Abstract
Circulating tumor cells (CTCs) are rare cancer cells released from tumors into the bloodstream that are thought to have a key role in cancer metastasis. The presence of CTCs has been associated with worse prognosis in several major cancer types, including breast, prostate and colorectal cancer. There is considerable interest in CTC research and technologies for their potential use as cancer biomarkers that may enhance cancer diagnosis and prognosis, facilitate drug development, and improve the treatment of cancer patients. This review provides an update on recent progress in CTC isolation and molecular characterization technologies. Furthermore, the review covers significant advances and limitations in the clinical applications of CTC-based assays for cancer prognosis, response to anti-cancer therapies, and exploratory studies in biomarkers predictive of sensitivity and resistance to cancer therapies.
Keywords: circulating tumor cells, CTC, cancer biomarker, prognostic biomarker, predictive biomarker
1. Introduction
The process of cancer metastasis by which tumor cells detach from a primary site, spread through the circulatory system, and form distant secondary tumors is responsible for the majority of cancer deaths (Hanahan and Weinberg, 2011). Since they were first described by Thomas Ashworth in 1869, the presence of circulating tumor cells (CTCs) has been suggested to be associated with cancer by various early studies (Ashworth, 1869; Carey et al., 1976; Gallivan and Lokich, 1984; Myerowitz et al., 1977). Through a proposed process known as epithelial-mesenchymal transition (EMT), epithelial cells of solid tumors undergo cellular changes that enable them to escape their structural confines via increased mobility and invasiveness, to enter into the blood stream, and to adhere and develop into distant metastases (Steeg, 2003; Thiery, 2002). Thus, it is very attractive to isolate and characterize CTCs, as they may represent both the phenotypic and genetic compositions of the primary tumors and potentially serve as a “liquid biopsy” for any metastatic tumors.
A validated CTC enrichment and enumeration technology has been established in which CTC counts above a known threshold is a prognostic marker and predictor of patient outcome in metastatic breast (Hayes et al., 2006), prostate (Danila et al., 2007), and colon cancers (Cohen et al., 2008). Based on these clinical trials, the US Food and Drug Administration (FDA) cleared the CellSearch® technology (Veridex, LLC, Raritan, NJ, USA) for CTC enrichment and enumeration for the above indicated cancers. The success of CellSearch® proves that enumeration of CTCs is indeed a surrogate for active disease and that increased CTC numbers are predictive of worse prognosis. Also, by demonstrating the successful isolation of clinically relevant cells from the blood of cancer patients, it revealed the potential for further analysis of CTCs beyond enumeration.
There is great interest in obtaining molecular information from CTCs, as they may constitute a read-out for both primary and metastatic tumors. Success in CTC-based analysis has the potential to provide real-time and non-invasive surrogates for diagnosis and prognosis, predictive biomarkers for making treatment decisions, and samples for monitoring drug resistance. The majority of conventional cancer treatments have had limited success in curing metastatic disease. As tumors evolve, even an effective response to therapy is typically short lived, and patients often relapse within 12-24 months of therapeutic intervention (Cristofanilli et al., 2005; Lacy et al., 1998; Ushijima, 2009). CTCs may provide a source for longitudinal molecular analysis of tumors during the clinical management of patients that could facilitate both clinical investigations and cancer patient care.
The fact that CTCs occur at extremely low levels in the circulation and are obscured by billions of peripheral blood cells has hindered their isolation and molecular characterization (Alix-Panabières and Pantel, 2013). There have been numerous efforts, and many technologies developed to enrich and analyze CTCs, many of which have been explored and evaluated with samples from cancer patients. This review will mainly focus on CTC enrichment technologies, studies, and applications that have been successfully tested or evaluated with clinical samples. We will review the recent advances that have been made towards applying CTC assays to clinical practice, discuss the substantial challenges facing the field, and elaborate on future prospects.
2. Isolation of CTCs: current advances
CTC isolation techniques must be sensitive enough to capture the rare and heterogeneous population of CTCs, while also being sufficiently specific for substantial enrichment against blood cells. It is also important for the isolation to be repeatable, reliable, rapid, cost-effective, capable of processing clinically-relevant volumes of blood, and compatible with process automation and downstream CTC analysis. Further, it is desirable for some analyses that isolated cells will maintain their viability and that they experience minimal disturbance caused by the isolation process that might alter their status or phenotype.
Various approaches that have been developed for CTC isolation from blood are discussed below. The technologies are grouped by their principle of CTC enrichment as illustrated in Figure 1, and summarized in Table 1. These technologies are typically evaluated using cell line model systems for multiple performance parameters (i.e. capture efficiency/recovery, enrichment against leukocytes, cell viability, processing speed, blood sample capacity) and then validated through testing with clinical samples. The optimal isolation approach may require a compromise among performance parameters, and is likely to depend on the intended downstream application.
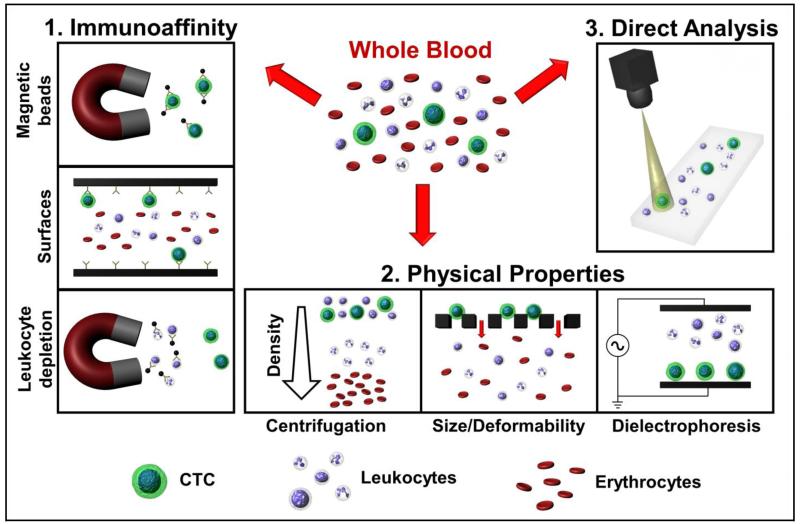
Fig. 1.
Approaches for CTC isolation from whole blood. 1: Immunoaffinity based techniques target specific markers to selectively enrich CTCs or deplete leukocytes. 2: Physical properties may be exploited to separate CTCs from blood cells based on differences in density, size, deformability and electrical properties. 3: Direct analysis is achieved by high throughput assaying of all cells in blood after erythrocyte lysis.
Table 1. Summary of technologies for CTC isolation.
| Isolation method | Cell line | Capture efficiency | Enrichment / (Purity) | Throughput* | Sample volume |
Clinical detection | Reference |
|---|---|---|---|---|---|---|---|
| 1. Immunoaffinity: | |||||||
| anti-EpCAM-coated beads and magnetic sweeper |
MCF7 | 62 ± 7% | 10^8 / (51 ± 18%) | 9 mL/hr | 9 mL | IF: 100% (17/17) of metastatic breast cancer patients | Talasaz et al., 2009 |
| anti-EpCAM-coated beads and magnetic sweeper |
- | - | − / − | 9 mL/hr | 9 mL | RT-PCR: 70% (35/50) ofprimary and metastatic breast cancer patients |
Powell et al., 2012 |
| anti-EpCAM-coated beads and magnetic sifter |
H-1650 | 91.4% | − / (17.7 ± 9.3%) | 10 mL/hr | 0.9 - 3.3 mL | IF: 100% (6/6) of lung cancer patients | Earhart et al., 2013 |
| AdnaTest® BreastCancer | - | - | − / − | - | 10 mL | RT-PCR: 52% (22/42) ofmetastatic breast cancer patients | Tewes et al., 2009 |
| AdnaTest® BreastCancer | - | - | − / − | - | 10 mL | RT-PCR: 19% (16/86) ofpre-surgery and 27% (19/70) of post-chemotherapy ovarian cancer patients |
Aktas et al., 2011a |
| AdnaTest® BreastCancer | - | - | − / − | - | 10 mL | RT-PCR: 19% (97/502) of primary breast cancer patients | Kasimir-Bauer et al., 2012 |
| anti-EpCAM-coated microposts | NCI-H1650 | > 60% | **10^6 / (> 47%) | 1 - 2 mL/hr | 0.9 - 5.1 mL | IF: 99% (122/123) of metastatic cancer and early prostate cancer patients |
Nagrath et al., 2007 |
| anti-PSMA-coated microposts | LNCaP | 85 ± 5% | 10^9 / (68 ± 6%) | 1 mL/hr | 1 mL | IF: 90% (18/20) of prostate cancer patients | Gleghorn et al., 2010 |
| anti-EpCAM-coated herringone chip | PC3 | up to 91.8 ± 5.2% | − / (14.0 ± 0.1%) | 1.5 - 2.5 mL/hr | ~4 mL | IF: 93% (14/15) of metastatic prostate cancer patients | Stott et al., 2010a |
| anti-EpCAM or leukocyte depletion microchip |
Various | up to 98.6 ± 4.3% | **10^3.5 / (0.02 - 43%) | 8 mL/hr | 6 - 12 mL | IF: 88% (37/42) of metastatic cancer patients | Ozkumur et al., 2013 |
| anti-EpCAM-coated silicon nanopillar array | MCF7 | > 95% | − / − | 1 mL/hr | 1 mL | IF: 77% (20/26) of prostate cancer patients | Wang et al., 2011 |
| selectin and anti-EpCAM or anti-PSMA in microtubes |
KG1a | ~50% | − / (66.0 ± 3.9%) | 4.8 mL/hr | 7.5 mL | IF: 100% (14/14) of metastatic cancer patient samples | Hughes et al., 2012 |
| in vivo sampling with anti-EpCAM coated needle |
SK-Br-3 | - | − / − | 3 - 6 L/hr | 1.5 - 3 L | IF: 92% (22/24) of breast and lung cancer patients | Saucedo-Zeni et al., 2012 |
| anti-CD45 leukocyte depletion | MCF-7 | 46% | ~1.5 × 10^5 / − | - | - | - | Lara et al., 2004 |
| 2. Physical properties: | |||||||
| Ficoll-Paque® centrifugation | HT 29 | - | − / − | - | 10 mL | RT-PCR: 41% (24/58) of colorectal cancer patients | Weitz et al., 1998 |
| Ficoll-Paque® centrifugation | HT-29 | 84% | **3.8 / − | - | - | - | Rosenberg et al., 2002 |
| OncoQuick® centrifugation | HT-29 | 87% | **632 / − | - | 10 - 30 mL | RT-PCR: 30% (11/37) of gastrointestinal carcinoma patients | Rosenberg et al., 2002 |
| OncoQuick® centrifugation | MCF7 | 70.6% | − / − | - | 20 mL | IF: 24% (30/123) of primary and metastatic breast cancer patients |
Muller et al., 2005 |
| OncoQuick® centrifugation | - | - | − / − | - | 15 mL | IF: 23% (14/61) of metastatic carcinoma patients | Balic et al., 2005 |
| Diagnostic leukapheresis and CellSearch® | SK-BR-3 | - | − / − | - | ~ 4.5 L | IF: 72% (21/29) of carcinoma patient samples | Fischer et al., 2013 |
| round pore track-etched microfilter | - | - | − / − | - | 6 mL | Cytomorphology: 52% (23/44) of primary liver cancer patients |
Vona et al., 2004 |
| round pore track-etched microfilter | - | - | − / − | - | 7.5 mL | Cytomorphology: 95% (57/60) of metastatic cancer patients | Farace et al., 2011 |
| round pore parylene microfilter | LNCaP | 89.0 ± 9.5% | >10^6 / − | - | - | - | Zheng et al., 2007 |
| round pore parylene microfilter | Various | 92 ± 14% | 10^7/ − | 225 mL/hr | 7.5 mL | IF: 89% (51/57) of metastatic cancer patients | Lin et al., 2010 |
| electroformed nickel microcavity array | Various | 68 - 100% | − / − | 12 mL/hr | 3 - 7.5 mL | IF: 88% (37/42) of metastatic lung cancer patients | Hosokawa et al., 2013 |
| 3D parylene microfilter | MCF-7 | 86.5 ± 5.3% | **10^3/ − | 12 - 20 mL/hr | - | - | Zheng et al., 2011 |
| slot pore parylene microfilter | PC3 | ~70% | **1.5 × 10^3 / − | 12 mL / hr | 1 mL | Telomerase assay: 46% (6/13) of metastatic prostate cancer patients |
