Abstract
Semaphorin 7A (Sema7A) plays a major role in TGF-β1-induced lung fibrosis. Based on the accumulating evidence that eosinophils contribute to fibrosis/remodeling in the airway, we hypothesized that airway eosinophils may be a significant source of sema7A. In vivo, sema7A was expressed on the surface of circulating eosinophils and upregulated on bronchoalveolar lavage eosinophils obtained after segmental bronchoprovocation with allergen. Based on mRNA levels in unfractionated and isolated bronchoalveolar cells, eosinophils are the predominant source of sema7A. In vitro, among the members of the IL-5-family cytokines, sema7A protein on the surface of blood eosinophils was increased more by IL-3 than by GM-CSF or IL-5. Cytokine-induced expression of cell surface sema7A required translation of newly synthetized protein. Finally, a recombinant sema7A induced alpha-smooth muscle actin production in human bronchial fibroblasts. Semaphorin 7A is a potentially important modulator of eosinophil profibrotic functions in the airway remodeling of patients with chronic asthma.
Keywords: Eosinophil, semaphorin 7A, IL-3, fibrosis, translation
1. INTRODUCTION
The semaphorins are a family of soluble and membrane-associated proteins characterized by a conserved “sema” domain [1]. Semaphorins were initially identified as axonal growth and guidance proteins [2]. More than 20 types of semaphorins have been described that have a variety of functions including angiogenesis, vasculogenesis, and immune regulation. Unlike the other semaphorins, which are integral membrane proteins, semaphorin 7A (sema7A) is associated with the membrane by a glycophosphatidylinositol (GPI)-linked sequence [3]. While some sema7A functions, such as axon growth and monocyte activation, are mediated through β1-integrins [1], the transmembrane protein plexin C1 binds exclusively to sema7A [4]. Sema7A is highly expressed on activated T lymphocytes [3] and its localization in the immunological synapse amplifies pro-inflammatory cytokine expression by antigen-presenting cells [5]. Recently, sema7A was found to play a critical function as an inducer of pulmonary fibrosis and liver fibrogenesis [6–8]. These observations suggest a role for sema7A in fibrotic lung diseases and potentially asthma where increase subepithelial fibrosis is well described [9].
Eosinophils (EOS) are a hallmark of allergic asthma. Although the role of airway EOS has been controversial, recent trials with anti-EOS therapies such as mepolizumab and reslizumab (both anti-IL-5 monoclonal antibodies) demonstrated reduction in asthma exacerbations [10–13]. EOS are capable of releasing cytokines that direct the T lymphocyte response [14, 15], and enhance airway remodeling [16]. Genetic ablation of EOS attenuated submucosal matrix deposition and reduced airway smooth muscle hyperplasia [17, 18]. Using genome-wide expression analyses, we recently reported that sema7A mRNA was among the transcripts that were increased in parallel with the increase of EOS in bronchoalveolar lavages (BAL) and sputum of mild allergic asthmatics following an in vivo allergen challenge [19]. While airway remodeling has been linked at least in part to the presence of EOS, their expression of sema7A has not yet been reported. Our present study aims to examine the expression of the profibrotic factor sema7A on airway EOS and to determine its regulation.
2. MATERIALS AND METHODS
2.1 Subjects and cell preparations
The study protocol was approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board. Informed written consent was obtained from subjects prior to participation. All subjects were atopic, with at least one positive skin prick test. For the bronchoscopy study to obtain airway EOS, subjects had a history of mild asthma with airway reversibility to albuterol. None of the subjects were using inhaled or oral corticosteroids.
Detailed methods for bronchoscopy, segmental antigen challenge (SBP-Ag), and BAL cell preparation have previously been described [20]. Blood EOS were purified by negative selection as previously described [14]. More details are provided in the online supplement.
2.2 Real-time PCR
Total RNA preparation, real-time quantitative PCR (RT-qPCR) using SYBR Green Master Mix, and calculation of mRNA fold change using the comparative cycle threshold (ΔΔCT) method, have been described previously [15], and are described in the online supplement.
2.3 ELISA
Unlabeled and biotinylated anti-IL-3 mAbs and corresponding recombinant protein standards for ELISA were from BD Biosciences. Unlabeled and biotinylated anti-GM-CSF mAb and recombinant protein standard for ELISA were from R&D Systems. To measure cytokine concentrations, BAL fluid was concentrated 20-fold at 4°C using a low protein-binding Centriprep centrifugal filter unit (Millipore) with a molecular mass cutoff limit of 3 kDa. A sensitive two-step sandwich ELISA was used as described [21]. BAL fluid was diluted in LowCross-Buffer (Boca Scientific, Boca Raton, FL, USA) to reduce non-specific low-affinity binding. The assay sensitivities were below 3 pg/ml for GM-CSF, and 12 pg/ml for IL-3.
2.4 Immunocytochemistry
EOS in suspension were fixed with 3.7% paraformaldehyde, incubated with 0.1 M glycine for 10 min and washed with PBS. Cells were onto poly-L-lysine-coated coverslips by cytocentrifugation and permeabilized with 0.5% SDS in PBS for 15 min at room temperature. Cells were washed three times with PBS and blocked with 10% BSA in PBS for 1 h at room temperature. Cells were incubated overnight at 4°C with 5 μg/ml of goat anti-human Sema7A antibody or control IgG (both from R&D Systems, Minneapolis, MN) diluted in PBS containing 2% BSA and 0.1% SDS. After 3 washes with PBS, cells were stained with 1:100 FITC-rabbit anti-goat antibody at room temperature for 1 h. Cells were washed 3 times with PBS. Nuclei were stained with diamidino-2-phenylindole (DAPI) and the coverslip was mounted to a slide. Pictures were taken using a Nikon A1R confocal microscope (Nikon Instruments INC, Melville, NY), and 100x/1.40 oil immersion objective lens with 1 or 1.2 AU pinhole and 0.15–1 μm z stack thickness. Images were obtained and exported by NIS Elements Advanced Research software (Nikon Instruments INC).
