Abstract
The rs1344706, an intronic SNP within the zinc-finger protein 804A gene (ZNF804A), was identified as one of the most compelling risk SNPs for schizophrenia (SZ) and bipolar disorder (BD). It is however not clear by which molecular mechanisms ZNF804A increases disease risk. We evaluated the role of ZNF804A in SZ and BD by genotyping the originally associated rs1344706 SNP and an exonic SNP (rs12476147) located in exon four of ZNF804A in a sample of 428 SZ, 385 BD, and 578 controls from the isolated population of the Costa Rica Central Valley. We also investigated the rs1344706 SNP for allelic specific expression (ASE) imbalance in the dorsolateral prefrontal cortex (DLPFC) of 46 heterozygous postmortem brains.
While no significant association between rs1344706 and SZ or BD was observed in the Costa Rica sample, we observed an increased risk of SZ for the minor allele (A) of the exonic rs12476147 SNP (p =0.026). Our ASE assay detected a significant over-expression of the rs12476147 A allele in DLPFC of rs1344706 heterozygous subjects. Interestingly, cDNA allele ratios were significantly different according to the intronic rs1344706 genotypes (p-value = 0.03), with the rs1344706 A allele associated with increased ZNF804A rs12476147 A allele expression (average 1.06, p-value = 0.02, for heterozygous subjects vs. genomic DNA).
In conclusion, we have demonstrated a significant association of rs12476147 with SZ, and using a powerful within-subjects design, an allelic expression imbalance of ZNF804A exonic SNP rs12476147 in the DLPFC. Although this data does not preclude the possibility of other functional variants in ZNF804A, it provides evidence that the rs1344706 SZ risk allele is the cis-regulatory variant directly responsible for this allelic expression imbalance in adult cortex.
Keywords: schizophrenia, bipolar disorder, ZNF804A, association study, rs1344706, allelic-specific expression
1. Introduction
The zinc finger protein 804A (ZNF804A) gene represents a robustly replicated locus for schizophrenia (SZ). The rs1344706 SNP, located in intron 2 of ZNF804A, was the first risk variant to achieve genome-wide significance for psychosis (O'Donovan et al., 2008). Since then ZNF804A has been extensively investigated, and the association of rs1344706 has been further confirmed by a number of independent replications studies in several European populations (Riley et al., 2010; Steinberg et al., 2011; Zaharie et al., 2012; Zhang et al., 2011a; Zhang et al., 2012). Furthermore, a meta-analysis of more than 55,000 subjects (Williams et al., 2011) found genome-wide significant association of rs1344706 with SZ (p = 2.5×10–11), and the result was strengthened when bipolar disorder (BD) samples were added into the meta-analysis (p = 4.1×10–13) even though the effect size remained relatively small (odds ratio = 1.10, 95% confidence interval = 1.07–1.14). This intronic SNP remained the most strongly associated marker in the gene, even after a fine-scale linkage disequilibrium (LD) mapping (Williams et al., 2011), and extensive re-sequencing of ZNF804A showed no evidence for a role of moderately rare non-synonymous coding variants in the association of ZNF804A with SZ (Williams et al., 2011).
Attempts to replicate this association using Asian samples have been mixed. Significant associations of rs1344706 with SZ risk were observed in Han Chinese and Indonesian samples (Schwab et al., 2013; Zhang et al., 2011b; Zhang et al., 2012). However, GWAS replication studies (Li et al., 2011; Yue et al., 2011) and recent meta-analyses (Li et al., 2012; Li et al., 2013) have shown that rs1344706 is not a risk SNP for SZ in the Chinese population, suggesting potential genetic heterogeneity of SZ susceptibility at this locus. Moreover, the rs4380187 SNP near ZNF804A almost met genome-wide significance (p = 5.66 × 10–8) in a recently published large-scale collaborative GWAS (Ripke et al., 2013).
Although the evidence for association of ZNF804A with SZ remains compelling, the function of the gene is still unknown. The gene is known to be expressed in the brain (Hawrylycz et al., 2012), and is predicted to encode a 1210 amino acids protein, containing a C2H2-type motif. This DNA-binding domain is characteristic of the zinc-finger protein family, thus suggesting a role of ZNF804A in the regulation of gene expression in brain (Donohoe et al., 2010). Moreover, the molecular mechanisms by which rs1344706 increases disease risk have yet to be determined. The SZ associated A allele of rs1344706 appears to correlate with reduced binding affinity for unidentified nuclear protein(s) in neural cells (Hill and Bray, 2011). The A-risk allele has been associated with a higher ZNF804A allelic expression in adult human brain, but does not seem to be the expression quantitative trait loci (eQTL) directly responsible. The association might be through additional cis-regulatory variants that are in high LD with the expressed SNPs rs4667001 or rs12476147 (Hill and Bray, 2012; Williams et al., 2011).
We investigated the role of ZNF804A plays in SZ and BD by: i) genotyping the rs1344706 and rs12476147 SNPs in a sample of 428 SZ, 385 BD, and 578 controls from an isolated population from Costa Rica (DeLisi et al., 2001; Morera et al., 2003); and ii) analyzing the rs1344706 allelic specific expression (ASE) in the postmortem dorsolateral prefrontal cortex (DLPFC).
