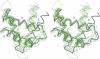Abstract
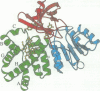
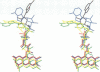
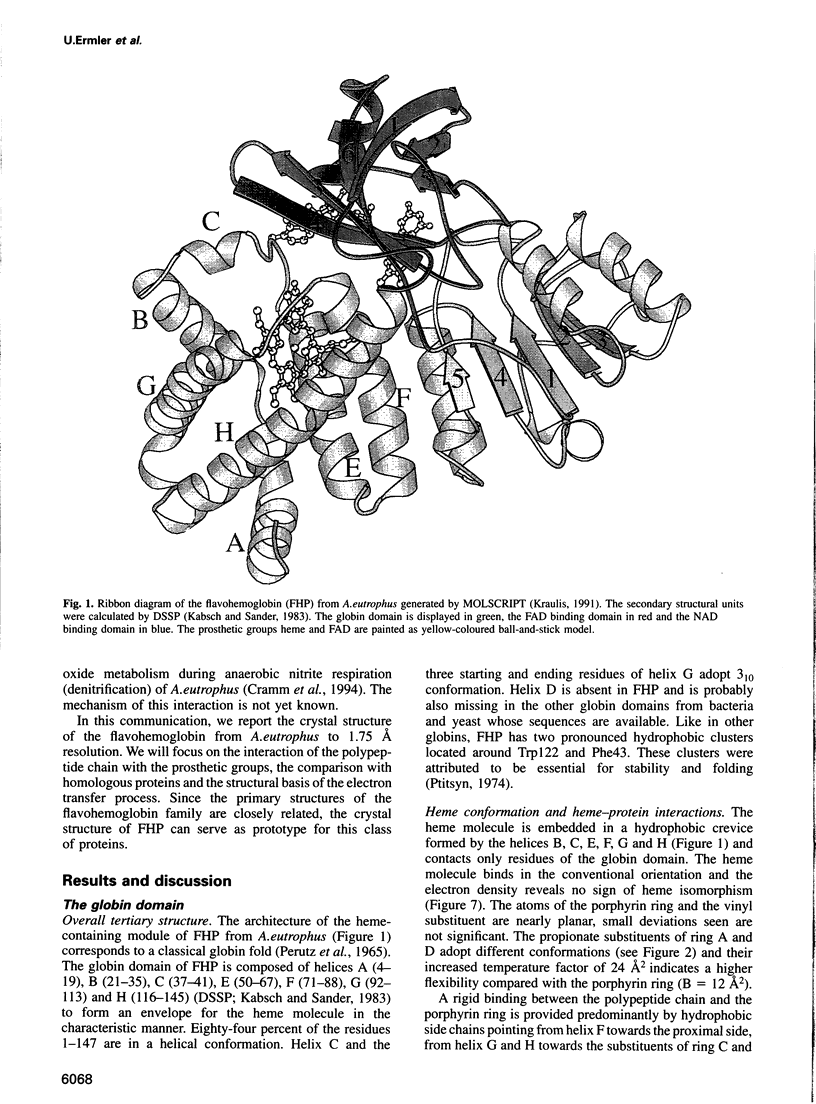
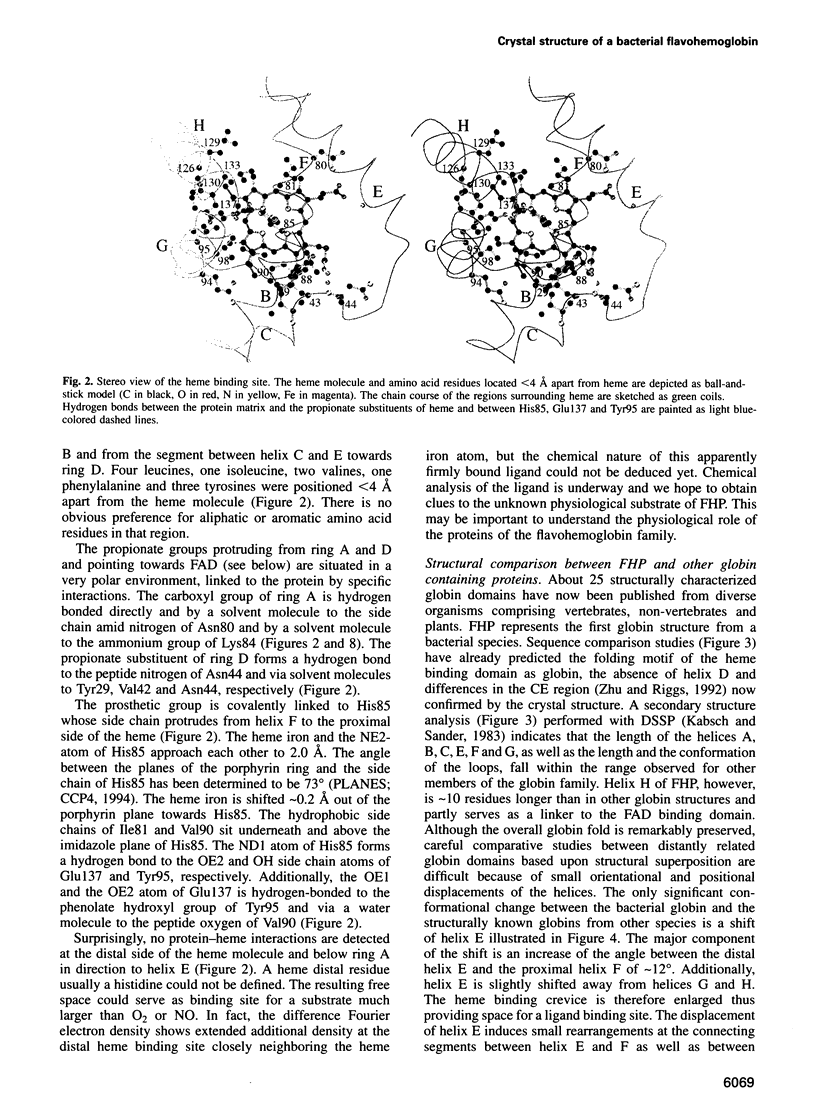
The molecular structure of the flavohemoglobin from Alcaligenes eutrophus has been determined to a resolution of 1.75 A and refined to an R-factor of 19.6%. The protein comprises two fused modules: a heme binding module, which belongs to the globin family, and an FAD binding oxidoreductase module, which adopts a fold like ferredoxin reductase. The most striking deviation of the bacterial globin structure from those of other species is the movement of helix E in a way to provide more space in the vicinity of the distal heme binding site. A comparison with other members of the ferredoxin reductase family shows similar tertiary structures for the individual FAD and NAD binding domains but largely different interdomain orientations. The heme and FAD molecules approach each other to a minimal distance of 6.3 A and adopt an interplanar angle of 80 degrees. The electron transfer from FAD to heme occurs in a predominantly polar environment and may occur directly or be mediated by a water molecule.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews S. C., Shipley D., Keen J. N., Findlay J. B., Harrison P. M., Guest J. R. The haemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal domain shares homology with ferredoxin NADP+ reductases. FEBS Lett. 1992 May 18;302(3):247–252. doi: 10.1016/0014-5793(92)80452-m. [DOI] [PubMed] [Google Scholar]
- Arents G., Love W. E. Glycera dibranchiata hemoglobin. Structure and refinement at 1.5 A resolution. J Mol Biol. 1989 Nov 5;210(1):149–161. doi: 10.1016/0022-2836(89)90297-0. [DOI] [PubMed] [Google Scholar]
- Aronson H. E., Royer W. E., Jr, Hendrickson W. A. Quantification of tertiary structural conservation despite primary sequence drift in the globin fold. Protein Sci. 1994 Oct;3(10):1706–1711. doi: 10.1002/pro.5560031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D., Chothia C., Lesk A. M. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987 Jul 5;196(1):199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- Bolognesi M., Onesti S., Gatti G., Coda A., Ascenzi P., Brunori M. Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution. J Mol Biol. 1989 Feb 5;205(3):529–544. doi: 10.1016/0022-2836(89)90224-6. [DOI] [PubMed] [Google Scholar]
