Abstract
SWI/SNF chromatin remodeling complexes are pleomorphic multi-subunit cellular machines that utilize the energy of ATP hydrolysis to modulate chromatin structure. The complexes interact with transcription factors at promoters and enhancers to modulate gene expression and contribute to lineage specification, differentiation and development. Initial clues to a role in tumor suppression for SWI/SNF complexes came over a decade ago when the gene encoding the SMARCB1/SNF5 core subunit was found specifically inactivated in nearly all pediatric rhabdoid tumors. In the last 3 years, cancer genome sequencing efforts have revealed an unexpectedly high mutation rate of SWI/SNF subunit genes, which are collectively mutated in 20% of all human cancers and approach the frequency of p53 mutations. Here we provide a background on these newly recognized tumor suppressor complexes, discuss mechanisms implicated in the tumor suppressor activity, and highlight findings that may lead to potential therapeutic targets for SWI/SNF mutant cancers.
Background
Chromatin and SWI/SNF complexes
Nuclear DNA wrapped around a histone octamer constitutes the nucleosome, the basic unit of chromatin. Further compaction of DNA through progressive coiling provides an organizational structure for the two meters of DNA contained within each cell but also presents an access barrier to the transcriptional machinery. Numerous chromatin-modifying complexes exist in mammalian cells and these are intimately involved in processes that require DNA access such as transcription, replication and repair. These complexes can be grouped into two classes: those that covalently modify nucleosomes and those, like the SWI/SNF complex, that consume ATP to mobilize nucleosomes and modulate chromatin compaction.
SWI/SNF complexes are evolutionarily conserved and were originally identified in yeast. Genes encoding SWI/SNF subunits were revealed in screens of yeast for defects in mating type SWItching and in sucrose metabolism (Sucrose Non-Fermentable, SNF)(1). Mammalian SWI/SNF complexes are also referred to as BAF (BRG1 associated factors) complexes in recognition that mammalian complexes contain additional subunits not found in the yeast complex and therefore the extent to which activities are conserved remains unclear(2). Mammalian SWI/SNF complexes (herein referred to simply as SWI/SNF complexes) are enriched at promoters and enhancers of active genes and have been shown to contribute to regulation of differentiation and proliferation across many lineages(3-5). SWI/SNF complexes are large, ~2 MDa, and composed of 12-15 subunits(2). These complexes are comprised of one of two mutually exclusive catalytic ATPase subunits: SMARCA2 (Brahma or BRM) or SMARCA4 (BRM/SWI2 related gene 1, or BRG1), and a set of widely expressed core subunits that include SMARCB1 (SNF5, INI-1 or BAF47), SMARCC1 (BAF155) and SMARCC2 (BAF170)(2). In addition, SWI/SNF complexes also contain a large number of lineage-restricted subunits often encoded by multi-gene families (Table 1). Recent work has demonstrated that these complexes may further contain additional subunits not previously appreciated(6). Considering the large number of variant subunits, it has been estimated that several hundred versions of SWI/SNF complexes may exist, each with a conserved core of subunits but containing distinct combinations of variant subunits(7).
Table 1.