2.5 Flow cytometry
For the in vivo study, unfractionated EDTA-treated blood (100 μl) or BAL cells (250,000) were analyzed by flow cytometric analysis as previously described [20]. Cells were stained with PE-conjugated anti-CD108 (BD Pharmingen™, BD Biosciences, San Jose, CA) along with a cocktail of FITC-conjugated anti-CD16 and anti-CD14 to discriminate EOS from neutrophils and monocytes, respectively. RBCs were lysed (BD-lysing solution) and 5–10,000 gated events (EOS) were acquired on a FACSCalibur (BD Biosciences). EOS were identified by forward and side scatter and then histograms were based on CD16− CD14− populations within the scatter gate. As a positive control for sema7A, CD108+ CD4+ T cells were assessed in unfractionated EDTA-treated blood or BAL cells. An isotype control antibody was used to set the negative gate to yield 1% positive cells thus allowing analysis of the percentage of positive cells. Data were analyzed with FlowJo (TreeStar Inc, Ashland, OR, USA) and expressed as the percentage of positive cells within the CD4+ lymphocyte population.
For in vitro analyses of cultured purified blood EOS, cells were stained with PE-conjugated anti-CD108 (sema7A), anti-CD125 (IL-5Rα), or anti-CD123 (IL-3Rα) or appropriate isotype control antibodies (all purchased from BD Biosciences, San Jose, CA). Data for all EOS experiments are expressed as geometric mean of specific stain minus that of the isotype control antibody.
2.6 Polyribosome preparation
Blood EOS were activated with either IL-3 or GM-CSF (2ng/ml) for 14 h. Cells (6×106) were washed with PBS and polyribosomes were prepared as previously described [22] with some changes. All steps of the protocol were performed on ice or at 4°C. Briefly, cell pellets were suspended in a lysis buffer composed of 10 mM Tris hydrochloride, pH 8.0, 1 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol, 0.5 % Triton X100, 0.05 % NP-40, 100 μg/ml of cycloheximide (Calbiochem,), 800 U/ml of RNasin® (Promega, Madison, WI) and a cocktail of protease inhibitor for mammalian cells (Sigma, St. Louis, MO). Cells were briefly sonicated to resuspend the pellet, and were incubated for 30 min on ice with 15 passages through a 29-gauge needle. After centrifugation to remove the nuclei and cell debris, the supernatant was layered on 30% sucrose and centrifuged at 130,000 x g for 2 h. The polyribosome pellet was then washed once with the lysis buffer without detergent and then lysed in TRIzol® (Sigma, St. Louis, MO) for RNA extraction. RT-qPCR for sema7A was performed as described in the supplemental information (Real-time PCR). Ribosomes and polyribosomes were quantified by the 18S RT-qPCR using the TaqMan human 18S rRNA endogenous control primers (reference sequence: X03205.1) and hydrolysis probe (VICR/MGB probe, Life Technologies).
2.7 EOS adhesion assay
EOS adhesion was assessed using the EOS peroxidase (EPO) assay as previously described [23]. A 96-well Immulon™ plate was coated overnight with 100 μl of plexin C1 (R&D Systems) or VCAM-7d both at 10 μg/ml in bicarbonate buffer (pH 9.6) and then wells were aspirated and blocked with 0.1% gelatin in HBSS for 1.5 hours at 37°C. After an overnight culture of the EOS at 37°C with IL-3 or IL-5 (2 ng/ml) or in complete medium only, EOS were suspended (1×105 cells/ml) in HBSS containing 0.2% BSA and 100 μl per well was added to the precoated plates. EOS were incubated at 37°C in a CO2 incubator for 60 minutes and then the plate was washed three times with 200 μL of HBSS (pH of 8.0) to remove non-adherent cells. One hundred μl of the original EOS suspension were used as a measure of total EPO activity (maximum absorbance at 490 nm). 100 μl of substrate reagent (1mM o-phenylenediamine in 55 mM Tris buffer at pH 8 containing 1mM H2O2 and 0.1% Triton X100) was then added to the wells. After a 30 min incubation at room temperature, 50 μl of 4 M H2S04 was added to stop the reaction. Absorbance was measured at 490 nm in a plate reader (Biotec, Winoski, VT, USA). Adherence was calculated as follows: % adherence = [(experimental absorbance at 490 nm/ max absorbance at 490 nm) x 100]. Each condition was performed in quadruplicate.
2.8 Expression of α-smooth muscle actin (α-SMA) by primary human bronchial fibroblasts
The preparation of primary fibroblasts has been previously described [24]. Bronchial fibroblasts were propagated in FBM (Clonetics, Walkersville, MD) supplemented with 10% FCS. Recombinant human sema7A (R&D Systems, Minneapolis, MN USA) was coated in PBS onto a 48-well plate (0.7 μg per well). Trypsinized fibroblasts were seeded on sema7A for 7 h in medium containing 10 % FCS before a 3.5 day starvation using DMEM F12, 0.1% FBS. Anti-α-SMA and anti-tubulin antibodies (Sigma, St Louis, MO USA) were used for western-blot analysis performed as previously described [25].
2.9 Statistical analyses
To compare expression of genes in total BAL cells and purified BAL EOS by RT-qPCR, data were analyzed using a Student’s paired t-test. In the in vitro studies of purified EOS, a one way ANOVA or Student’s paired t test was used to compare levels of sema7A mRNA, geometric means of flow cytometric analyses and α-SMA production. Log transformation was performed if data were not normally distributed. Statistical analyses were performed using SigmaPlot 11.0 software package.
3. RESULTS
3.1 Airway EOS express sema7A mRNA
Sema7A was assessed by RT-qPCR of total unfractionated BAL cells before and 48 h after SBP-Ag and BAL EOS purified 48 h after SBP-Ag (Table 1). Sema7A mRNA level increased 163-fold in BAL cells after SBP-Ag compared to BAL cells obtained from the same subjects at baseline (before allergen challenge). The main cell population (76 % of the total) recruited into the airway 48 h after SBP-Ag was EOS (Table 1). Sema7A mRNA level was ~10 fold higher in purified BAL EOS compared to unfractionated BAL cells (Table 1). Therefore, the elevated levels of sema7A mRNA in BAL cells after antigen challenge are likely attributable to infiltrating EOS.
Table 1.
