Abstract
Most patients with BRAFV600 metastatic melanoma develop resistance to selective RAF kinase inhibitors. The spectrum of clinical genetic resistance mechanisms to RAF inhibitors and options for salvage therapy are incompletely understood. We performed whole exome sequencing on formalin-fixed, paraffin embedded (FFPE) tumors from 45 patients with BRAFV600 metastatic melanoma who received vemurafenib or dabrafenib monotherapy. Genetic alterations in known or putative RAF inhibitor resistance genes were observed in 23 of 45 patients (51%). Besides previously characterized alterations, we discovered a “long tail” of new MAPK pathway alterations (MAP2K2, MITF) that confer RAF inhibitor resistance. In three cases, multiple resistance gene alterations were observed within the same tumor biopsy. Overall, RAF inhibitor therapy leads to diverse clinical genetic resistance mechanisms, mostly involving MAPK pathway reactivation. Novel therapeutic combinations may be needed to achieve durable clinical control of BRAFV600 melanoma. Integrating clinical genomics with preclinical screens may model subsequent resistance studies.
Keywords: Melanoma, resistance, RAF, inhibitor, genetic
INTRODUCTION
RAF inhibitors (vemurafenib, dabrafenib) administered alone or in combination with MEK inhibitors have improved progression-free and overall survival in patients with BRAFV600-mutated metastatic melanoma1–4. Although the vast majority of patients experience clinical benefit, almost all develop resistance to these agents5. Furthermore, some of these patients shows either intrinsic resistance or short-lived responses (e.g. disease progression (PD) in less than 12 weeks).
Several mechanisms of RAF inhibitor resistance have been identified. Genetic causes of resistance identified thus far include NRAS6 and MEK15,7 mutations, BRAF amplification8, and NF1 loss9. Non-genetic causes of RAF inhibitor resistance include activation of COT10 or EGFR11, suppression of BIM expression via PTEN loss12, alternative splicing of BRAF RNA transcripts13, disrupted feedback regulation14, receptor tyrosine kinase dysregulation6,15, and stromal secretion of growth factors like HGF16,17.
Most previously described resistance mechanisms were identified using preclinical models and confirmed in limited numbers of clinical specimens. A detailed understanding of the somatic genetic causes of RAF inhibitor resistance may have longstanding clinical impact, since these mechanisms may inform future clinical development priorities. However, our understanding of the spectrum of genetic resistance mechanisms is incomplete. Therefore, we performed whole exome sequencing of BRAFV600 melanoma tissue obtained before treatment and after the development of resistance to RAF inhibitors to characterize the clinical spectrum of genetic resistance for this patient population.
RESULTS
A spectrum of genetic alterations is associated with clinical resistance to RAF inhibition
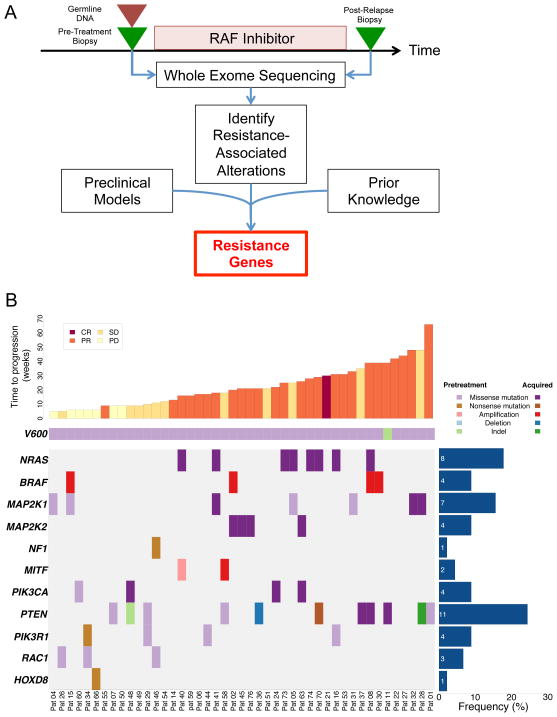
Among the 45 patients in this cohort (Fig. 1A), 14 (31%) had early resistance (on therapy for less than 12 weeks) and 31 (68.9%) developed acquired resistance (Table 1). Among the early resistance patients, 7 (50%) had progressive disease as best response, 6 (43%) had short-lived stable disease, and one (7%) had a brief partial response. The mean target coverage for tumor samples was 200X and 92X for germline DNA (Supplementary Table S1). BRAF mutations were detected in all pre-treatment biopsy specimens by WES, of which 44 of 45 were missense mutations affecting codon V600. Patient 11 had an in-frame deletion event predicted to generate a functional effect similar to V600E (Val600_Lys601delinsGlu).
Figure 1. Genetic alterations in the context of RAF inhibitor therapy.
(A) Schematic overview of tumor biopsy collection in the context of RAF inhibitor therapy, followed by whole exome sequencing and analysis. (B) Spectrum of putative resistance genes, including known genes (NRAS, BRAF, MEK1) and new genes (MEK2, MITF). Additional recurrently altered pathways (PI3K pathway) or genomic correlates of early resistance (HOXD8, RAC1) are also shown. Results are sorted by duration of therapy (weeks). CR = Complete response, PR = partial response, SD = stable disease, PD = progressive disease.
Table 1. Clinical Characteristics.
The RAF inhibitor used, duration of response, and best clinical response data for 45 patients are detailed here, further stratified by whether the patient exhibited early or acquired resistance to therapy. Age is defined as the age at time of starting therapy.
| Total | Early | Acquired | |
|---|---|---|---|
|
|
|||
| Number of Patients | 45 | 14 | 31 |
| Age (years) | 51.0 | 48.8 | 52.0 |
| Gender (% Female) | 48.9 | 50 | 48.3 |
| Medication | |||
| Vemurafenib (%) | 66.6 | 64.3 | 67.7 |
| Dabrafenib (%) | 33.3 | 35.7 | 32.3 |
| Median Duration of therapy (weeks) | 20 (SD 14.1) | 9 (SD 2.3) | 25 (SD 12.4) |
| Best Response (%) | |||
| CR | 2.2 | 0 | 3.2 |
| PR | 55.6 | 7.1 | 80.6 |
| SD | 24.4 | 42.9 | 16.1 |
| PD | 17.8 | 50 | 0 |
|
|
|||
All values are medians across the cohort. CR = Complete response, PR = partial response, SD = stable disease, PD = progressive disease.
Mutational analysis of resistant tumors revealed several genes shown previously to confer resistance to RAF inhibition (Supplementary Table S2–4). These include somatic mutations in NRAS (17.8%; seven involving the Q61 loci and one involving T58), amplifications of BRAF (8.9%), and mutations in MAP2K1 (15.6%), although MAP2K1 mutations did not universally preclude clinical response (Fig. 1B). As expected, acquired NRAS mutations occurred exclusively in patients on therapy for more than 12 weeks (P = 0.04). We also observed multiple additional putative resistance drivers that occurred at low frequencies across the cohort (Fig. 1B).