2. Materials and methods
2.1 Subjects and DNA samples
The association study cohort is composed of Costa Rican subjects: i) 3 extended families with SZ and BD (N = 107); ii) unrelated individuals with diagnoses of SZ (n = 390) and BD (n = 368); and iv) unrelated controls (N = 507) with no family history of SZ, BD, suicide or hospitalization for psychiatric reasons, with no self-reports of suicide attempts, psychosis, diagnosis of SZ, BD, or use of medications for depression or psychiatric conditions. A total of 1372 subjects participated, including 422 SZ (44.1% male), 382 BD (42.1% male) and 507 control subjects (56.9% males). All subjects were ascertained in the Costa Rica Central Valley (CRCV) by a psychiatrist, and only subjects whose four grandparents were of Spanish descent were included in this cohort. At the time of blood sampling, for all study participants structured interviews, summaries of hospital records, and family history information was obtained by a trained interviewer. Details about enrolment criteria and clinical definitions have been described elsewhere (Moon et al., 2011). The study was approved by the Ministry of Health of Costa Rica and the ethics committee for the Hospital Nacional Psiquiatrico and by the Institutional Review Board at the University of California at Irvine. Written informed consent was obtained from all subjects.
Blood samples were collected from participants in Costa Rica and sent to the laboratory at UC Irvine. Genomic DNA was purified from venous blood by using the salting-out method. DNA samples were quantified using the SpectraMax Plus spectrophotometer (Molecular Devices, Sunnyvale, CA), standardized for concentration, and arrayed into 96-deep-well plates.
2.2 Genotyping
DNA samples were genotyped for ZNF804A SNPs rs12476147 by a pre-validated TaqMan assay (Applied Biosystems, Foster City, CA) and for rs1344706 using an in-house developed allele-specific PCR assay. Both assays were validated in house by Sanger sequencing of random subjects.
For rs12476147 genotyping PCR reactions were performed on a 7900HT real-time sequence detection system using the TaqMan Genotyping Master Mix and the TaqMan SNP genotyping assay (Applied Biosystems). Optimal conditions were as follows: Step 1, 95°C for 10 min; Step 2, 92°C for 15 s, 63°C for 60 s; repeated for 45 cycles. The genotype of each sample was determined by measuring allelic-specific fluorescence using SDS 2.3 software for allelic discrimination (Applied Biosystems). PCR reactions for rs1344706 genotyping were carried out using the LightCycler 480 HRM Master Mix (Roche Applied Science, Indianapolis, IN, USA) and a common forward primer (5′-ggattgggacgaggagaaa-3′) and allele-specific reverse primers (5′-aaacactgaaacaaagaatcaaaaac[T/G]-3′). PCR cycling was performed on a LightCycler 480 (Roche), under the following thermal conditions: Step 1, 95°C for 10 min; Step 2, 95°C for 10 s, 48°C for 15 s, 72°C for 20 s; repeated for 45 cycles. After amplification, samples were kept at 97°C for 1 min and 40°C for 1 min, then melting curves were generated by ramping from 65 to 95°C at 0.02°C/s with continuous fluorescence acquisition. The genotype of each sample was determined by measuring allelic-specific fluorescence using the LightCycler® Software Version 1.5 (Roche).
2.3 Statistical analysis
All procedures were performed using the R program (http://www.r-project.org/) and the software package PLINK v1.07 {Purcell, 2007 #46}. Deviations from Hardy-Weinberg equilibrium (HWE) were tested using the exact test {Wigginton, 2005 #45} implemented in PLINK software. For each SNP, a DFAM (family-based association for disease traits) analysis was performed. This procedure implements the sib-TDT and also allows for unrelated individuals to be included (via a clustered-analysis using the Cochran-Mantel-Haesnzel); asymptotic p-values were provided for minor alleles, and presented as non-corrected. The level of statistical significance was set at 5%. Statistical power analysis was examined using software using the genetic power calculator (Purcell et al., 2003) (http://pngu.mgh.harvard.edu/~purcell/gpc/).
2.4 Postmortem Brain Samples
DLPFC samples were obtained from the University of California, Irvine Brain Bank (UCI BB) and assayed in the allelic imbalance study. This cohort includes postmortem brain tissues of 91 subjects (34 CTR, 17 SZ, 19 BD, 21 MDD; 70.3% male, mean age=50.0 ± 13.2 ). Written informed consent was obtained from the next of kin for each subject and the study was reviewed and approved by UCI Institutional Review Board. A psychological autopsy was conducted on all subjects consisting of an extensive review of the medical examiner's conclusions, coroner's investigation, medical and psychiatric records, toxicology results, interviews of the decedents’ next-of-kin, and results of gross neuropathology examinations to exclude possible neurodegenerative disorders. All subjects that were clinically characterized with the psychological autopsy method died suddenly without prolonged agonal state. The human brain dissection and freezing protocol is described in detail elsewhere (Jones et al., 1992). RNA was prepared from 80-100 mg of frozen tissue samples, with Omni Prep Multi-Sample Homogenizer (Omni International, Kennesaw, GA). Total RNA isolation was performed with TRIZOL™ reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. RNA was quantified by OD260/280 with a UV spectrophotometer and treated with RNase-free DNase using the RNeasy MinElute columns (Qiagen, Valencia, CA). The quality of the total RNA was finally evaluated using the Agilent 2100 Bioanalyser RNA Chip (Santa Clara, CA). Genomic DNA was extracted from tissues using the DNeasy Blood & Tissue Kits (Qiagen), following the manufacturer's instructions.