Eosinophils are a major source of sema7A mRNA in BAL cells after an allergen challenge*
| Total cells × 106 | Mono/Mac (%) | Neut (%) | Lymph (%) | Eos (%) | Sema7A mRNA | |
|---|---|---|---|---|---|---|
| Unfractionated BAL cells before SBP-Ag | 11±2 | 92.5±2.9 | 0.8±0.4 | 6.3±2.6 | 0.5±0.1 | 1 |
| Unfractionated BAL cells after SBP-Ag | 238±43 | 16.3±5.1 | 1.9±0.8 | 6.4±3.7 | 75.7±3.7 | 163±84† |
| Purified BAL EOS after SBP-Ag | not applicable | <1 | <1 | <1 | >99 | 1617±865‡ |
BAL cells were obtained before and 48 h after SBP-Ag. Total cell counts and differential cell counts for monocytes (Mono), macrophages (Macs), neutrophils (Neuts), and EOS are expressed as mean±SD. Sema7A mRNA was analyzed by qPCR in unfractionated BAL cells and purified BAL EOS. Sema7A mRNA was normalized to the housekeeping gene (GUSB) and fold change was calculated compared to sema7A mRNA level in unfractioned BAL cells before SBP-Ag. Sema7A mRNA fold change was log transformed and a student’s t test was performed. Data are expressed as mean ± SE,
indicates statistical difference (p<0.05) compared to BAL cells before SBP-Ag, and
indicates difference (p<0.05) compared to total BAL cells after SBP-Ag, n=4.
3.2 Airway EOS display high levels of sema7A on their membrane
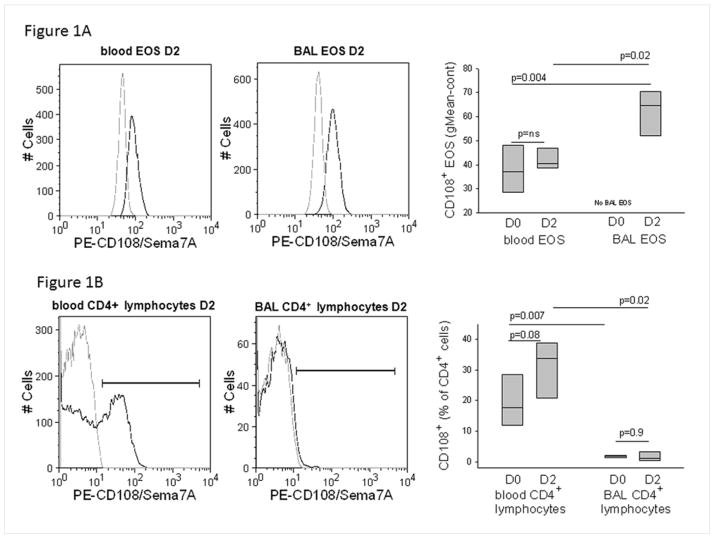
Cell-surface sema7A was determined by flow cytometric analysis of gated EOS in whole blood and unfractionated BAL. Blood and BAL EOS expressed sema7A (Fig. 1A, left and central panels). The fluorescence intensity on BAL EOS (geometric mean fluorescence intensity ± SE of 62 ± 10, Fig. 1A, right panel) was significantly greater than that of blood EOS either before (D0, 38 ± 10) or after challenge (D2, 42 ± 5).
Figure 1.
Sema7A expression in blood and airway cells. A/ Representative flow cytometric histograms showing staining of Sema7A protein (solid black line) on EOS in blood or BAL fluid 48 h after a SBP-Ag (D2). The isotype control is indicated by the gray line. On the right panel, box plots are composite data from 4 subjects showing Sema7A surface staining intensity of blood and BAL EOS before (D0) and 48 h after (D2) SBP-Ag. BAL EOS were not analyzed before SBP-Ag due to their low percentage in BAL cells. B/ Representative flow cytometric histograms showing staining of Sema7A protein (solid black line) on CD4+ lymphocytes and EOS in blood or BAL fluid 48 h after a SBP-Ag (D2). The isotype control is indicated by the gray line. Box plots depict the median and the interquartile range between the 25th and 75th percentiles for 4 subjects. P values are indicated on the graphs.
Unlike EOS that were uniformly positive, CD4+ lymphocytes fell into distinct sema7A negative and sema7A positive subpopulations (Fig. 1B, left panel). The percentage of blood CD4+ lymphocytes that expressed sema7A was 19 ± 9 (mean ± SD) before and 31 ± 10 after allergen challenge (p=0.08) (Figure 1B, right panel). In marked contrast to BAL EOS, the percentage of sema7A+ CD4+ BAL lymphocytes either before or after challenge (Fig. 1B, central and right panels) was low to undetectable.
3.3 Regulation of EOS Sema7A mRNA by IL-5 family cytokines
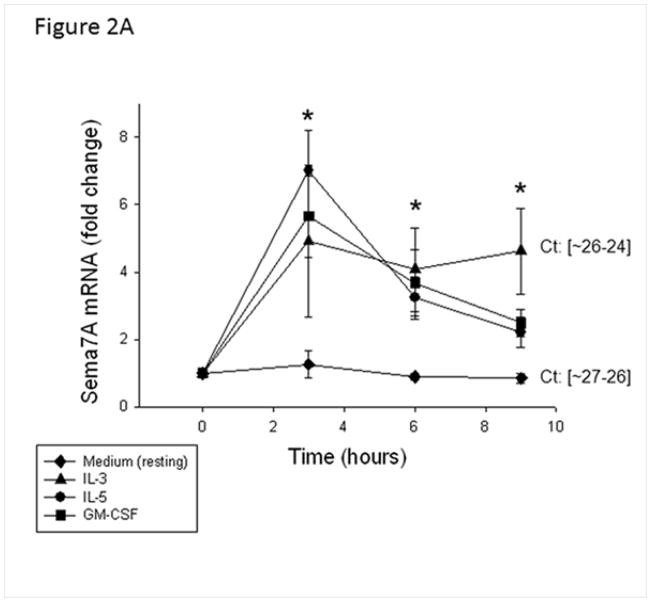
IL-5 family cytokines that signal through receptors comprising a specific α chain (IL-5Rα, GM-CSFRα, and IL-3Rα) and a common β chain are important for EOS survival and function. These cytokines are elevated in BAL fluid obtained after allergen challenge [26]. Therefore, we evaluated the effect of these cytokines on sema7A mRNA expression by blood EOS in vitro. All three cytokines equally induced sema7A mRNA expression compared to medium alone (Fig. 2A).
Figure 2.
Effect of IL-5 family cytokines on sema7A in EOS. A/ EOS were activated with IL-5, GM-CSF, or IL-3 (2 ng/ml) for the indicated times. Data are calculated as mRNA fold changes compared to before culture (Time = 0) are shown and represent the average ± SD of EOS from 3 different donors. * indicates that all three cytokines induced greater levels of sema7A mRNA compared to medium alone (resting). The range of cycle thresholds (Ct) for sema7A obtained with resting or IL-3-activated EOS at 9 h is shown and indicates resting EOS expressed abundant amount of sema7A (similar Ct as for the housekeeping transcript, GUSB). B/ Sema7A (CD108) was analyzed by flow cytometry; geometric means were calculated by subtracting values for isotype control. I/ and II/ EOS were incubated for 20 h with IL-5, GM-CSF or IL-3 (2 ng/ml). A representative histogram for sema7A is shown in panel I with the mean ± SD for EOS from 3 different donors depicted in panel II. III/ and IV/ EOS were activated for 20 h with increasing doses of IL-3 from 0.1 to 10 ng/ml as indicated. A representative histogram is shown in panel III with the mean ± SD for EOS from 4 different donors depicted in panel IV. *p<0.05 compared to resting EOS.
