Abstract
Purpose
Radiotherapy remains a primary treatment modality for pancreatic carcinoma, a tumor characterized by aberrant mTOR activity. Given mTOR's regulatory role in gene translation, in this study we defined the effects of the clinically relevant, ATP-competitive mTOR inhibitor, INK128 on the radiosensitivity of pancreatic carcinoma cell lines.
Experimental Design
Clonogenic survival was used to determine the effects of INK128 on in vitro radiosensitivity on 3 pancreatic carcinoma cell lines and a normal fibroblast cell line with mTOR activity defined using immunoblots. DNA double strand breaks were evaluated according to γH2AX foci. The influence of INK128 on radiation-induced gene translation was determined by microarray analysis of polysome-bound mRNA. Leg tumor xenografts grown from pancreatic carcinoma cells were evaluated for mTOR activity, eIF4F cap complex formation and tumor growth delay.
Results
INK128, while inhibiting mTOR activity in each of the cell lines, enhanced the in vitro radiosensitivity of the pancreatic carcinoma cells, but had no effect on normal fibroblasts. The dispersal of radiation-induced γH2AX foci was inhibited in pancreatic carcinoma cells by INK128 as were radiation-induced changes in gene translation. Treatment of mice with INK128 resulted in an inhibition of mTOR activity as well as cap-complex formation in tumor xenografts. Whereas INK128 alone had no effect of tumor growth rate, it enhanced the tumor growth delay induced by single and fractionated doses of radiation.
Conclusion
These results indicate that mTOR inhibition induced by INK128 enhances the radiosensitivity of pancreatic carcinoma cells and suggest that this effect involves the inhibition of DNA repair.
Keywords: Radiosensitization, mTOR, pancreatic carcinoma, INK128, xenograft
Introduction
Radiation regulates gene expression primarily via translational control, a process that operates independently from changes in the transcriptome (1). Moreover, radiation-induced gene translation profiles have revealed a set of genes and networks that are unique to tumor or normal cells (2), suggesting that translation control processes may provide a source of tumor-selective targets for radiosensitization. Along these lines, we recently showed that knockdown of the eukaryotic initiation factor 4E (eIF4E), the rate-limiting protein in cap-dependent translation initiation, enhanced the radiosensitivity of tumor but not normal cell lines (3). These results suggested that eIF4E targeting strategies in combination with radiotherapy may be of benefit in cancer treatment. One approach to targeting eIF4E is through inhibition of the mechanistic target of rapamycin (mTOR) kinase, which regulates mRNA translation in response to a wide variety of environmental signals (4). Specifically as part of the multi-protein complex mTORC1, mTOR phosphorylates eIF4E-binding protein 1 (4E-BP1) resulting in its release of eIF4E, which then initiates the formation of the eIF4F cap-complex leading to cap-dependent translation (5). Inhibiting the mTOR-mediated phosphorylation of 4E-BP1 severely limits eIF4E availability and thus eIF4F complex formation, essentially mimicking the knockdown of eIF4E.
To target mTORC1 activity most studies have focused on rapamycin or its analogs (rapalogs). Whereas these agents effectively inhibit S6 kinase phosphorylation, the other primary substrate of mTORC1, they only partially inhibit phosphorylation of 4E-BP1 (5). In addition, these drugs do not inhibit mTORC2 (6), an mTOR containing complex that, although not as well defined as mTORC1, has been reported to phosphorylate several AGC kinases including AKT, SGK, and PKC. As single agents or in combination protocols, rapamycin/rapalogs have only modest clinical activity against a variety of tumor types (7), which has been attributed to their incomplete inhibition of 4E-BP1 phosphorylation and/or the lack of mTORC2 inhibition (6, 8). In comparison to the allosteric inhibition by rapamycin, the recently developed ATP-competitive inhibitors of mTOR inhibit mTORC1 output more completely and inhibit mTORC2 (9-12). We recently showed that in contrast to rapamycin, the competitive mTOR inhibitor PP242 enhanced the radiosensitivity of a glioma and breast carcinoma cell line, while having no effect on the radiosensitivity of a normal cell line (13). Whereas these results are consistent with mTOR as a target for tumor selective radiosensitization, PP242 has unfavorable pharmacokinetics in humans (14). Thus, to extend investigations to a clinically applicable compound, we have evaluated the radiosensitizing potential of the PP242 analog INK128 (14), which is currently undergoing clinical trials (15).
We have focused these initial INK128 studies on pancreatic carcinoma cell lines. Clinical results indicate that although the combination of radiation and gemcitabine significantly prolongs survival as compared to gemcitabine alone (16), the prognosis for patients with pancreatic carcinoma remains poor with an overall 5 year survival rate of approximately 5% (17-18). Thus, this tumor site would seem likely to benefit from an effective radiosensitizing agent. Pancreatic carcinomas have high incidences of K-RAS mutations with the associated increased signaling through the PI3K/AKT/mTOR pathway (19) and approximately 70% of these tumors have constitutive mTOR activation (20) suggesting mTOR kinase as a target for radiosensitization. The data presented here indicate that the competitive mTOR inhibitor INK128 enhances the radiosensitivity of pancreatic carcinomas cells in vitro and when grown as tumor xenografts. Furthermore, this radiosensitization was associated with an inhibition of DNA double strand break repair as well as the suppression of radiation-induced translation of functionally related mRNAs.