2.5 Allele specific expression assay
Total RNA (500 ng) was reverse transcribed with oligo (dT) primers and MultiScribe Reverse Transcriptase, using the TaqMan Gold RT-PCR Kit (Invitrogen), according to the manufacturer's instructions. Allele specific expression for rs12476147 coding variant was performed in cDNA from heterozygous individuals using a pre-validated TaqMan 5′-allele discrimination assays (Applied Biosystems, Assay ID), see Section 2.2 Genotyping for assay details. A 1.5 μl sample of the cDNA synthesized in the RT reaction was used in a real-time PCR reaction (12.5 μl total assay volume). To investigate whether this assay could be utilized to detect the allele difference accurately, genomic DNA from two homozygous individuals were mixed in the following ratios: 8:1, 4:1, 2:1, 1:1, 1:2, 1:4, 1:8. These two allele mixes were quantified by the same method. For normalization, the same assay was also performed on 45 heterozygous genomic DNA samples. The average allele ratio from all genomic DNA samples was used as a correction factor for all genomic DNA and cDNA allele ratios, since this could be assumed to reflect a perfect 1:1 ratio of the two alleles and could therefore be used to correct for any inequalities in allelic representation specific to the assay (Bray et al., 2003). T-tests were performed using the average corrected allele ratio for each genomic DNA and cDNA sample. The t-tests were two-tailed with unequal variance, and p values <0.05 were considered to be significant. The real time PCR was performed in triplicate for each cDNA sample and the allelic imbalance ratio was expressed as Aquantity / Tquantity.
3. Results
3.1 Association study
The rs1344706 and rs12476147 SNPs were genotyped in the entire cohort to investigate their association with SZ and BD in the Costa Rica population. The genotyping success rate exceeded 98%, and the accuracy was >99%, according to random duplicated genotyping of 5% of samples. No significant deviation from Hardy–Weinberg equilibrium was observed either in cases, or in controls, or in both. The association analysis results of the genotyped SNPs are presented in Table 1. The minor A allele of the rs12476147 SNP, located in the exon 4 of the ZNF804A gene, showed association with an increased risk of SZ (p = 0.026, one tailed) using DFAM. In this analysis, we used a one-tailed test, since rs12476147 had been previously associated, so there was a prior direction of effect to assume. No significant association between rs1344706 and SZ or BD was observed using DFAM.
Table 1.
Association analysis results
|
SZ |
BD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CHR | SNP | A1 | A2 | OBS | EXP | CHISQ | p-value | OBS | EXP | CHISQ | p-value |
| 2 | rs1344706 | C | A | 285 | 293.5 | 0.70 | 0.402 | 272 | 276.8 | 0.23 | 0.630 |
| 2 | rs12476147 | A | T | 226 | 208.1 | 3.75 | 0.053* | 184 | 182.4 | 0.03 | 0.868 |
A1: minor allele, A2: major allele; OBS/EXP: Number of observed/expected minor alleles.
P < 0.026 one-tailed test.
3.2 Allelic Imbalance assay
The effect of the intronic risk SNP rs1344706 genotypes on ZNF804A allelic expression was analyzed. The allele-specific expression differences can be identified by comparing the relative levels of exonic SNP alleles (A and T) in RNA samples from subjects heterozygous for rs12476147. We measured the allelic specific expression (ASE) of the rs12476147 coding variant, located in the last exon of the full-length ZNF804A mRNA (NM_194250), and in strong LD with rs1344706. To make sure the ASE assay was accurate, we first quantified the two alleles at SNP rs12476147 in genomic DNA mixes with known ratios. The observed ratio presented a highly linear relation with the expected ratio (r2 = 0.99), thus confirming that our assay could be used to differentiate two alleles, in the range of 0.125 – 8 A/T ratio (data not shown).
Genomic DNA from the DLPFC samples was genotyped for both the SZ risk SNP rs1344706 and the exonic SNP rs12476147. The LD between these two SNPs was calculated using PLINK, and pairwise LD analysis revealed substantial LD between the two SNPs (r2 = 0.343, D′ = 0.952) (Figure 1).