3.4 EOS cell surface sema7A protein is increased by IL-5 family cytokines
After 20 h of stimulation with IL-5 family cytokines, the expression of sema7A protein by blood EOS was significantly upregulated with IL-3>GM-CSF>IL-5 (Fig. 2BI and II). Fig. 2BIII and IV show that IL-3 induced sema7A/CD108 in a dose dependent manner. Kinetic studies demonstrated the presence of sema7A on the surface of blood EOS before culture (T0). IL-3 (2 ng/ml) increased the level of sema7A on the cell surface after 20 h incubation compared to T0 (Fig. S1A) while IL-5 had little effect compared to the level of sema7A observed at T0. Differential response to IL-3 vs IL-5 may reflect loss of IL-5Rα on IL-5-activated EOS (Fig. S1B) [27] and the increase in IL-3Rα on IL-3-activated EOS (Fig S1b) [28].
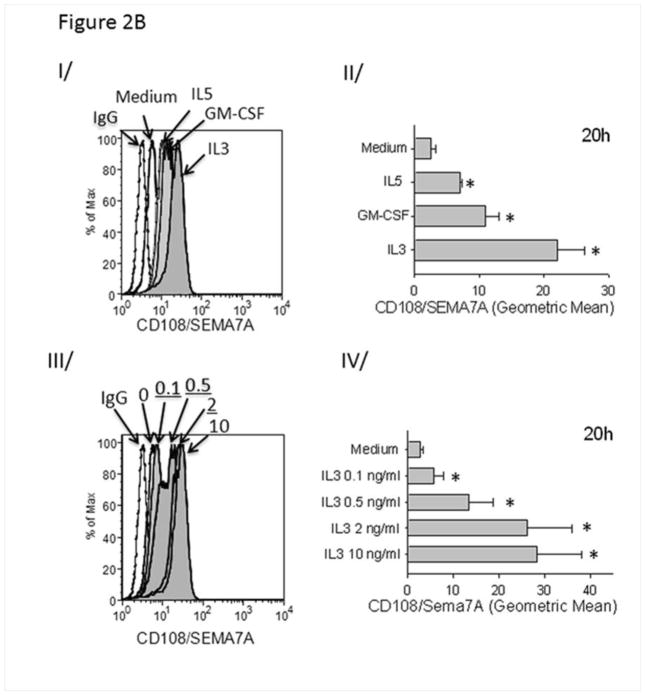
The biological relevance of IL-3 in allergic asthma is demonstrated by an increased amount of IL-3 in BAL fluid 48 h after SBP-Ag (84 pg/ml; Fig. 3). This amount is likely an underestimated of the concentration in bronchial epithelial lining fluid due to the dilution factor of the lavages [29].
Figure 3.
Accumulation of IL-3 protein the BAL fluid 48 h after SBP-Ag. IL-3 and GM-CSF protein levels were measured by ELISA before (Pre-Ag) and after (Post-Ag) SBP-Ag. Box plots depict the median and the interquartile range between the 25th and 75th percentiles for 3 subjects. Means are presented in parentheses. *p<0.05 indicates statistical significant differences between groups.
3.5 IL-3 induces EOS synthesis of sema7A protein
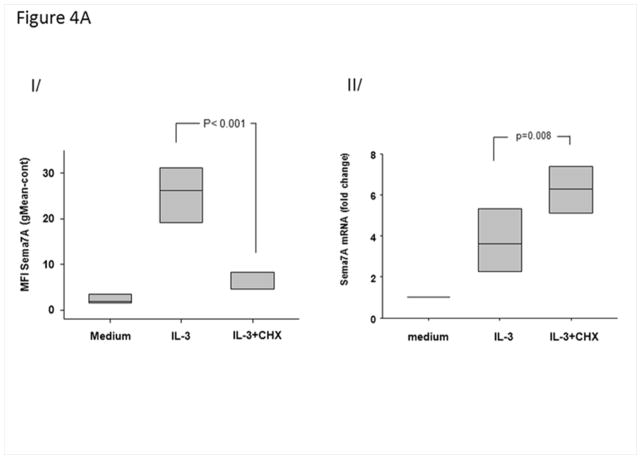
Fig. 4AI shows that inhibition of translation with cycloheximide (10−6 M) significantly attenuated (>80%) IL-3-induced increases of sema7A on the surface of EOS. Conversely, sema7A mRNA levels were increased (Fig. 4AII), consistent with a cessation of mRNA turnover due to the absence of translation.
Figure 4.