Summary of the SWI/SNF complex subunits and mutations in cancers
| Subunit | Aliases | Function | Mutated in human cancer |
|---|---|---|---|
| ARID1A | BAF250A | Variant subunit, BAF complex only | Ovarian; hepatocellular; bladder; gastric; endometrioid; pancreatic; colon; lung; neuroblastoma; Burkitt lymphomas |
| ARID1B | BAF250B | Variant subunit, BAF complex only | Melanoma; neuroblastoma; hepatocellular; pancreatic; liver |
| PBRM1 | BAF180 | Variant subunit, PBAF complex only | renal cell carcinomas; breast, gastric and pancreatic |
| ARID2 | BAF200 | Variant subunit, PBAF complex only | Melanoma; hepatocellular; pancreatic |
| SMARCA2 | BRM | Catalytic ATPase subunit | Lung, colon, breast |
| SMARCA4 | BRG1 | Catalytic ATPase subunit | Lung, medulloblastoma; Burkitt lymphomas |
| SMARCB1 | SNF5, INI1 | Core subunit | Rhabdoid tumor; Familial schwannomatosis |
| SMARCC1 | BAF155 | Core subunit | |
| SMARCC2 | BAF170 | Core subunit | |
| SMARCD1/2/3 | BAF60A/B/C | Variant subunit | Breast |
| SMARCE1 | BAF57 | Variant subunit | Spinal mengioma |
| ACTL6A/B | BAF53A/B | Variant subunit | |
| PHF10 | BAF45A | Variant subunit | |
| DPF1/2/3 | BAF45B/C/D | Variant subunit | |
| ACTB | Beta-Actin |
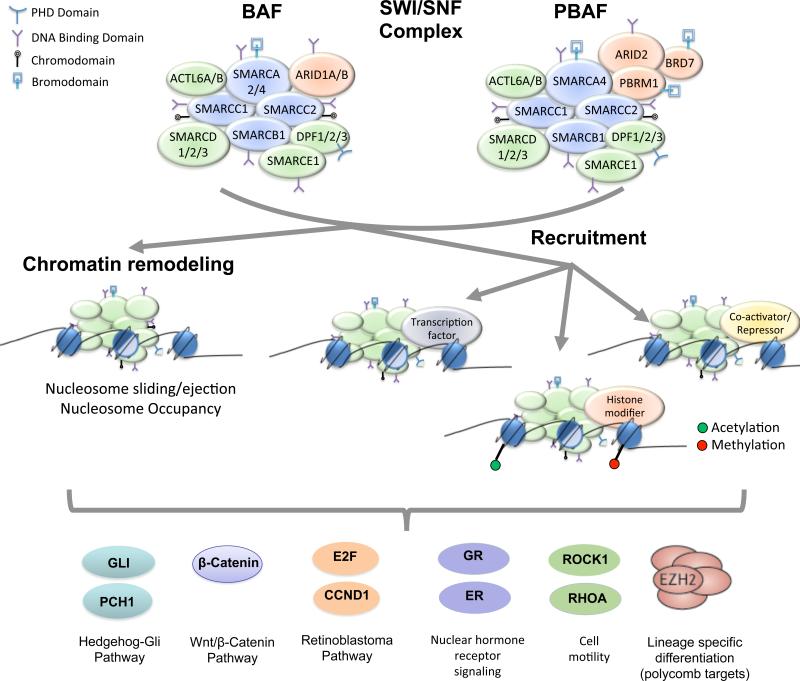
The precise biochemical function of SWI/SNF complexes remains somewhat unclear. In vitro assays have clearly demonstrated that the complexes are capable of mobilizing and ejecting histone octamers on DNA(8). Functional studies performed to evaluate biochemical activity of SWI/SNF complexes in living cells have implicated the complexes in the establishment of nucleosome occupancy and phasing at promoters and enhancers at a subset of active genes(3-5), as well as in DNA repair processes(9-11). Recent studies have begun to demonstrate that while SWI/SNF complexes may be ubiquitously expressed, individual cells contain a select set of variant subunits that contribute to lineage-specific targeting and determination of cell fate(3-5). Perhaps some of the clearest evidence has come from studies on neural differentiation, which showed SWI/SNF complex composition undergoes an essential subunit switch during the progression from neural progenitors to post-mitotic neurons(12). Similarly, embryonic stem (ES) cells have been shown to contain a distinctive assembly of SWI/SNF subunits essential for ES cell maintenance and pluripotency(13). Such interaction with and recruitment of lineage-specific transcriptional regulators appear to be a central mechanism by which SWI/SNF complexes contribute to lineage specification. For example, MyoD, the muscle determination factor, can be directly incorporated into SWI/SNF complexes, which then results in transcription of MyoD-target genes(14). Similarly, Olig2 has been shown to physically associate with SWI/SNF complex at oligodendrocyte specific enhancers during differentiation(15). SWI/SNF complexes may contribute specificity to transcription factor interactions and targeting via subunits that contain DNA-binding and histone binding domains, including HMG, ARID, HSA, PHD, and Bromodomains, which are found in both core and variant SWI/SNF subunits(7). Thus, regulation of gene expression and cell lineage specification via direct recruiting or interaction with key transcription factors may be a central function of SWI/SNF complexes (Figure 1).