Globally, these events could be aggregated based on the cellular pathways or mechanisms implicated by the resistance-associated genes. Resistance alterations predominantly involved the MAPK pathway or downstream effectors (NRAS, BRAF, MAP2K1, MAP2K2, MITF, NF1), representing 44.4% (20/45) of the patient cohort. Additional alterations with less clear resistance relationships were observed in the phosphoinositide 3-kinase (PI3K) pathway (PIK3CA, PTEN, PIK3R1), and in HOXD8 or RAC1 (Fig. 1B).
MEK2 mutations confer resistance to RAF and MEK inhibitors
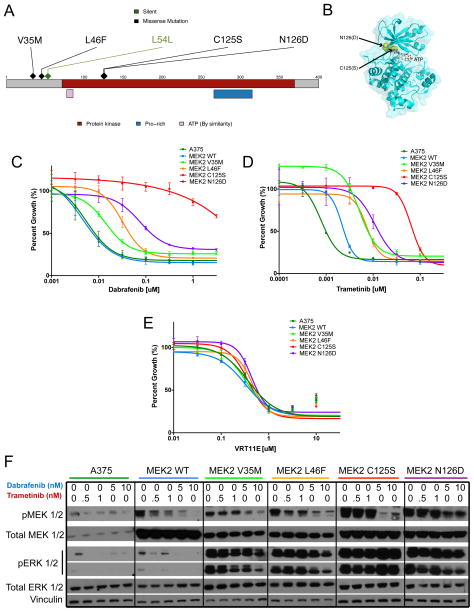
We identified four mutations involving the MAP2K2 gene (which encodes the MEK2 kinase) in drug-resistant melanoma specimens (Fig. 2A–B). Like its homologue MEK1, MEK2 is situated immediately downstream of RAF proteins in the MAPK pathway. MEK2 forms a heterodimer with MEK1 that promotes extracellular signal-related kinase (ERK) phosphorylation18. One of these mutations (MEK2C125S) is homologous to a previously described MEK1C121S mutation that confers cross-resistance to RAF and MEK inhibitors in vitro7.
Figure 2. MEK2 mutations confer resistance to RAF/MEK but not ERK inhibition.
(A) A stick plot of MAP2K2 (which encodes the MEK2 kinase); the location of putative resistance-associated mutations observed in the patient cohort are indicated. (B) The crystal structure for MEK2. The locations of somatically mutated bases are denoted in yellow; the first stretch of amino acids are missing from the MEK2 structure in PDB, so the V35M and L46F mutations cannot be shown on the structure. (C–E) Growth inhibition curves are shown for MEK2 mutants in the context of RAF (C), MEK (D), or ERK (E) inhibitors. (F) The effect of dabrafenib or trametinib on ERK1/2 phosphorylation (pERK 1/2) in wild-type A375 cells (BRAFV600E) and those expressing wildtype MEK2 (WT) or mutant constructs for MEK2. The levels of pERK1/2, total ERK1/2, pMEK1/2, MEK1/2, and vinculin are shown for A375 cells expressing novel MEK2 mutations after a 16-hour incubation at various drug concentrations as indicated.
To verify the predicted resistance phenotypes conferred by MEK2 mutations, MEK2 mutant constructs were cloned into a doxycycline-inducible vector and expressed in A375 melanoma cells – which harbor BRAFV600E mutation and are sensitive to RAF inhibition – and treated with increasing concentrations of MAP kinase pathway inhibitors. Compared to the effects of wild type MEK2, cells expressing resistance-associated MEK2 mutations were less sensitive to both RAF (dabrafenib; Fig. 2C) and MEK (trametinib; Fig. 2D) inhibition. As with the homologous and previously-reported MEK1C121S resistance mutation7, MEK2C125S conferred profound resistance to both RAF and MEK inhibition, with fold change in GI50 (half-maximal inhibitor concentration) greater than 100. The MEK2V35M, MEK2L46F, and MEK2N126D mutants also engendered resistance to RAF and MEK inhibition, although their effect were not as pronounced as those of MEK2C125S. In contrast, all resistance-associated MEK2 mutations remained sensitive to ERK inhibition using the tool compound VRT11E (Fig. 2E). All MEK2 mutant alleles examined conferred sustained MEK and ERK phosphorylation in the context of RAF inhibitor treatment (Fig. 2F).
MEK1 mutations confer resistance to RAF and MEK inhibitors when expressed inducibly
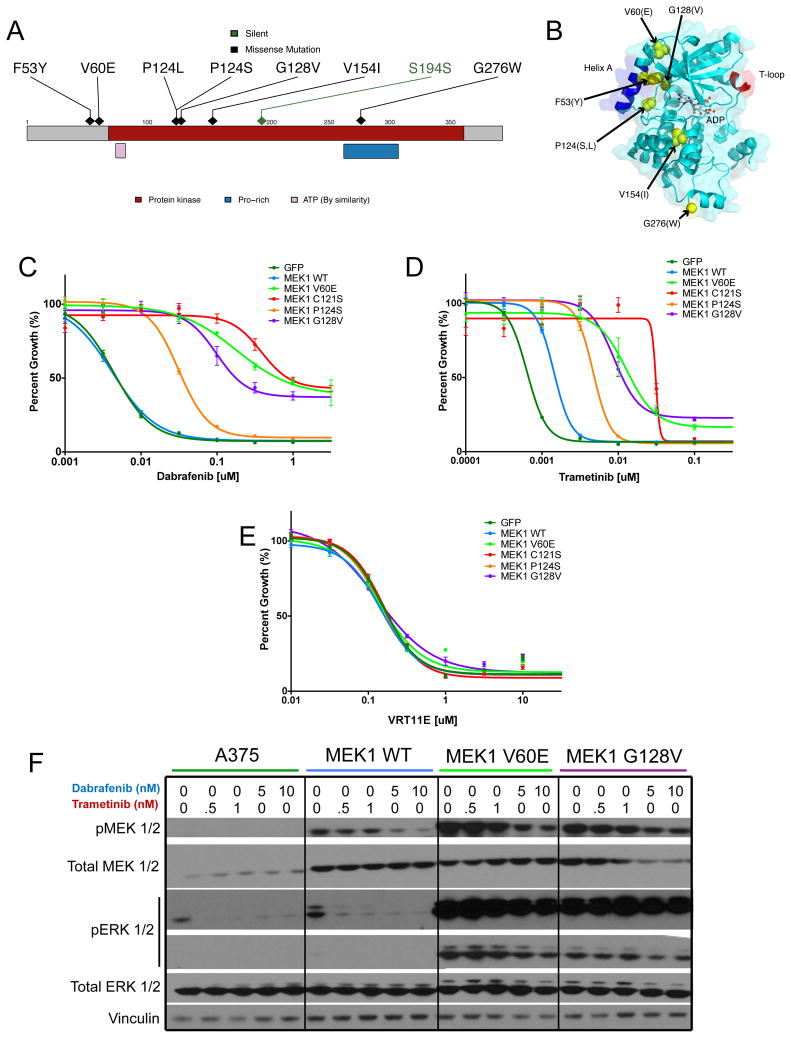
Five MAP2K1 gene mutations (encodes the MEK1 kinase) were detected in either drug-resistance specimens (3 mutations; MEK1V60E, MEK1G128V, and MEK1V154I in Patients 41, 32, and 28, respectively) or pre-treatment tumors that progressed rapidly in the face of clinical RAF inhibition (2 mutations; MEK1P124S and MEK1P124L in Patients 4 and 15, respectively) (Fig. 1B). Two additional MEK1 mutations (MEK1G276W and MEK1F53Y) occurred in pre-treatment tumor samples from patients (Patient 5 and 31) who experienced a clinical benefit from RAF inhibitors based on time on therapy (25–33 weeks; Fig. 1B). This finding was consistent with prior studies indicating that the presence of MEK1 mutations does not necessarily preclude a clinical benefit from RAF inhibitors5,19. Nonetheless, several MEK1 mutations involved residues predicted to cause RAF inhibitor resistance based on mutagenesis screens and experimental studies reported previously7 (Fig. 3A–B), including MEK1V60E and MEK1G128V. These results raised the possibility that somatic MAP2K1 mutations might promote resistance to RAF inhibition in some contexts but not others.