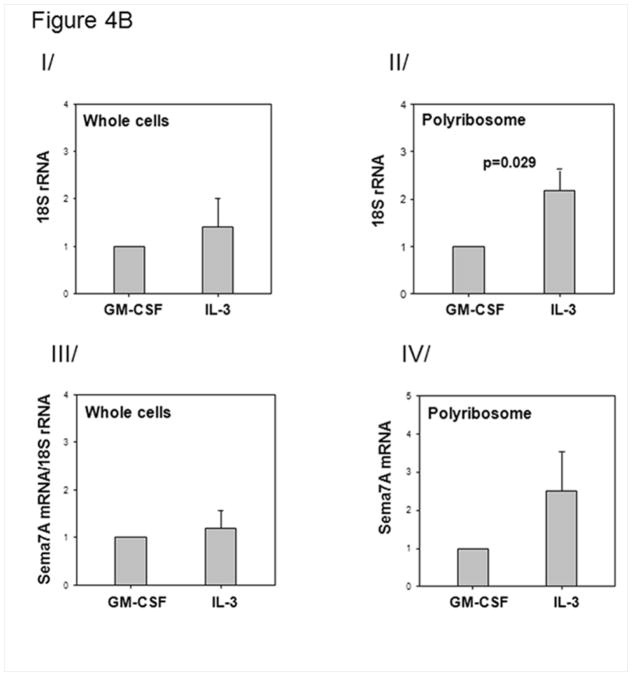
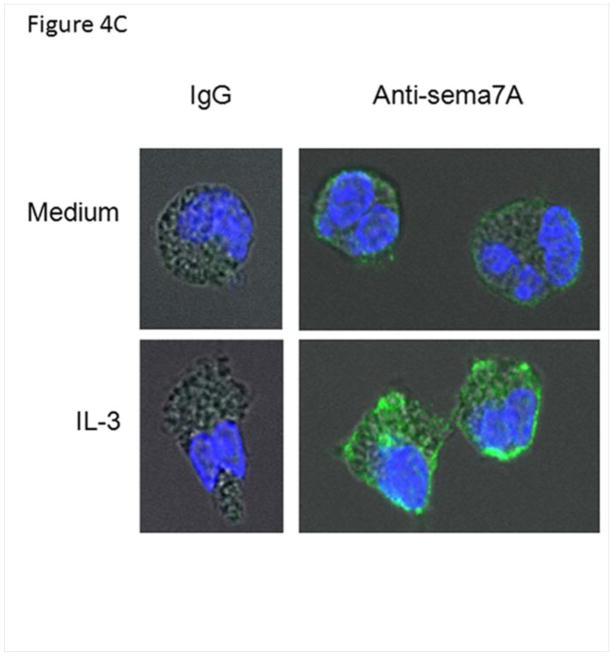
IL-3 induces newly translated sema7A. A/ Effect of a protein translation inhibitor on sema7A cell surface protein of IL-3-activated EOS. EOS were treated simultaneously with IL-3 (2 ng/ml) and cyclohexamide (10−6 M). I/ mean fluorescence intensities (MFI) were determined by flow cytometry 20 h after culture of resting, IL-3-activated or IL-3-activated cells treated with cyclohexamide (IL-3+CHX). II/ at 4 h, sema7A mRNA levels were measured by RT-qPCR. Box plots depict the median and the interquartile range between the 25th and 75th percentiles for 5 subjects. p<0.05 indicates statistical significant differences between groups. B/ Amount of polyribosomes in IL-3- versus GM-CSF-stimulated EOS. EOS were cultured with IL-3 or GM-CSF (2 ng/ml) for 14 h. The amount of ribosome was determined by quantifying 18S rRNA using RT-qPCR. I/ Steady state level of 18S in whole cells indicate similar ribosome amounts in IL-3- and GM-CSF-activated cells. II/ RT-qPCR for 18S using the polyribosome preparations from IL-3- or GM-CSF-activated EOS. III/ Total sema7A mRNA in unfractionated IL-3- and GM-CSF-activated EOS normalized to total levels of 18S rRNA. IV/ RT-qPCR for sema7A using the polyribosome preparations from IL-3- or GM-CSF-activated EOS. Graphs shown are an average ± SE using EOS from 4 different donors, and p<0.05 indicates statistical significant differences. C/ Localization of Sema7A in IL-3-activated and resting EOS. EOS cultured in medium (resting) or treated with IL-3 (2 ng/ml) for 20 h, and were permeabilized and stained with an anti-sema7A antibody or its IgG control. Pictures using a confocal microscope show an increase of green fluorescence for the presence of sema7A protein in IL-3-activated EOS.
In addition, Fig. 4B shows enhanced presence of sema7A mRNA with polyribosomes in IL-3- versus GM-CSF-activated EOS (Fig. 4BIV). However, we noted a similar increase in the amount of polyribosomes in IL-3-activated EOS compared to GM-CSF (Fig. 4BII). These results suggest that IL-3 may increase the general translation rate and presumably upregulates the production of additional proteins.
Confocal immunofluorescence microscopy was performed to corroborate further de novo synthesis of sema7A protein. Substantial increase of total sema7A protein (membrane and cytoplasmic) was appreciated after EOS stimulation with IL-3 (Fig. 4C).
3.6 IL-3-activated EOS are strongly adherent to plexin C1
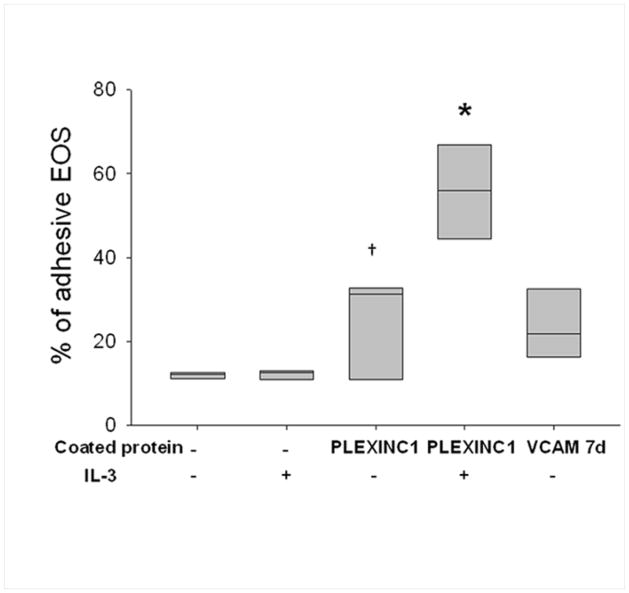
To test the potential functionality of sema7A, we assessed the ability of IL-3-activated EOS to bind to plexin C1. Of note, sema7A is the only ligand known to bind plexin C1 [4, 30]. The adherence of IL-3-activated EOS to plate-bound plexin C1 compared to unactivated cells was doubled from ~30% to ~60% (Fig. 5), which indicates that the cytokine-induced sema7A protein expressed on the EOS cell surface is likely functional.
Figure 5.
Effect of IL-3 on EOS binding to plexin C1. EOS were pre-activated or not with IL-3 (2 ng/ml) for 20 h. EOS were then seeded on the indicated protein, plexin C1 or VCAM 7d (positive control), coated at 10 μg/ml and blocked with gelatin. Percentage of EOS bound to coated protein was determined with the EPO assay. *indicates % adhesive EOS is higher than either EOS on gelatin alone after treatment with IL-3, or untreated cells on plexin C1 (P<0.05, one way analysis of variance, n=3). †indicates adhesion of untreated EOS to plexin C1 is higher than adhesion to gelatin (P<0.05, paired t test, n=3).
3.7 Sema7A increases production of α-SMA in primary human bronchial fibroblasts
Finally, we asked if sema7A could induce intracellular signaling into fibroblasts. To start answering this question and to avoid confounding variables due to lack of blocking antibodies against sema7A, human fibroblasts were cultured on recombinant sema7A rather than in co-culture with EOS. Consistent with productive signaling, α-SMA was elevated by sema7A (Fig. 6).