Methods
Cell lines and treatments
The human pancreatic carcinoma cell lines Miapaca-2 and Panc1 were obtained from American Type Culture Collection (ATCC); PSN1 was provided by Deborah Citrin (National Cancer Institute, Bethesda, Maryland). The human normal fibroblast cell line MRC9 was obtained from American Type Culture Collection (ATCC). Cells were maintained in RPMI (Miapaca-2, Panc1, and PSN1) or MEM (MRC9) media supplemented with 10% FBS (Invitrogen). Cells were cultured less than 3 months after resuscitation and were maintained in an atmosphere of 5% CO2/95% air at 37°C. INK128 (Chemietek) was dissolved in DMSO. Cell cultures were irradiated using 320kV X-ray source at 2.3 Gy/min (Precision XRay Inc.).
Clonogenic Survival Assay
To evaluate radiosensitivity cells were plated in six well plates and treated the next day. 10 to 14d after seeding, plates were stained with 0.5% crystal violet, the number of colonies determined, and the surviving fractions were calculated. Radiation survival curves were generated after normalizing for the cytotoxicity induced by INK128 treatment alone..
Immunoblotting and antibodies
Cells were lysed as described (13). Protein was quantified using BCA protein assay (Thermo Scientific); separated by SDS-PAGE; transferred to PVDF (Bio-Rad) and probed with the indicated antibodies. Bands were visualized using Pierce ECL Western Blotting Substrate (Thermo Scientific). Anti-AKT, anti-phospho-AKT S473, anti-4E-BP-1, anti-phospho-4E-BP-1 T37/46, anti-eIF4E, and anti-phospho-mTOR S2481 were purchased from Cell Signaling Technology. Anti-β-actin was obtained from Sigma-Aldrich. Anti-eIF4G was purchased from BD Biosciences. Donkey-anti-rabbit and sheep-anti-mouse Horseradish Peroxidase conjugated secondary antibodies were purchased from GE Healthcare.
Polysome isolation and microarray analysis
Isolation of polysome-bound RNA was performed as described (1-2, 21) in biological triplicate. The isolated RNA was amplified with GeneChip 3’ IVT Express Kit (Affymetrix) and hybridized to GeneChip PrimeView Human Gene Expression Array chips (Affymetrix) per manufacturer's protocol. Using Partek Genomics suite, RMA normalization and log2 transformation was performed. Statistical significance was determined by ANOVA testing with Partek Genomics suite. Probesets that were modified by at least ± 1.2 fold with a p < 0.1 (vs. control) were identified and submitted to Ingenuity Pathway Analysis (IPA) as described in the results. The data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibues (GEO) and are accessible through GEO Series accession number GSE48098.
Immunofluorescent analysis of γH2AX foci
Cells grown in chamber slides, were fixed, permeabilized, and blocked as described (13). The slides were incubated with antibody to phospho-H2AX (Millipore) followed by goat-anti-mouse-Alexa488 (Invitrogen) and mounted with Prolong gold anti-fade reagent containing DAPI (Invitrogen) to visualize nuclei. Cells were analyzed on a Leica upright fluorescent microscope.
Immunohistochemistry
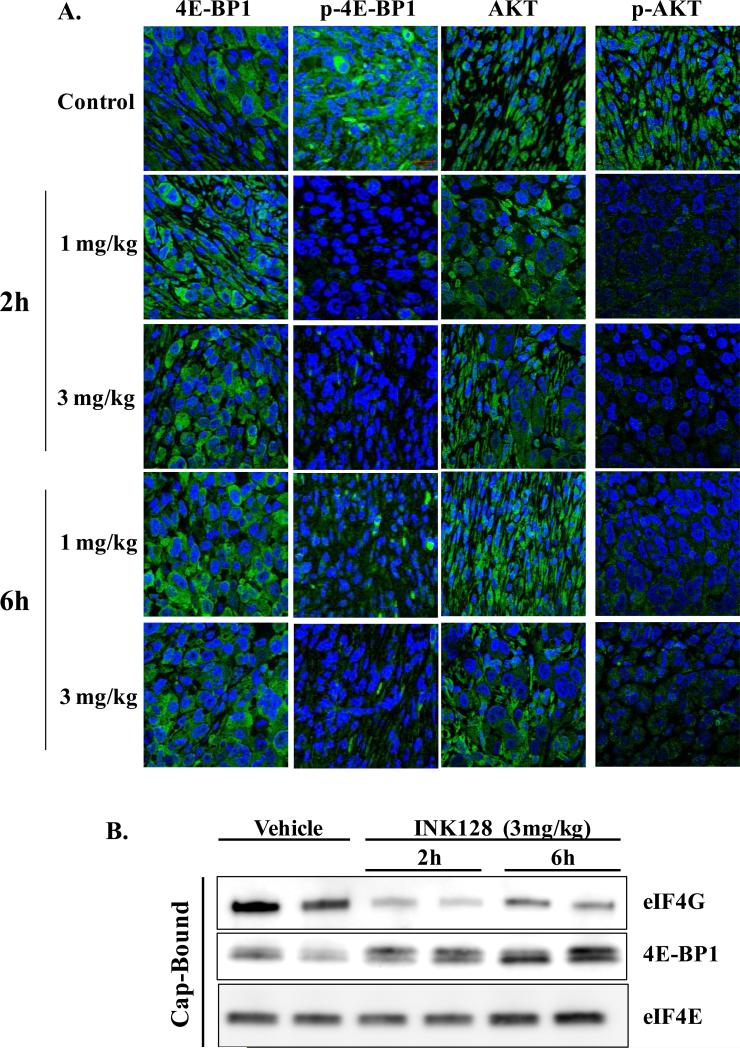
Tumors were excised and fixed in 10% buffered formalin. Paraffin sections were deparaffinized in xylene and rehydrated in decreasing amounts of alcohol. Sections were boiled in citrate buffer and incubated in 1% BSA containing 10% goat serum (Sigma-Aldrich). Anti-phospho-4E-BP1 T37/46, anti-phospho-AKT S473, anti-4E-BP1, and anti-AKT were incubated overnight at 4°C followed by incubation with FITC-labeled secondary antibody (Invitrogen), and finally mounted with mounting media containing DAPI (Vector) to visualize nuclei. Images were generated from two individual tumors per treatment group using a Zeiss confocal microscope with a 63x objective with representative images shown in Figure 5.