Figure 3. MEK1 mutations confer varying degrees of resistance to RAF/MEK but not ERK inhibition.
(A) A stick plot of MAP2K1 (which encodes the MEK1 kinase); the location of putative resistance-associated mutations observed in the patient cohort are indicated. (B) The crystal structure for MEK1. The locations of somatically mutated bases are denoted in yellow. (C-E) Growth inhibition curves are shown for MEK1 mutants in the context of RAF (C), MEK (D), or ERK (E) inhibitors. (F) The effect of dabrafenib or trametinib on ERK1/2 phosphorylation (pERK 1/2) in wild-type A375 cells (BRAFV600E) and those expressing wildtype MEK1 (WT) or mutant constructs for MEK1. The levels of pERK1/2, total ERK1/2, pMEK1/2, MEK1/2, and vinculin are shown for A375 cells expressing novel MEK2 mutations after a 16-hour incubation at various drug concentrations as indicated.
To explore this possibility, we cloned several MEK1-mutant cDNAs identified herein into a doxycycline-inducible expression vector, and expressed the mutant proteins alongside wild-type MEK1 in A375 melanoma cells. Cell growth inhibition curves were generated in the presence of dabrafenib (RAF inhibitor), trametinib (MEK inhibitor), or VRT11E (ERK inhibitor), as shown in Fig. 3 (see Methods for additional details). All MEK1 mutations examined conferred robust resistance to both RAF and MEK inhibition following doxycycline induction, with fold change in GI50 (half-maximal inhibitory concentration) of 10–80 fold for dabrafenib (Fig. 3C) and 3–20 fold for trametinib (Fig. 3D) as compared to wild-type MEK1, noting that only 50% growth inhibition was maximally achieved. Of note, doxycycline induction of wild-type MEK1 produced modest (approximately 3 fold) resistance to MEK1 inhibition but had no effect on the dabrafenib GI50 value (Figs. 3C–D). In contrast, doxycycline induction of these MEK1 mutants had no effect on BRAFV600E-mutant melanoma cell sensitivity to ERK inhibition using VRT11E (Fig. 3E). As seen with the somatic MEK2 mutations described above, doxycycline-inducible expression of MEK1 mutations resulted in elevated MEK and ERK phosphyorylation, which was sustained in the presence of drug concentrations that inhibited MAPK signaling in A375 melanoma cells that overexpressed wild-type MEK1 (Fig. 3F). Together, these results indicated that inducible expression of somatic MEK1 mutations can confer resistance to RAF/MEK inhibition, and suggested that dynamic regulation of mutant MEK1 protein may conceivably modulate clinical sensitivity to these agents in melanoma tumors.
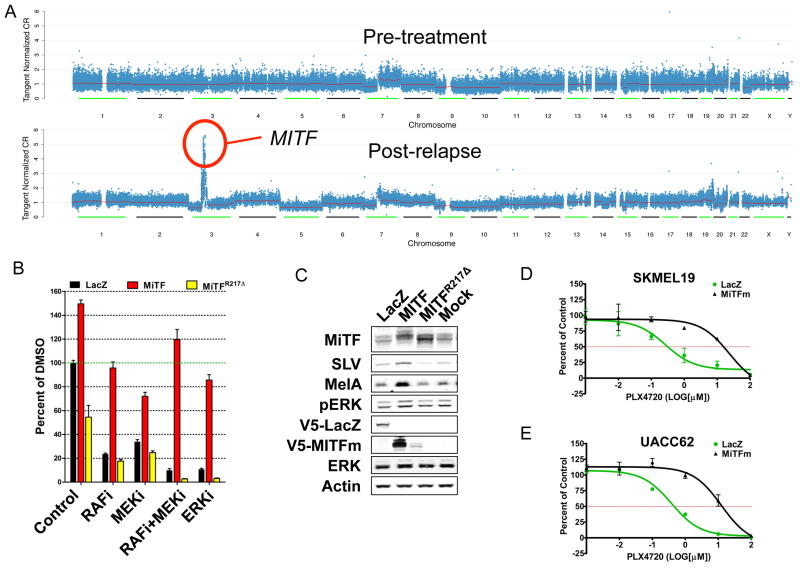
MITF amplification is associated with resistance to MAPK inhibition
In one patient whose melanoma tumor lacked a previously described genetic resistance mechanism, we identified a relapse-associated focal amplification of MITF (Fig. 4A). This gene encodes a master lineage transcription factor that governs melanocyte development, and is also an amplified oncogene within the melanocyte lineage20. While paired RNA or immunohistochemistry analysis was not possible with this clinical sample, the highly focal nature of the amplification and its specificity for the relapsed tumor suggested that this amplicon in general, and MITF in particular, might contribute to the resistance phenotype. To test this hypothesis, we overexpressed wild type MITF alongside a DNA-binding impaired MITF mutant (MITFR217Δ), or negative control (LacZ) in WM266.4 (BRAFV600E) melanoma cells. The resulting cells were cultured in the presence of RAF (PLX4720), MEK (AZD6244), or ERK (VRT11E) inhibitors at fixed concentrations shown previously to cause resistance. Indeed, forced MITF overexpression rendered these BRAFV600E melanoma cells resistance to RAF, MEK, and ERK inhibition (Fig. 4B–C).
Figure 4. Acquired MITF amplification confers resistance to MAP kinase pathway inhibition.
(A) Focal genomic amplification of MITF occurs in a post-relapse sample in the absence of additional, known resistance mechanisms. (B) Relative viability of BRAFV600E-mutant WM266.4 cells following overexpression of wild type MITF, a DNA-binding impaired MITF mutant (MITF-mR217Δ) or control (LacZ) in the presence of RAF, MEK, combined RAF/MEK, or ERK inhibitors. (C) Western blot analysis of WM266.4 expressing the constructs used in (B). Half-maximal drug response curves in BRAFV600E-mutant melanoma cell lines SKMEL19 (D) and UACC62 (E) expressing either LacZ or the melanocyte-specific isoform of MITF (MITFm) in the presence of increasing concentrations of PLX4720.