Figure 6.
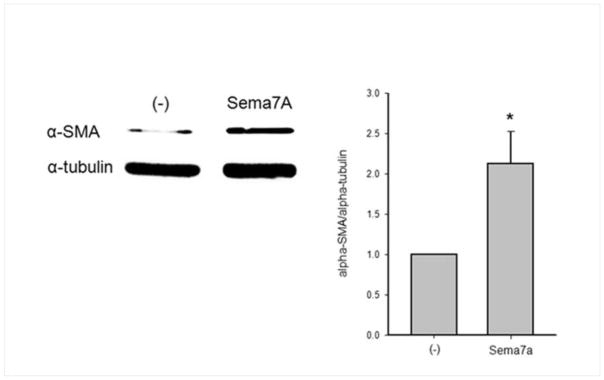
Effect of recombinant human sema7A on α-SMA production in human bronchial fibroblasts. Fibroblasts were seeded on coated sema7A or plastic (−) for 7 h in medium containing 10 % FCS and for 3.5 days in starvation medium. Cellular α-SMA was quantified by western-blot. Tubulin was used as a loading control. A representative western-blot is shown on the left panel. The ratios sema7A/tubulin were calculated and the right panel shows mean ± SD for 3 experiments (*P<0.05, t test, n=3).
4. DISCUSSION
Our study localizes expression of sema7A mRNA to airway EOS in total BAL cells obtained from atopic subjects with mild asthma after an in vivo allergen challenge. Previous studies have implicated sema7A in immunity via its expression on T lymphocytes [5, 8]. In our model, we confirmed that CD4+ blood lymphocytes express surface sema7A but airway T lymphocytes that are considered to have an activated phenotype, had little or no detectable sema7A on their cell surface. Thus, surface sema7A on human airway T lymphocytes is either lost during transit from blood into the airway lumen or the sema7A-expressing blood T lymphocytes are specifically not recruited into the airway. In contrast to CD4+ T lymphocytes, membrane sema7A levels were increased on BAL compared to blood EOS. While other BAL cells, such as alveolar macrophages may express sema7A, our observation that the amount of sema7A mRNA in purified EOS was greater than that in the total cell population indicates that in our model of SBP-Ag, EOS are a predominant source of sema7A. Expression of sema7A on other hematopoietic BAL cells and lung parenchymal cells merit future investigations.
Little is known about regulation of sema7A at the protein and gene level. In an animal model, expression of sema7A in lung tissue was dependent on TGF-β1 [6], but the identification of the direct cell activators was not reported. Based on the observation that the spontaneous level of sema7A tended to decrease over time in vitro, the TGF-β1 present in the fetal bovine serum used in cell culture medium in our study does not appear to increase sema7A on EOS. At the transcriptional level, the hypoxia-inducible factor (HIF1A) increases sema7A mRNA through its hypoxia-response element (HRE) in endothelial cells [31]. We found here that in vitro, sema7A mRNA and protein are upregulated by the IL-5-family cytokines, particularly IL-3. IL-3-mediated augmentation of sema7A protein on the surface of EOS was preceded by mRNA accumulation. However, the incongruence between the relatively small increase of sema7A mRNA expression by all three IL-5 family cytokines and the great induction of membrane sema7A protein by IL-3 suggests translational regulation of sema7A protein.
Translational regulation of EOS sema7A was further implicated by the low level of sema7A protein and high level of sema7A mRNA in resting EOS. The use of a translational inhibitor (cycloheximide) and visualization of EOS sema7A by confocal microscopy demonstrated that IL-3 induced de novo synthesis of sema7A protein. Additional support for translational regulation was provided by the observation that IL-3 led to further recruitment of sema7A mRNA into the translational machinery (polyribosomes) compared to GM-CSF. Although specific enrichment of sema7A mRNA into the polyribosomes in IL-3- versus GM-CSF-activated cells was not observed, the presence of more polyribosomes in IL-3-activated cells suggested greater general translatability of mRNAs.
Upstream of the translational events, the differences in sema7A protein induction among IL-5, GM-CSF, and IL-3 could originate from differential regulation of their respective receptors. We, and others, have previously shown [27, 28] that IL-5 activation leads to a rapid loss of membrane IL-5Rα while GM-CSF induced no change in membrane GM-CSFRα over a period of 24 h. Conversely, as previously described [28], we confirm here that IL-3 strongly induces its own receptor, IL-3Rα. It is possible that EOS expression of GPI-coupled proteins such as sema7A, and as recently described by Munitz et al [32], CD48, are particularly responsive to IL-3 compared to IL-5 or GM-CSF.
The mediators responsible for the augmentation of sema7A in vivo on airway EOS after an allergen challenge are unknown. Possible candidates are IL-3 and/or GM-CSF. We have confirmed our earlier finding that airway allergen challenge leads to increased levels of IL-3 in BAL fluid [26]. In support for a role of IL-3 in asthma, serum IL-3 levels are higher in poorly controlled asthmatics [33]. Moreover, IL-3-positive cells are more abundant in BAL cells from subjects with asthma compared to control subjects, and their numbers escalate with asthma symptoms [34]. Finally, IL-3 levels are heightened in ex vivo-activated T lymphocytes from severe asthmatic subjects [35].
EOS expression of sema7A is yet another avenue by which EOS may drive fibroblasts toward a more profibrotic phenotype. We have showed here that fibroblasts in contact with sema7A produce more alpha-smooth muscle actin. However, a deeper analysis of EOS function on fibroblasts via sema7A would require co-cultures, and the use of blocking anti-sema7A or anti-plexin C1 antibodies, which are to date unavailable. Sema7A is considered a requirement for TGF-β1-induced fibrosis and accumulation of extracellular matrix proteins (ECM) in the lung [6, 36]. Importantly, EOS themselves have been reported to be the main source of TGF-β1 in the airways of asthmatic subjects [37], and both airway eosinophilia and TGF-β1 correlate with asthma severity [38]. In addition, the presence of EOS in the airway associates with deposition of ECM in both human and mouse models of allergen-induced airway inflammation models [16–18].
Finally, the interaction of EOS with plexin C1 and the potential interaction with β1 integrins by sema7A will bring new insight toward understanding EOS biology. We showed here that IL-3-activated EOS strongly bind to plexin C1. Through the sema7A/plexin C1 interaction, EOS may affect the function of other hematopoietic cell types. For instance, sema7A interaction with plexin C1 expressed on dendritic cells is known to impair their migration [30]. Also, sema7A should allow functional interaction of EOS with other β1 integrin-expressing cells such as epithelial cells, endothelial cells, or fibroblasts.