Figure 5.
The effects of INK128 treatment on mTOR activity in pancreatic tumor xenografts. Mice bearing PSN1 xenografts were treated with vehicle or the indicated dose of INK128 (oral gavage). Tumors were collected 2, or 6 hours later and prepared for immunohistochemical staining or affinity chromatography. A). Sections were probed with an antibody specific to the indicated protein followed by staining with a FITC coupled secondary antibody (green). Nuclei were visualized with DAPI (blue). Each image is of representative of at least two mice per treatment group. B) eIF4F complex formation. m7-GTP affinity chromatography was performed on PSN1 tumor xenografts with m7-GTP bound proteins resolved via SDS-PAGE followed by immunoblot analysis. Each lane represents the tumor from an individual mouse.
In vivo cap-binding assay
eIF4F cap complex formation in PSN1 xenografts was measured using m7-GTP batch chromatography (22). Briefly, tumors were homogenized and lysed in 20mM Tris-HCl (pH 7.4), 150mM NaCl, 1mM EDTA, 1mM EGTA, 1mM β-glycerophosphate, 1mM sodium orthovanadate, 1% Triton X-100, 0.2mM PMSF, 1x phosphatase inhibitor cocktails II and III (Sigma-Aldrich), and 1x HALT protease inhibitor cocktail (Thermo Scientific) for 15m on ice. 400μg of lysate were pre-cleared for 1h at 4°C then incubated with m7-GTP Sepharose 4B beads (GE Healthcare) overnight at 4°C. Beads were washed three times with lysis buffer; bound protein was eluted, denatured, and then separated using SDS-PAGE followed by immunoblotting.
In vivo tumor growth delay
Eight to ten-week-old female athymic nude mice (NCr nu/nu; NCI Animal Production Program, Frederick, MD) were used in these studies. Animals are caged in groups of 5 or less and fed animal chow and water ad libitum. A single cell suspension PSN1 cells (5 × 106) was injected subcutaneously into the right hind leg. INK128 was dissolved in 5% n-methylpyrrolidone, 15% polyvinylpyrrolidone, and 80% water and delivered by oral gavage. When tumors grew to a mean volume of approximately 180 mm3 mice were randomized into four groups: vehicle treated controls, drug (INK128, oral gavage), radiation, or drug/radiation combination. The specific treatment protocols are described in the results. Radiation was delivered locally using a Pantak X Ray source with animals restrained in a custom designed lead jig. To obtain tumor growth curves, perpendicular diameter measurements of each tumor were measured 2 to 3 times per week with a digital caliper and volumes were calculated using a formula (L × W2) / 2. Data are expressed as mean ± SEM tumor volume. Each experimental group contained between 5-7 mice. All animal studies were conducted in accordance with the NIH Guide for Care and Use of Animals. Statistical analysis was performed using one-way ANOVA and Tukey's post hoc analysis.
Results
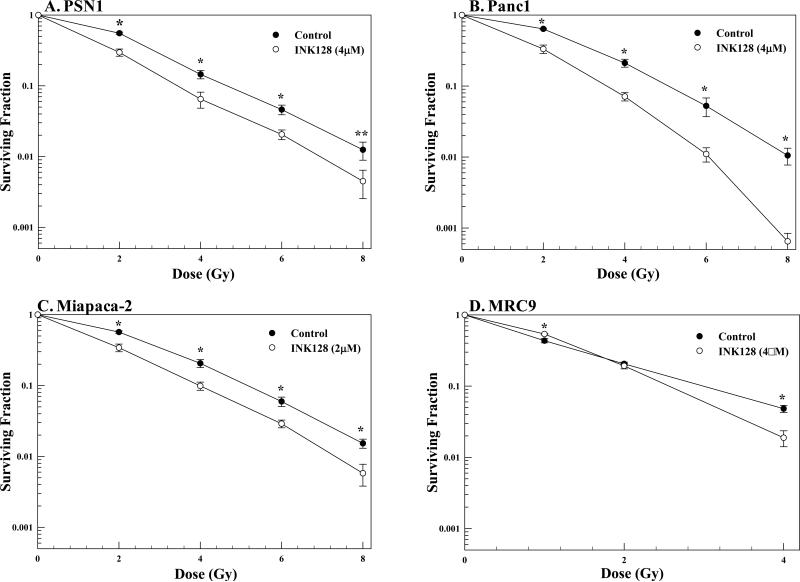
The effects of INK128 on pancreatic cancer cell radiosensitivity as measured by clonogenic survival analysis are shown in Figure 1A-C. For this study cells were plated at clonogenic density, allowed to attach overnight, irradiated, followed immediately by adding the specified concentration of INK128. The treatment protocol was chosen based upon our recent study in which maximum radiosensitization induced by the ATP-competitive mTOR inhibitor PP242 occurred when drug was added immediately after radiation (13); concentrations of INK128 used are similar to those achievable in vivo (14). Twenty-four hours after irradiation media was removed, fresh drug-free media was added and colonies determined 10-14 days later. INK128 treatment alone had no effect on the surviving fraction of PSN1 or Panc1 cells. Because Miapaca-2 cells were more sensitive to INK128-induced cytotoxicity, to maintain a minimal level of drug-induced cell killing similar to the other tumor cell lines, the concentration was decreased to 2 μM, which reduced surviving fraction of MiaPaca-2 cells to 0.85 ± 0.02. In all 3 pancreatic tumor cell lines treatment with INK128 immediately after irradiation resulted in an increase in radiosensitivity with dose-enhancement factors at a surviving fraction of 0.1 (DEFs) of 1.32 ± 0.06, 1.45 ± 0.01 and 1.30 ± 0.09 for PSN1, Panc1 and Miapaca-2, respectively. As an initial investigation into the potential for tumor selective actions, the effect of INK128 on the radiosensitivity of the normal lung fibroblast line MRC9 was determined. INK128 treatment alone reduced the MRC9 surviving fraction to 0.73 ± 0.05; in contrast to the 3 tumor cell lines INK128 had no consistent effect on the radiosensitivity of MRC9 cells (Figure 1D).
