Abstract
Purpose
Suppression of P53 transcriptional function mediates poor therapeutic response in cancer patients. AURKA and HDM2 are negative regulators of P53. Herein, we examined the role of AURKA in regulating HDM2 and its subsequent effects on P53 apoptotic function in gastric cancer (GC).
Experimental design
Primary tumors and in vitro GC cell models with overexpression or knockdown of AURKA were used. The role of AURKA in regulating HDM2 and cell survival coupled with P53 expression and activity were investigated.
Results
Overexpression of AURKA enhanced HDM2 protein level; conversely, knockdown of endogenous AURKA decreased expression of HDM2 in AGS and SNU-1 cells. Dual co-immunoprecipitaion assay data indicated that AURKA was associated with HDM2 in a protein complex. The in vitro kinase assay using recombinant AURKA and HDM2 proteins followed by co-immunoprecipitaion revealed that AURKA directly interacts and phosphorylates HDM2 protein in vitro. The activation of HDM2 by AURKA led to induction of P53 ubiquitination and attenuation of cisplatin-induced activation of P53 in gastric cancer cells. Inhibition of AURKA using an investigational small molecule specific inhibitor, alisertib, decreased HDM2 protein level and induced P53 transcriptional activity. These effects markedly decreased cell survival in vitro and xenograft tumor growth in vivo. Notably, analysis of immunohistochemistry on tissue microarrays revealed significant overexpression of AURKA and HDM2 in human GC samples (P<0.05).
Conclusion
Collectively, our novel findings indicate that AURKA promotes tumor growth and cell survival through regulation of HDM2-induced ubiquitination and inhibition of P53.
Keywords: AURKA, HDM2, Stomach, Cancer, P53, Ubiquitination, Esophagus
Introduction
Gastric cancer (GC) exhibits poor patient survival rates due to intrinsic resistance to chemotherapeutic drugs (1, 2). According to current estimates, GC is a frequently diagnosed disease world-wide with an estimated incidence rate of approximately 1 million cases and mortality rate of 740,000 cases, respectively (3, 4). Multiple oncogenic signaling mechanisms have been shown to mediate cancer cell survival and drug resistance against several chemotherapeutics in GCs (5–9). Unfortunately, the survival rate for patients with GC has shown only marginal improvement (10). Therefore, investigations specifically aimed at further understanding of the mechanisms regulating cell death and drug resistance in GC are essential for the development of novel effective anti-cancer therapeutic regimens.
AURKA (Aurora Kinase A), a serine/threonine cell cycle kinase, has been mapped to the 20q13 chromosomal region, which is frequently amplified in GC (11). Frequent amplification and/or overexpression of AURKA have been reported in breast, colon, esophageal, gastric, liver, ovarian, and pancreatic cancers (8, 12–18). AURKA plays an important role in facilitating mitosis and its expression is tightly regulated in normal cells. However, deregulated overexpression of AURKA causes genetic instability, dedifferentiated morphology and oncogenic transformation (6, 13). Overexpression of AURKA promotes pro-growth and anti-apoptotic signaling pathways resulting in cancer cell proliferation, drug resistance, and poor patient prognosis in GCs (13, 19). The potent DCF (Docetaxel, Cisplatin and 5-Fluorouracil) chemotherapy regimen is frequently used against GCs (20, 21). Nonetheless, AURKA overexpression has been shown to mediate resistance against Taxol and Cisplatin and can thereby undermine the therapeutic outcome of the DCF regimen in GCs (22, 23). The efficacy of chemotherapeutic drugs is dependent on induction of apoptosis by P53 (Tumor Protein 53) family of pro-apoptotic proteins. We and others have previously reported that overexpression of AURKA can attenuate both P53 and P73 (Tumor Protein 73) protein expression and function (24, 25). These findings indicate that constitutive overexpression of AURKA can mediate poor response to chemotherapy in GCs. Therefore, further investigation of mechanism(s) by which AURKA suppresses P53 expression and function can aid in the development of novel and effective therapeutic strategies against GCs.