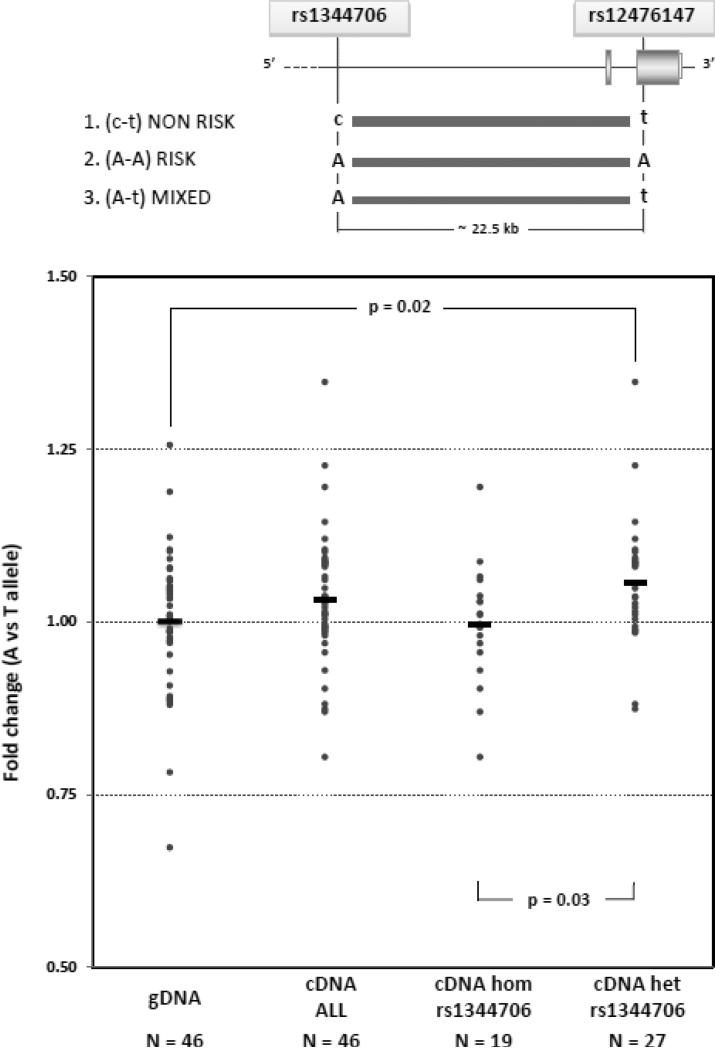
Figure 1. ZNF804A allelic specific expression in DLPFC.
TOP: The structure of the 3′ end of the ZNF804A gene is shown (drawn to scale; exons are represented by boxes, introns by horizontal lines). The two genotyped SNPs are listed, and their location is indicated by lines relative to the ZNF804 gene. Below the gene scheme, the three identified haplotypes are represented as horizontal lines, the thickness of which is proportional to the haplotype frequency. Risk alleles are indicated by uppercase letters. BOTTOM: Individual data points (grey dots) represent the A/T allele ratio at the expressed rs12476147 SNP. gDNA ratios, cDNA ratios, and cDNA ratios divided into rs1344706 homozygotes (hom) and rs1344706 heterozygotes (het) are shown. The mean value for each distribution is also indicated by a horizontal bar. cDNA allele ratios are significantly higher than gDNA allele ratios in the rs1344706 heterozygotes (p-value = 0.02). Moreover, there is a significant difference in cDNA allele ratios between homozygotes and heterozygotes for the rs1344706 SNP (p-value = 0.03).
The rs12476147 ASE was then evaluated in the DLPFC of the 46 heterozygous subjects. ASE analyses were performed on all heterozygous subjects from the 4 diagnostic groups merged together. A possible effect of diagnosis on rs12476147 ASE was tested, and there was no difference among diagnostic groups (one-way ANOVA p-value=0.99).
Figure 1 illustrates ASE of ZNF804A at the expressed SNP rs12476147 in the DLPFC. A non-significant over-expression of the rs12476147 A allele was detected in cDNA A/T allele ratios compared to A/T allele ratios of rs12476147 observed in genomic DNA (average 1.03, 95% CI 1.00-1.06; p-value < 0.12, after correction for genomic DNA A/T assay).
Since the rs12476147 SNP is in strong, but not perfect, LD with the rs1344706 SNP, we were able to test the effect of the rs1344706 genotype on ZNF804A allelic expression of rs12476147. The 46 rs12476147 heterozygotes were further divided into 2 groups based on the rs1344706 homozygous AA (N=19) and heterozygous CA (N=27) genotypes. cDNA allele A/T ratios were significantly different depending on the intronic rs1344706 genotypes (AA vs CA, p-value = 0.03). In the rs1344706 heterozygous group the cDNA allele ratio was significantly higher than the genomic DNA allele ratio (average 1.06, 95% CI 1.02-1.09; p-value = 0.02), whereas in rs1344706 homozygotes, cDNA allele ratios did not differ from the genomic DNA allele ratios (average 1.00, 95% CI 0.96-1.03; p-value=0.87).
4. Discussion
The involvement of ZNF804A in SZ and BD was evaluated by a combined approach: i) we genotyped the rs1344706 and rs12476147 SNPs in an independent sample of 428 SZ, 385 BD and 578 controls from the demographically well characterized population of the CVCR; and ii) we analyzed the rs12476147 ASE in the DLPFC of 46 heterozygous postmortem brains.
Genetically and geographically isolated populations provide several advantages for studying genes related to complex diseases. The CVCR population is a recently founded population, which displays similar features to other genetic isolates with few founders, therefore presenting a homogenous population suitable for studying genes underlying complex genetic disorders. In fact, chromosomal areas of interest have already been identified in this cohort for both SZ (Cooper-Casey et al., 2005; DeLisi et al., 2002; Walss-Bass et al., 2006) and BD (Freimer et al., 1996).