Figure 1. SWI/SNF functions in chromatin modulation and tumor suppression.
Two subclasses, BAF and PBAF, of many variants of SWI/SNF complexes are shown. BAF and PBAF differ in composition of the subunits shown in orange. Core subunits are shown in blue. Further diversity is derived from the subunits shown in green as these are all encoded by multi gene families, each yielding a slightly different amino acid sequence. These diverse complexes then make specific contributions to targeting of chromatin remodeling and transcriptional control. Several of the pathways that have been implicated in tumor suppression are shown at bottom.
SWI/SNF complex mutations in cancer
In 1998, specific biallelic inactivating mutations of the core SWI/SNF subunit SMARCB1 (SNF5/INI1/BAF47) were identified in the vast majority of cases of rhabdoid tumors (RT), highly aggressive and lethal cancers arising in the kidney, brain, and soft tissues of young children (16, 17). The bona fide tumor suppressor properties of SMARCB1 were subsequently demonstrated in genetically engineered mouse models: while homozygous loss results in embryonic lethality, heterozygous mice are viable and thirty percent develop cancers that closely resemble human RT(18-20). In these tumors, the wildtype allele has been spontaneously inactivated so, as in humans, the tumors completely lack SMARCB1. In order to bypass embryonic lethality associated with homozygous loss, conditional SMARCB1 alleles were generated. Conditional homozygous inactivation in post-natal mice results in cancer in 100% of mice at a median onset of only 11 weeks, a remarkably rapid rate of onset for inactivation of a single gene(21). In comparison, biallelic inactivation of p53 leads to cancer at a median of 22 weeks.
While rhabdoid tumors served as the initial clue linking mutation of genes encoding SWI/SNF subunits to cancer, recent cancer genome sequencing studies have revealed a much broader role(22-24). Only 3 years ago, two papers reported that more than 50% of ovarian clear cell carcinomas contain inactivating mutations in ARID1A(25, 26), which encodes a variant subunit of the complexes. Subsequently, frequent mutations of SWI/SNF complex subunits have been identified in a variety of human cancers (Table 1) (Selected references and summary reviews: 6, 22-24, 27-46). So far, at least eight genes encoding SWI/SNF complex subunits have been found to be recurrently mutated in a variety of cancers including subsets of lung, breast, stomach, liver, pancreas, kidney, bladder, skin and brain cancers. These mutations are predominantly loss of function(6, 22-24, 27). Interestingly, while SMARCB1 is always biallelically mutated, other subunits are often heterozygously mutated, raising the possibility of haploinsufficiency for these subunits in tumor suppression(6, 27). Thus far SMARCB1 and SMARCA4 have been validated to have bona fide tumor suppressor function using genetically engineered mouse models(18-21, 47), while others remain to be tested. Collectively, genes encoding SWI/SNF complex subunits have now been recognized as one of the most commonly mutated targets in human cancer, being thus far detected in 20% of human cancers(6, 48).
Genetics or Epigenetics?
Despite the prevalence of SWI/SNF complex mutations in cancer, insight into the mechanisms underlying tumor suppression and the reason for the differing cancer spectrum associated with each subunit is still in its infancy. In addition to a role in transcriptional regulation, several studies have linked the complex to DNA repair including nucleotide excision repair, double-strand break repair and DNA decatenation(9-11, 49). Consequently, it has been a question of great interest whether the tumor suppressor activity of the complex arises via a role in the control of transcriptional programs or whether it is derived from a role for the complex in protecting genome integrity. Perhaps providing substantial insight, RT are diploid, chromosomally stable and contain a marked paucity of aberrations detectable by SNP arrays, other than loss of SMARCB1 itself(50). More recently, exome sequencing of human RT revealed that despite their highly aggressive nature these cancers contain an extremely low rate of mutations(51) – one of the lowest ever measured(52). Across 35 of these SMARCB1-deficient cancers, there were essentially no other recurrent mutations and in two of the cancers there were no other identified mutations at all. The same conclusion was reached in other studies as well(53, 54). Given the paucity of other mutations and the extremely rapid onset of cancer in mouse models, these findings seem to suggest that SMARCB1-deficiency may not cause cancer via defects in DNA repair but rather due to epigenetic alterations such as disruption of chromatin-based contributions to control of cell fate. Given that at least eight subunits of the SWI/SNF complex are recurrently mutated in cancer, it is tempting to speculate that the general mechanism is likely shared. However, while evidence seems to suggest SMARCB1 loss driving cancer via an epigenetic mechanism, whether a similar mechanism underlies cancer associated with ARID1A, SMARCA4, PBRM1 and other subunit mutations remains to be determined. Interestingly mutations in more than one SWI/SNF subunit gene can occur in primary tumors perhaps reflecting both haploinsufficiency and compound heterozygous effects (6, 47).