Figure 1.
Effects of INK128 on cellular radiosensitivity. A) PSN1, B) Panc1, C) Miapaca-2 or D) MRC9 cells were plated, allowed to attach overnight, irradiated and the indicated concentration of inhibitor was added immediately after radiation. Twenty-four hours after irradiation media was removed and fresh drug-free media was added. Colony-forming efficiency was determined 10-14 days later and survival curves were generated after normalizing for cell killing from drug alone. Values shown represent the means ± SEM for 3 independent experiments, *p < 0.05, **p < 0.1 according to Student's t test (INK128 vs. control).
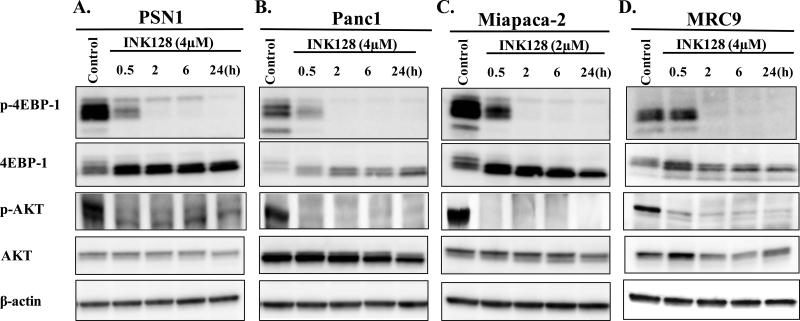
Immunoblot analysis was performed to determine the effects of INK128 treatment on mTOR activity using the concentrations from Figure 1. mTORC1 and mTORC2 activity was determined in each cell line after specified times of INK128 exposure (Figure 2A-D). Towards this end, the level of p-4E-BP1 (t37/46) was used as a marker of mTORC1 activity (5) and p-AKT (s473) was used as a marker of mTORC2 activity (10). INK128 exposure in all three tumor cell lines reduced activity of mTORC1 reaching a maximum inhibition by approximately 2h as judged by a decrease in p-4E-BP1 as well as the observed mobility shift from the slower migrating hyperphosporylated bands to the faster migrating hypophosphorylated bands seen in total-4E-BP1 (23). Treatment with INK128 also reduced mTORC2 activity reaching a maximum inhibition by approximately 2h as indicated by the decrease in p-AKT levels. The results presented in Figure 2 are thus consistent with an inhibitory effect of INK128 on both mTORC1 and mTORC22 activity as reported for other tumor cell lines (14). Of note, whereas treatment with INK128 did not enhance the radiosensitivity of normal MRC9 cells (Figure 1D), it did inhibit mTORC1 and mTORC2 activities to similar degrees as detected in the tumor cell lines (Figure 2D).
Figure 2.
Effects of INK128 on mTORC1/2 activity. A) PSN1, B) Panc1, C) Miapaca-2 or D) MRC9 cells were treated with the specified dose of inhibitor. Cells were collected at the specified time points and subjected to immunoblot analysis. Actin was used as a loading control.
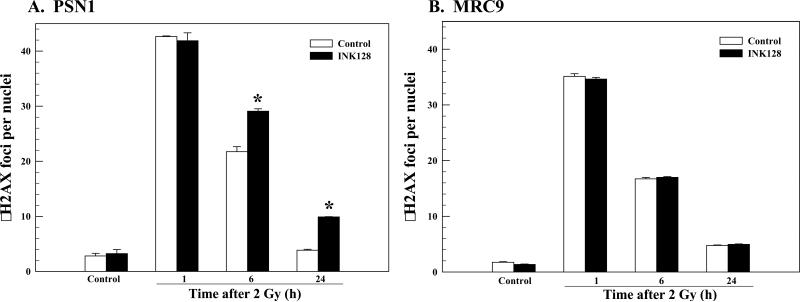
To begin to investigate the mechanisms mediating INK128-induced radiosensitization we focused on PSN1 cells. The critical lesion responsible for radiation-induced cell death is the DNA double strand break (DSB). Because γH2AX foci correspond to radiation-induced DSBs and their dispersal correlates with DSB repair (24-25), the effects of INK128 on radiation-induced γH2AX were evaluated in PSN1 cells (Figure 3A). In this study the same treatment protocol used for the clonogenic survival assays shown in Figure 1 was used: INK128 was added immediately after irradiation (2 Gy) with γH2AX nuclear foci determined at times out to 24h. No difference in foci levels was detected between control (vehicle) and INK128 treated cells at 1 hour after irradiation, suggesting that mTOR inhibition has no effect on the initial levels of radiation-induced DSBs. However, at 6 and 24h after irradiation the number of γH2AX foci remaining was significantly greater in the INK128 treated cells as compared to control cells. These results suggest that INK128-induced radiosensitization involves the inhibition of DNA DSB repair, at least as reflected by γH2AX foci dispersal. Although INK128 inhibited mTOR activity in MRC9 cells (Figure 2D), following the same treatment protocol as for PSN1 cells, it had no effect on radiation-induced γH2AX foci or their dispersal in these normal fibroblasts (Figure 3B), consistent with the lack of an enhancement of radiation-induced cell killing (Figure 1D).