Xu et al., 2010 |
| crescent-shaped microfuidic traps | Various | > 80% | − / (> 80%) | 0.7 mL/hr | - | - | Tan et al., 2009 |
| crescent-shaped microfuidic traps | - | - | − / − | - | 1 - 3 mL | IF: 100% (5/5) ofmetastatic lung cancer patients | Tan et al., 2010 |
| inertial microfluidic trapping reservoirs | MCF-7 | ~ 80% | **1.2 × 10^4 / − | 1.2 mL/hr | - | - | Bhagat et al., 2011 |
| inertial microfluidic trapping reservoirs | HeLa, MCF-7 | 10 - 23% | **5.5 - 7.1 / − | - | - | - | Hur et al., 2011 |
| inertial microfluidic trapping reservoirs | MCF7 | 20.7% | **3.5×10^4 / (89%) | 22.5 mL/hr | 7.5 mL | IF: 100% (12/12) of breast and lung cancer patients | Sollier et al., 2013 |
| inertial microfluidic spiral channel | Various | > 85% | **~10^3 / (~10%) | 3 mL/hr | ~ 6 mL | IF: 100% (20/20) of metastatic lung cancer patients | Hou et al., 2013 |
| dielectrophoretic flow separation | SKOV3, MDA- MB-231 |
71.2 - 75.4% | **~10^2 / − | 7.5 mL/hr | - | - | Gupta et al., 2013 |
| 3. Direct analysis: | |||||||
| fiber-optic array scanning of nucleated cells | SKBR3 | ~ 100% | − / − | - | - | IF: 68% (46/68) of prostate, breast and pancreatic cancer patients |
Marrinucci et al., 2012 |
| fiber-optic array scanning of nucleated cells | - | - | − / − | - | - | IF: 73% (57/78) of early and late stage lung cancer patients | Wendel et al., 2012 |
| fiber-optic array scanning of nucleated cells | - | - | − / − | - | - | IF: 68% (45/66) of samples from 28 lung cancer patients | Nieva et al., 2012 |
| Hall-effect sensing after antibody magnetic labeling |
MDA-MB-453 | - | − / − | 3.75 mL/hr | 7.5 mL | Hall-effect sensing: 100% (20/20) of ovarian cancer patients | Issadore et al., 2012 |
Due to differences in the underlying principles of isolation, the cells acquired by different methods are likely to be overlapping CTC subpopulations. Thus, it is important to fully characterize isolated CTCs and to establish clinical correlation and usability as in the case of the CellSearch® clinical trials. It is also important to compare various CTC isolation approaches to fully appreciate the benefits and drawbacks of each method. Practically, this may be achieved through blind comparison with the CellSearch® instrument using duplicate clinical samples.
2.1. Immunoaffinity
Immunoaffinity-based CTC isolation takes advantage of highly specific affinity reactions between capture antibodies and target antigens present on cells of interest. The following antibody capture approaches have been developed:
2.1.1 Magnetic beads
The CellSearch® instrument is the only FDA-cleared technology that is clinically applied for CTC enrichment. Its enrichment process involves the binding of antibody functionalized magnetic beads to the epithelial cell adhesion molecule (EpCAM) antigen on CTCs, and the subsequent isolation of these beads with a magnet. Enumeration of CellSearch®-enriched CTCs has been established as a prognostic marker and predictor of patient outcome in metastatic breast (Hayes et al., 2006), prostate (Danila et al., 2007), and colon cancers (Cohen et al., 2008). A similar approach has been developed using a prototype magnetic sweeper device to improve the capture of cells bound to anti-EpCAM-coated magnetic beads. This technology has demonstrated a capture efficiency of 62%, a purity of 51% from whole blood, and a throughput of 9 mL/hour (Talasaz et al., 2009). This “MagSweeper” device identified CTCs in 14 of 20 primary and 21 of 30 metastatic breast cancer patient blood samples using single cell level reverse transcription polymerase chain reaction (RT-PCR) based detection (Powell et al., 2012). A recent magnetic sifter device generates extremely high magnetic field gradients around the edges of magnetic pores in a microarray format to enhance capture efficiency to 91.4% or higher and performs enrichment in a vertical flow configuration to improve the processing speed, optimized at 10 mL/hour (Sieuwerts et al., 2009). AdnaTest® (Adnagen AG, Langenhagen, Germany) is a commercialized series of assays that employs magnetic beads functionalized with cocktails of antibodies specific to either breast, prostate, colon, ovarian or EMT/stem cell markers to improve enrichment. AdnaTest BreastCancer™ coupled with multiplexed RT-PCR based CTC detection has been demonstrated to correlate with patient outcome in metastatic breast (Tewes et al., 2009) and ovarian cancer (Aktas et al., 2011a). The assay was also used to identify a subset of CTCs expressing stem cell and EMT markers in primary breast cancer (Kasimir-Bauer et al., 2012). In a comparative study AdnaTest BreastCancer™ was positive for 29 of 55 metastatic breast cancer patients compared with the detection of ≥2 CTCs in 26 of 55 patients by CellSearch® (Andreopoulou et al., 2012).
2.1.2 Microfluidic flow
Nagrath, Toner and colleagues developed a microchip consisting of an array of 78,000 silicon micropillars functionalized with antibodies targeting EpCAM, allowing the direct processing of whole blood. The micropillar geometry provides an abundant total surface area for potential contact (970 mm2), resulting in a capture efficiency of ≥60% and a final sample purity of about 50% when processing at a throughput of 2.5 mL/hour. This “CTC-chip” was used to enrich and identify CTCs in 115 of 116 tested blood samples from patients with various metastatic cancer types (Nagrath et al., 2007). A similar micropillar approach was implemented on a “geometrically enhanced differential immunocapture” microchip coated with antibodies targeting prostate-specific membrane antigen (PSMA). This microchip achieved a capture efficiency of 85% and purity of 68%, and identified CTCs from 18 of 20 prostate cancer patient samples (Gleghorn et al., 2010). To overcome fabrication challenges with the first generation CTC-chip, Stott and Toner et al have reported an improved second generation “herringbone chip” that encouraged microfluidic mixing through the generation of microvortices (Stott et al., 2010a). Here the specific capture antibodies were conjugated to the herringbone-shaped grooves along the bottom surface of the device, and the flow patterns resulted in increased cell to surface contact. This improved capture efficiency to 91.8% using antibodies for EpCAM, and CTCs and microclusters were detected in samples from 14 of 15 prostate cancer patients (Stott et al., 2010a). More recently, to facilitate the retrieval of CTCs for further analysis, Ozkumur and Toner et al developed a “CTC-iChip” that enables either positive anti-EpCAM CTC selection or leukocyte depletion after an initial size-based enrichment step and hydrodynamic focusing. They reported high capture efficiencies of up to 98.6%, with varying purities in the range of 0.02 – 42%. CTCs were detected in 37 of 42 metastatic cancer patient samples as compared with 29 of 42 using CellSearch® (Ozkumur et al., 2013).
2.1.3 Nanostructured substrate
Wang et al employed nanostructured substrates to take advantage of an extremely high contact surface area for immunoaffinity due to roughness at the nano-scale (Wang et al., 2009). They used anti-EpCAM conjugated silicon nanopillars and chaotic micromixing to achieve a capture efficiency of >95% from blood at an optimal throughput of 1 mL/hour. This approach detected CTCs in 20 of 26 prostate cancer patient blood samples as compared with only 8 of 26 using the CellSearch® system (Wang et al., 2011). The chip was further modified with electrospun polymer nanofibers and incorporated with laser capture microdissection to isolate single prostate cancer CTCs for amplification and whole exome sequencing (Zhao et al., 2013).
2.1.4 Microtubes
Hughes et al employed a biomimetic approach to simulate the process of selectin-mediated cell adhesion in blood vessels for CTC capture. Selectin-coated microtubes induce cell attachment and rolling, which encourages CTC binding with anti-EpCAM and anti-PSMA antibodies, even at a high flow rate of 4.8 mL/hour. The device achieved a capture efficiency of ~50% and an average purity of 66%, while successfully detecting CTCs in 14 of 14 tested patient blood samples as compared with 9 of 14 with the CellSearch® system (Hughes et al., 2012).
2.1.5 In vivo sampling
A novel approach for in vivo sampling was developed by functionalizing a medical wire with EpCAM antibodies. The medical wire is injected through a cannula into the patient’s cubital vein for a duration of 30 minutes to allow direct continuous sampling of large volumes of blood (1.5 - 3 liters). This approach successfully enriched CTCs in 22 of 24 patients diagnosed with either breast or lung cancer (Saucedo-Zeni et al., 2012).
2.1.6 Leukocyte depletion
An alternate approach to positive immunoaffinity based CTC selection is to use monoclonal antibodies targeting leukocyte antigens (i.e., CD45, CD14) to deplete cells of hematopoietic origin. Some strategies include antibody labeling of leukocytes for removal through immunomagnetic separation (Lara et al., 2004; Yang et al., 2009), or through centrifugation with the RosetteSep™ kit (StemCell Technologies, Vancouver, Canada) (Baccelli et al., 2013; He et al., 2008). These approaches are capable of high recovery rates with minimal disturbance to CTCs, but may achieve relatively low sample purities.
Summary
The specificity of the antibody-antigen reaction allows the isolation of CTCs with a very high level of purity. However, CTC enrichment results will be heavily dependent on the performance of the particular antibody employed. There is currently no known ideal CTC antigen target that would allow capture of all CTCs at the exclusion of all hematopoietic cells. Recent reports have indicated that anti-EpCAM approaches may miss out on significant populations of CTCs that do not exhibit an epithelial phenotype, presumably due to EMT, in addition to the variable expression of EpCAM on certain phenotypes of epithelial CTCs (Sieuwerts et al., 2009; Yu et al., 2013). Capture rates may be increased by using combined cocktails of antibodies that also target antigens specific to a particular cancer type, though this may then result in reduced specificity and lower sample purity. Leukocyte depletion approaches have the benefit of not disturbing the CTCs which may minimize phenotypic alterations caused by the isolation process, but may lose CTCs that are attached to or interacting with leukocytes. Leukocyte depletion methods result in purities that are typically at least one order of magnitude lower than positive CTC targeting. In general, an immune-affinity approach requires long incubation/interaction times to optimize CTC recovery, which could be a bottleneck for processing speed. For methods using positive selection, it might be challenging to reversibly remove CTCs from the immune-affinity tag or surface.