HDM2 (Human Double Minute 2), an E3-ubiquitin ligase, is one of the critical negative regulators of P53 protein function and expression. HDM2 has been shown to inhibit P53 by blocking P53 transcriptional activity, promoting cytosolic translocation of P53 from the nucleus and tagging it with ubiquitin for proteasomal degradation (26). HDM2 gene amplification has been reported in various cancers and when overexpressed HDM2 becomes a bona-fide oncogene that can promote malignant transformation and drug resistance (27, 28). Given the fact that HDM2 plays a critical role in regulating P53, a function shared with AURKA, we postulated that activation of HDM2 by AURKA regulates P53 in GC cells. In this study, we investigated the role of AURKA in regulating HDM2-mediated P53 inhibition affecting gastric tumor growth and cell survival. We also examined the role of an investigational small molecule specific inhibitor of AURKA, alisertib, in suppressing HDM2 and promoting cell death in vitro and in vivo.
Materials and Methods
Cell culture and pharmacologic reagents
The gastric adenocarcinoma cell lines (AGS, SNU-1, SNU-16, MKN28, MKN45, MKN75, Kato III and RF-1) were obtained from American Tissue Culture Collection (ATCC, Manassas, VA) and RIKEN BioResource Center (Ibaraki, Japan). The cell lines were maintained in F-12 or DMEM (Gibco, CA) medium supplemented with 10% (v/v) fetal bovine serum (Gibco, CA) (29). All cell lines were ascertained to conform to in vitro morphological characteristics.
Alisertib (Millenium Pharmaceuticals, Inc.) solutions for in vitro and in vivo studies were prepared as described previously (8). Nutlin3A (Cayman Chemicals, MI) solution was prepared in dimethyl sulfoxide (Sigma-Aldrich, MO). Cisplatin (CDDP, APP Pharmaceuticals, LLC., IL) solution (3.3 mmol/L) was prepared in sterile water. AKT, p-AKT (Ser473), p-AURKA (Thr288), AURKA, HDM2, p-HDM2 (Ser166), P53, P21, and β-Actin primary antibodies were obtained from Cell Signaling Technology, MA.
AURKA and HDM2 expression and plasmids
The AURKA expression plasmid was generated as described previously (30). The HDM2 expression plasmid was purchased from Addgene. Transient transfection of GC cells was performed using X-tremeGENE HP (Roche Applied Sciences, IN). The recombinant adenovirus expressing AURKA or control was generated as described previously (31).
Clonogenic cell survival assay
GC cells were seeded at 5000 cells/well in a six well plate and treated with vehicle (dimethyl sulfoxide, DMSO) or alisertib (0.25–5.0 μmol/L) for 24 hours. Next, cells were cultured for 10 days, and colonies were stained and quantified as described previously (8).
Western blot analysis
Cells were lysed in lysis-buffer (50 mmol/L Tris-HCl buffer, pH 7.4, 150 mmol/L NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) supplemented with 1x Halt protease inhibitor cocktail (Pierce, Rockford, IL). Proteins were analyzed by Western blot as described previously (32).
Dual-immunofluorescence
GC cells plated in 8-chamber slides (BD Falcon, NJ) were permeabilized and fixed in 2% paraformaldehyde. Cells were then incubated in a mixture of Rabbit AURKA (1:100) and Mouse HDM2 (1:100) primary antibodies for 3 hours. After washing with PBS (phosphate buffered saline), cells were stained with Alexa-Fluor-488 Anti-Rabbit and Alexa-Fluor-568 Anti-Mouse secondary antibodies. The cells were washed and mounted with DAPI (4,6-diamidino-2-phenylindole) and examined by fluorescence microscopy (Olympus America Inc.).
Immunoprecipitation
Immunoprecipitation (IP) was performed as described previously (33). Briefly, cells were lysed in lysis buffer and proteins were immunoprecipitated at room temperature with primary antibodies previously bound to 50 μl Dyna-beads Protein G (Invitrogen).
In vitro Kinase activity assay
Active human recombinant AURKA (Cell Sciences, MA) and HDM2 (Thermo Scientific, IL) proteins were used for an in vitro kinase assay. Briefly, increasing concentrations of AURKA (0.2–1.0 μg) were added to a fixed concentration of HDM2 (0.5 μg) in the assay buffer. The reaction mixtures were incubated at 37°C for 30 minutes to initiate kinase activity, and the protein samples were subjected to Western blot analysis.