Next, we overexpressed MITF in two additional BRAFV600E-mutant melanoma cell lines (SKMEL19, UACC62) and performed cell growth inhibition studies using the RAF inhibitor tool compound (PLX4720). In both cell lines, MITF overexpression conferred a 30–80 fold increase in the PLX4720 GI50 values relative to control (LacZ) gene expression (Fig. 4D–E). Since MITF activity is regulated by MAPK signaling in melanocytes and melanoma, these results suggest that restoration of a MITF-driven transcriptional output (by genomic amplification or other means) comprises a newly recognized clinical resistance mechanism (Johannessen, et al, in press, Nature).
Intra-tumor heterogeneity of resistance mechanisms
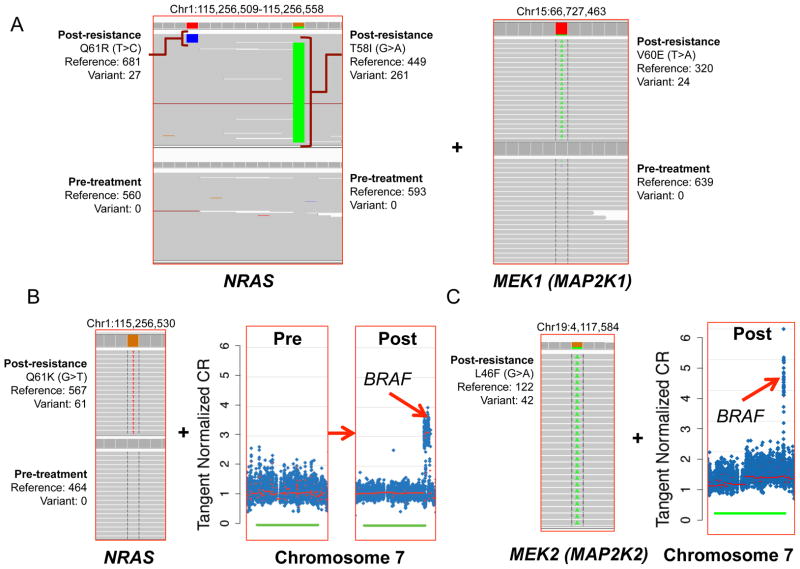
In three patients, multiple independent resistance mechanisms were evident within the same resistant tumor biopsy. For example, one tumor biopsied at progression after 18 weeks on treatment contained resistance-associated mutations in both NRAS and MEK1 (Patient 41, Fig. 5A). Moreover, two distinct somatic NRAS mutations were observed exclusively in the resistant tumor: a validated resistance alteration (Q61R), and a second alteration (T58I) that is homologous to germline KRAS alterations that cause Noonan syndrome21. These alterations occurred on mutually exclusive reads that spanned both loci, suggesting either that the two alterations occur in trans in the same resistant tumor cells or that they represent separate subclonal resistant populations harboring different NRAS mutations (Fig. 5A).
Figure 5. Intra-tumor resistance heterogeneity.
(A) Co-occuring NRAS (Integrated Genomics Viewer [IGV] compressed window) and MEK1V60E (IGV regular window) missense mutations in a relapsed tumor sample (Patient 41). NRASQ61R occurs in mutually exclusive reads compared to a neighboring NRAST58I somatic missense mutation in this patient. (B) Acquired NRASQ61K missense mutation together with BRAF amplification in the same tumor (Patient 8). (C) A MEK2L46F missense mutation coincident with BRAF amplification in a resistant tumor specimen (Patient 02 – no pre-treatment tumor sample was available in this case).
Another post-relapse tumor (Patient 08) harbored an acquired NRASQ61K missense mutation together with focal BRAF amplification (Fig. 5B). The resistant tumor from a third patient harbored both a MEK2 mutation and BRAF amplification (Patient 02, Fig. 5C). While each of these events has been shown to confer a RAF inhibitor resistance phenotype, the pretreatment tumor was unavailable for this case. Thus, we cannot formally exclude the possibility that these mutations were present prior to treatment with RAF inhibition. Nonetheless, these results further indicate that BRAF-mutant melanoma tumors may elaborate multiple resistance mechanisms simultaneously, as suggested by an earlier study involving a single case22. Given that “non-genetic” resistance mechanisms such as BRAF alternative splicing or COT overexpression could not be assessed in this cohort, these findings may vastly undersample the actual clinical incidence of intra-tumor resistance heterogeneity.
PI3K pathway activation may contribute to clinical RAF inhibitor resistance
In contrast to MAPK pathway activation, the clinical importance of PI3K pathway dysregulation as a RAF inhibitor resistance mechanism has been less clear12,23,24. Prior studies have demonstrated that PI3K pathway activation through multiple mechanisms (e.g. PTEN loss, AKT activation) is associated with RAF inhibitor resistance in some preclinical models12,25, and that patients with PTEN loss trend towards shorter progression free survival to dabrafenib26. However, as is the case with MEK1 (above), PI3K pathway alterations did not necessarily preclude clinical response in tumors characterized herein. For example, Patient 58 harbored a pre-treatment PTENK128T missense mutation but was on therapy for 18 weeks with stable disease before developing an acquired MITF amplification. Patient 1 had a PTENH93D missense mutation in the pre-treatment tumor but was on therapy for 66 weeks and achieved a partial response.
We noted that several resistant tumors contained PI3K pathway gene alterations in the absence of known MAPK resistance mechanisms (Fig. 1B, Supplementary Table S5). For example, one patient (Patient 48) whose pre-treatment tumor harbored PTENY86fs frameshift exhibited a new PIK3CAH1047R mutation in the resistant tumor biopsy (PIK3CAH1047R is a well-characterized oncogenic mutation in the catalytic PI3 kinase alpha subunit27). The resistant tumor from Patient 36 harbored a PTEN deletion (Fig. 6A), although the pre-treatment tumor was unavailable for this case. In 9 of the 11 patients with PTEN missense mutations, the mutation occurred in the phosphatase domain of the protein (Supplementary Table S5). Such mutations have been shown previously to confer a loss of function phenotype28,29. Other patients with known resistance mechanisms also harbored post-resistant somatic variants of uncertain significance in the PI3K pathway (predominantly PTEN missense mutations, but also PIK3R1 alterations) that lack experimental validation. For instance, Patient 70 was on therapy for 29 weeks and achieved a partial response before acquiring an NRASQ60H mutation and a PTENR233* nonsense mutation that is may be inactivating but requires experimental confirmation.