Figure 3.
Influence of INK128 on radiation-induced γH2AX foci. A) PSN1 or B) MRC9 cells were exposed to the INK128 (4 μM) immediately after irradiation (2 Gy). Cells were collected at the specified time; γH2AX foci were counted in at least 50 nuclei per condition. Values shown represent the means ± SEM for 3 independent experiments, *p < 0.05 according to Student's t test (INK128 vs. control).
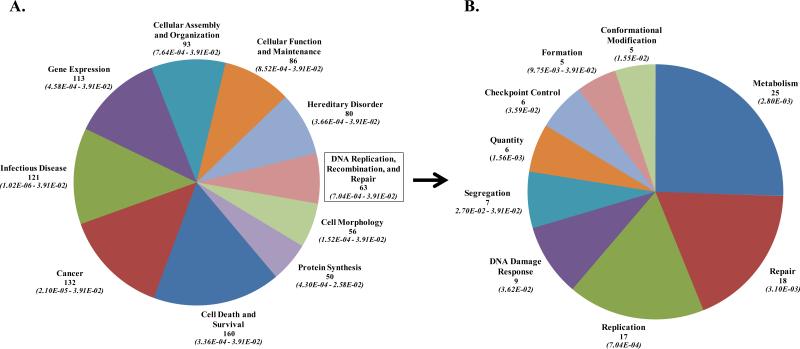
Given mTOR's role in regulating eIF4E availability and thus gene translation in response to environmental signals (5), we determined the effects of INK128 on radiation-induced gene translation profiles (1-2). For this study, PSN1 cells were irradiated (2 Gy) followed by the immediate addition of INK128 (4 μM), 6h later cells were collected for polysome isolation and the polysome-bound mRNA subjected to microarray analysis. INK128 treatment alone decreased the polysome abundance of 4741 genes and increased the polysome abundance of 4119 transcripts as compared to control cells, changes consistent with previous reports and indicating that inhibition of mTORC1/2 has a substantial effect on gene translation (14, 26). Radiation alone increased the abundance of 943 and decreased the abundance of 1238 transcripts in the polysome fraction compared to control cells. However, in cells receiving radiation followed by INK128, 808 of the 943 genes were no longer increased by radiation and 1114 of the 1238 genes were no longer decreased by radiation. These data suggest that mTORC1/2 plays a significant role in radiation-induced control of gene translation. To generate insight into potential functional consequences, the mRNAs no longer increased by radiation in the presence of INK128 (808) were subjected to IPA, which distributes genes into networks defined by known interactions and then matches these networks with specific biologically significant pathways. The top 10 biological functions of the mRNAs whose radiation-induced up-regulation was inhibited by INK128 (Figure 4A) included Cell Death and Survival, Cancer, Gene Expression and, of perhaps most relevance, DNA replication, recombination, and repair, which when further subdivided (Figure 4B) is shown to encompass a number of critical aspects of the DNA damage response, including DSB repair and checkpoint control. The specific genes comprising the DNA replication, recombination and repair biological function whose radiation-induced up-regulation was inhibited by INK128 are listed in Supplemental Table S1. The 10 networks identified by IPA and their associated functions are shown in Supplemental Table S2. Whereas there are a number of functions associated with these networks, of particular interest with respect to radiosensitivity are Networks 6 and 8, which again include genes associated with DNA replication, recombination, and repair. The data presented in Figure 4 and Supplemental Tables S1-2 indicate that mTORC1/2 inhibition suppresses the radiation-induced translation of functionally related mRNAs, many of which are related to DNA replication, recombination, and repair, consistent with the radiosensitizing effect shown in Figure 1. The 1114 genes that were no longer decreased by radiation after INK128 treatment were also subjected to IPA and the top 10 networks are shown in Supplemental Table S3. However, whereas these genes distributed to biological functions and networks, there was no obvious relationship to radiosensitivity.
Figure 4.
The effects of INK128 on radiation-induced gene translation. PSN1 cells were irradiated followed by addition of INK128 (4 μM), 6 hours later polysome-bound mRNA was isolated, subjected to microarray analysis and genes whose radiation-induced increase was prevented by INK128 were submitted to IPA. A.) Top 10 biological functions (containing 50 or more genes) of the 808 genes whose radiation-induced increase was prevented by INK128and B) the biological functions of the mRNAs (with more than 5 genes) within the DNA replication, recombination, and repair category are further delineated. For each biological function the number of genes whose radiation-induced increase was prevented by INK128 is listed (top number) along with the range in p-values (italicized in parentheses).
To evaluate the potential of INK128 to enhance radiosensitivity under in vivo conditions PSN1 cells were grown as leg tumor xenografts in nude mice. Initially, we determined the effects of INK128 on mTORC1 and mTORC2 activity according to immunohistochemical analysis of p-4E-BP1 (t37/46) and p-AKT (s473), respectively (Figure 5A). INK128 (1 or 3 mg/kg) was delivered by oral gavage; PSN1 tumors were collected 2 or 6h after treatment and processed for immunohistochemical analysis. As compared to controls, 2h after INK128 treatment (1 and 3 mg/kg) p-4E-BP1 and p-AKT staining were substantially reduced indicative of mTORC1 and mTORC2 inhibition, respectively. At 6h, although remaining below control levels, p-4E-BP1 and p-AKT staining began to return to untreated levels suggesting a recovery of mTOR activity. These results indicate that INK128 inhibits mTOR activity in PSN1 tumor xenografts.