In vivo tumor xenograft
AGS cells (4×106) suspended in 200 μl of DMEM matrigel mixture (50% DMEM supplemented with 10% FBS and 50% matrigel) were injected into the flank regions of female athymic nude - Foxn1 nu/nu mice (Harlan Laboratories Inc., IN). The tumors were allowed to grow until 200 mm3 in size before starting the treatment with a daily alisertib (30 mg/kg, orally) for 21 days. Tumor xenografts were measured every four days and tumor size was calculated according to the following formula: where Tvol is tumor volume, L is tumor length and W is tumor width (34). At the end of treatment, the xenograft tumors were collected and processed for qRT-PCR (PUMA, NOXA, P21, and BAX) or IHC (P53; SC-126, and HDM2; SMP14, Santa Cruz Biotechnology Inc.) antibodies as described previously (31).
Tissue microarray and immunohistochemistry
Immunohistochemistry analysis (IHC) was performed on tissue microarrays (TMA) containing 94 de-identified archival cases of GC and 113 cases of esophageal adenocarcinomas (EAC) along with 71 normal gastric epithelial tissue samples and 26 normal esophageal tissue samples were included. All tissue samples were obtained in accordance with the IRB (Institutional Review Board) approved protocols at Vanderbilt University. 5 μm thick TMA sections of normal and tumor tissue samples were used for IHC staining of AURKA and HDM2 proteins. The intensity and frequency of staining was scored as described previously (35).
AKT and HDM2 silencing by small interfering RNA (siRNA)
AGS cells overexpressing AURKA or control vector were transfected with AKT siRNA, HDM2 siRNA or control siRNA for 48 hours, purchased from Cell Signaling (Cell Signaling Technologies, MA) and IDT (Integrated DNA technologies Inc., IL), respectively. The Cell lysates were analyzed by Western blot analysis.
Statistical analysis
Data are presented as means ± SEM. All in vitro experiments were performed in triplicates. One-way analysis of variance (ANOVA) with Tukey post hoc analysis was used to evaluate statistical difference between groups. Statistical analyses were carried out using GraphPad Prism 5 software (GraphPad Software Inc., CA). The correlation between two parameters was determined by the Spearman correlation and kappa test. The P value of ≤0.05 was considered statistically significant.
Results
AURKA regulates HDM2 and cell survival in gastric adenocarcinoma cells
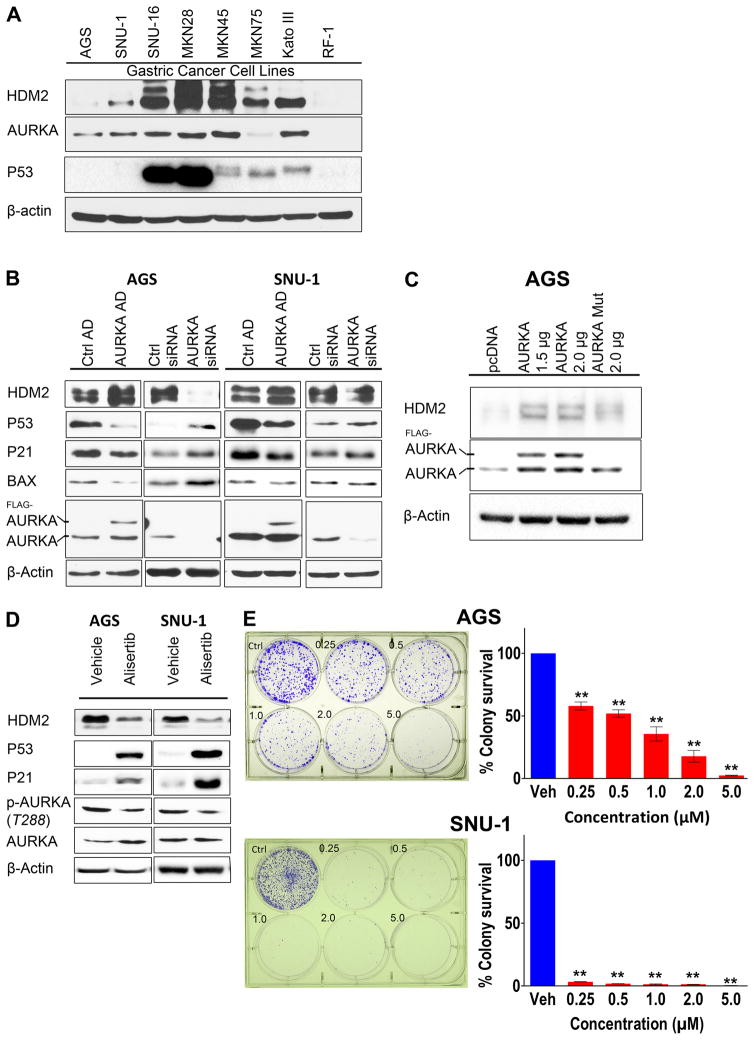
