan M, Siebel CW, Maillard I. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J Clin Invest. 2013 doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]