- Bruns C. M., Karplus P. A. Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states. J Mol Biol. 1995 Mar 17;247(1):125–145. doi: 10.1006/jmbi.1994.0127. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chen Z. W., Koh M., Van Driessche G., Van Beeumen J. J., Bartsch R. G., Meyer T. E., Cusanovich M. A., Mathews F. S. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994 Oct 21;266(5184):430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- Correll C. C., Batie C. J., Ballou D. P., Ludwig M. L. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science. 1992 Dec 4;258(5088):1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- Correll C. C., Ludwig M. L., Bruns C. M., Karplus P. A. Structural prototypes for an extended family of flavoprotein reductases: comparison of phthalate dioxygenase reductase with ferredoxin reductase and ferredoxin. Protein Sci. 1993 Dec;2(12):2112–2133. doi: 10.1002/pro.5560021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramm R., Siddiqui R. A., Friedrich B. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J Biol Chem. 1994 Mar 11;269(10):7349–7354. [PubMed] [Google Scholar]
- Drennan C. L., Huang S., Drummond J. T., Matthews R. G., Lidwig M. L. How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science. 1994 Dec 9;266(5191):1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- Ermler U., Siddiqui R. A., Cramm R., Schröder D., Friedrich B. Crystallization and preliminary X-ray diffraction studies of a bacterial flavohemoglobin protein. Proteins. 1995 Apr;21(4):351–353. doi: 10.1002/prot.340210408. [DOI] [PubMed] [Google Scholar]
- Eschenbrenner M., Coves J., Fontecave M. Ferric reductases in Escherichia coli: the contribution of the haemoglobin-like protein. Biochem Biophys Res Commun. 1994 Jan 14;198(1):127–131. doi: 10.1006/bbrc.1994.1018. [DOI] [PubMed] [Google Scholar]
- Favey S., Labesse G., Vouille V., Boccara M. Flavohaemoglobin HmpX: a new pathogenicity determinant in Erwinia chrysanthemi strain 3937. Microbiology. 1995 Apr;141(Pt 4):863–871. doi: 10.1099/13500872-141-4-863. [DOI] [PubMed] [Google Scholar]
- Fermi G., Perutz M. F., Shaanan B., Fourme R. The crystal structure of human deoxyhaemoglobin at 1.74 A resolution. J Mol Biol. 1984 May 15;175(2):159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- Goodin D. B., McRee D. E. The Asp-His-Fe triad of cytochrome c peroxidase controls the reduction potential, electronic structure, and coupling of the tryptophan free radical to the heme. Biochemistry. 