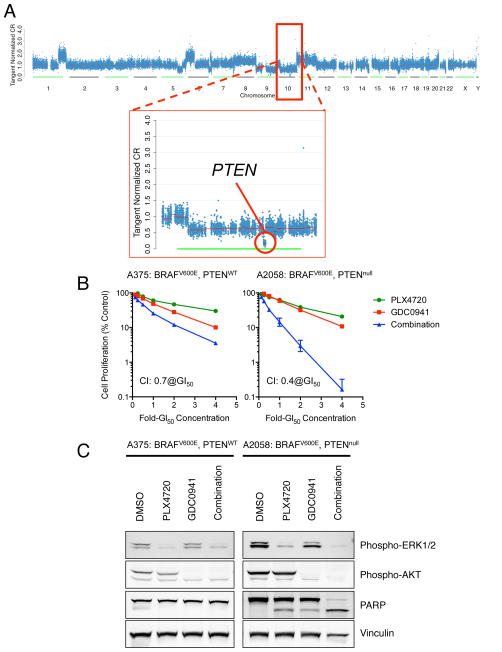
Figure 6. Combined inhibition of BRAF and PI3K can confer synergistic activity.
(A) Copy number profile from a resistant tumor sample (Patient 36 – no pre-treatment tumor sample was available in this case) demonstrates a focal homozygous deletion in chromosome 10 that includes PTEN. (B) A375 (BRAFV600E, PTENWT) and A2058 (BRAFV600E, PTENnull) cells were treated with multiples of their respective GI50s for PLX4720 or GDC0941 alone and in combination for 4 d. Cell proliferation was assessed using the CellTiter-Glo reagent. Combined BRAF and PI3K inhibition demonstrated synergistic activity in A2058 cells but to a lesser extent in A375 cells. The combination index (CI) was determined at the GI50 concentration using the Chou and Talalay method and Calcusyn (Biosoft)50. (C) A375 and A2058 cells were treated with 4xGI50 concentrations of PLX4720, GDC0941 or a combination of both inhibitors for 20 h. Cell lysates were analyzed by Western blotting for the indicated proteins.
In light of these observations, we sought to probe the possible therapeutic implications of PI3K pathway alterations in BRAF-mutant melanoma. Accordingly, we treated BRAF-mutant melanoma cells that were either wild type or mutant for PTEN with both a RAF inhibitor and a PI3 kinase inhibitor in vitro. Combined MAPK (PLX4720) and PI3K (GDC0941) inhibition had a modest but discernible impact on cell proliferation in BRAFV600E/PTENWT cells (A375), but in the BRAFV600E/PTENnull cell line (A2058) the combination has a marked effect on cell proliferation and apoptosis (Figure 6B–C), consistent with prior reports23,30–32. Despite these results (which are limited to only one cell line per condition), the cellular contexts in which PI3K pathway activation may promote cell viability in the setting of RAF inhibition remains unclear5,9,10. Some in vitro studies argue against a strong proliferative effect (Johannessen, et al, in press, Nature), therefore implying that PI3K pathway dysregulation may augment cell survival in some contexts and allow such tumor cells the opportunity to elaborate additional (non-genetic) adaptive resistance mechanisms. This model is consistent with the observation that many, though not all, tumors with resistance-associated PI3K pathway mutations lack genetic resistance mechanisms linked to MAPK pathway activation (Fig. 1B).
Genetic correlates of early resistance
Among the 14 patients exhibiting early disease progression in the setting of RAF inhibition (disease progression within 12 weeks of initial RAF inhibitor therapy), three pre-treatment tumor biopsies harbored RAC1P29S mutations. Patients 26 and 46 had transient stable disease, and Patient 34 had immediate progressive disease. RAC1P29S was previously identified as a gain-of-function oncogene mutation in melanoma33,34. No patients who demonstrated a sustained response to therapy (≥ 12 weeks on therapy) exhibited this mutation (P = 0.026) (Fig. 1B). Conceivably, the RAC1P29S mutations may mark a tumor cell population that is partially resistant to single-agent RAF inhibition, although experimental evidence in support of this notion is necessary.
Finally, a nonsense mutation in the HOXD8 gene was observed in a single resistant tumor from a patient with early resistance (Fig. 1B). HOXD8 is a homeobox transcription factor35 that has been shown to be dysregulated in multiple cancers36,37, although it has not previously been associated with melanoma, nor is there a known functional relationship between this gene and RAF signaling. Interestingly, HOXD8 suppression was implicated in RAF inhibitor resistance following a genome-scale RNA interference suppressor screen reported previously9. The presence of a HOXD8 nonsense mutation in the absence of other known resistance associated alterations raises the possibility that inactivation of this homeobox transcription factor may represent another new resistance mechanism. However, this gene has not previously been interrogated in drug-resistant clinical specimens.
DISCUSSION
Whole-exome sequencing of BRAFV600-mutant melanoma tumors obtained prior to treatment with selective class I RAF inhibitors and following resistance has revealed a landscape of putative clinical resistance gene mutations linked to this therapy. This study utilized clinical FFPE tumor samples rather than research-grade frozen tissue. The regular use of FFPE material for massively parallel sequencing studies should greatly expand the specimens available for systematic studies of response and resistance to anticancer agents. In addition to known alterations, we found multiple new gene mutations that may implicate novel resistance mechanisms. Consequently, this work has extended knowledge of potential genetic avenues through which BRAFV600-mutant melanomas can achieve resistance to RAF inhibition.
This study was performed with patient samples longitudinally obtained from a nationwide clinical consortium (the Dermatologic Cooperative Oncology Group of Germany [DeCOG]). Whole exome sequencing was completed using genomic DNA obtained from archival (e.g. formalin-fixed, paraffin-embedded) tumor material. This study therefore demonstrates the feasibility of comprehensive clinical resistance studies that unite the potentially expansive clinical access of consortium groups with the production capabilities of a large-scale genome center. Such efforts may guide future cancer genomics studies that incorporate multiple biopsies obtained from patients for clinical purposes to effectively study therapeutic resistance and other salient clinical questions. The use of FFPE tumor samples for whole exome sequencing did not limit this study; indeed, sequencing metrics for this cohort met or exceeded standards that have been employed for The Cancer Genome Atlas (TCGA) and other cancer genome projects of similar scale. This technical success may therefore provide a framework for many new clinical studies that allow insights into the molecular basis of therapeutic response and resistance in cancer.
Computational analysis of the tumor genomic data in this study was enhanced by integration with available systematic functional screening data to prioritize candidate RAF inhibitor resistance-associated genes. Since the pattern or resistance genes may approximate a “long tail” distribution (similar to the distribution of cancer genes38), many nominated resistance-associated genes may not reach statistical significance without exceedingly large clinical cohorts, as was the case with this study. Genome-wide resistance screens that comprehensively assess for genes that, through loss or gain, promote resistance in vitro promote new resistance-associated genes, but in the absence of such events observed in patients may lack clinical validation. To better assess the functional significance of the genomics data generated for this study, we integrated it with existing and emerging preclinical biological studies that provide a biological foundation the potential importance of the alterations we observed in the clinical resistance setting. By analyzing genomics data in the context of genome-wide resistance screens, rare resistance-associated events that have clear functional roles may be characterized for future studies. Generally, the convergence of in vitro genome-wide resistance screens with in vivo comprehensive genomic profiling may enrich for robust resistance mechanisms, which demonstrates a potential model for resistance studies involving targeted therapies across tumor types.