Clinical–Translational Advances
As increasing evidence has linked mutation of the SWI/SNF complex to a variety of human cancers, there has been a growing desire to identify vulnerabilities created by SWI/SNF mutation. Unlike some activating oncogene mutations such as BCR-ABL fusion or EGFR mutation, which can be directly therapeutically targeted, the majority of SWI/SNF complex mutations are inactivating. Thus the proteins are absent and cannot be directly targeted. Thus a goal should be to achieve a better understanding of the mechanisms underlying transformation and dependencies created by mutation of SWI/SNF subunits. Indeed, early studies on the basic function of SWI/SNF complexes have revealed potential vulnerabilities currently being translated.
Pathways affected by SWI/SNF mutations
Some of the earliest studies of cancer associated gene mutations, identification of genes activated by chromosomal translocation in acute leukemias, revealed the theme that these genes were often transcription factors that were master regulators of lineage development such that their forced overexpression resulted in perturbation of lymphoid development and predisposition to transformation. In its role of interacting with transcription factors to facilitate lineage specific development, it is conceivable that mutation of SWI/SNF complexes predispose to cancer in much the same way – via disruption of development, proliferation and differentiation pathways. However, relevant target pathways are only beginning to be characterized.
One of the challenges for the field is that while SMARCB1 was recognized as a tumor suppressor 15 years ago, other subunits have only been linked to cancer recently. Consequently, mechanisms underlying the role of SMARCB1 in tumor suppression have been more extensively investigated. However, it remains to be determined the extent to which these mechanisms are shared with cancers driven by other SWI/SNF subunit mutations. Collectively, a shared mechanism of perturbation of SWI/SNF function seems likely. However, as the spectrum of cancer types associated with each subunit varies, the specific pathways affected may depend upon the subunit mutated. Ultimately, additional studies will be needed to determine the extent to which vulnerabilities caused by mutation of one SWI/SNF subunit gene extend to others.
The tumor suppressor activity of SWI/SNF complexes was first linked to the RB tumor suppressor pathway when inactivation of SMARCB1 was shown to down-regulate p16INK4A expression, a cyclin-dependent kinase (CDK) inhibitor that regulates the RB pathway (55-59). SWI/SNF complexes can also bind to RB itself and facilitate the repression of RB target genes, such as E2Fs and CCND1(60-62), suggesting a contribution of SWI/SNF complexes to cell cycle regulation. Some, but not all, studies have found high level CyclinD1 expression in RT, and pharmacological inhibitors of Cyclin D1 have been reported effective in reducing the growth of RT cell lines in vitro and in vivo (50, 63-66). Collectively, these findings have lead to a Phase I/II clinical trial of RT using CDK4 inhibitors (http://clinicaltrials.gov/show/NCT01747876).