1993 Apr 6;32(13):3313–3324. [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Iwaasa H., Takagi T., Shikama K. Amino acid sequence of yeast hemoglobin. A two-domain structure. J Mol Biol. 1992 Oct 5;227(3):948–954. doi: 10.1016/0022-2836(92)90236-d. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- Lu G., Campbell W. H., Schneider G., Lindqvist Y. Crystal structure of the FAD-containing fragment of corn nitrate reductase at 2.5 A resolution: relationship to other flavoprotein reductases. Structure. 1994 Sep 15;2(9):809–821. doi: 10.1016/s0969-2126(94)00082-4. [DOI] [PubMed] [Google Scholar]
- Mathews F. S., Chen Z. W., Bellamy H. D., McIntire W. S. Three-dimensional structure of p-cresol methylhydroxylase (flavocytochrome c) from Pseudomonas putida at 3.0-A resolution. Biochemistry. 1991 Jan 8;30(1):238–247. doi: 10.1021/bi00215a034. [DOI] [PubMed] [Google Scholar]
- May A. C., Johnson M. S. Protein structure comparisons using a combination of a genetic algorithm, dynamic programming and least-squares minimization. Protein Eng. 1994 Apr;7(4):475–485. doi: 10.1093/protein/7.4.475. [DOI] [PubMed] [Google Scholar]
- Moser C. C., Keske J. M., Warncke K., Farid R. S., Dutton P. L. Nature of biological electron transfer. Nature. 1992 Feb 27;355(6363):796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Ornston L. N., Bairoch A., Rekik M., Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991 Sep;173(17):5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H., Inaka K., Yamanaka M., Kaida S., Kobayashi K., Miki K. Crystal structure of NADH-cytochrome b5 reductase from pig liver at 2.4 A resolution. Biochemistry. 1995 Mar 7;34(9):2763–2767. doi: 10.1021/bi00009a004. [DOI] [PubMed] [Google Scholar]
- Phillips S. E. Structure and refinement of oxymyoglobin at 1.6 A resolution. J Mol Biol. 1980 Oct 5;142(4):531–554. doi: 10.1016/0022-2836(80)90262-4. [DOI] [PubMed] [Google Scholar]
- Pittsyn O. B. Invariant features of globin primary structure and coding of their secondary structure. J Mol Biol. 1974 Sep 15;88(2):287–300. doi: 10.1016/0022-2836(74)90482-3. [DOI] [PubMed] [Google Scholar]
- Poole R. K., Ioannidis N., Orii Y. Reactions of the Escherichia coli flavohaemoglobin (Hmp) with oxygen and reduced nicotinamide adenine dinucleotide: evidence for oxygen switching of flavin oxidoreduction and a mechanism for oxygen sensing. Proc Biol Sci. 1994 Mar 22;255(1344):251–258. doi: 10.1098/rspb.1994.0036. [DOI] [PubMed] [Google Scholar]