2.2. Physical properties
Physical properties may be exploited to effectively separate CTCs from peripheral blood cells. The following technologies have been developed based on differences in density, size, deformability and electrical properties:
2.2.1. Density gradient centrifugation
Centrifugation is a cheap and efficient method for separating CTCs in the mononucleocyte fraction of blood away from erythrocytes and granulocytes based on cell density. Centrifugation with Ficoll-Paque® solution (Pharmacia-Fine Chemicals, Uppsala, Sweden) was used to detect CTCs using an RT-PCR based assay for cytokeratin 20 expression with a resolution of 1 cell/mL of blood in model systems, successfully identifying CTCs in 24 of 58 colorectal cancer patients undergoing surgical resection (Weitz et al., 1998). OncoQuick® (Grenier BioOne, Frickenhausen, Germany) is a novel technology that incorporates a porous barrier for size-based separation of CTCs in conjunction with density-based centrifugation. Rosenberg et al reported a vastly improved enrichment of 632-fold against leukocytes with OncoQuick® compared to 3.8-fold with Ficoll-Paque® (Rosenberg et al., 2002). OncoQuick® has identified CTCs in blood samples obtained from 11 of 37 gastrointestinal cancer patients with RT-PCR (Rosenberg et al., 2002), 5 of 60 primary breast cancer patients and 25 of 63 advanced breast cancer patients by immunofluorescence (IF) (Muller et al., 2005). In another clinical study CTCs were detected in 14 of 61 patients using cytospins prepared after OncoQuick® enrichment, compared to 33 of 61 with CellSearch® (Balic et al., 2005). Recently, leukapheresis has been shown as a centrifugation technique to sample a much larger volume of patient blood for CTC analysis, resulting in improved sensitivity for downstream detection and analysis. CTCs were detected in 21 of 29 carcinoma patient samples using the CellSearch® system after concentration by leukapheresis, compared with only 8 of 29 using the conventional 7.5 mL assay (Fischer et al., 2013).
2.2.2. Microfiltration
Microfiltration operates on the principle of retaining larger CTCs while allowing smaller leukocytes to pass through pores of varying geometries. Vona, Paterlini-Brechot, and colleagues developed the “isolation by size of epithelial tumor cells” (ISET) technique using randomly track-etched polycarbonate filters with 8 μm diameter circular pores for CTC enrichment and cytological detection from fixed blood samples (Vona et al., 2000). Track-etched microfilters have been used to enrich and characterize CTCs in studies involving liver cancer (Vona et al., 2004), melanoma (De Giorgi et al., 2010), lung cancer (Hofman et al., 2011a; Lecharpentier et al., 2011), prostate cancer (Chen et al., 2012) and various other cancers (Hofman et al., 2011b). These microfilters were demonstrated to be more sensitive than CellSearch®, detecting CTCs in 57 of 60 metastatic patients with breast, prostate and lung cancer compared to 42 of 60 with CellSearch® (Farace et al., 2011). Zheng, Tai, and colleagues used deterministic photolithography to develop an improved microfilter with circular- or oval-shaped pores fabricated from parylene polymer, reporting a capture efficiency of 89% (Zheng et al., 2007). This parylene microfilter obtained CTCs in 51 of 57 cancer patients while CellSearch® was positive for only 26 (Lin et al., 2010). Similar microfilters have been developed from silicon substrates (Lim et al., 2012) and electroformed nickel (Hosokawa et al., 2010) detecting CTCs in 37 of 42 lung cancer patient samples compared with 19 of 43 with CellSearch® (Hosokawa et al., 2013). To encourage viable CTC capture a three-dimensional microfilter was designed out of two layers of parylene to incorporate support structures that mitigate cell damage (Zheng et al., 2011). Xu et al. employed a slot pore microfilter designed by Tai’s group to detect telomerase activity from viable enriched CTCs filtered from Ficoll-Paque®-isolated buffy coats of metastatic prostate cancer patients (Xu et al., 2010).
2.2.3. Microfluidics
Microfluidic devices have been developed to achieve size and deformability-based sorting of CTCs in a more controlled fashion. Tan, Lim, and colleagues designed crescent-shaped trap arrays with a fixed 5 μm gap width to enrich CTCs from whole blood, reporting a capture efficiency and purity of over 80% (Tan et al., 2009a). This microdevice successfully detects CTCs in 1-3 mL blood samples obtained from metastatic lung cancer patients (Tan et al., 2010). Higher throughput microfluidic approaches apply hydrodynamic forces to select for cells of different sizes by inertial flow fractionation. This principle was incorporated through contraction and expansion reservoirs developed for pinch alignment of tumor cells by Bhagat et al (Bhagat et al., 2011) and tumor cell trapping in microscale vortices by Hur et al (Hur et al., 2011). An improved form of the latter approach has been applied to detect cancer cells from a cohort of 12 breast and lung cancer patients (Sollier et al., 2013). These devices allow vastly improved throughput and are capable of processing larger sample volumes compared to previous microfluidic approaches, but with potential reductions to cell recovery rate and enrichment against leukocytes. Sun et al developed a double spiral microfluidic channel to hydrodynamically separate tumor cells using drag forces, reporting a recovery rate of 88.5% from diluted blood (Sun et al., 2012; Sun et al., 2013). Lim and colleagues incorporated a spiral microfluidic channel to successfully enrich CTCs and microclusters from 20 metastatic lung cancer patients (Hou et al., 2013).
2.2.4. Dielectrophoresis
Electrical properties of CTCs may be exploited to discriminate them from blood cells by applying a non-uniform electric field through the phenomenon of dielectrophoresis (DEP). Interdigitated gold electrodes were used by Becker, Gascoyne, and colleagues to isolate leukemia (Becker et al., 1994) and breast cancer cell lines (Becker et al., 1995) from spiked healthy donor blood. The application of an electric field generated by the electrodes attracts tumor cells by positive DEP, while other cells flow past. Upon removal of the electric field the tumor cells can be collected with a capture efficiency of 95% (Becker et al., 1995). Based on the success of this method, Huang et al proposed a DEP field flow fractionation approach to allow continuous processing of samples that did not require intermittent application of the electrical field for cell recovery (Huang et al., 1997). Gupta et al developed the ApoStream™ instrument for DEP field flow fractionation, demonstrating viable capture with an efficiency of greater than 70% from cell lines spiked into whole blood (Gupta et al., 2012). Thus far, only preliminary efforts of the application of this technology to clinical samples have been reported (Shim et al., 2013).
Summary
Physical separation is achieved through simple approaches that isolate CTCs independent of cell antigen expression. They are therefore not vulnerable to antigen expression variability or loss of epithelial markers that may occur due to EMT, and typically achieve high capture efficiencies of greater than 80%. Density gradient centrifugation is a robust technique that is very effective as a first enrichment step. Leukapheresis is a promising yet somewhat more invasive approach that allows the sampling of much more blood, improving detection sensitivity in cancer patients. However, density based approaches result in insufficient purity for most downstream analyses and typically require further enrichment. Microfiltration enables extremely high throughput processing of full tubes of blood within minutes. However, the overlap in size distributions between CTCs and large leukocytes results in sample purities of less than 10%. It is also possible that smaller CTCs or CTC fragments may be missed. Improved control in microfluidic chambers can achieve high sample purities of greater than 80%, but at the expense of reducing throughput to approximately 1 mL/hour. Higher throughputs may be achieved using hydrodynamic separation in microfluidic channels, but with reduced capture efficiency and/or purity. The use of DEP to exploit cell electrical properties is a novel approach that has demonstrated excellent viability and minimal disruption to captured cells in testing with cell lines. DEP based technology has yet to be thoroughly evaluated with clinical samples, so its clinical utility must be determined.
2.3. Direct analysis
An alternative approach to overcoming the challenges of isolating rare CTCs is to carry out high-throughput assaying of the entire population of cells in blood.
2.3.1. Fiber-optic array scanning
Krivacic, Kuhn, and colleagues developed a high-throughput “fiber-optic array scanning technology” system capable of analyzing 300,000 cells per second (Krivacic et al., 2004). Whole blood is first treated for erythrocyte lysis, and then all remaining cells are plated onto several glass slides. This approach has demonstrated efficient enumeration of CTCs in breast, prostate, pancreatic, lung and ovarian cancers (Cho et al., 2012; Lazar et al., 2012; Marrinucci et al., 2012; Nieva et al., 2012; Phillips et al., 2012; Wendel et al., 2012).
2.3.2. Micro-Hall sensor
Issadore et al used microfabricated Hall effect sensors to detect CTCs with an assay throughput of ~107 cells/min. CTCs are labeled with magnetic nanoparticles conjugated to antibodies targeting various antigens of interest (ie. EpCAM, HER2, EGFR, MUC1), then flowed past an array of sensors that measure Hall voltages induced by the magnetic flux of each labeled cell. This approach demonstrated improved sensitivity over the CellSearch® system when evaluated with a cohort of 20 ovarian cancer patients (Issadore et al., 2012).
Summary
Direct analysis approaches are promising for CTC detection as they are less vulnerable to cell loss. They involve minimal enrichment of blood samples, typically only requiring erythrocyte lysis. Fiber-optic scanning has enabled a “high definition-CTC” assay that allows effective CTC detection with particularly high quality imaging and optical analysis. The lack of concentration of CTCs or enrichment against leukocytes does however pose challenges for other desirable CTC analysis applications. The development of simple on-chip electrical detection with Hall effect sensing does not require optical instrumentation and therefore has great potential for portability and point-of-care application, though the approach is essentially limited to CTC enumeration.
3. Analytic tools for CTCs
The successful clinical application of CTCs requires the development of analytic tools to exploit their potential for affecting clinical outcomes. Various analytic tools have been deployed for the cellular and molecular characterization of CTCs (Figure 2). These tools are developed in conjunction with CTC enrichment technologies as each downstream application will have its own requirements regarding sensitivity, sample purity, cell viability, and ability to recover cells after enrichment. Different isolation technologies may, therefore, be more suitable for specific downstream applications. As any CTC isolation and analysis process applies certain criteria for enrichment and detection, it is important to characterize the false-positive and false-negative rates of the overall CTC process, which can be performed with cell line models. For clinical samples, a common practice to establish a new method is to compare results from duplicate samples with the CellSearch®.