The majority of resistance alterations involve reactivation of the MAPK pathway. Experimental assessment of a subset of resistance mutations further stratified these events into discrete categories that may inform future therapeutic or clinical trial options. Several MEK1 and MEK2 resistance mutations confer cross-resistance to MEK inhibitor therapy and raise the possibility that combined RAF/MEK inhibition may also have limited efficacy in this patient subpopulation (Wagle et al., co-submission). In contrast, NRAS mutations and BRAF amplifications may still prove responsive to subsequent MEK inhibitor-based regimens, although the existing clinical data suggests that patients who progress following single-agent RAF inhibition are less likely to benefit from MEK inhibitors39. Most of these mutations remain sensitive to ERK inhibitors in vitro, suggesting that such agents40 may represent a high-priority avenue for new targeted therapeutic development.
Alterations in genes encoding transcription factors may also implicate a rare category of clinical resistance mechanisms that involves restoration (MITF amplification) of an oncogenic transcriptional output downstream of the MAPK pathway. Unlike signaling based resistance, transcriptional effectors may confer cross-resistance to all MAPK pathway inhibitors (including ERK inhibitors) and therefore require alternative therapeutic approaches. HOXD8, another transcriptional effector that emerged in an RNAi screen and was subsequently observed to be mutated in a patient with early resistance, may also contribute to RAF inhibitor resistance, although further functional studies are necessary to confirm this model. Moreover, the identification of alterations that activate PI3K signaling suggests that novel combinations of MAPK and PI3K pathway inhibitors may merit clinical evaluation in BRAF-mutant melanoma.
Comprehensive cancer genome characterization studies have revealed a “long tail” of mutated genes that drive carcinogenesis38,41. The present study suggests that a similar long tail of genetic effectors may mediate resistance to RAF inhibitors. Additionally, multiple resistance mechanisms may arise within the same tumor, and the mechanisms observed in this study differ from those reported in case report form22. As noted earlier, given we could not interrogate non-somatic mechanisms of resistance, such as BRAF alternative splicing, this result likely underestimates the true incidence of intra-tumor resistance heterogeneity observed clinically. Therapeutically, this finding implies that multiple pathways may need to be targeted either in parallel if not limited by toxicity, or in series as part of an intermittent dosing schedule42.
Although informative, this study is limited in several aspects. For example, multiple known resistance mechanisms cannot be identified by whole exome sequencing (including BRAF alternative splicing, COT up-regulation, and ligand/receptor tyrosine kinase overexpression); thus, this study undoubtedly underestimates the true prevalence of molecular changes that drive clinical resistance to RAF inhibition. Also, since serial biopsies cannot always be performed on the same tumor focus, it may be difficult to discern resistance-associated changes between pre-treatment and resistant tumor samples given pre-existing tumor heterogeneity. The advent of systematic approaches to functional evaluation of candidate resistance effectors may help address such challenges, which are inherent to many genomics studies using clinical samples.
Moreover, some genetic alterations observed in this cohort (e.g. RAC1, HOXD8) require further experimental validation and assessment in large clinical cohorts to determine their specific impact on mediating RAF inhibitor resistance. It also seems possible that, in certain settings, dysregulation of the PI3K pathway may help improve tumor cell survival during treatment with these agents until a proliferative resistance allele emerges, although experimental validation of many PI3K variants observed in this cohort is necessary.
In conclusion, this work demonstrates the promise of serial tumor biopsies coupled with systematic genetic characterization to reveal a wide spectrum of clinical resistance mechanisms. Widespread clinical application of similar approaches may illuminate new biological and therapeutic insights across many cancer types and inform clinical trial design. Furthermore, these findings underscore the future importance of novel therapeutic combinations (including both targeted agents and immunotherapy) to achieve more durable control of metastatic melanoma and other advanced cancers.
METHODS
Patients and Tumor Specimens
Biopsy samples were secured from 45 patients with metastatic melanoma under an IRB-approved protocol to obtain research biopsies before and after resistance developed to systemic therapy (University Hospital Essen 12-4961-BO). All patients provided written informed consent. The clinical characteristics of these patients are described in Table 1. Clinical responses to therapy were determined using RECIST criteria. Pre-treatment biopsies were obtained prior to starting therapy, and resistant biopsy samples were obtained upon discontinuation of vemurafenib or dabrafenib at disease progression (Fig. 1A). For patients where complete “trios” (germline sample, pre-treatment tumor, post-progression tumor) were not available (n = 13), pre-treatment biopsy results were included if the patient experienced rapid disease progression, and resistant biopsy results were included if an objective clinical response was achieved. Results from one responding patient (patient 65) were included to inform analyses related to predictors of early resistance, although this patient did not have a matched resistant tumor sample for analysis. Additional details regarding the patient cohort can be found in Supplementary Table S1.
FFPE DNA Extraction
After fixation and mounting, 5–10 10μm slices from formalin-fixed, paraffin-embedded (FFPE) tumor block were obtained and tumor-enriched tissue was macrodissected. Germline DNA was obtained from adjacent normal tissue or peripheral blood mononuclear cells. Paraffin was removed from FFPE sections and cores using CitriSolv™ (Fisher Scientific) followed by ethanol washes, then tissue was lysed overnight at 56°C. Samples were then incubated at 90°C to remove DNA crosslinks, and extraction was performed using Qiagen QiaAmp DNA Mini Kit (#51306).
Whole Exome Sequencing
We performed whole exome sequencing on the extracted DNA using the Illumina HiSeq platform43 (Van Allen, Wagle, et al, in press, Nature Medicine):
Library Construction
DNA libraries for massively parallel sequencing were generated as previously described43 with the following modifications: the initial genomic DNA input into the shearing step was reduced from 3μg to 10–100ng in 50μL of solution. For adapter ligation, Illumina paired-end adapters were replaced with palindromic forked adapters (purchased from Integrated DNA Technologies) with unique 8 base index molecular barcode sequences included in the adapter sequence to facilitate downstream pooling. With the exception of the palindromic forked adapters, all reagents used for end repair, A-base addition, adapter ligation, and library enrichment PCR were purchased from KAPA Biosciences in 96-reaction kits. In addition, during the post-enrichment solid phase reversible immobilization (SPRI) bead cleanup, elution volume was reduced to 20μL to maximize library concentration, and a vortexing step was added to maximize the amount of template eluted from the beads. Libraries with concentrations above 40ng/μl, as measured by a PicoGreen assay automated on an Agilent Bravo instrument, were considered acceptable for hybrid selection and sequencing.
Solution-phase hybrid selection
The exon capture procedure was performed as previously described43 with the following modifications: prior to hybridization, any libraries with concentrations >60ng/μL (as determined by PicoGreen) were brought to 60ng/μL, and 8.3μL of library was combined with blocking agent, bait, and hybridization buffer. Libraries with concentrations between 50 and 60ng/μL were normalized to 50ng/μL, and 10.3μL of library was combined with blocking agent, bait, and hybridization buffer. Libraries with concentrations between 40 and 50ng/μL were normalized to 40ng/μL, and 12.3μL of library was combined with blocking agent, bait, and hybridization buffer. Finally, the hybridization reaction was reduced to 17 hours, with no changes to the downstream capture protocol.