We were unable to confirm the association of rs1344706 with schizophrenia in our CVCR cohort. This is consistent with some recent GWAS replication studies, and recent meta-analyses conducted with Chinese population cohorts (Li et al., 2011; Li et al., 2013). The A allele frequency of rs1344706 in our control group was 0.60, which is similar to the frequencies reported for samples with European ancestry. However, we observe a significant association with an increase risk of SZ for the minor A allele of the exonic SNP rs12476147 (p-value = 0.026), which is in agreement with published data (Williams et al., 2011). In this analysis, we use a one-tailed test, since rs12476147 had been previously associated, so there was a prior direction of effect to assume. Further, given the extent of LD between rs1344706 and rs12476147, it can be argued that a specific allele at rs12476147 (the allele most commonly found with the A risk allele at rs1344706) could be identified a priori.
One possibility to explain our non-significant association with SZ of rs1344706 is insufficient power to detect a significant difference. In fact, the statistical power sufficient to replicate findings with relatively low relative risk is a critical determinant of the success of any replication study. An odds ratio of 1.38 and 1.23 was reported for association of SZ with rs1344706 and rs12476147, respectively (O'Donovan et al., 2008; Williams et al., 2011), and odds ratios are usually expected to be lower in replication studies. Our post-hoc power analysis shows that with the present sample size, the study has 35% and 44% power for the rs1344706 and rs12476147 SNPs respectively to detect a significant association.
There is growing evidence that allele-specific differences in gene expression, arising from cis-acting regulatory polymorphisms, are an important source of phenotypic variability, including susceptibility to complex disorders (Bray et al., 2003; Sadee, 2009). Allelic imbalance is a robust and accurate tool to identify cis-regulatory variation while minimizing confounding trans-acting factors, including tissue preparation methods and post-mortem interval as well as trans-acting genetic events (Bray and O'Donovan, 2006; Pastinen and Hudson, 2004; Stamatoyannopoulos, 2004; Yan and Zhou, 2004). Allelic differences in expression appear to be context specific, for example, with regard to tissue-type, and the effects of cis-regulatory polymorphism on gene expression can differ widely between tissues (Sun et al., 2010; Wilkins et al., 2007; Zhang et al., 2009). Moreover, differential allelic expression across multiple regions of the adult human brain has been reported (Buonocore et al., 2010). Therefore, it's important that ASE analysis focuses on relevant target tissues.
We used ASE to assess cis-regulatory variation of ZNF804A expression, and to what extent the SZ risk SNP rs1344706 accounts for ASE, in the DLPFC of postmortem human brains. The DLPFC was chosen since this brain region has been consistently implicated in the pathogenesis of SZ. Moreover, functional imaging genetic studies have shown an association between rs1344706 and altered connectivity of DLPFC with other cortical regions (Esslinger et al., 2009; Paulus et al., 2013; Rasetti et al., 2011). We observed significant allelic expression imbalance of ZNF804A, indicating heterozygous cis effects on ZNF804A expression in the DLPFC. In contrast with previously published results (Hill and Bray, 2012; Williams et al., 2011), genotype at rs1344706 was found to have a significant effect on ZNF804A allelic expression in adults, with the SZ risk allele associated with increased ZNF804A allelic expression. Our data provide evidence for a possible functional role of the rs1344706 SNP, thus suggesting that this variant may be the cis-regulatory element directly responsible for ZNF804A allelic expression imbalance. Our results fit reasonably well with the previously published study showing rs1344706 as a functional polymorphism, with potential effects on ZNF804A expression through altered DNA-protein interactions (Hill and Bray, 2011), and are also consistent with the previously reported association between the rs1344706 risk allele and higher total ZNF804A expression in the adult DLPFC (Riley et al., 2010).
It should be noted that the observed ZNF804A allelic imbalance is small; therefore it's crucial to use a large sample size. However, since ASE assays can only be performed on heterozygous subjects for a coding SNP, the sample sizes in published allelic imbalance studies using human brain are typically modest (Bray et al., 2003; Fukuda et al., 2006; Sun et al., 2010). Because small sample sizes reduce the statistical power, and since no effect of diagnosis on rs12476147 ASE was observed, we analyzed the ASE on the rs12476147 heterozygotes from four diagnostic groups merged together, thus reaching a sample size of 46 subjects. Furthermore, the majority of the approaches that have been used to detect ASE are end-point readings of PCR, and therefore they may not be accurate enough in quantifying the ratio between the two alleles. In order to have a higher sensitivity, in our ASE assay we used real time PCR with Taqman probes to distinguish the two alleles (Chen et al., 2008).