A number of other target pathways and functions have been implicated in cancer driven by SWI/SNF mutations. For example, SWI/SNF complexes have been implicated in the control of cell motility via regulation of RhoA (67), and actin cytoskeletal organization (68). SWI/SNF complexes can also directly interact with MYC, an oncogene frequently overexpressed in cancer(69), and SMARCB1-deficient RTs show activation of MYC programs(70, 71). SWI/SNF complexes interact with nuclear hormone receptors (NHRs), such as glucocorticoid receptors, estrogen receptors, and retinoic acid receptors(72). Clinical studies found that decreased levels of SWI/SNF subunits SMARCB1/ SMARCA4/ARID1A correlate with steroid resistance in pediatric Acute Lymphoblastic Leukemia (ALL), with the underlying mechanism remaining unknown(73). SWI/SNF complexes have also been implicated in regulation of the Wnt/β-catenin pathway. BRG1 deletion results in down-regulation of the Wnt receptor family and degradation of β-catenin(74). The telomerase protein component TERT (telomerase reverse transcriptase) can also interact with BRG1, to modulate the Wnt/β-catenin pathway(75) and another study reported that loss of SMARCB1 results in β-catenin hyperactivation(76). SWI/SNF complexes also directly interact with Gli1, the effector of Hedgehog signaling and in RT cells, loss of SMARCB1 leads to an aberrant activation of the Hedgehog-Gli pathway(77). These results suggest that SWI/SNF complexes directly regulate the canonical Wnt/β-catenin and Hedgehog pathways. Collectively, SWI/SNF complexes have been implicated in the control of a number of cancer-related transcriptional pathways, some of which may provide therapeutic potential (Figure 1). Via such investigations a theme is also emerging that SWI/SNF mutations may directly affect genes and pathways at the level of chromatin and thus uncouple pathways from upstream canonical control. For example, cancers in which the Hedgehog pathway is activated by mutation of the PTCH receptor are sensitive to blockade of SMO downstream in the signaling cascade. In contrast SMO inhibitors are unable to block Hedgehog signaling in SMARCB1 mutant cancers. Similarly, the Wnt/β-catenin can be inhibited at multiple levels. However, none of these upstream inhibitors have an effect upon the Wnt pathway in SMARCB1 mutant cancers where a key contributor appears to be failure of the constitutive Wnt repressor TCF to bind target DNA in the absence of SMARB1 thus resulting in spontaneous Wnt signaling uncoupled from canonical pathway control. While still early in the understanding of mechanism, it is tempting to speculate that chromatin remodeler mutations, such as SWI/SNF, may provide an explanation for findings such as the discrepancy that EGFR mutant cancers are susceptible to EGFR blockade while cancers that show transcriptional activation of the EGFR pathway, but lack mutations in EGFR itself, are unresponsive. Consequently, disruption of chromatin structure may have the potential to directly alter pathway signaling. Therefore, gaining insight into the function of SWI/SNF complexes in transcriptional control may offer novel approaches modulation of cancer-related pathways.
Antagonism of SWI/SNF complex and polycomb complex – Targeting EZH2?
High mutation rate of SWI/SNF chromatin remodelers in a large variety of cancer suggests a chromatin-based epigenetic mechanism that drives cancer development by regulating chromatin structure. Therefore, mutations of SWI/SNF complexes likely lead to an aberrant chromatin landscape, providing the driving force of oncogenesis. Recent discoveries describing an epigenetic antagonism between Polycomb and the SWI/SNF complex supports this model(70, 78). Loss of SMARCB1 was found to cause elevated levels of EZH2 and H3K27me3 at PcG target genes thus leading to broad repression of these targets(70). Another study in ES cells found complex interactions between SWI/SNF and Polycomb complexes, at times antagonistic and at others cooperative(4). In a genetically engineered in vivo tumor model, inactivation of EZH2 completely blocked tumor onset driven by SMARCB1 loss(70). This discovery suggested EZH2 as a target for SMARCB1 mutant cancers. In contrast to these findings, another group found that knock down of SMARCB1 doesn't affect the level of EZH2(79), raising the possibility that antagonism between SMARCB1 and EZH2 might be complex and cell-context dependent(79, 80). Nonetheless, a recent study reported that targeted pharmacological inhibition of EZH2 results in durable regressions in genetically altered malignant rhabdoid tumors, suggesting this as a potential new treatment for RT(81), stimulating interest in therapeutic trials for RT. Whether antagonism between SMARCB1 and EZH2 extends to other SWI/SNF subunits and associated cancers needs to be further investigated.
Summary/Prospective
Cancer genome sequencing studies have identified genes encoding subunits of the SWI/SNF chromatin remodeling complexes as being frequently mutated across a wide variety of cancers. However, identification of these links is largely recent and the mechanisms by which these complexes act as tumor suppressors are only just beginning to be elucidated. While some evidence indicates that these complexes may contribute to DNA repair, involvement of these complexes in chromatin-based epigenetic regulation has been implicated in their tumor suppressor activity. Ultimately, understanding the fundamental biology of SWI/SNF complexes, such roles in the regulation of chromatin structure/landscape, the biochemical contributions of different subunits, and activity in modulating transcription, offer promise for novel therapeutic approaches to SWI/SNF mutant cancers.