Preparation of libraries for cluster amplification and sequencing
After post-capture enrichment, libraries were quantified using PicoGreen, normalized to equal concentration using a Perkin Elmer MiniJanus instrument, and pooled by equal volume on the Agilent Bravo platform. Library pools were then quantified using quantitative PCR (KAPA Biosystems) with probes specific to the ends of the adapters; this assay was automated using Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries were brought to 2nM and denatured using 0.2 N NaOH on the Perkin-Elmer MiniJanus. After denaturation, libraries were diluted to 20pM using hybridization buffer purchased from Illumina.
Cluster amplification and sequencing
Cluster amplification of denatured templates was performed according to the manufacturer’s protocol (Illumina). HiSeq v3 cluster chemistry and flowcells, as well as Illumina’s Multiplexing Sequencing Primer Kit. DNAs were added to flowcells and sequenced using the HiSeq 2000 v3 Sequencing-by-Synthesis method, then analyzed using RTA v.1.12.4.2 or later. Each pool of whole exome libraries was subjected to paired 76bp runs. An 8-base index sequencing read was performed to read molecular indices, across the number of lanes needed to meet coverage for all libraries in the pool.
Sequencing Analysis and Interpretation
Sequence data processing
Exome sequence data processing was performed using established analytical pipelines at the Broad Institute. A BAM file was produced with the Picard pipeline (http://picard.sourceforge.net/), which aligns the tumor and normal sequences to the hg19 human genome build using Illumina sequencing reads. The BAM was uploaded into the Firehose pipeline (http://www.broadinstitute.org/cancer/cga/Firehose), which manages input and output files to be executed by GenePattern44.
Sequencing quality control
Quality control modules within Firehose were applied to all sequencing data for comparison of the origin for tumor and normal genotypes and to assess fingerprinting concordance. Cross-contamination of samples was estimated using ContEst45. Where 5–15% contamination was observed (n = 2), SNP fingerprints from each lane of a tumor/normal pair were cross-checked to confirm concordance; those without global concordance were excluded from subsequent analysis.
Somatic alterations
MuTect46 was applied to identify somatic single-nucleotide variants. Indelocator (http://www.broadinstitute.org/cancer/cga/indelocator) was applied to identify small insertions or deletions. Artifacts introduced by DNA oxidation during sequencing were computationally removed using a filter-based method47. Annotation of identified variants was done using Oncotator (http://www.broadinstitute.org/cancer/cga/oncotator). Copy ratios were calculated for each captured target by dividing the tumor coverage by the median coverage obtained in a set of reference normal samples. The resulting copy ratios were segmented using the circular binary segmentation algorithm48. Genes in copy ratio regions with segment means of greater than log2(4) were evaluated for focal amplifications, and genes in regions with segment means of less than log2(0.5) were evaluated for deletions. For cases where we had pre-treatment and post-resistance tumors (n = 32), alterations that we present in the post-resistance tumor only were selected; in other cases we included all non-synonymous alterations (Supplementary Table S2–3). We then integrated this data with preclinical functional models of resistance and existing literature on melanoma oncogenesis7,9,33,34,49. The analyses were performed using the R statistical software. All somatic mutations and short insertion/deletions are provided in Supplementary Table S4.
Experimental Analysis
Physical and biologic containment procedures for recombinant DNA followed institutional protocols in accordance with the National Institutes of Health Guidelines for Research Involving Recombinant DNA Molecules.
Plasmids and site-directed mutagenesis
The tet-inducible construct, pCW57.1, was a generous gift of Dr. David Root and Dr. John Doench (The Broad Institute of Harvard and MIT). The V5-tagged tet-inducible construct, pLIX_403, was obtained from addgene (41395). MEK1 cDNA was described previously7 and MEK2 cDNA was obtained from addgene (23555). MITF (m isoform) was previously described20. Site-directed mutagenesis was performed in the pDONR (Invitrogen) construct using Quick-Change II (Stratagene) according to the manufacturer’s instructions. MEK cDNA was then transferred from pDONR to pCW57.1 or pLIX_403 using LR Clonase II (Invitrogen). Arginine 217 of MITF-m36 was deleted using the QuikChange Lightning Mutagenesis Kit (Agilent), performed in pDonor223 (Invitrogen). MITF-mR217Δ was transferred into pLX304 using LR Clonase (Invitrogen) per manufacturer’s recommendation.
Viral infections
293T cells (70% confluent) were transfected with expression vectors (pCW57.1 MEK1 (tet-inducible), pLIX_403 MEK2 (V5 tagged, tet-inducible), or pLX_304 MIF-m (V5 tagged), together with packaging vectors Δ8.91 and VSVG, using X-treme Gene 9 (Roche). Viral supernatants were passed through a 0.45μm syringe. A375 cells were infected for 16 hours with virus in the presence of polybrene (4 ug/ml, Sigma); puromycin was introduced 48 hours post infection to create stable cell lines. For MITF expression in WM266.4, cells were infected at a 1:10–1:20 dilution of virus in 6-well plates (2.0 × 105 cells/well, for immunoblot assays) or 96-well plates (3.0 × 103, for cell growth assays) in the presence of 5.5 μg/ml polybrene and centrifuged at 2250 RPM for 60 min. at 373 C followed immediately by removal of media and replacement with complete growth media.
Cell lines
A375, SKMEL28, A2058 and WM266.4 cells were purchased from ATCC and were maintained in DMEM or RPMI-1640 with 10% heat inactivated FBS. A375 cells stable expressing tet-inducible MEK1 (pCW57.1 MEK1) or tet-inducible MEK2-V5 (pLIX_403 MEK2) were maintained in 10% tet-approved FBS (Clontech) and 2 μg/mL puromycin. Cell lines obtained from ATCC, which verify identity by short tandem repeat profiling, were passaged less than 6 months following receipt.
In vitro pharmacologic growth inhibition assays
Dabrafenib (RAF inhibitor), trametinib (MEK inhibitor) and VRT11E (ERK inhibitor) were purchased from ChemieTek. PLX4720 (RAF inhibitor), GDC0941 (PI3K inhibitor) and selumetinib (MEK inhibitor) were purchased from Selleck Chemicals. For growth inhibition analysis, A375 cells expressing MEK1 or MEK2 WT and mutants were seeded in 96-well plates (2000 cells/well for A375 cells) and allowed to adhere for 16 hours. Afterwards, media containing serial dilutions of inhibitor were added, ensuring that the final volume of DMSO did not exceed 0.1%. Doxycycline (1μg/mL) was added to the media at the time of drug treatment to induce the protein expression. Cells were incubated for 72 hours in the presence of drug, and viability was measured by the CellTiter96 AQueous assay (Promega). For shoter-term culture growth inhibition analysis, cells were seeded in 96-well plates and treated the following day with serial dilutions of inhibitor. Cells were incubated for 72 hours in the presence of drug, and viability was measured by the CellTiter96 Aqueous assay (Promega). WM266.4 engineered to express MITF were seeded into 96-well, white-walled, clear bottom plates. Seventy-two hours after viral infection, dilutions of the relevant compound were prepared in DMSO to 1000x stocks. Drug stocks were then diluted 1:100 into appropriate growth media and added to cells at a dilution of 1:10(1x final). Cell viability was measured using CellTiterGlo viability assay (Promega) at a dilution of 1:6. Viability was calculated as a percentage of the control (untreated cells) after background subtraction. Six replicates were performed in each cell line and drug combination experiment, and the entire experiment was also repeated three times. Data from the pharmacologic growth-inhibition assays were modeled using a nonlinear regression curve fit with a sigmoidal dose response. These curves were displayed using GraphPad Prism 5 (GraphPad Software).