In conclusion, our study describes, for the first time, expression of sema7A on EOS. Furthermore, we demonstrate that IL-3 is a strong activator of sema7A protein production. In vivo, IL-3 is upregulated after an allergen challenge, and airway EOS display high levels of sema7A, a protein that is known to enhance pro-inflammatory cytokine production and fibrosis/remodeling in lung diseases.
Supplementary Material
Highlights.
Semaphorin 7A is known to be critical for the induction of lung remodeling.
We show that human airway eosinophils are a major carrier of semaphorin 7A.
IL-3 is a strong inducer of de novo semaphorin 7A protein on blood eosinophils.
Semaphorin 7A directly induces alpha-SMA in human bronchial fibroblasts.
We propose that eosinophils can increase tissue remodeling via semaphorin 7A.
Acknowledgments
The authors thank the subject volunteers who participated in this study, the staff of the Eosinophil Core facility for blood eosinophil purification, and our research nurse coordinators for subject recruitment and screening.
Funding Support: This work was supported in part by a Program Project Grant P01 HL088594 and Clinical and Translational Research Center Grant UL1 RR025011 and UL1 TR000427 from the National Institutes of Health.
Abbreviations
- BAL
bronchoalveolar lavage
- SBP-Ag
segmental bronchoprovocation with allergen
- EOS
eosinophils
- RT-qPCR
real-time quantitative PCR
- sema7A
semaphorin 7A
Footnotes
Conflict of Interest Statement
None of the authors has conflicts of interest with the present study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This article has online Supporting Information
References
- 1.Jongbloets BC, Ramakers GM, Pasterkamp RJ. Semaphorin7A and its receptors: pleiotropic regulators of immune cell function, bone homeostasis, and neural development. Semin Cell Dev Biol. 2013;24:129–138. doi: 10.1016/j.semcdb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- 3.Angelisova P, Drbal K, Cerny J, Hilgert I, Horejsi V. Characterization of the human leukocyte GPI-anchored glycoprotein CDw108 and its relation to other similar molecules. Immunobiology. 1999;200:234–245. doi: 10.1016/s0171-2985(99)80073-4. [DOI] [PubMed] [Google Scholar]
- 4.Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, Kitao T, Takagi J, Rennert PD, Kolodkin AL, Kumanogoh A, Kikutani H. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. 2007;446:680–684. doi: 10.1038/nature05652. [DOI] [PubMed] [Google Scholar]
- 6.Kang HR, Lee CG, Homer RJ, Elias JA. Semaphorin 7A plays a critical role in TGF-beta1-induced pulmonary fibrosis. J Exp Med. 2007;204:1083–1093. doi: 10.1084/jem.20061273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Minicis S, Rychlicki C, Agostinelli L, Saccomanno S, Trozzi L, Candelaresi C, Bataller R, Millan C, Brenner DA, Vivarelli M, Mocchegiani F, Marzioni M, Benedetti A, Svegliati-Baroni G. Semaphorin 7A Contributes to TGF-beta-Mediated Liver Fibrogenesis. Am J Pathol. 2013 doi: 10.1016/j.ajpath.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reilkoff RA, Peng H, Murray LA, Peng X, Russell T, Montgomery R, Feghali-Bostwick C, Shaw A, Homer RJ, Gulati M, Mathur A, Elias JA, Herzog EL. Semaphorin 7a+ regulatory T cells are associated with progressive idiopathic pulmonary fibrosis and are implicated in transforming growth factor-beta1-induced pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:180–188. doi: 10.1164/rccm.201206-1109OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al Muhsen S, Johnson JR, Hamid Q. Remodeling in asthma. J Allergy Clin Immunol. 2011;128:451–462. doi: 10.1016/j.jaci.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 10.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID. Mepolizumab and exacerbations of refractory eosinophilic asthma. New Eng J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro M, Mathur S, Hargreave F, Boulet LP, Xie F, Young J, Wilkins HJ, Henkel T, Nair P. Reslizumab for poorly controlled, eosinophilic asthma: A randomized, placebo-controlled study. Am J Respir Crit Care Med. 2011;184:1125–1132. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 12.Pavord ID, Korn S, Howarth P, Bleecker ER, Buhl R, Keene ON, Ortega H, Chanez P. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380:651–659. doi: 10.1016/S0140-6736(12)60988-X. [DOI] [PubMed] [Google Scholar]
- 13.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O’Byrne PM. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. New Eng J Med. 2009;360:985–993. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 14.Liu LY, Mathur SK, Sedgwick JB, Jarjour NN, Busse WW, Kelly EAB. Human airway and peripheral blood eosinophils enhance Th1 and Th2 cytokine secretion. Allergy. 2006;61:589–598. doi: 10.1111/j.1398-9995.2006.01060.x. [DOI] [PubMed] [Google Scholar]
- 15.Esnault S, Kelly EA, Nettenstrom LM, Cook EB, Seroogy CM, Jarjour NN. Human eosinophils release IL-1β and increase expression of IL-17A in activated CD4(+) T lymphocytes. Clin Exp Allergy. 2012;42:1756–1764. doi: 10.1111/j.1365-2222.2012.04060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flood-Page P, Menzies-Gow A, Phipps S, Ying S, Wangoo A, Ludwig MS, Barnes N, Robinson D, Kay AB. Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J Clin Invest. 2003;112:1029–1036. doi: 10.1172/JCI17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JJ, Dimina D, Macias MP, Ochkur SI, McGarry MP, O’Neill KR, Protheroe C, Pero R, Nguyen T, Cormier SA, Lenkiewicz E, Colbert D, Rinaldi L, Ackerman SJ, Irvin CG, Lee NA. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004;305:1773–1776. doi: 10.1126/science.1099472. [DOI] [PubMed] [Google Scholar]
- 18.Humbles AA, Lloyd CM, McMillan SJ, Friend DS, Xanthou G, McKenna EE, Ghiran S, Gerard NP, Yu C, Orkin SH, Gerard C. A critical role for eosinophils in allergic airways remodeling. Science. 2004;305:1776–1779. doi: 10.1126/science.1100283. [DOI] [PubMed] [Google Scholar]