Acknowledgments
The authors thank Katherine C. Helming for critical reading of the manuscript.
Grant Support
The work in Dr. Roberts’ laboratory is partly supported by R01CA172152 (CWMR) and R01CA113794 (CWMR), and a U01 NCI Mouse Models of Cancer Consortium Award (CWMR). Alex's Lemonade Stand Foundation, Hyundai Hope on Wheels, the Cure AT/RT Now fund, Garrett B. Smith Foundation, Miles for Mary, The Ellison Foundation and Cookies for Kids Cancer provided additional support. X.W. is supported by St. Baldrick's Foundation, the Rally Foundation and David Abraham Foundation.
Footnotes
Conflict of Interest statement:
CWMR, Commercial Research Grant and Consultant, Novartis Institutes for Biomedical Research
References
- 1.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–58. doi: 10.1093/genetics/108.4.845. Epub 1984/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. The EMBO journal. 1996;15:5370–82. Epub 1996/10/01. [PMC free article] [PubMed] [Google Scholar]
- 3.Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10165–70. doi: 10.1073/pnas.1302209110. Epub 2013/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5187–91. doi: 10.1073/pnas.0812888106. Epub 2009/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome research. 2011;21:1650–8. doi: 10.1101/gr.121145.111. Epub 2011/07/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. Epub 2013/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–6. doi: 10.1016/j.cell.2009.01.009. Epub 2009/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. Epub 1999/03/17. [DOI] [PubMed] [Google Scholar]
- 9.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Molecular and cellular biology. 2002;22:6779–87. doi: 10.1128/MCB.22.19.6779-6787.2002. Epub 2002/09/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. The EMBO journal. 2006;25:3986–97. doi: 10.1038/sj.emboj.7601291. Epub 2006/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nature structural & molecular biology. 2006;13:902–7. doi: 10.1038/nsmb1152. Epub 2006/10/03. [DOI] [PubMed] [Google Scholar]
- 12.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15. doi: 10.1016/j.neuron.2007.06.019. Epub 2007/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. Epub 2009/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nature genetics. 2001;27:187–90. doi: 10.1038/84826. Epub 2001/02/15. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–61. doi: 10.1016/j.cell.2012.12.006. Epub 2013/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. Epub 1998/07/22. [DOI] [PubMed] [Google Scholar]
- 17.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer research. 1999;59:74–9. Epub 1999/01/19. [PubMed] [Google Scholar]
- 18.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13796–800. doi: 10.1073/pnas.250492697. Epub 2000/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO reports. 2000;1:500–6. doi: 10.1093/embo-reports/kvd129. Epub 2001/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Molecular and cellular biology. 2001;21:3598–603. doi: 10.1128/MCB.21.10.3598-3603.2001. Epub 2001/04/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer cell. 2002;2:415–25. doi: 10.1016/s1535-6108(02)00185-x. Epub 2002/11/27. [DOI] [PubMed] [Google Scholar]
- 22.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. Epub 2013/04/02. [DOI] [PubMed] [Google Scholar]
- 23.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews Cancer. 2011;11:481–92. doi: 10.1038/nrc3068. Epub 2011/06/10. [DOI] [PubMed] [Google Scholar]
- 24.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. Epub 2013/04/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. The New England journal of medicine. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. Epub 2010/10/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. Epub 2010/09/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer discovery. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. Epub 2012/12/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature genetics. 2012;44:760–4. doi: 10.1038/ng.2291. Epub 2012/05/29. [DOI] [PubMed] [Google Scholar]
- 29.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nature genetics. 2012;44:1117–21. doi: 10.1038/ng.2391. Epub 2012/08/28. [DOI] [PubMed] [Google Scholar]
- 30.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nature genetics. 2013;45:12–7. doi: 10.1038/ng.2493. Epub 2012/12/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith MJ, O'Sullivan J, Bhaskar SS, Hadfield KD, Poke G, Caird J, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nature genetics. 2013;45:295–8. doi: 10.1038/ng.2552. Epub 2013/02/05. [DOI] [PubMed] [Google Scholar]