There are some differences between our results and previously published studies (Hill and Bray, 2012; Schultz et al., 2013; Williams et al., 2011), which can be explained for some of the above reasons. A recently published paper (Schultz et al., 2013) showed significant difference in general ZNF804A expression mediated by the rs1344706 SNP, and the A risk allele was associated with lower prefrontal ZNF804A expression in patients (Schultz et al., 2013), whereas the opposite effect in controls has been observed by prior analyses (Riley et al., 2010). The expression profile of ZNF804A appears to be different between patients and healthy controls in terms of an opposite effect of the risk allele. No effect of rs1344706 genotype on ZNF804A allelic expression was reported in adult brain regions (Hill and Bray, 2012; Williams et al., 2011), leading to the conclusion that the observed allelic expression imbalance was not directly attributable to rs1344706, at least in the adult brain. In fact, rs1344706 genotypes were found to have a significant effect on ZNF804A allelic expression in second-trimester fetal brain tissue, however with the SZ risk allele associated with a reduced expression (Hill and Bray, 2012). This is in contrast with our findings, but one possible explanation could be the reversal of fetal expression trajectories. It has been shown that fetal expression changes are negatively correlated with those in other stages of life, thus suggesting that select fetal expression changes are reversed at different times across the lifespan (Colantuoni et al., 2011), in fact ZNF804A general expression appears to be increased in fetal brain compared to levels found in adult prefrontal cortex. Finally, it is possible, but not likely, that rs1344706 might be indirectly related to the ASE we have observed, as it could be in LD with the unknown causal variant, or could be related to general ZNF804A expression, these possibilities will require further analysis in independent cohorts.
In conclusion, using a powerful within-subjects technique that detects cis-acting influences on gene expression, we have demonstrated an imbalanced allelic expression of ZNF804A in DLPFC. Our ASE assay detected a significant excess of the rs12476147 A allele in heterozygous subjects for the SZ risk SNP rs1344706. Further we confirmed a prior association of the SZ risk SNP rs12476147 in the CVCR isolated population. Although our data do not exclude the possibility of other functional variants in ZNF804A, they provide support for rs1344706 risk allele as the cis-regulatory variant directly responsible for ZNF804A allelic expression imbalance.
Acknowledgments
We acknowledge members of the Pritzker Neuropsychiatric Disorders Research Consortium for collecting DNA samples, and we appreciate the Costa Rica staff for subject ascertainment. We appreciate the assistance of Preston Cartagena, Psy.D., David Walsh, Ph.D., and Richard Stein, Ph.D. of the University of California, Irvine (UCI) Brain Bank for their contributions to postmortem clinical characterization of subjects, Kathleen Burke (UCI), and Claudia M. Cervantes (UCI) for procurements of brain tissue, as well as Chief Deputy Coroner Jacque Berndt, and the staff of Orange County Coroners’ Office. Neuropathological evaluation of the postmortem brains was performed by F. Warren Lovell, M.D. Tissue specimens were processed and stored at the Human Brain and Spinal Fluid Resource Center, Veteran's Medical Center, Los Angeles under the direction of Wallace W. Tourtellotte, M.D., Ph.D.
Funding body agreements and policies
The present work has been in part supported by NIMH Grants R01MH085801, R01MH085548, and R01MH 099440 (MPV), Della Martin Foundation and the William Lion Penzner Foundation (UCI Department of Psychiatry and Human Behavior). The authors of this paper are members of the Pritzker Neuropsychiatric Disorders Research Consortium currently supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. A shared intellectual property agreement exists between the Fund and all the universities involved, in order to encourage the development of appropriate findings for research and clinical applications. The academic and philanthropic entities involved in this research program are jointly filing patent applications related to the present findings.
The funding source had no other role other than financial support. They had no involvement in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributors
IG designed and carried out the laboratory experiments, analyzed the data, interpreted the results and wrote the paper. LM performed the SNP genotyping for the CVCR cohort. AS and BR carried out the brain dissection. RMM, SJW, HA and WEB were involved in the conception of the study and revising the manuscript. LED and WB contributed in the study design, clinical characterizations, and in the discussion of the results. MPV participated in the conception of the study, interpretation of the results, in writing the manuscript, and supervised the entire study. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare no conflict of interest.
References
- Bray NJ, Buckland PR, Owen MJ, O'Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113(2):149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- Bray NJ, O'Donovan MC. Investigating cis-acting regulatory variation using assays of relative allelic expression. Psychiatr Genet. 2006;16(4):173–177. doi: 10.1097/01.ypg.0000218612.35139.84. [DOI] [PubMed] [Google Scholar]