Western blot analysis
Immunoblot studies were performed using standard procedures. Briefly, melanoma cells were lysed with RIPA or 1% NP40 lysis buffer containing both protease and phosphatase inhibitor cocktails (Roche). Lysates were quantified (Bradford assay), denatured (95°C), and resolved by SDS gel electrophoresis. Protein was transferred to PVDF or nitrocellulose membranes and probed with primary antibodies recognizing p-ERK1/2, p-MEK1/2 (Ser217/221), MEK1/2, pan-AKT, pAKT Ser473, PARP, actin and α-tubulin (Cell Signaling Technology; 1:1000 dilution) or MITF (NeoMarkers, C5), Silver, pERK (Sigma), V5 (Invitrogen), ERK2 and Melan-A (Santa Cruz). After incubation with the appropriate secondary antibody (anti-rabbit or anti-mouse IgG, HRP-linked; 1:1000 dilution) (Cell Signaling Technology), proteins were detected using chemiluminescence (Pierce).
Statistical analysis
Analyses of clinical parameters were performed using the R statistical package. Significance between two means was calculated with the two-tailed Mann-Whitney test. The two-tailed Fisher’s exact test was used to test the statistical significance of the contingency table represented by NRAS or RAC1 mutation status in early resistance and responding cohorts. P < 0.05 was considered significant.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
The use of RAF inhibitors for BRAFV600-mutant metastatic melanoma improves patient outcomes, but most patients demonstrate early or acquired resistance to this targeted therapy. We reveal the genetic landscape of clinical resistance mechanisms to RAF inhibitors from patients using whole exome sequencing, and experimentally assess new observed mechanisms to define potential subsequent treatment strategies.
Acknowledgments
We thank the patients that were enrolled in this study. We also thank the funding sources that support this effort, which include the National Cancer Institute (Award #P01 CA163222 01A1, P50 CA 93683; L.A.G.), National Human Genome Research Institute (Award #5U54HG003067-11; L.A.G., G.G., S.A.G.), the Melanoma Research Alliance (L.A.G.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (L.A.G.), the Harvard Clinical and Translational Science Center NIH Training Grant Award (Award #UL1 RR 025758; C.S.P.), the National Institute of General Medical Sciences Grant T32GM07753 (E.H.), the Conquer Cancer Foundation (E.M.V. and N.W.), the Dana-Farber Leadership Council (E.M.V.), and the American Cancer Society (E.M.V.).
Footnotes
CONFLICTS OF INTEREST
Dr. Garraway and Dr. Wagle are equity holders in and consultants to Foundation Medicine. Dr. Garraway is a consultant to Novartis, Millenium/Takeda, and Boehringer Ingelheim, and a recipient of a grant from Novartis. Dr. Schadendorf is a consultant for GlaxoSmithKline, Bristol-Myers Squibb, Merck, Amgen, Novartis, and Roche, and a recipient of grant funding from Merck.
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. The New England journal of medicine. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:1767–74. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 6.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer discovery. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer discovery. 2013;3:158–67. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer research. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalanotti F, Reyes G, Jesenberger V, Galabova-Kovacs G, de Matos Simoes R, Carugo O, et al. A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nature structural & molecular biology. 2009;16:294–303. doi: 10.1038/nsmb.1564. [DOI] [PubMed] [Google Scholar]
- 19.Shi H, Moriceau G, Kong X, Koya RC, Nazarian R, Pupo GM, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer discovery. 2012;2:414–24. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 21.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, et al. Germline KRAS mutations cause Noonan syndrome. Nature genetics. 2006;38:331–6. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 22.Romano E, Pradervand S, Paillusson A, Weber J, Harshman K, Muehlethaler K, et al. Identification of Multiple Mechanisms of Resistance to Vemurafenib in a Patient with BRAFV600E-Mutated Cutaneous Melanoma Successfully Rechallenged after Progression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013 doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]
- 23.Deng W, Gopal YN, Scott A, Chen G, Woodman SE, Davies MA. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment cell & melanoma research. 2012;25:248–58. doi: 10.1111/j.1755-148X.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 24.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, et al. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Molecular cancer therapeutics. 2012;11:909–20. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 25.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31:446–57. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nathanson KL, Martin AM, Wubbenhorst B, Greshock J, Letrero R, D’Andrea K, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor Dabrafenib (GSK2118436) Clinical cancer research: an official journal of the American Association for Cancer Research. 2013 doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer research. 2013;73:276–84. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Escudero I, Oliver MD, Andres-Pons A, Molina M, Cid VJ, Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Human molecular genetics. 2011;20:4132–42. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 29.Han SY, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer research. 2000;60:3147–51. [PubMed] [Google Scholar]
- 30.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer research. 2010;70:8736–47. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoeflich KP, Herter S, Tien J, Wong L, Berry L, Chan J, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer research. 2009;69:3042–51. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 32.Hoeflich KP, Merchant M, Orr C, Chan J, Den Otter D, Berry L, et al. Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer research. 2012;72:210–9. doi: 10.1158/0008-5472.CAN-11-1515. [DOI] [PubMed] [Google Scholar]
- 33.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nature genetics. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spitz F, Gonzalez F, Peichel C, Vogt TF, Duboule D, Zakany J. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes & development. 2001;15:2209–14. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinawi T, Hill VK, Krex D, Schackert G, Gentle D, Morris MR, et al. DNA methylation profiles of long- and short-term glioblastoma survivors. Epigenetics: official journal of the DNA Methylation Society. 2013;8:149–56. doi: 10.4161/epi.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116:1025–34. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Kefford R, Miller WH, Jr, Tan D, Sullivan RJ, Long GV, Dienstmann R, et al. Preliminary results from a phase Ib/II, open-label, dose-escalation study of the oral BRAF inhibitor LGV818 in combination with the oral MEK1/2 inhibitor MEK162 in BRAF V600-dependent advanced solid tumors. J Clin Oncol. 2013;31:2013. (suppl; abstr 9029) [Google Scholar]
- 40.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 2013;3:742–50. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 41.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome biology. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2. 0. Nature genetics. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 45.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013 doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic acids research. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 49.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.