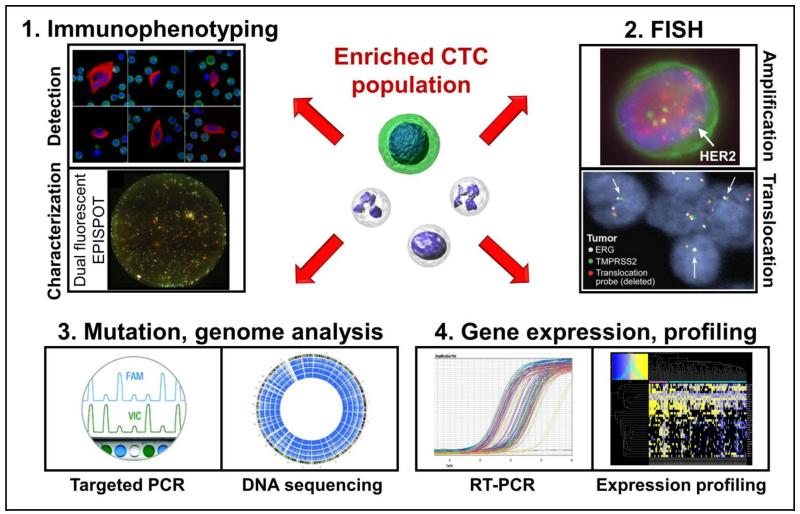
Fig. 2.
Analytic tools for analysis of CTCs after isolation. 1: Immunophenotyping employs antibodies to label proteins for CTC detection and characterization. 2: Fluorescence in situ hybridization detects aberrant amplification and translocation of specific genes. 3: PCR and DNA sequencing techniques identify oncogenic mutations and alterations to the genome. 4: RT-PCR and RNA based profiling characterize CTC gene expression.
Listed images are reproduced with permission and with all rights reserved from the following sources:
Detection: IOP Publishing (Marrinucci et al., 2012)
Characterization: BioMed Central (Alix-Panabieres et al., 2009)
Amplification: Public Library of Science (Punnoose et al., 2010)
Translocation: The American Association for the Advancement of Science (Stott et al., 2010b)
DNA sequencing: John Wiley and Sons (Zhao et al., 2013)
Expression profiling: Public Library of Science (Powell et al., 2012)
3.1. Immunophenotyping
Immunostaining is the current gold standard technique for CTC detection and enumeration as established by the FDA-cleared CellSearch® system. The defined criteria for identifying CTCs includes (1) positive expression of cytokeratins (CK), a class of intermediate filaments in epithelial cells; (2) negative expression of the leukocyte common antigen, CD45; and (3) positive staining of the nucleus with DAPI (Cristofanilli et al., 2004). Markers specific to cancer types, such as PSA or PSMA in prostate cancer, have also been used for positive CTC identification (Kirby et al., 2012; Stott et al., 2010b; Wang et al., 2000).
Recently, attempts have been made to investigate the expression of other proteins to further characterize CTCs. The presence of stem cell markers, such as CD44+/CD24−, CD133+, ALDH1+, have been identified in CTCs (Aktas et al., 2009; Armstrong et al., 2011; Baccelli et al., 2013; Hou et al., 2013; Theodoropoulos et al., 2010). Studies attempting to investigate the EMT status of CTCs have reported high expression levels of mesenchymal markers such as TWIST, AKT2, PIK3α, N-cadherin and vimentin (Aktas et al., 2009; Armstrong et al., 2011; Kallergi et al., 2011; Lecharpentier et al., 2011). While the direct clinical relevance of such markers has yet to be established, they are of significant research interest for better characterizing CTCs and understanding their roles in cancer metastasis.
Immunophenotyping of CTCs may be applied to the clinic in ways that could more directly influence therapy selection. In conjunction with other methods, human epidermal growth factor receptor 2 (HER2) expression status of breast cancer CTCs was determined through IF (Ignatiadis et al., 2011; Pestrin et al., 2009; Riethdorf et al., 2010). In prostate cancer, CTCs have been assayed for epidermal growth factor receptor (EGFR) expression (Shaffer et al., 2007), as well as androgen receptor (AR) status (Lazar et al., 2012). A functional EPISPOT assay was developed to distinguish apoptotic versus viable CTCs based on IF detection of proteins secreted by CTCs after short-term culturing (Alix-Panabières et al., 2005). In preclinical studies of M1 stage breast cancer patients, this technology shows that the secretion of CK-19 is associated with worse outcomes (Markou et al., 2011). In M1 stage prostate patients, a large fraction of CTCs secrete fibroblast growth factor-2, a stem cell growth factor (Alix-Panabieres et al., 2007).
Since CTCs are rare and heterogeneous, it is desirable to multiplex marker analysis for comprehensive single cell characterization. Currently, multiplexing is severely limited due to fluorescence spectral overlap. This is exacerbated by the fact that at least three channels are required for definitive CTC detection (i.e. CK, DAPI, and CD45). The implementation of quantum dots or other advanced imaging technologies will improve capabilities in the future. The subjectivity of IF detection must also be addressed through a greater emphasis on quantitative imaging and process automation.
3.2. Fluorescence In Situ Hybridization
Somatic alterations to gene copy numbers are a hallmark of many cancers, and can be an important prognostic marker. They can further provide valuable predictive information on therapeutic efficacy. Meng et al employed a multicolor fluorescence in-situ hybridization (FISH) assay on CTCs isolated from breast cancer patients to determine the amplification status of the HER2 oncogene (Meng et al., 2004). They reported some level of non-concordance between HER2 status determined by the analysis of primary tumors and with their novel CTC assay. Other studies have confirmed this observation with HER2 assays based on IF (Pestrin et al., 2009) and RT-PCR (Fehm et al., 2007). This suggests that a sub-set of patients with initially HER2-negative disease may develop HER2-positive CTCs over the course of progression. CTC analysis provides a minimally-invasive avenue for repeated screening for the development of genomic aberrations. It is interesting to note that CTC HER2 amplification screening has been successfully demonstrated in even primary breast cancer patients (Fehm et al., 2009; Wulfing et al., 2006). In castration-resistant prostate cancer, multicolor FISH has shown potential as a surrogate for status determination of AR and MYC status in CTCs after immunomagnetic enrichment with CellSearch® (Leversha et al., 2009; Shaffer et al., 2007).
Oncogenic translocation is an important pathogenic mechanism for some cancers. TMPRSS2-ERG translocation occurs in 30-70% of therapy-naïve prostate cancer patients (Tomlins et al., 2005). Using FISH analysis on CTCs, it has been shown that TMPRSS2-ERG fusion can be identified in CTCs of prostate cancer patients (Attard et al., 2009; Mao et al., 2008; Stott et al., 2010b). Further, there appears to be a consistency in TMPRSS2-ERG translocation between therapy-naïve prostate cancer from core biopsy specimens and from CTCs at the time of castration-resistance from the same patients (Attard et al., 2009). In a recent study using FISH, it has been shown that ALK-rearrangement can be identified in non-small cell lung cancer patients harboring ALK-translocations (Pailler et al., 2013).
There is, therefore, much promise through CTC analysis for minimally-invasive tumor profiling and improved prognosis related to metastatic disease. However, the low sensitivity of current enrichment technologies remains a significant hurdle to routine clinical application. In addition, the reliability and clinical relevance of these FISH-based assays must first be thoroughly validated.
3.3 Oncogene mutation detection and genomic analysis
One of the key attractions for isolating CTCs is to enable the detection of cancer gene mutations and genomic aberrations in these metastatic tumor cells that are associated with the worst prognosis for survival, reflect drivers of active disease, and predict drug sensitivity and resistance. Using CTCs isolated with microchips, it has been shown that activating EGFR mutations can be effectively detected in patients with non-small cell lung cancer using an allele-specific PCR amplification technique (Maheswaran et al., 2008). Further, the EGFR T790M mutation that confers drug resistance can also be detected from patients who have received EGFR kinase inhibitor treatment. While allele-specific PCR is a fast and cost-effective approach to determine specific mutations, prior knowledge of the mutations are necessary for the design of amplicon primer sets.
For genes with a broader distribution of pathogenic mutations, targeted regions can be amplified from genomic DNA or cDNA of CTCs and sequenced. Mutations in multiple genes can also be examined following whole genome amplification and mutation-specific PCR in which mutations in KRAS and PIK3CA have been detected from CTCs of patients with colorectal cancer (Gasch et al., 2013). It has also been reported in castration-resistant prostate cancer that silent mutations, and small deletions and insertions in the AR gene can be identified from CTCs and then confirmed with biopsy or autopsy specimens, some of which were associated with resistance to androgen-directed therapies (Jiang et al., 2010).
With the ability to perform single-cell whole genome amplification, it has been shown that one can perform next-generation exome sequencing with CTCs to uncover cancer-specific mutations, both pathogenic and passenger, which are present at very different levels of prevalence in primary and metastatic colon cancer (Heitzer et al., 2013). Once amplified, the DNA from a single CTC can also be used for array-comparative genomic hybridization analysis to uncover genomic aberrations, such as large deletions and insertions that cannot be detected with exome DNA sequence analysis (Heitzer et al., 2013). Although single-cell DNA sequencing is tedious, such studies confirm the feasibility of applying CTCs to discovery research in clinical oncology studies. Technologies for cancer gene exome analysis such as the Ion Torrent AmpliSeq™ Cancer Panels (Life Technologies, Carlsbad, CA) and the TruSeq® Amplicon-Cancer Panel (Illumina, San Diego, CA) would facilitate the analysis of cancer mutations in CTCs.
3.4 Gene expression analysis of CTCs
While the analysis of mRNA from CTCs presents significant technical challenges due to the molecule’s labile nature, it is desirable due to: (1) specificity to amplicons that are highly selectively expressed in tumors but not in white blood cells; (2) the ability to detect gene translocations and alternative splice variants; (3) quantitative high-throughput technologies such as multiplex digital PCR and expression arrays for cell profiling; (4) potential application to next generation sequencing analysis.
One study on mRNA analysis explains the low abundance of EpCAM expression associated with increased vimentin in basal-like breast cancer that is poorly captured with EpCAM-based CTC purification schemes (Punnoose et al., 2010). Another study further demonstrate dynamic changes in epithelial and mesenchymal composition in circulating breast cancer cells with mixed probes using RNA in situ hybridization, particularly in CTC clusters (Yu et al., 2013). It also suggests a more mesenchymal phenotype for the basal-like molecular subtype of breast cancer. Other studies have looked at oncogenic drivers with CTC analysis. TMPRSS2-ERG translocation is the most prevalent pathogenic event in prostate cancer, and various attempts have been made to detect this translocation in CTCs with some modest success in correlating it with primary tumors (Danila et al., 2011a; Stott et al., 2010b). Previous studies have also shown that it is possible to detect the expression of PSA from patients with castration-resistant prostate cancer and its expression is associated with worse prognosis in the patients, while the predictive value is yet to be established (Ghossein et al., 1997; Helo et al., 2009). Such a method of analysis, once matured, would allow investigators to better detect the cancer driver in individual patients and to correlate it with clinical response. Expression analysis of estrogen receptor (ER) and progesterone receptor (PR), or HER2 signature has been explored, and studies show the feasibility of such analysis (Aktas et al., 2011a; Aktas et al., 2011b; Sieuwerts et al., 2011). As biomarkers such as HER2 in breast cancer and AR in castration-resistant prostate cancer change during the course of hormone therapies, assays that can quantify their levels as well as their signaling activities would be extremely useful to correlate with drug response and to understand the development of resistance.