We examined AURKA and HDM2 protein expression in gastric adenocarcinoma (GC) cell lines. The Western blot analysis data indicated frequent concomitant overexpression of AURKA and HDM2 proteins in 5/8 GC cell lines with RF-1 cell line exhibiting relatively low expression of AURKA and HDM2 (Figure 1A). Similarly, the data showed that 4/5 esophageal adenocarcinoma (EAC) cell lines concomitantly overexpressed AURKA and HDM2 proteins, and 3/3 un-transformed immortalized esophageal cells were negative for AURKA and HDM2 expression (Supplemental Figure 1). We have previously reported that AURKA suppresses P53 protein function in GC cells (36). HDM2 is an E3-ubiquitin ligase closely involved in regulating P53 protein expression and stability. Since AURKA and HDM2 are frequently overexpressed in GC cell lines, we hypothesized that AURKA regulates HDM2 expression in gastric cancer. To test this hypothesis, we employed P53 wild-type AGS and SNU-1 GC cell in vitro models. The Western blot analysis indicated that transient overexpression of AURKA using adenovirus system enhanced HDM2 protein levels in AGS and SNU-1 cells (Figure 1B). Following AURKA overexpression, P53 and its downstream targets (P21 and BAX) were down-regulated in AGS cells. Conversely, siRNA mediated knockdown of endogenous AURKA increased P53, P21, and BAX expression in AGS cells. AURKA overexpression and knockdown exhibited similar effects on HDM2, P53, and P21 in SNU-1 cells (Figure 1B). However, unlike AGS cells, we did not observe a change in BAX protein expression in SNU-1 cells. To investigate whether AURKA-mediated increase in HDM2 protein level was dependent on its serine/threonine kinase activity, we utilized kinase-dead mutant AURKA (D274A). The Western blot analysis revealed that, unlike wild-type AURKA, kinase-dead mutant AURKA (D274A) did not significantly affect HDM2 expression in the AGS cells (Figure 1C). To validate the role of AURKA in regulating HDM2 and examine its impact on cell survival, we pharmacologically inhibited AURKA with alisertib, an investigational AURKA specific inhibitor, in AGS and SNU-1 cells. The Western blot data confirmed that inhibition of AURKA led to a significant decrease in HDM2 protein level coupled with upregulation of P53 and P21 expression in both cell lines (Figure 1D). The clonogenic survival assay data indicated that alisertib-mediated inhibition of AURKA activity significantly suppressed AGS (P<0.01) and SNU-1 (P<0.01) cell survival (Figure 1E).
Figure 1. AURKA promotes cell survival through regulation of HDM2 and P53 expression in GC cell lines.