- 19.Esnault S, Kelly EA, Schwantes EA, Liu LY, Delain LP, Hauer JA, Bochkov YA, Denlinger LC, Malter JS, Mathur SK, Jarjour NN. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PloS one. 2013;8:e67560. doi: 10.1371/journal.pone.0067560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, Kelly EAB. Decreased expression of membrane IL-5R alpha on human eosinophils: I. Loss of membrane IL-5 alpha on eosinophils and increased soluble IL-5R alpha in the airway after antigen challenge. J Immunol. 2002;169:6452–6458. doi: 10.4049/jimmunol.169.11.6452. [DOI] [PubMed] [Google Scholar]
- 21.Kelly EA, Rodriguez RR, Busse WW, Jarjour NN. The effect of segmental bronchoprovocation with allergen on airway lymphocyte function. Am J Resp Crit Care Med. 1997;156:1421–1428. doi: 10.1164/ajrccm.156.5.9703054. [DOI] [PubMed] [Google Scholar]
- 22.Rajagopalan LE, Malter JS. Modulation of granulocyte-macrophage colony-stimulating factor mRNA stability in vitro by the adenosine-uridine binding factor. J Biol Chem. 1994;269:23882–23888. [PubMed] [Google Scholar]
- 23.Sedgwick JB, Calhoun WJ, Vrtis RF, Bates ME, McAllister PK, Busse WW. Comparison of airway and blood eosinophil function after in vivo antigen challenge. J Immunol. 1992;149:3710–3718. [PubMed] [Google Scholar]
- 24.Nakamura Y, Esnault S, Maeda T, Kelly EA, Malter JS, Jarjour NN. Ets-1 regulates TNF-alpha-induced matrix metalloproteinase-9 and tenascin expression in primary bronchial fibroblasts. J Immunol. 2004;172:1945–1952. doi: 10.4049/jimmunol.172.3.1945. [DOI] [PubMed] [Google Scholar]
- 25.Kelly EA, Liu LY, Esnault S, Quinchia-Rios BH, Jarjour NN. Potent synergistic effect of IL-3 and TNF on matrix metalloproteinase 9 generation by human eosinophils. Cytokine. 2012;58:199–206. doi: 10.1016/j.cyto.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson MW, Kelly EA, Busse WW, Jarjour NN, Mosher DF. Up-regulation and activation of eosinophil integrins in blood and airway after segmental lung antigen challenge. J Immunol. 2008;180:7622–7635. doi: 10.4049/jimmunol.180.11.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, Kelly EAB. Decreased expression of membrane IL-5R alpha on human eosinophils: II. IL-5 down-modulates its receptor via a proteinase-mediated process. J Immunol. 2002;169:6459–6466. doi: 10.4049/jimmunol.169.11.6459. [DOI] [PubMed] [Google Scholar]
- 28.Gregory B, Kirchem A, Phipps S, Gevaert P, Pridgeon C, Rankin SM, Robinson DS. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor alpha-chain expression by cytokines: IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor alpha expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor alpha expression. J Immunol. 2003;170:5359–5366. doi: 10.4049/jimmunol.170.11.5359. [DOI] [PubMed] [Google Scholar]
- 29.Rennard SI, Basset G, Lecossier D, O’Donnell KM, Pinkston P, Martin PG, Crystal RG. Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of dilution. J Appl Physiol. 1986;60:532–538. doi: 10.1152/jappl.1986.60.2.532. [DOI] [PubMed] [Google Scholar]
- 30.Walzer T, Galibert L, Comeau MR, De Smedt T. Plexin C1 engagement on mouse dendritic cells by viral semaphorin A39R induces actin cytoskeleton rearrangement and inhibits integrin-mediated adhesion and chemokine-induced migration. J Immunol. 2005;174:51–59. doi: 10.4049/jimmunol.174.1.51. [DOI] [PubMed] [Google Scholar]
- 31.Morote-Garcia JC, Napiwotzky D, Kohler D, Rosenberger P. Endothelial Semaphorin 7A promotes neutrophil migration during hypoxia. Proc Natl Acad Sci U S A. 2012;109:14146–14151. doi: 10.1073/pnas.1202165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munitz A, Bachelet I, Eliashar R, Khodoun M, Finkelman FD, Rothenberg ME, Levi-Schaffer F. CD48 is an allergen and IL-3-induced activation molecule on eosinophils. J Immunol. 2006;177:77–83. doi: 10.4049/jimmunol.177.1.77. [DOI] [PubMed] [Google Scholar]
- 33.Patil SP, Wisnivesky JP, Busse PJ, Halm EA, Li XM. Detection of immunological biomarkers correlated with asthma control and quality of life measurements in sera from chronic asthmatic patients. Ann Allergy Asthma Immunol. 2011;106:205–213. doi: 10.1016/j.anai.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson DS, Ying S, Bentley AM, Meng Q, North J, Durham SR, Kay AB, Hamid Q. Relationships among numbers of bronchoalveolar lavage cells expressing messenger ribonucleic acid for cytokines, asthma symptoms, and airway methacholine responsiveness in atopic asthma. J Allergy Clin Immunol. 1993;92:397–403. doi: 10.1016/0091-6749(93)90118-y. [DOI] [PubMed] [Google Scholar]
- 35.Lai CK, Ho SS, Chan CH, Leung R, Lai KN. Gene expression of interleukin-3 and granulocyte macrophage colony-stimulating factor in circulating CD4+ T cells in acute severe asthma. Clin Exp Allergy. 1996;26:138–146. doi: 10.1111/j.1365-2222.1996.tb00072.x. [DOI] [PubMed] [Google Scholar]
- 36.Gan Y, Reilkoff R, Peng X, Russell T, Chen Q, Mathai SK, Homer R, Gulati M, Siner J, Elias J, Bucala R, Herzog E. Role of semaphorin 7a signaling in transforming growth factor beta1-induced lung fibrosis and scleroderma-related interstitial lung disease. Arthritis Rheum. 2011;63:2484–2494. doi: 10.1002/art.30386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohno I, Nitta Y, Yamauchi K, Hoshi H, Honma M, Woolley K, O’Byrne P, Tamura G, Jordana M, Shirato K. Transforming growth factor β1 (TGFβ1) gene expression by eosinophils in asthmatic airway inflammation. Am J Resp Cell Mol Biol. 1996;15:404–409. doi: 10.1165/ajrcmb.15.3.8810646. [DOI] [PubMed] [Google Scholar]
- 38.Halwani R, Al Muhsen S, Al Jahdali H, Hamid Q. Role of transforming growth factor-beta in airway remodeling in asthma. Am J Resp Cell Mol Biol. 2011;44:127–133. doi: 10.1165/rcmb.2010-0027TR. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.