- Probst I., Schlegel H. G. Respiratory components and oxidase activities in Alcaligenes eutrophus. Biochim Biophys Acta. 1976 Aug 13;440(2):412–428. doi: 10.1016/0005-2728(76)90075-x. [DOI] [PubMed] [Google Scholar]
- Probst I., Wolf G., Schlegel H. G. An oxygen-binding flavohemoprotein from Alcaligenes eutrophus. Biochim Biophys Acta. 1979 Feb 26;576(2):471–478. doi: 10.1016/0005-2795(79)90422-7. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Venkatachalam C. M., Krimm S. Stereochemical criteria for polypeptide and protein chain conformations. 3. Helical and hydrogen-bonded polypeptide chains. Biophys J. 1966 Nov;6(6):849–872. doi: 10.1016/s0006-3495(66)86699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer W. E., Jr High-resolution crystallographic analysis of a co-operative dimeric hemoglobin. J Mol Biol. 1994 Jan 14;235(2):657–681. doi: 10.1006/jmbi.1994.1019. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Steigemann W., Weber E. Structure of erythrocruorin in different ligand states refined at 1.4 A resolution. J Mol Biol. 1979 Jan 25;127(3):309–338. doi: 10.1016/0022-2836(79)90332-2. [DOI] [PubMed] [Google Scholar]
- Vasudevan S. G., Armarego W. L., Shaw D. C., Lilley P. E., Dixon N. E., Poole R. K. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol Gen Genet. 1991 Apr;226(1-2):49–58. doi: 10.1007/BF00273586. [DOI] [PubMed] [Google Scholar]
- Vinogradov S. N., Walz D. A., Pohajdak B., Moens L., Kapp O. H., Suzuki T., Trotman C. N. Adventitious variability? The amino acid sequences of nonvertebrate globins. Comp Biochem Physiol B. 1993 Sep;106(1):1–26. doi: 10.1016/0305-0491(93)90002-m. [DOI] [PubMed] [Google Scholar]
- Wakabayashi S., Matsubara H., Webster D. A. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. 1986 Jul 31-Aug 6Nature. 322(6078):481–483. doi: 10.1038/322481a0. [DOI] [PubMed] [Google Scholar]
- Warshel A., Naray-Szabo G., Sussman F., Hwang J. K. How do serine proteases really work? Biochemistry. 1989 May 2;28(9):3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]
- Xia Z. X., Mathews F. S. Molecular structure of flavocytochrome b2 at 2.4 A resolution. J Mol Biol. 1990 Apr 20;212(4):837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]
- Zhu H., Riggs A. F. Yeast flavohemoglobin is an ancient protein related to globins and a reductase family. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5015–5019. doi: 10.1073/pnas.89.11.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]