Various technological advances would undoubtedly facilitate such expression analysis. BioMark digital PCR (Fluidigm, South San Francisco, CA) has been used for high degree multiplexed analysis of gene expression from single CTCs after immunomagnetic enrichment (Powell et al., 2012). The expression profiles showed a high degree of heterogeneity among CTCs, but also minimal concurrence between metastatic breast cancer patients and a panel of breast cancer cell lines, which warrants further investigation and validation (Powell et al., 2012). Further analysis is also needed to cluster CTC expression with that of known subtypes of breast cancer for both validation and clinical relevance. Strati et al performed a comparison of three RT-PCR based molecular characterization techniques in breast cancer, showing a concordance level of approximately 70% (Strati et al., 2013). Markou, Lianidou and colleagues developed a novel liquid bead array hybridization assay for CTCs based on the simultaneous detection of 6 gene targets, demonstrating feasibility in early and late stage breast cancer (Markou et al., 2011). Next generation RNA sequencing has been performed with CTCs isolated from patients with pancreatic cancer using a microfluidic chip (Yu et al., 2012). RNA sequencing results revealed the activation of WNT signaling in the CTCs, which may contribute to tumor metastasis. Taken together, while it is very attractive to analyze mRNA for gene expression and pathway profiling, its quantitative analysis requires a high quality source of mRNA that is consistently well preserved.
4. Clinical applications of CTC assays
Due to the minimally invasive nature of obtaining blood from cancer patients and its potential implications for cancer diagnosis, there is a great interest in developing methods to extract and analyze CTCs for diagnosis, prognosis, and clinical management of cancer patients. There are several potential benefits for successful CTC or blood-based diagnostics. With the ability to obtain multiple blood draws from cancer patients throughout the duration of clinical management, the analysis of CTCs may potentially provide clinicians with information on biomarkers predictive of drug sensitivity (Maheswaran et al., 2008; Miyamoto et al., 2012), with an independent surrogate biomarker of response and patient prognosis (Cristofanilli et al., 2004; Cristofanilli et al., 2005; de Bono et al., 2008; Hayes et al., 2006), with an indicator for early relapse (Pachmann et al., 2008), as well as with material for the analysis of drug resistance (Scher et al., 2009). Because of the extreme rarity of CTCs in the circulation, while their enumeration has been widely accepted as a strong and independent prognostic biomarker for several major epithelial tumors, obtaining information on other types of biomarkers, i.e., predictive biomarkers that may affect treatment decisions, have been highly elusive and challenging to implement.
4.1 CTC enumeration as a prognostic biomarker for overall survival
Veridex’s CellSearch® assay is the only FDA-cleared method for CTC enumeration. It is considered the sole CTC methodology that has been analytically validated for sensitivity, accuracy, and reproducibility (Allard et al., 2004). Using the CellSearch® assay, CTC enumeration has been established in many prospective studies to be a prognostic marker for metastatic breast (Budd et al., 2006; Cristofanilli et al., 2004; Cristofanilli et al., 2005; Hayes et al., 2006), prostate (de Bono et al., 2008; Scher et al., 2009), and colon cancer (Cohen et al., 2008; Cohen et al., 2009). The CellSeach® assay has also been shown to have prognostic utility in some lung cancer patients, particularly in those with small cell lung cancer who present with higher CTC counts in the majority of cases (Hou et al., 2012; Krebs et al., 2011). While CTC enumeration with the CellSearch® assay is the most established method, it has limitations, particularly the sensitivity of detection and information that may influence treatment decisions as well as clinical outcomes for patients. For instance, the CellSearch® assay detects ≥5 CTCs per 7.5 ml blood in only about 10% of patients with metastatic non-small cell lung cancer, which is associated with worse prognosis (Krebs et al., 2011), a sensitivity which is far less than desirable to be effective in patient management. Further, while CellSearch® Profile kits can be obtained from Veridex for IF and FISH analysis of CTCs, their performance and effectiveness have not been established for clinical usage for the analysis of predictive biomarkers (www.veridex.com).
An early stage anti-EpCAM antibody-coated microchip-based technology has been shown to be much more effective in capturing CTCs with a near 100% success rate and high CTC counts in a wide range of cancers, including all of the 55 lung cancer patient samples used in one study (Nagrath et al., 2007). Subsequent work revealed that biopsy-matching mutations in EGFR can be effectively detected from the CTCs isolated from non-small cell lung cancer patients with this anti-EpCAM antibody-coated microchip (Maheswaran et al., 2008). However, the analysis of two commercial versions of this CTC-capturing microchip in comparison with the CellSearch® method done by Genentech Inc. fails to support such microchip specific claims, both analytically and with clinical specimens (Punnoose et al., 2010). In fact, the amount of CTCs captured with microchips was comparable to that with the CellSearch® assay. While the microchip technology has promise, the isolation technology and subsequent assays need to be sufficiently mature to be used reproducibly in different diagnostic labs. Further, associations between the number of CTCs isolated by microchips and clinical endpoints need to be established.
Various other technologies, including microchips of different isolation mechanisms (Tan et al., 2009b), microfilters (Hofman et al., 2011a; Lin et al., 2010), and methods using physical and electrical properties (Gupta et al., 2012; Hou et al., 2013; Ozkumur et al., 2013) of CTCs have been invented and evaluated for CTC isolation with various success. However, they need to be vigorously validated analytically for accuracy, precision, stability, and reproducibility. Further, prognostic or other predictive biomarkers need to be established with prospective clinical studies for these technologies to be useful (Danila et al., 2011c; Parkinson et al., 2012).
The direct analysis of CTCs by assaying all nucleated cells in blood may have some advantages due to potentially significant variations between CTC isolation technologies that may result in different rates of recovery and loss of tumor cells for different types of tumors. This approach, therefore, may be less susceptible to the variations in physical and biological properties of different types of tumors and within heterogeneous CTC populations. In fact, the recent advances in high-definition cell image analysis revealed that multiple fluorescence images of very high resolution can be obtained and analyzed from millions of cells on slides to enable CTC identification (Nieva et al., 2012; Wendel et al., 2012). This assay detects ≥ 5 CTCs per ml of blood from all stages of non-small cell lung cancer patients (Wendel et al., 2012), which initially has been associated with worse overall survival (Nieva et al., 2012). The technology is suitable for detection by IF of cancer-specific proteins and by FISH for genomic aberrations in cancer cells and thus, has a potential to integrate the analysis of one additional biomarker for correlative studies in treatment trials.
While the enumeration of CTCs with various isolation and detection technologies, particularly the CellSearch® assay, may provide valuable prognostic information, its limited utilities have restricted a broader adoption in oncology practice. In a recommendation by the American Society of Clinical Oncology, it was suggested that there were no data generated to prove that the use of this CTC test would lead to a longer survival time or improved quality of life for cancer patients, and additional studies are necessary to determine the utility of CTCs (Harris et al., 2007). The identification and analysis of biomarkers predictive of response to therapeutic agents are likely needed for such advancement and, indeed, are being intensely investigated.
4.2 CTC enumeration as an indicator of response
Strong associations between reduced CTC counts and progression-free or overall survival have also been established, in which the conversion of high to low CTC counts for patients on therapies indicates good prognosis for breast (Cristofanilli et al., 2004; Cristofanilli et al., 2005), prostate (Danila et al., 2007; de Bono et al., 2008), and colon cancer (Cohen et al., 2008). In contrast, patients whose low CTC counts at the baseline convert to high CTC counts during treatment do significantly worse. In metastatic breast cancer, CTC analysis has been evaluated as a response marker for chemo- and hormone-therapies, in which reduced CTC counts at weeks 3-5 have been correlated with radiographic response (Liu et al., 2009). It was further shown that the patients with <5 CTCs at weeks 3-5 following treatment have much improved progression-free survival, whereas the CTC count at the baseline is not informative (Liu et al., 2009). Thus, CTC enumeration may be suitable as an adjunct to the standard methods for monitoring disease status of certain cancer patients during therapy.
CTC analysis has been incorporated into phase I and II clinical trials for the development of novel targeted therapies. In the development of abiraterone for castration-resistant prostate cancer, the analysis of CTCs was performed in two phase II studies. Between 60% and 70% of castration-resistant prostate cancer patients enrolled had unfavorable initial CTC counts (CTCs ≥ 5) of which nearly half converted to favorable counts following abiraterone treatment (Danila et al., 2010; Reid et al., 2010), providing evidence for drug activity in the patients. Subsequently, a phase III study showed that abiraterone is effective in improving overall survival for castration-resistant prostate cancer patients (de Bono et al., 2011). When performed properly, CTC enumeration can be a good response biomarker for anti-cancer therapies and could certainly have a role in the clinical development of novel therapeutics. Further, with CTCs as the only potential source of tumor material from the patients in some studies, the added ability of reliably obtaining information on cancer genomics and other molecular markers may facilitate the clinical identification and validation of predictive biomarkers for new therapeutic agents.
4.3 Exploratory studies on predictive biomarkers with CTCs
The arguments for more-extensive molecular analysis of CTCs are very compelling. Primary or metastatic cancer samples may not be available or readily obtainable. In addition, CTCs may be a source for longitudinal molecular analysis of the tumor to provide necessary molecular information during clinical management. Also, CTCs may facilitate response monitoring, residual disease assessment, and early relapse detection. Further, they may serve as a means to address issues such as inter-tumor and intra-tumor heterogeneity. While challenging, there has been consistent interest in the analysis of molecular markers of CTCs in various clinical studies (Danila et al., 2011b; Devriese et al., 2011). As the development of targeted cancer therapies will likely need companion diagnostics for the selection of appropriate patients, CTC isolation may have an edge in providing continuous access to tumor specimens without a biopsy procedure. The utility of CTCs is particularly recognizable in cases where relapsed tumors are very different from the original primary ones and when the relapsed tumors are very difficult to obtain, such as in castration-resistant prostate cancer. Early exploratory studies have been performed with CTCs for detecting specific predictive biomarkers in various cancer, including breast, prostate, colon, and lung cancer.
In the case of breast cancer, ER, PR, and HER2 status primarily inform treatment selection. There is a body of evidence that hormone receptors and HER2 status can change during relapse or disease progression (Liedtke et al., 2009; Meng et al., 2004; Simmons et al., 2009). However, some early analyses of molecular markers on CTCs presented some technical challenges. In one study with 213 preoperative breast cancer patients, CTCs were detected in 22% of the patients with the CellSearch® assay, and HER2 over-expression was detected by IF in 14 of 58 CTC-positive patients. Thus, the percentages of both CTCs and HER2-positive cells were too low to be effective (Riethdorf et al., 2010). Also, the correlation between HER2 status in primary tumors and CTCs appears to be relatively weak (Riethdorf et al., 2010), and the assay may require further validation. In another study of 254 metastatic breast cancer patients, HER2 status was analyzed with both HER2 IF with the CellSearch® assay and HER2 mRNA with RT-PCR from AdnaTest BreastCancer™. While both assays were able to detect HER2-positive CTCs in 40-50% of patients, the correlation between the assays was weak (Fehm et al., 2010). Similar results with low concordance rates were also observed with ER and PR in CTCs and primary tumors (Aktas et al., 2011b). It was concluded that a gold standard for HER2 evaluation in CTCs is required to implement HER2 status as a stratification marker of HER2 targeted therapy in patients where repeated biopsies are not feasible (Fehm et al., 2010).