- 32.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nature genetics. 2011;43:1219–23. doi: 10.1038/ng.982. Epub 2011/11/01. [DOI] [PubMed] [Google Scholar]
- 33.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. Epub 2012/07/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. Epub 2013/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. Epub 2011/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E252–9. doi: 10.1073/pnas.1114817109. Epub 2012/01/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. Epub 2012/07/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. Epub 2012/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature genetics. 2011;43:875–8. doi: 10.1038/ng.907. Epub 2011/08/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature genetics. 2012;44:694–8. doi: 10.1038/ng.2256. Epub 2012/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nature genetics. 2012;44:1310–5. doi: 10.1038/ng.2455. Epub 2012/10/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Streppel MM, Lata S, Delabastide M, Montgomery EA, Wang JS, Canto MI, et al. Next- generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus. Oncogene. 2013 doi: 10.1038/onc.2012.586. Epub 2013/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. Epub 2012/10/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–4. doi: 10.1182/blood-2012-06-437624. Epub 2012/10/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome research. 2012;22:2109–19. doi: 10.1101/gr.145144.112. Epub 2012/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome research. 2012;22:2120–9. doi: 10.1101/gr.137596.112. Epub 2012/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–8. doi: 10.1038/sj.onc.1210664. Epub 2007/07/20. [DOI] [PubMed] [Google Scholar]
- 48.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PloS one. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. Epub 2013/01/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–7. doi: 10.1038/nature12146. Epub 2013/05/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna ES, Sansam CG, Cho YJ, Greulich H, Evans JA, Thom CS, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Molecular and cellular biology. 2008;28:6223–33. doi: 10.1128/MCB.00658-08. Epub 2008/08/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. The Journal of clinical investigation. 2012;122:2983–8. doi: 10.1172/JCI64400. Epub 2012/07/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. Epub 2013/06/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasselblatt M, Isken S, Linge A, Eikmeier K, Jeibmann A, Oyen F, et al. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes, chromosomes & cancer. 2013;52:185–90. doi: 10.1002/gcc.22018. Epub 2012/10/18. [DOI] [PubMed] [Google Scholar]
- 54.Hoell JI, Gombert M, Bartenhagen C, Ginzel S, Husemann P, Felsberg J, et al. Whole-genome paired-end analysis confirms remarkable genomic stability of atypical teratoid/rhabdoid tumors. Genes, chromosomes & cancer. 2013 doi: 10.1002/gcc.22092. Epub 2013/08/10. [DOI] [PubMed] [Google Scholar]
- 55.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LM, Mohd-Sarip A, et al. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. The Journal of biological chemistry. 2004;279:3807–16. doi: 10.1074/jbc.M309333200. Epub 2003/11/08. [DOI] [PubMed] [Google Scholar]
- 56.Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17745–50. doi: 10.1073/pnas.0509014102. Epub 2005/11/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staehling-Hampton K, Ciampa PJ, Brook A, Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–87. doi: 10.1093/genetics/153.1.275. Epub 1999/09/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strobeck MW, Knudsen KE, Fribourg AF, DeCristofaro MF, Weissman BE, Imbalzano AN, et al. BRG-1 is required for RB-mediated cell cycle arrest. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7748–53. doi: 10.1073/pnas.97.14.7748. Epub 2000/07/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–203. doi: 10.1038/sj.onc.1205706. Epub 2002/08/01. [DOI] [PubMed] [Google Scholar]
- 60.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11268–73. doi: 10.1073/pnas.94.21.11268. Epub 1997/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Versteege I, Medjkane S, Rouillard D, Delattre O. A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle. Oncogene. 2002;21:6403–12. doi: 10.1038/sj.onc.1205841. Epub 2002/09/13. [DOI] [PubMed] [Google Scholar]