A biochemical consequence of mTOR inhibition is a decrease in cap-dependent translation, which can be evaluated by eIF4F cap-complex formation. mTOR controls eIF4F cap-complex formation primarily by phosphorylation of the translation inhibitor 4E-BP1. Upon phosphorylation, 4E-BP1 is released from the 5’ mRNA cap followed by binding of eIF4G and subsequently translation initiation (27). Thus, to extend the immunohistochemical analysis of INK128-mediated mTOR inhibition to a functional endpoint, m7-GTP batch chromatography was used to evaluate eIF4F cap-complex formation (22, 28) in PSN1 tumor xenografts. Mice were treated with INK128 (3 mg/kg) and tumors collected 2 or 6 hours later (Figure 5B). At 2h after INK128 treatment eIF4F cap-complex formation was reduced in PSN1 tumors as indicated by the decrease in bound eIF4G and increase in bound 4E-BP1. Consistent with the immunohistochemical analyses of mTOR activity (Figure 5A) bound eIF4G begins to return to control levels at 6 h after treatment. These results indicate that INK128 inhibits eIF4F cap-complex formation in PSN1 xenografts. To our knowledge, this is the first data indicating that mTOR inhibition reduces eIF4F cap-complex formation in tumor xenografts.
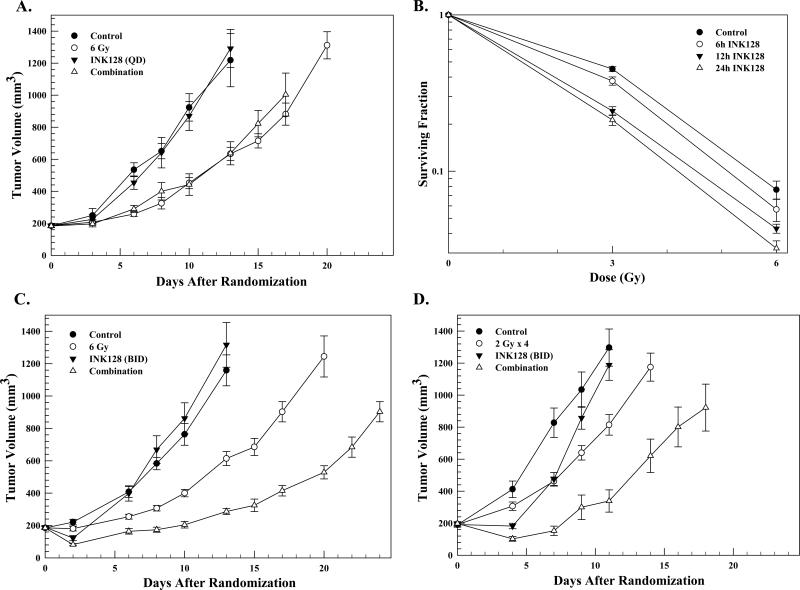
Using the data in Figure 5, a combination protocol was designed aimed at determining whether the INK128-mediated radiosensitization in vitro could be extended to an in vivo tumor xenograft model. Specifically, mice bearing PSN1 leg tumors (~180mm3) were randomized into four groups: vehicle, INK128 (3 mg/kg, oral gavage), radiation (6 Gy), and the combination of radiation and INK128 (INK128 delivered immediately after radiation). The growth rates of PSN1 tumors corresponding to each treatment are shown in Figure 6A. Whereas INK128 alone had no effect as compared to controls, radiation resulted in a significant decrease in tumor growth rate. However, there was no difference in tumor growth rates between the radiation only and combination treatment group indicating no enhancement of in vivo tumor radiosensitivity.
Figure 6.
A). PSN1 xenografts were locally irradiated (6 Gy) followed by a single dose of INK128 (3 mg/kg.) Each group contained six mice. Values represent the mean tumor volumes ± SEM. B.) PSN1 cells were plated in vitro at clonal density and allowed to attach overnight, irradiated and INK128 (4μM) was added immediately after irradiation. At the specified times after irradiation media was removed and fresh drug-free media was added; colonies were counted 10-14 days later. Values represent the means ± SEM of 3 independent experiments. C) Mice bearing PSN1 xenografts were treated with INK128 (1.5 mg/kg) 1h prior to tumor irradiation (6Gy) followed by a second drug dose 6h later; the next day INK128 (1.5 mg/kg) was delivered with each dose separated by 7h. INK128 alone contained 5 mice and all other groups contained 6 mice. Values represent the mean tumor volumes ± SEM. D) Mice bearing PSN1 xenografts were treated with INK128 (1.5 mg/kg) 1h before and 6h after each tumor irradiation (2 Gy) for 4 consecutive days; INK128 only was delivered on the fifth day (twice with each dose separated by 7h). The INK128/radiation combination group contained 6 mice and all other group contained 7 mice. Values represent the mean tumor volumes ± SEM.
mTOR activity in tumor xenografts begins to return as early as 6h after INK128 treatment (Figure 5), suggesting that the duration of mTOR inhibition after irradiation may be a determinant of INK128-induced radiosensitization. Under the in vitro conditions (Figure 1) that established INK128-mediated enhancement of tumor cell radiosensitivity, drug was added immediately after irradiation and not removed from the culture media until 24h later. To determine whether the duration of post-irradiation INK128 exposure was a variable in the radiosensitization induced under in vitro conditions, clonogenic survival analysis was performed using a treatment protocol in which INK128 was added to PSN1 culture media immediately after irradiation and cultures rinsed and fed drug-free media 6, 12, or 24h later (Figure 6B). In each of the 3 treatment protocols INK128 alone had no effect on the surviving fraction. Whereas removing INK128 12 or 24h after irradiation resulted in an increase in PSN1 radiosensitivity with DEFs of 1.23 and 1.33, respectively, removal of drug at 6h had no effect on radiosensitivity.