There are enormous interests in the analysis of CTCs for castration-resistant prostate cancer. The cancer generally occurs a few years after the removal of the primary cancer and tends to metastasize to bone, making them generally inaccessible to biopsy. There are further extensive changes in the tumors during the development of castration-resistance, with half of them having amplifications or mutations of the AR gene (Grasso et al., 2012). The development of second generation anti-androgen therapies and the absence of any companion diagnostics (Rathkopf and Scher, 2013) leave castration-resistant prostate cancer as the only remaining major cancer without methods of identifying appropriate patients for the available targeted therapies (Scher et al., 2013). While CTC enumeration has been extensively evaluated as a potential efficacy (surrogate) biomarker of survival for AR-targeted therapies, much effort was made to perform molecular profiling of CTCs, including the analysis of AR status, TMPRSS2-ERG translocation, PTEN deletion and MYC amplification with FISH-based analysis (Attard et al., 2009; Leversha et al., 2009). A study of the TMPRSS2-ERG fusion gene in CTCs suggests that the presence of the translocation may indicate susceptibility to the CYP17 inhibitor, abiraterone acetate (Attard et al., 2009). Using CTCs isolated with the CellSearch® instrument, there are some examples of detecting PSA expression and AR mutation with RT-PCR-based approaches (Helo et al., 2009; Jiang et al., 2010). Using herringbone-chip based technology, CTCs isolated from prostate cancer patients have been evaluated for TMPRSS2-ERG and AR signaling with IF and RT-PCR (Miyamoto et al., 2012; Stott et al., 2010a; Stott et al., 2010b) and recently, for the exploration of single cell RNA expression analysis (Ozkumur et al., 2013). While some of the technologies and exploratory studies show great promise, none of them have been clinically validated, and thus, significant advances in analytic validation and clinical qualification are required for their use in prostate cancer (Danila et al., 2011c; Scher et al., 2013).
In metastatic colorectal cancer, the final major cancer with FDA-cleared use of CellSearch, patients with ≥3 CTCs per 7.5 ml blood have significantly worse overall survival (Cohen et al., 2008). Efforts were made to explore biomarkers from CTCs in patients who had undergone curative surgery. Using total blood, mRNA and cDNA were generated and analyzed. A positive expression signature of CEA/CK/CD133 was identified in a training set and then validated in a second validation set of patients to have worse prognosis (Iinuma et al., 2011). Successful attempts were reported in analyzing biomarkers that may predict drug sensitivity by detecting mutations in KRAS and PIK3CA from CTCs isolated with CellSearch® followed by micromanipulation (Gasch et al., 2013; Heitzer et al., 2013). Further, proof-of-principle single-cell whole genome analysis with DNA array-comparative genomic hybridization and exome sequencing have been performed with CTCs isolated with the CellSearch® instrument (Heitzer et al., 2013). Deep-sequencing analysis reveals that the method is capable of detecting mutations in APC, KRAS, and PIK3CA from multiple CTCs, which are also present in both the primary and metastatic tumors of the same patient (Heitzer et al., 2013). Such whole genome analysis from CTCs may have the potential to uncover biomarkers predictive of sensitivity or resistance when tumor tissues are not available.
Lung cancer is the only major cancer without an FDA-cleared CTC application. A recent study shows that in the case of non-small cell lung cancer, the most prevalent lung cancer type, only about 20% of stage III and IV patients have ≥ 2 CTCs per 7.5 ml of blood (Krebs et al., 2011). Only about 10% of these patients have ≥ 5 CTCs, which is a worse prognostic marker for overall survival. In contrast, the detection sensitivity for small cell lung cancer is far better at 85%, and a high CTC count is a significantly worse prognostic biomarker (Hou et al., 2012). The CTCs isolated by CTC-chip have been used for EGFR mutation detection with mutation-specific PCR analysis (Maheswaran et al., 2008). While such oncogene mutations can also be detected by methods utilizing plasma tumor DNA (ptDNA), the isolation of CTCs has certain advantages in detecting gene rearrangements, cancer-specific expression, and more-extensive whole genome analysis for discovery research. For the clinical application of CTC-based assays, a recent study shows that by using the ISET filtration method for CTC enrichment, followed by in-situ hybridization, ELM4-ALK translocation can be detected in all 18 ALK-rearrangement positive non-small cell lung cancer patients, but not in the ALK rearrangement-negative patients (Pailler et al., 2013). If confirmed, such a method may provide a non-invasive method for the analysis of predictive biomarkers for lung cancer.
Various cell surface marker-independent technologies have the potential to isolate live CTCs, including a microfluidic chip (Tan et al., 2009b), dielectrophoretic field flow fractionation (Gupta et al., 2012), and Dean flow fractionation (Hou et al., 2013). Such methods may allow the isolation and ex vivo expansion of the isolated tumor cells from circulating blood for genomic analysis as well as molecular pharmacology studies. Although difficult, a recent report demonstrates the possibility of establishing rare mouse xenografts with cells from breast cancer patients who have very high CTC counts that help identify metastasis-initiating cells (Baccelli et al., 2013). It is also worth noting a recent publication in which a medium containing ROCK kinase inhibitor and feeder cells was capable of reprograming epithelial cells to enable long-term ex vivo culturing and expansion of a wide range of normal and tumor cells (Liu et al., 2012). Such a method could be used in conjunction with CTC isolation technology to evaluate the potential to establish CTCs ex vivo. The ability to grow tumor cells isolated from blood offers a very attractive option for both cancer research and patient care.
4.4 Alternative technology to CTCs, ptDNA and its analysis
As an alternative to CTCs, ptDNA has been intensively investigated to determine its feasibility as a form of ‘liquid biopsy’ for the analysis of cancer biomarkers, including particular mutations in genes that are predictive of drug sensitivity or resistance. Very significant progress has been made in this area that may represent an alternative approach to CTC-based analysis. Similar to CTC enumeration that is prognostic for cancer patients, the relative quantification of ptDNA can be determined by analyzing plasma DNA using an emulsion bead-based single-molecular amplification that is capable of determining the relative abundance of tumor DNA in plasma. Detectable ptDNA has been shown to be a better predictive marker for recurrence-free survival when compared with that of plasma carcinoembryonic antigen for colorectal cancer patients (Diehl et al., 2008). Moreover, the analysis of ptDNA from metastatic breast cancer patients has been shown to be a more sensitive biomarker with better correlation to tumor burden when compared with circulating tumor antigen CA 15-3 (Dawson et al., 2013). By using targeted deep DNA sequencing analysis, mutation in selective oncogenes and tumor suppressors can be identified, and their allele frequencies determined (Forshew et al., 2012). KRAS mutation is predictive of non-responsiveness to anti-EGFR therapies. Mutations in KRAS have been seen to emerge in ptDNA in patients that have been treated with the EGFR antibodies, cetuximab and panitumumab, indicating that developing KRAS mutations is a mechanism to escape EGFR blockage (Diaz et al., 2012; Misale et al., 2012). Similarly, mutations in a large number of genes can be identified with increased allele frequency in ptDNA from cancer patients who progressed on a number of targeted or cytotoxic chemotherapies using whole genome exon analysis (Murtaza et al., 2013). It is very likely that the analysis of ptDNA will have value in monitoring tumor burden and in detecting gene mutations for clinical research and for cancer patient management. Recent advances in digital PCR technologies such as Biomark PCR from Fluidigm and Droplet PCR from BioRad will simplify the assays and speed up the adoption in the cancer research community.
5. Conclusions and future prospects
The CellSearch® Circulating Tumor Cell test is the only FDA-cleared in vitro CTC diagnostic in which a positive enumeration is associated with decreased progression-free survival and overall survival for metastatic breast, prostate and colon cancer patients. It is currently the gold standard method for CTC enumeration and the only one that has been both analytically and clinically validated. This test has also been shown to be useful in monitoring these patients during clinical management. However, the CellSearch® test has limitations. It has limited sensitivity and is not applicable to all types of cancer. Assays with improved sensitivity could not only be applicable to other types of cancers, but also may detect cancers at early stages when patients are more likely to benefit from therapies. Also, while extensive work has been performed with both IF and FISH to examine biomarkers that may predict drug sensitivity, these predictive biomarker studies are still at the research and development stage. Further, the CellSearch® tests have not yet been shown to lead to improved survival or quality of life.
CTCs have the promise of serving as liquid biopsies for tumors with the potential for generating information on biomarkers predictive of response and resistance. CTC isolation and analysis is one of the most active areas for translational cancer research. Various methods have been shown to be able to isolate more CTCs from a higher percentage of cancer patients. Moreover, many assays on the analysis of potentially predictive biomarkers of therapies have been explored, including IF, FISH, RT-PCR, cancer gene mutation detection, and genomic analysis. Some of them have shown success in early clinical exploratory studies. However, for them to be used for cancer management, assays and technologies need to be better validated analytically, and methods need to be standardized and reproduced by others. Furthermore, commercial success of a CTC technology requires a robust technology that is cost effective with a fast turn-around time. The eventual integration of CTC technology into clinical patient care requires the demonstration of specific utilities in treatment decisions, such as the analysis of biomarkers predictive of response or the development of CTC-based companion diagnostics for new targeted therapies.