- Buonocore F, Hill MJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C, Hortobagyi T, Williams BP, Cooper JD, Bray NJ. Effects of cis-regulatory variation differ across regions of the adult human brain. Hum Mol Genet. 2010;19(22):4490–4496. doi: 10.1093/hmg/ddq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Weaver J, Bove BA, Vanderveer LA, Weil SC, Miron A, Daly MB, Godwin AK. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum Mol Genet. 2008;17(9):1336–1348. doi: 10.1093/hmg/ddn022. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Casey K, Mesen-Fainardi A, Galke-Rollins B, Llach M, Laprade B, Rodriguez C, Riondet S, Bertheau A, Byerley W. Suggestive linkage of schizophrenia to 5p13 in Costa Rica. Mol Psychiatry. 2005;10(7):651–656. doi: 10.1038/sj.mp.4001640. [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K. Clinical characteristics of schizophrenia in multiply affected Spanish origin families from Costa Rica. Psychiatr Genet. 2001;11(3):145–152. doi: 10.1097/00041444-200109000-00006. [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K, Relja M, Byerley W, Sherrington R. Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet. 2002;114(5):497–508. doi: 10.1002/ajmg.10538. [DOI] [PubMed] [Google Scholar]
- Donohoe G, Morris DW, Corvin A. The psychosis susceptibility gene ZNF804A: associations, functions, and phenotypes. Schizophr Bull. 2010;36(5):904–909. doi: 10.1093/schbul/sbq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, Haddad L, Mier D, Opitz von Boberfeld C, Raab K, Witt SH, Rietschel M, Cichon S, Meyer-Lindenberg A. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324(5927):605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- Freimer NB, Reus VI, Escamilla M, Spesny M, Smith L, Service S, Gallegos A, Meza L, Batki S, Vinogradov S, Leon P, Sandkuijl LA. An approach to investigating linkage for bipolar disorder using large Costa Rican pedigrees. Am J Med Genet. 1996;67(3):254–263. doi: 10.1002/(SICI)1096-8628(19960531)67:3<254::AID-AJMG3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Koga M, Arai M, Noguchi E, Ohtsuki T, Horiuchi Y, Ishiguro H, Niizato K, Iritani S, Itokawa M, Arinami T. Monoallelic and unequal allelic expression of the HTR2A gene in human brain and peripheral lymphocytes. Biol Psychiatry. 2006;60(12):1331–1335. doi: 10.1016/j.biopsych.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SG, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Allelic differences in nuclear protein binding at a genome-wide significant risk variant for schizophrenia in ZNF804A. Mol Psychiatry. 2011;16(8):787–789. doi: 10.1038/mp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am J Psychiatry. 2012;169(12):1301–1308. doi: 10.1176/appi.ajp.2012.11121845. [DOI] [PubMed] [Google Scholar]
- Jones EG, Hendry SH, Liu XB, Hodgins S, Potkin SG, Tourtellotte WW. A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J Neurosci Methods. 1992;44(2-3):133–144. doi: 10.1016/0165-0270(92)90006-y. [DOI] [PubMed] [Google Scholar]
- Li M, Luo XJ, Xiao X, Shi L, Liu XY, Yin LD, Diao HB, Su B. Allelic differences between Han Chinese and Europeans for functional variants in ZNF804A and their association with schizophrenia. Am J Psychiatry. 2011;168(12):1318–1325. doi: 10.1176/appi.ajp.2011.11030381. [DOI] [PubMed] [Google Scholar]
- Li M, Shi CJ, Shi YY, Luo XJ, Zheng XB, Li ZQ, Liu JJ, Chong SA, Lee J, Wang Y, Liu XY, Yin LD, Pu XF, Diao HB, Xu Q, Su B. ZNF804A and schizophrenia susceptibility in Asian populations. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(7):794–802. doi: 10.1002/ajmg.b.32084. [DOI] [PubMed] [Google Scholar]
- Li M, Zhang H, Luo XJ, Gao L, Qi XB, Gourraud PA, Su B. Meta-Analysis Indicates That the European GWAS-Identified Risk SNP rs1344706 within ZNF804A Is Not Associated with Schizophrenia in Han Chinese Population. PLoS One. 2013;8(6):e65780. doi: 10.1371/journal.pone.0065780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon E, Rollins B, Mesen A, Sequeira A, Myers RM, Akil H, Watson SJ, Barchas J, Jones EG, Schatzberg A, Bunney WE, DeLisi LE, Byerley W, Vawter MP. Lack of association to a NRG1 missense polymorphism in schizophrenia or bipolar disorder in a Costa Rican population. Schizophr Res. 2011;131(1-3):52–57. doi: 10.1016/j.schres.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera B, Barrantes R, Marin-Rojas R. Gene admixture in the Costa Rican population. Ann Hum Genet. 2003;67(Pt 1):71–80. doi: 10.1046/j.1469-1809.2003.00010.x. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306(5696):647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Paulus FM, Krach S, Bedenbender J, Pyka M, Sommer J, Krug A, Knake S, Nothen MM, Witt SH, Rietschel M, Kircher T, Jansen A. Partial support for ZNF804A genotype-dependent alterations in prefrontal connectivity. Hum Brain Mapp. 2013;34(2):304–313. doi: 10.1002/hbm.21434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry. 2011;68(12):1207–1217. doi: 10.1001/archgenpsychiatry.2011.103. [DOI] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O'Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15(1):29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O'Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Levinson DF, Gejman PV, Laurent C, Mowry BJ, O'Donovan MC, Pulver AE, Schwab SG, Wildenauer DB, Dudbridge F, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Van Os J, Walshe M, Weisbrod M, Wiersma