Based on these in vitro data suggesting that maintaining mTOR inhibition beyond 6h is critical for INK128-induced radiosensitization along with the in vivo data (Figure 5) indicating that mTOR activity in PSN1 tumors begins to return 6h after INK128 treatment, a tumor growth delay experiment was performed using a modified combination protocol. In this experiment, INK128 (1.5 mg/kg) was delivered 1h before and again 6h after radiation (6 Gy), followed the next day by two additional INK128 doses separated by 7h. It should be noted that the 1.5 mg/kg INK128 dose used in this experiment is half that used in Figure 6A, yet sufficient to reduce mTOR activity (Figure 5A). As shown in Figure 6C, whereas INK128 alone again had no effect on tumor growth rate, the INK128/radiation combination slowed tumor growth rate to a greater extent than radiation alone. For each group the time to grow from 180mm3 (volume of tumors at initiation of treatment) to 1000mm3 was calculated using the tumor volumes from the individual mice in each group (mean ± SEM). These data were then used to determine the absolute growth delays (the time in days for tumors in treated mice to grow from 180 to 1000 mm3 minus the time in days for tumors to reach the same size in vehicle treated mice). For PSN1 tumors the absolute growth delay for radiation alone was 6.3± 0.7 days. INK128 treatment alone had no effect on tumor growth delay. For tumors treated with the combination of INK128 and radiation the absolute growth delay was 12.3 ± 0.4 days, which is greater than the sum of the growth delays from the individual treatments and indicative of an enhancement of tumor radiosensitivity (p < 0.01 versus drug and radiation alone). The dose enhancement factor (DEF) obtained using this INK128 treatment protocol was 2.0.
The single radiation dose study was then extended to a fractionated radiation protocol. In this experiment, INK128 (1.5 mg/kg) was delivered 1h before and again 6h after each dose of radiation (2 Gy) delivered on 4 consecutive days with two additional INK128 doses separated by 7h on the fifth day. As shown in Figure 6D, this INK128 treatment protocol initially slowed tumor growth when delivered alone and increased tumor growth delay when combined with radiation as compared to radiation alone. The absolute growth delays for INK128 alone and radiation alone were 1.0 ± 0.6 and 3.7 ± 0.6 days, respectively. For tumors treated with the combination of INK128 and radiation the absolute growth delay was 10.1 ± 1.8 days, which is greater than the sum of the growth delays from the individual treatments and indicative of enhanced tumor radiosensitivity (p < 0.01 versus drug and radiation alone). The normalized tumor growth delay for the combination treatment, which accounts for the growth delay induced by drug alone, was 9.1 resulting in a DEF of 2.5. These results indicate that the combination of INK128 with single dose or fractionated radiation enhances tumor radiosensitivity. Moreover, during this study there was no appreciable weight loss or detectable skin toxicities.
Discussion
mTOR kinase serves as a central node integrating a number of signaling pathways to affect gene translation and ultimately regulate cell proliferation and survival (4). Given that these pathways are frequently activated or dysregulated in tumors, mTOR is considered a target for cancer therapy. In addition, because components of these pathways (e.g. RTKs, Ras, PI3K) have been associated with tumor cell radioresistance, mTOR has been suggested as a target for radiosensitization. Data indicating that eIF4E availability, which is regulated by mTOR via 4E-BP1 phosphorylation (7), can be a determinant of tumor cell radiosensitivity (3) also implicates mTOR as a target for radiosensitization. We recently reported that the competitive mTOR inhibitor PP242, in contrast to the allosteric inhibitor rapamycin, enhanced tumor cell radiosensitivity (13). To address the potential clinical relevance of this initial observation, we have extended this investigation to the competitive mTOR inhibitor INK128, which is currently undergoing clinical trials as a single agent and in combination with paclitaxel and/or trastuzumab (15). As shown, INK128 inhibited mTORC1 and mTORC2 activity in 3 pancreatic carcinoma cell lines, which was accompanied by an enhancement in radiosensitivity. Although INK128 also inhibited mTOR activity in a normal fibroblast cell line, there was no corresponding enhancement in radiation-induced cell death. These results are thus consistent with eIF4E knock down preferentially enhancing the radiosensitivity of tumor over normal cell lines (3) and the role of mTOR as a downstream hub for oncogenic signaling processes (29), a number of which can also influence radiosensitivity.
Based on the analysis of radiation-induced γH2AX foci, the mechanism of INK128-induced radiosensitization appears to involve an inhibition of DNA DSB repair. Furthermore, clonogenic survival analysis showing that maintenance of mTOR inhibition for greater than 6h was required for radiosensitization suggests that INK128 inhibits a later stage of DNA DSB repair. Although the direct interaction of mTOR or one of its substrates with a component of the DNA repair machinery cannot be eliminated, the role of mTOR as a critical regulator of gene translation may play a role in the inhibition of DSB repair. Consistent with this hypothesis, the gene translation profiles generated from polysome-bound RNA indicate that INK128 exposure suppresses the radiation-induced translation of specific mRNA subpopulations; a major subset of these mRNAs corresponded to those coding for proteins involved in DNA replication, recombination, and repair. These results are in line with a recent report showing that the knockdown of eIF4G (another component of the eIF4F complex) inhibits the radiation-induced translation of a number of mRNAs related to DNA damage and repair (30). Thus, whereas additional studies are required, INK128-induced radiosensitization appears to involve an inhibition of radiation-induced gene translation leading to an inhibition of DSB repair.