- 62.Biegel JA, Kalpana G, Knudsen ES, Packer RJ, Roberts CW, Thiele CJ, et al. The role of INI1 and the SWI/SNF complex in the development of rhabdoid tumors: meeting summary from the workshop on childhood atypical teratoid/rhabdoid tumors. Cancer research. 2002;62:323–8. Epub 2002/01/10. [PubMed] [Google Scholar]
- 63.Alarcon-Vargas D, Zhang Z, Agarwal B, Challagulla K, Mani S, Kalpana GV. Targeting cyclin D1, a downstream effector of INI1/hSNF5, in rhabdoid tumors. Oncogene. 2006;25(5):722–34. doi: 10.1038/sj.onc.1209112. Epub 2005/11/23. [DOI] [PubMed] [Google Scholar]
- 64.Smith ME, Cimica V, Chinni S, Challagulla K, Mani S, Kalpana GV. Rhabdoid tumor growth is inhibited by flavopiridol. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(2):523–32. doi: 10.1158/1078-0432.CCR-07-1347. Epub 2008/01/29. [DOI] [PubMed] [Google Scholar]
- 65.Gadd S, Sredni ST, Huang CC, Perlman EJ. Rhabdoid tumor: gene expression clues to pathogenesis and potential therapeutic targets. Laboratory investigation; a journal of technical methods and pathology. 2010;90(5):724–38. doi: 10.1038/labinvest.2010.66. Epub 2010/03/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venneti S, Le P, Martinez D, Eaton KW, Shyam N, Jordan-Sciutto KL, et al. p16INK4A and p14ARF tumor suppressor pathways are deregulated in malignant rhabdoid tumors. Journal of neuropathology and experimental neurology. 2011;70(7):596–609. doi: 10.1097/NEN.0b013e31822146ca. Epub 2011/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caramel J, Quignon F, Delattre O. RhoA-dependent regulation of cell migration by the tumor suppressor hSNF5/INI1. Cancer research. 2008;68(15):6154–61. doi: 10.1158/0008-5472.CAN-08-0115. Epub 2008/08/05. [DOI] [PubMed] [Google Scholar]
- 68.Rosson GB, Bartlett C, Reed W, Weissman BE. BRG1 loss in MiaPaCa2 cells induces an altered cellular morphology and disruption in the organization of the actin cytoskeleton. Journal of cellular physiology. 2005;205(2):286–94. doi: 10.1002/jcp.20397. Epub 2005/05/12. [DOI] [PubMed] [Google Scholar]
- 69.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nature genetics. 1999;22(1):102–5. doi: 10.1038/8811. Epub 1999/05/13. [DOI] [PubMed] [Google Scholar]
- 70.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer cell. 2010;18(4):316–28. doi: 10.1016/j.ccr.2010.09.006. Epub 2010/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Werneck MB, Wilson BG, Kim HJ, Kluk MJ, Thom CS, et al. TCR-dependent transformation of mature memory phenotype T cells in mice. The Journal of clinical investigation. 2011;121(10):3834–45. doi: 10.1172/JCI37210. Epub 2011/09/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nuclear receptor signaling. 2008;6:e004. doi: 10.1621/nrs.06004. Epub 2008/02/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pottier N, Yang W, Assem M, Panetta JC, Pei D, Paugh SW, et al. The SWI/SNF chromatin- remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. Journal of the National Cancer Institute. 2008;100(24):1792–803. doi: 10.1093/jnci/djn416. Epub 2008/12/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(6):2282–7. doi: 10.1073/pnas.1013751108. Epub 2011/01/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460(7251):66–72. doi: 10.1038/nature08137. Epub 2009/07/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mora-Blanco EL, Mishina Y, Tillman EJ, Cho YJ, Thom CS, Pomeroy SL, et al. Activation of beta-catenin/TCF targets following loss of the tumor suppressor SNF5. Oncogene. 2013 doi: 10.1038/onc.2013.37. Epub 2013/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nature medicine. 2010;16(12):1429–33. doi: 10.1038/nm.2251. Epub 2010/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Molecular and cellular biology. 2008;28(10):3457–64. doi: 10.1128/MCB.02019-07. Epub 2008/03/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You JS, De Carvalho DD, Dai C, Liu M, Pandiyan K, Zhou XJ, et al. SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS genetics. 2013;9(4):e1003459. doi: 10.1371/journal.pgen.1003459. Epub 2013/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell research. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. Epub 2011/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7922–7. doi: 10.1073/pnas.1303800110. Epub 2013/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]