Acknowledgment
We thank Jennifer Crawford for editing the manuscript. This work is directly supported by the Intramural Research Program of the US National Cancer Institute (NCI) of the National Institutes of Health (Z.K. and L.C.) and by the NCI (R21CA161835, DP2CA174508, R.H. and S.Z.) in preparing the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- AR
androgen receptor
- CK
cytokeratin
- CTC
circulating tumor cell
- DEP
dielectrophoresis
- EGFR
epidermal growth factor receptor
- EMT
epithelial-mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- ER
estrogen receptor
- FDA
US Food and Drug Administration
- FISH
fluorescence in situ hybridization
- HER2
human epidermal growth factor receptor 2
- IF
immunofluorescence
- ISET
isolation by size of epithelial tumor cells technique
- PR
progesterone receptor
- PSA
prostate specific antigen
- PSMA
prostate-specific membrane antigen
- ptDNA
plasma tumor DNA
- RT-PCR
reverse transcription polymerase chain reaction
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aktas B, Kasimir-Bauer S, Heubner M, Kimmig R, Wimberger P. Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int J Gynecol Cancer. 2011a;21:822–830. doi: 10.1097/IGC.0b013e318216cb91. [DOI] [PubMed] [Google Scholar]
- Aktas B, Muller V, Tewes M, Zeitz J, Kasimir-Bauer S, Loehberg CR, Rack B, Schneeweiss A, Fehm T. Comparison of estrogen and progesterone receptor status of circulating tumor cells and the primary tumor in metastatic breast cancer patients. Gynecol Oncol. 2011b;122:356–360. doi: 10.1016/j.ygyno.2011.04.039. [DOI] [PubMed] [Google Scholar]
- Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabières C, Brouillet J-P, Fabbro M, Yssel H, Rousset T, Maudelonde T, Choquet-Kastylevsky G, Vendrell J-P. Characterization and enumeration of cells secreting tumor markers in the peripheral blood of breast cancer patients. Journal of immunological methods. 2005;299:177–188. doi: 10.1016/j.jim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clinical Chemistry. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres C, Vendrell JP, Pelle O, Rebillard X, Riethdorf S, Muller V, Fabbro M, Pantel K. Detection and characterization of putative metastatic precursor cells in cancer patients. Clinical Chemistry. 2007;53:537–539. doi: 10.1373/clinchem.2006.079509. [DOI] [PubMed] [Google Scholar]
- Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- Andreopoulou E, Yang LY, Rangel KM, Reuben JM, Hsu L, Krishnamurthy S, Valero V, Fritsche HA, Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. International journal of cancer Journal international du cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ, Garcia-Blanco MA. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Molecular Cancer Research. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1869;14:146–149. [Google Scholar]
- Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bäuerle T, Wallwiener M. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nature Biotechnology. 2013 doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- Balic M, Dandachi N, Hofmann G, Samonigg H, Loibner H, Obwaller A, van der Kooi A, Tibbe AGJ, Doyle GV, Terstappen L, Bauernhofer T. Comparison of two methods for enumerating circulating tumor cells in carcinoma patients. Cytometry Part B-Clinical Cytometry. 2005;68B:25–30. doi: 10.1002/cyto.b.20065. [DOI] [PubMed] [Google Scholar]
- Becker F, Wang X-B, Huang Y, Pethig R, Vykoukal J, Gascoyne P. The removal of human leukaemia cells from blood using interdigitated microelectrodes. Journal of Physics D: Applied Physics. 1994;27:2659. [Google Scholar]
- Becker FF, Wang X-B, Huang Y, Pethig R, Vykoukal J, Gascoyne P. Separation of human breast cancer cells from blood by differential dielectric affinity. Proceedings of the National Academy of Sciences. 1995;92:860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat AAS, Hou HW, Li LD, Lim CT, Han J. Pinched flow coupled shear-modulated inertial microfluidics for high-throughput rare blood cell separation. Lab On A Chip. 2011;11:1870–1878. doi: 10.1039/c0lc00633e. [DOI] [PubMed] [Google Scholar]
- Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, Matera J, Repollet M, Doyle GV, Terstappen LW, Hayes DF. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- Carey RW, Taft PD, Bennett JM, Kaufman S. Carcinocythemia (carcinoma cell leukemia): An acute leukemia-like picture due to metastatic carcinoma cells. The American journal of medicine. 1976;60:273–278. doi: 10.1016/0002-9343(76)90437-x. [DOI] [PubMed] [Google Scholar]
- Chen CL, Mahalingam D, Osmulski P, Jadhav RR, Wang CM, Leach RJ, Chang TC, Weitman SD, Kumar AP, Sun L. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. The Prostate. 2012 doi: 10.1002/pros.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Physical Biology. 2012;9:016001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse MA, Mitchell E, Miller MC, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009;20:1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- Danila DC, Anand A, Sung CC, Heller G, Leversha MA, Cao L, Lilja H, Molina A, Sawyers CL, Fleisher M, Scher HI. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol. 2011a;60:897–904. doi: 10.1016/j.eururo.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila DC, Fleisher M, Scher HI. Circulating tumor cells as biomarkers in prostate cancer. Clin Cancer Res. 2011b;17:3903–3912. doi: 10.1158/1078-0432.CCR-10-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- Danila DC, Morris MJ, de Bono JS, Ryan CJ, Denmeade SR, Smith MR, Taplin ME, Bubley GJ, Kheoh T, Haqq C, et al. Phase II multicenter study of abiraterone acetate plus prednisone therapy in patients with docetaxel-treated castration-resistant prostate cancer. J Clin Oncol. 2010;28:1496–1501. doi: 10.1200/JCO.2009.25.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila DC, Pantel K, Fleisher M, Scher HI. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 2011c;17:438–450. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr., Saad F, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- De Giorgi V, Pinzani P, Salvianti F, Panelos J, Paglierani M, Janowska A, Grazzini M, Wechsler J, Orlando C, Santucci M. Application of a filtration-and isolation-by-size technique for the detection of circulating tumor cells in cutaneous melanoma. Journal of Investigative Dermatology. 2010;130:2440–2447. doi: 10.1038/jid.2010.141. [DOI] [PubMed] [Google Scholar]
- Devriese LA, Voest EE, Beijnen JH, Schellens JH. Circulating tumor cells as pharmacodynamic biomarker in early clinical oncological trials. Cancer Treat Rev. 2011;37:579–589. doi: 10.1016/j.ctrv.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Diaz LA, Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. British Journal of Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehm T, Becker S, Duerr-Stoerzer S, Sotlar K, Mueller V, Wallwiener D, Lane N, Solomayer E, Uhr J. Determination of HER2 status using both serum HER2 levels and circulating tumor cells in patients with recurrent breast cancer whose primary tumor was HER2 negative or of unknown HER2 status. Breast Cancer Research. 2007;9 doi: 10.1186/bcr1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehm T, Hoffmann O, Aktas B, Becker S, Solomayer EF, Wallwiener D, Kimmig R, Kasimir-Bauer S. Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. 2009;11:R59. doi: 10.1186/bcr2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehm T, Muller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, Lattrich C, Lohberg CR, Solomayer E, Rack B, et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat. 2010;124:403–412. doi: 10.1007/s10549-010-1163-x. [DOI] [PubMed] [Google Scholar]
- Fischer JC, Niederacher D, Topp SA, Honisch E, Schumacher S, Schmitz N, Zacarias Fohrding L, Vay C, Hoffmann I, Kasprowicz NS, et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- Gallivan MV, Lokich JJ. Carcinocythemia (carcinoma cell leukemia). Report of two cases with English literature review. Cancer. 1984;53:1100–1102. doi: 10.1002/1097-0142(19840301)53:5<1100::aid-cncr2820530514>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Gasch C, Bauernhofer T, Pichler M, Langer-Freitag S, Reeh M, Seifert AM, Mauermann O, Izbicki JR, Pantel K, Riethdorf S. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem. 2013;59:252–260. doi: 10.1373/clinchem.2012.188557. [DOI] [PubMed] [Google Scholar]
- Ghossein RA, Rosai J, Scher HI, Seiden M, Zhang ZF, Sun M, Chang G, Berlane K, Krithivas K, Kantoff PW. Prognostic significance of detection of prostate-specific antigen transcripts in the peripheral blood of patients with metastatic androgen-independent prostatic carcinoma. Urology. 1997;50:100–105. doi: 10.1016/S0090-4295(97)00127-1. [DOI] [PubMed] [Google Scholar]
- Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab On A Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, Davis DW. ApoStream(), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV, Terstappen LW. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- He W, Kularatne SA, Kalli KR, Prendergast FG, Amato RJ, Klee GG, Hartmann LC, Low PS. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. International Journal of Cancer. 2008;123:1968–1973. doi: 10.1002/ijc.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- Helo P, Cronin AM, Danila DC, Wenske S, Gonzalez-Espinoza R, Anand A, Koscuiszka M, Vaananen RM, Pettersson K, Chun FK, et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Flejou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res. 2011a;17:827–835. doi: 10.1158/1078-0432.CCR-10-0445. [DOI] [PubMed] [Google Scholar]
- Hofman VJ, Ilie MI, Bonnetaud C, Selva E, Long E, Molina T, Vignaud JM, Fléjou JF, Lantuejoul S, Piaton E. Cytopathologic Detection of Circulating Tumor Cells Using the Isolation by Size of Epithelial Tumor Cell Method Promises and Pitfalls. American journal of clinical pathology. 2011b;135:146–156. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Hayata T, Fukuda Y, Arakaki A, Yoshino T, Tanaka T, Matsunaga T. Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells. Analytical Chemistry. 2010;82:6629–6635. doi: 10.1021/ac101222x. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Kenmotsu H, Koh Y, Yoshino T, Yoshikawa T, Naito T, Takahashi T, Murakami H, Nakamura Y, Tsuya A. Size-Based Isolation of Circulating Tumor Cells in Lung Cancer Patients Using a Microcavity Array System. Plos One. 2013;8:e67466. doi: 10.1371/journal.pone.0067466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, Lim WT, Han J, Bhagat AA, Lim CT. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang X-B, Becker FF, Gascoyne P. Introducing dielectrophoresis as a new force field for field-flow fractionation. Biophysical Journal. 1997;73:1118–1129. doi: 10.1016/S0006-3495(97)78144-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AD, Mattison J, Western LT, Powderly JD, Greene BT, King MR. Microtube Device for Selectin-Mediated Capture of Viable Circulating Tumor Cells from Blood. Clinical Chemistry. 2012;58:846–853. doi: 10.1373/clinchem.2011.176669. [DOI] [PubMed] [Google Scholar]
- Hur SC, Mach AJ, Di Carlo D. High-throughput size-based rare cell enrichment using microscale vortices. Biomicrofluidics. 2011;5 doi: 10.1063/1.3576780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis M, Rothe F, Chaboteaux C, Durbecq V, Rouas G, Criscitiello C, Metallo J, Kheddoumi N, Singhal SK, Michiels S. HER2-positive circulating tumor cells in breast cancer. Plos One. 2011;6:e15624. doi: 10.1371/journal.pone.0015624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iinuma H, Watanabe T, Mimori K, Adachi M, Hayashi N, Tamura J, Matsuda K, Fukushima R, Okinaga K, Sasako M, Mori M. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol. 2011;29:1547–1555. doi: 10.1200/JCO.2010.30.5151. [DOI] [PubMed] [Google Scholar]
- Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci Transl Med. 2012;4:141ra192. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Palma JF, Agus DB, Wang Y, Gross ME. Detection of androgen receptor mutations in circulating tumor cells in castration-resistant prostate cancer. Clin Chem. 2010;56:1492–1495. doi: 10.1373/clinchem.2010.143297. [DOI] [PubMed] [Google Scholar]
- Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimir-Bauer S, Hoffmann O, Wallwiener D, Kimmig R, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012;14:R15. doi: 10.1186/bcr3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, Gleghorn JP, Santana SM, Liu H, Smith JP, et al. Functional Characterization of Circulating Tumor Cells with a Prostate-Cancer-Specific Microfluidic Device. Plos One. 2012;7:e35976. doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, et al. A rare-cell detector for cancer. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy A, Delgado S, Garcia-Valdecasas J, Castells A, Pique J, Grande L, Fuster J, Targarona E, Pera M, Visa J. Port site metastases and recurrence after laparoscopic colectomy. Surgical endoscopy. 1998;12:1039–1042. doi: 10.1007/s004649900776. [DOI] [PubMed] [Google Scholar]