A critical aspect in the preclinical evaluation of a putative radiosensitizing agent is the extension of in vitro studies to an in vivo tumor xenograft model. Such studies typically involve not only defining tumor radioresponse, but also determining the ability of the agent to target the molecule in question under in vivo conditions. However, despite clearly establishing target inhibition in vivo at the time of irradiation (Figure 5), a single dose of INK128 was found to have no effect on the radiation-induced growth delay of PSN1 xenografts. This suggested that in contrast to in vitro conditions, that there was an additional requirement in terms of mTOR inhibition that was not satisfied in this initial in vivo experiment. Along these lines, because in vitro data indicated that INK128-induced radiosensitization was mediated via processes operative for 6-12h after irradiation (Figure 6B), we tested the hypothesis that mTOR inhibition had to be maintained in vivo for longer than the approximately 6h achieved by a single dose (Figure 5). Accordingly, when mice were treated with INK128 before and 6h after irradiation there was a significant enhancement of tumor radioresponse. This study thus illustrates not only the significance of understanding target dynamics in terms of the mechanisms of radiosensitization, but also suggests that this is a critical variable in the design of any potential clinical protocol combining INK128 and radiotherapy.
Supplementary Material
Translational Relevance.
INK128 is a clinically applicable, competitive inhibitor of mTOR kinase. In contrast to allosteric inhibitors (e.g. rapamycin), INK128 more completely inhibits mTORC1 and inhibits mTORC2, events that may lead to enhanced tumor radiosensitivity. Radiotherapy is a primary treatment modality for pancreatic carcinoma, tumors associated with hyperactivation of the mTOR pathway. Towards determining the potential of mTOR to serve as a target for radiosensitization in this tumor type, we initially showed that INK128 enhances the in vitro radiosensitivity of human pancreatic carcinoma cell lines. Treatment of mice bearing pancreatic carcinoma xenografts with INK128 was then shown to decrease tumor mTORC1 and mTORC2 activities along with eIF4F cap-complex formation. Whereas INK128 alone had no effect on tumor growth rate, it significantly increased the tumor growth delay induced by single and fractionated irradiation. These data suggest that the combination of INK128 with radiotherapy may be of benefit in the treatment of pancreatic carcinoma.
Footnotes
Conflict of Interest: none
References
- 1.Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–61. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- 2.Kumaraswamy S, Chinnaiyan P, Shankavaram UT, Lu X, Camphausen K, Tofilon PJ. Radiation-induced gene translation profiles reveal tumor type and cancer-specific components. Cancer Res. 2008;68:3819–26. doi: 10.1158/0008-5472.CAN-08-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayman TJ, Williams ES, Jamal M, Shankavaram UT, Camphausen K, Tofilon PJ. Translation initiation factor eIF4E is a target for tumor cell radiosensitization. Cancer Res. 2012;72:2362–72. doi: 10.1158/0008-5472.CAN-12-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 6.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–80. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16:4914–20. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle. 2009;8:567–72. doi: 10.4161/cc.8.4.7659. [DOI] [PubMed] [Google Scholar]
- 9.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–61. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K, Shi C, Toral-Barza L, Lucas J, Shor B, Kim JE, et al. Beyond rapalog therapy: preclinical pharmacology and antitumor activity of WYE-125132, an ATP-competitive and specific inhibitor of mTORC1 and mTORC2. Cancer Res. 2010;70:621–31. doi: 10.1158/0008-5472.CAN-09-2340. [DOI] [PubMed] [Google Scholar]
- 13.Hayman TJ, Kramp T, Kahn J, Jamal M, Camphausen K, Tofilon PJ. Competitive but Not Allosteric mTOR Kinase Inhibition Enhances Tumor Cell Radiosensitivity. Transl Oncol. 2013;6:355–62. doi: 10.1593/tlo.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ClinicalTrials.gov . [database on the Internet] National Library of Medicine (US); Bethesda (MD): 2000. [2013 June 1]. Available from: http://clinicaltrials.gov/ [Google Scholar]
- 16.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, et al. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72:282–93. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 18.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009;6:699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 19.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 20.Bellizzi AM, Bloomston M, Zhou XP, Iwenofu OH, Frankel WL. The mTOR pathway is frequently activated in pancreatic ductal adenocarcinoma and chronic pancreatitis. Appl Immunohistochem Mol Morphol. 2010;18:442–7. doi: 10.1097/PAI.0b013e3181de115b. [DOI] [PubMed] [Google Scholar]
- 21.Bae H, Gray JS, Li M, Vines L, Kim J, Pestka JJ. Hematopoietic cell kinase associates with the 40S ribosomal subunit and mediates the ribotoxic stress response to deoxynivalenol in mononuclear phagocytes. Toxicol Sci. 2010;115:444–52. doi: 10.1093/toxsci/kfq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Yue P, Deng X, Ueda T, Fukunaga R, Khuri FR, et al. Protein phosphatase 2A negatively regulates eukaryotic initiation factor 4E phosphorylation and eIF4F assembly through direct dephosphorylation of Mnk and eIF4E. Neoplasia. 2010;12:848–55. doi: 10.1593/neo.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–9. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–9. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 26.Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, et al. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A. 2012;109:8977–82. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh AC, Truitt ML, Ruggero D. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer. 2011;105:329–36. doi: 10.1038/bjc.2011.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–56. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badura M, Braunstein S, Zavadil J, Schneider RJ. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci U S A. 2012;109:18767–72. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.