D, Donnelly P, Barroso I, Blackwell JM, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, McCarthy MI, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013 doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadee W. Measuring cis-acting regulatory variants genome-wide: new insights into expression genetics and disease susceptibility. Genome Med. 2009;1(12):116. doi: 10.1186/gm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CC, Nenadic I, Riley B, Vladimirov VI, Wagner G, Koch K, Schachtzabel C, Muhleisen TW, Basmanav B, Nothen MM, Deufel T, Kiehntopf M, Rietschel M, Reichenbach JR, Cichon S, Schlosser RG, Sauer H. ZNF804A and Cortical Structure in Schizophrenia: In Vivo and Postmortem Studies. Schizophr Bull. 2013 doi: 10.1093/schbul/sbt123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SG, Kusumawardhani AA, Dai N, Qin W, Wildenauer MD, Agiananda F, Amir N, Antoni R, Arsianti T, Asmarahadi A, Diatri H, Djatmiko P, Irmansyah I, Khalimah S, Kusumadewi I, Kusumaningrum P, Lukman PR, Mustar L, Nasrun MW, Naswati S, Prasetiyawan P, Semen GM, Siste K, Tobing H, Widiasih N, Wiguna T, Wulandari WD, Benyamin B, Wildenauer DB. Association of rs1344706 in the ZNF804A gene with schizophrenia in a case/control sample from Indonesia. Schizophr Res. 2013;147(1):46–52. doi: 10.1016/j.schres.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA. The genomics of gene expression. Genomics. 2004;84(3):449–457. doi: 10.1016/j.ygeno.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, Mortensen PB, Andreassen OA, Sigurdsson E, Thorgeirsson TE, Bottcher Y, Olason P, Ophoff RA, Cichon S, Gudjonsdottir IH, Pietilainen OP, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Athanasiu L, Suvisaari J, Lonnqvist J, Paunio T, Hartmann A, Jurgens G, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Breuer R, Moller HJ, Giegling I, Glenthoj B, Rasmussen HB, Mattheisen M, Bitter I, Rethelyi JM, Sigmundsson T, Fossdal R, Thorsteinsdottir U, Ruggeri M, Tosato S, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Walshe M, Bramon E, Vassos E, Li T, Fraser G, Walker N, Toulopoulou T, Yoon J, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Peltonen L, Rujescu D, Collier DA, Stefansson H, St Clair D, Stefansson K. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry. 2011;16(1):59–66. doi: 10.1038/mp.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Southard C, Witonsky DB, Olopade OI, Di Rienzo A. Allelic imbalance (AI) identifies novel tissue-specific cis-regulatory variation for human UGT2B15. Hum Mutat. 2010;31(1):99–107. doi: 10.1002/humu.21145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walss-Bass C, Montero AP, Armas R, Dassori A, Contreras SA, Liu W, Medina R, Levinson D, Pereira M, Atmella I, NeSmith L, Leach R, Almasy L, Raventos H, Escamilla MA. Linkage disequilibrium analyses in the Costa Rican population suggests discrete gene loci for schizophrenia at 8p23.1 and 8q13.3. Psychiatr Genet. 2006;16(4):159–168. doi: 10.1097/01.ypg.0000218616.27515.67. [DOI] [PubMed] [Google Scholar]
- Wilkins JM, Southam L, Price AJ, Mustafa Z, Carr A, Loughlin J. Extreme context specificity in differential allelic expression. Hum Mol Genet. 2007;16(5):537–546. doi: 10.1093/hmg/ddl488. [DOI] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O'Donovan MC. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16(4):429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhou W. Allelic variations in gene expression. Curr Opin Oncol. 2004;16(1):39–43. doi: 10.1097/00001622-200401000-00008. [DOI] [PubMed] [Google Scholar]
- Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX, Li WQ, Zhang YL, Zhang Y, Ma CC, Du B, Wang LF, Ren YQ, Yang YF, Hu XF, Wang Y, Deng W, Tan LW, Tan YL, Chen Q, Xu GM, Yang GG, Zuo XB, Yan H, Ruan YY, Lu TL, Han X, Ma XH, Cai LW, Jin C, Zhang HY, Yan J, Mi WF, Yin XY, Ma WB, Liu Q, Kang L, Sun W, Pan CY, Shuang M, Yang FD, Wang CY, Yang JL, Li KQ, Ma X, Li LJ, Yu X, Li QZ, Huang X, Lv LX, Li T, Zhao GP, Huang W, Zhang XJ, Zhang D. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011;43(12):1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- Zaharie A, Ergul E, Ozel MD, Miclutia IV, Stanculete MF, Sazci A. ZNF804A rs1344706 variant and schizophrenia in a Romanian population from Cluj Napoca. Genet Test Mol Biomarkers. 2012;16(9):1135–1137. doi: 10.1089/gtmb.2012.0066. [DOI] [PubMed] [Google Scholar]
- Zhang F, Chen Q, Ye T, Lipska BK, Straub RE, Vakkalanka R, Rujescu D, St Clair D, Hyde TM, Bigelow L, Kleinman JE, Weinberger DR. Evidence of sex-modulated association of ZNF804A with schizophrenia. Biol Psychiatry. 2011a;69(10):914–917. doi: 10.1016/j.biopsych.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, Eggan K, Church GM. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods. 2009;6(8):613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lu SM, Qiu C, Liu XG, Gao CG, Guo TW, Valenzuela RK, Deng HW, Ma J. Population-based and family-based association studies of ZNF804A locus and schizophrenia. Mol Psychiatry. 2011b;16(4):360–361. doi: 10.1038/mp.2010.55. [DOI] [PubMed] [Google Scholar]
- Zhang R, Yan JD, Valenzuela RK, Lu SM, Du XY, Zhong B, Ren J, Zhao SH, Gao CG, Wang L, Guo TW, Ma J. Further evidence for the association of genetic variants of ZNF804A with schizophrenia and a meta-analysis for genome-wide significance variant rs1344706. Schizophr Res. 2012;141(1):40–47. doi: 10.1016/j.schres.2012.07.013. [DOI] [PubMed] [Google Scholar]