- Lara O, Tong X, Zborowski M, Chalmers JJ. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Experimental Hematology. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Lazar DC, Cho EH, Luttgen MS, Metzner TJ, Uson ML, Torrey M, Gross ME, Kuhn P. Cytometric comparisons between circulating tumor cells from prostate cancer patients and the prostate-tumor-derived LNCaP cell line. Physical Biology. 2012;9:016002. doi: 10.1088/1478-3975/9/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria J, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. British Journal of Cancer. 2011;105:1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leversha MA, Han J, Asgari Z, Danila DC, Lin O, Gonzalez-Espinoza R, Anand A, Lilja H, Heller G, Fleisher M, Scher HI. Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res. 2009;15:2091–2097. doi: 10.1158/1078-0432.CCR-08-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Broglio K, Moulder S, Hsu L, Kau SW, Symmans WF, Albarracin C, Meric-Bernstam F, Woodward W, Theriault RL, et al. Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol. 2009;20:1953–1958. doi: 10.1093/annonc/mdp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LS, Hu M, Huang MC, Cheong WC, Gan ATL, Looi XL, Leong SM, Koay ES-C, Li M-H. Microsieve lab-chip device for rapid enumeration and fluorescence in situ hybridization of circulating tumor cells. Lab On A Chip. 2012;12:4388–4396. doi: 10.1039/c2lc20750h. [DOI] [PubMed] [Google Scholar]
- Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, Rao SB, Eng-Wong J, Seillier-Moiseiwitsch F, Noone AM, Isaacs C. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27:5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Shaw G, James SY, Purkis P, Kudahetti SC, Tsigani T, Kia S, Young BD, Oliver RTD, Berney D. Detection of TMPRSS2: ERG fusion gene in circulating prostate cancer cells. Asian journal of andrology. 2008;10:467–473. doi: 10.1111/j.1745-7262.2008.00401.x. [DOI] [PubMed] [Google Scholar]
- Markou A, Strati A, Malamos N, Georgoulias V, Lianidou ES. Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin Chem. 2011;57:421–430. doi: 10.1373/clinchem.2010.154328. [DOI] [PubMed] [Google Scholar]
- Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Physical Biology. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, Beitsch P, Khan A, Euhus D, Osborne C, et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proc Natl Acad Sci U S A. 2004;101:9393–9398. doi: 10.1073/pnas.0402993101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller V, Stahmann N, Riethdorf S, Rau T, Zabel T, Goetz A, Janicke F, Pantel K. Circulating tumor cells in breast cancer: Correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clinical Cancer Research. 2005;11:3678–3685. doi: 10.1158/1078-0432.CCR-04-2469. [DOI] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- Myerowitz RL, Edwards PA, Sartiano GP. Carcinocythemia (carcinoma cell leukemia) due to metastatic carcinoma of the breast. Report of a case. Cancer. 1977;40:3107–3111. doi: 10.1002/1097-0142(197712)40:6<3107::aid-cncr2820400653>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J, Wendel M, Luttgen MS, Marrinucci D, Bazhenova L, Kolatkar A, Santala R, Whittenberger B, Burke J, Torrey M, et al. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: a longitudinal analysis. Phys Biol. 2012;9:016004. doi: 10.1088/1478-3975/9/1/016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachmann K, Camara O, Kavallaris A, Krauspe S, Malarski N, Gajda M, Kroll T, Jorke C, Hammer U, Altendorf-Hofmann A, et al. Monitoring the response of circulating epithelial tumor cells to adjuvant chemotherapy in breast cancer allows detection of patients at risk of early relapse. J Clin Oncol. 2008;26:1208–1215. doi: 10.1200/JCO.2007.13.6523. [DOI] [PubMed] [Google Scholar]
- Pailler E, Adam J, Barthelemy A, Oulhen M, Auger N, Valent A, Borget I, Planchard D, Taylor M, Andre F, et al. Detection of Circulating Tumor Cells Harboring a Unique ALK Rearrangement in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- Parkinson DR, Dracopoli N, Petty BG, Compton C, Cristofanilli M, Deisseroth A, Hayes DF, Kapke G, Kumar P, Lee J, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. 2012;10:138. doi: 10.1186/1479-5876-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestrin M, Bessi S, Galardi F, Truglia M, Biggeri A, Biagioni C, Cappadona S, Biganzoli L, Giannini A, Di Leo A. Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Research And Treatment. 2009;118:523–530. doi: 10.1007/s10549-009-0461-7. [DOI] [PubMed] [Google Scholar]
- Phillips KG, Velasco CR, Li J, Kolatkar A, Luttgen M, Bethel K, Duggan B, Kuhn P, McCarty OJ. Optical quantification of cellular mass, volume, and density of circulating tumor cells identified in an ovarian cancer patient. Front Oncol. 2012;2:72. doi: 10.3389/fonc.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et al. Single Cell Profiling of Circulating Tumor Cells: Transcriptional Heterogeneity and Diversity from Breast Cancer Cell Lines. Plos One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS, Lackner MR. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathkopf D, Scher HI. Androgen receptor antagonists in castration-resistant prostate cancer. Cancer J. 2013;19:43–49. doi: 10.1097/PPO.0b013e318282635a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Attard G, Danila DC, Oommen NB, Olmos D, Fong PC, Molife LR, Hunt J, Messiou C, Parker C, et al. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J Clin Oncol. 2010;28:1489–1495. doi: 10.1200/JCO.2009.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S, Muller V, Zhang L, Rau T, Loibl S, Komor M, Roller M, Huober J, Fehm T, Schrader I, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Gertler R, Friederichs J, Fuehrer K, Dahm M, Phelps R, Thorban S, Nekarda H, Siewert JR. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- Saucedo-Zeni N, Mewes S, Niestroj R, Gasiorowski L, Murawa D, Nowaczyk P, Tomasi T, Weber E, Dworacki G, Morgenthaler NG. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. International Journal of Oncology. 2012;41:1241. doi: 10.3892/ijo.2012.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Morris MJ, Larson S, Heller G. Validation and clinical utility of prostate cancer biomarkers. Nat Rev Clin Oncol. 2013;10:225–234. doi: 10.1038/nrclinonc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, et al. Circulating Tumor Cell Analysis in Patients with Progressive Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- Shim S, Stemke-Hale K, Noshari J, Becker FF, Gascoyne PR. Dielectrophoresis has broad applicability to marker-free isolation of tumor cells from blood by microfluidic systems. Biomicrofluidics. 2013;7:011808. doi: 10.1063/1.4774307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts AM, Mostert B, Bolt-de Vries J, Peeters D, de Jongh FE, Stouthard JM, Dirix LY, van Dam PA, Van Galen A, de Weerd V, et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res. 2011;17:3600–3618. doi: 10.1158/1078-0432.CCR-11-0255. [DOI] [PubMed] [Google Scholar]
- Simmons C, Miller N, Geddie W, Gianfelice D, Oldfield M, Dranitsaris G, Clemons MJ. Does confirmatory tumor biopsy alter the management of breast cancer patients with distant metastases? Ann Oncol. 2009;20:1499–1504. doi: 10.1093/annonc/mdp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier E, Go DE, Che J, Gossett D, O’Byrne S, Weaver WM, Kummer N, Rettig M, Goldman J, Nickols N. Size-Selective Collection of Circulating Tumor Cells using Vortex Technology. Lab Chip. 2013 doi: 10.1039/c3lc50689d. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010a;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010b;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati A, Kasimir-Bauer S, Markou A, Parisi C, Lianidou ES. Comparison of three molecular assays for the detection and molecular characterization of circulating tumor cells in breast cancer. Breast Cancer Res. 2013;15:R20. doi: 10.1186/bcr3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li M, Liu C, Zhang Y, Liu D, Liu W, Hu G, Jiang X. Double spiral microchannel for label-free tumor cell separation and enrichment. Lab On A Chip. 2012;12:3952–3960. doi: 10.1039/c2lc40679a. [DOI] [PubMed] [Google Scholar]
- Sun J, Liu C, Li M, Wang J, Xianyu Y, Hu G, Jiang X. Size-based hydrodynamic rare tumor cell separation in curved microfluidic channels. Biomicrofluidics. 2013;7:011802. doi: 10.1063/1.4774311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh K-H, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proceedings of the National Academy of Sciences. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Yobas L, Lee G, Ong C, Lim C. Microdevice for the isolation and enumeration of cancer cells from blood. Biomedical Microdevices. 2009a;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- Tan SJ, Lakshmi RL, Chen PF, Lim WT, Yobas L, Lim CT. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosensors & Bioelectronics. 2010;26:1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009b;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- Tewes M, Aktas B, Welt A, Mueller S, Hauch S, Kimmig R, Kasimir-Bauer S. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat. 2009;115:581–590. doi: 10.1007/s10549-008-0143-x. [DOI] [PubMed] [Google Scholar]
- Theodoropoulos PA, Polioudaki H, Agelaki S, Kallergi G, Saridaki Z, Mavroudis D, Georgoulias V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer letters. 2010;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Ushijima K. Treatment for recurrent ovarian cancer—at first relapse. Journal of Oncology. 2009;2010 doi: 10.1155/2010/497429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona G, Estepa L, Beroud C, Damotte D, Capron F, Nalpas B, Mineur A, Franco D, Lacour B, Pol S, et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology. 2004;39:792–797. doi: 10.1002/hep.20091. [DOI] [PubMed] [Google Scholar]
- Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, et al. Isolation by size of epithelial tumor cells - A new method for the immunomorphological and molecular characterization of circulating tumor cells. American Journal Of Pathology. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee Y-K, et al. Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angewandte Chemie International Edition. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angewandte Chemie (International ed in English) 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Eisenberger MA, Carducci MA, Partin AW, Scher HI, Ts’o PO. Identification and characterization of circulating prostate carcinoma cells. Cancer. 2000;88:2787–2795. doi: 10.1002/1097-0142(20000615)88:12<2787::aid-cncr18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Weitz J, Kienle P, Lacroix J, Willeke F, Benner A, Lehnert T, Herfarth C, von Knebel Doeberitz M. Dissemination of tumor cells in patients undergoing surgery for colorectal cancer. Clinical Cancer Research. 1998;4:343–348. [PubMed] [Google Scholar]
- Wendel M, Bazhenova L, Boshuizen R, Kolatkar A, Honnatti M, Cho EH, Marrinucci D, Sandhu A, Perricone A, Thistlethwaite P, et al. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys Biol. 2012;9:016005. doi: 10.1088/1478-3967/9/1/016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfing P, Borchard J, Buerger H, Heidl S, Zanker KS, Kiesel L, Brandt B. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clinical Cancer Research. 2006;12:1715–1720. doi: 10.1158/1078-0432.CCR-05-2087. [DOI] [PubMed] [Google Scholar]
- Xu T, Lu B, Tai Y-C, Goldkorn A. A Cancer Detection Platform Which Measures Telomerase Activity from Live Circulating Tumor Cells Captured on a Microfilter. Cancer Research. 2010;70:6420–6426. doi: 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnology and Bioengineering. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, Ouyang WH, et al. High-Purity Prostate Circulating Tumor Cell Isolation by a Polymer Nanofiber-Embedded Microchip for Whole Exome Sequencing. Advanced materials; Deerfield Beach, Fla: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Lin HK, Liu J-Q, Balic M, Datar R, Cote RJ, Tai Y-C. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. Journal of Chromatography A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- Zheng S, Lin HK, Lu B, Williams A, Datar RH, Cote RJ, Tai YC. 3D Microfilter Device for Viable Circulating Tumor Cell (CTC) Enrichment from Blood. Biomedical Microdevices. 2011;13:203. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]