A, Protein extracts from a panel of GC cell lines were subjected to Western blot analysis of AURKA, P53, and HDM2 proteins. B, Western blot analysis of HDM2, P53, P21, BAX, and AURKA proteins following adenovirus-mediated transient overexpression of AURKA, control adenovirus, siRNA- mediated knockdown of AURKA or control siRNA in AGS and SNU-1 cells is shown. Overexpression of AURKA led to an increase in the protein level of HDM2 and a decrease in P53. Conversely, inhibition of AURKA reduced the level of HDM2 with an increase in P53. C, AGS cells were transiently transfected with pcDNA3.1 empty vector, AURKA, or kinase-dead mutant AURKA D274A. Western blot analysis indicated that AURKA-mediated increase in the HDM2 protein level is dependent on AURKA kinase activity. D, AGS and SNU-1 cells were treated with alisertib (0.5 μmol/L) for 48 hours, and cell lysates were subjected to Western blot analysis. The data showed that treatment with alisertib reduced the level HDM2 and increased P53. E, Clonogenic cell survival analysis for AGS and SNU-1 cells was performed following treatment with alisertib (0.25–5.0 μmol/L). The data indicated a dose-dependent decrease in cell survival. Ctrl AD, Control Adenovirus; AURKA AD, Aurora Kinase A Adenovirus; AURKA Mut, Aurora Kinase A D274A Mutant; ** P<0.01.
AURKA directly interacts and phosphorylates HDM2 in gastric cancer
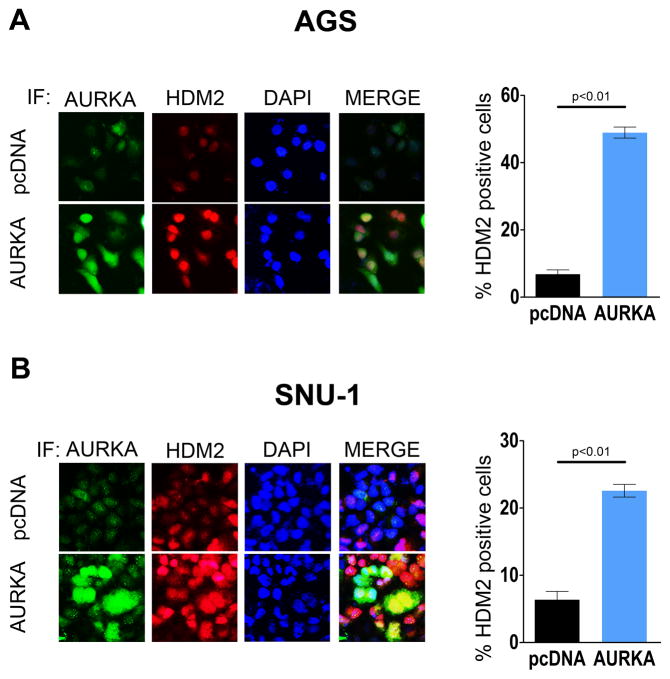
HDM2 localizes to the nucleus inducing ubiquitination and degradation of P53 protein in the proteosome (26). The dual immunofluorescence analyses results indicated that transient overexpression of AURKA significantly enhanced HDM2 protein expression, confirming the Western blot data (Figure 1B). The immunofluorescence data showed an overlap of AURKA and HDM2 signals in the merged image, suggesting co-localization of AURKA and HDM2 in both AGS and SNU-1 cells (Figure 2A & 2B). We have previously reported that AURKA phosphorylates AKT at Ser473 amino acid residue and active AKT has been shown to phosphorylate HDM2 at the Ser166 site (36, 37). Therefore, it is plausible that AURKA-mediated increase in HDM2 protein expression could be mediated by an AKT-dependent mechanism. To examine this hypothesis, we transiently overexpressed AURKA in AGS cells and evaluated HDM2 protein levels after treatment with an AKT inhibitor (AKTI) or genetic knockdown of AKT with siRNA. Overall, the Western blot data showed that HDM2 expression levels were not significantly affected by AKT inhibition or knockdown in AURKA-overexpressing cells (Supplemental Figure 2). Collectively, the data suggested that AURKA can upregulate HDM2 expression through an AKT-independent mechanism.
Figure 2. AURKA expression increases the HDM2 protein level in GC cell lines.
A & B, Dual immunofluorescence (IF) analysis for AURKA (Green) and HDM2 (Red) was performed on AGS and SNU-1 cells transiently transfected with pcDNA3.1 or AURKA. The nucleus was stained with DAPI (Blue). After merging AURKA, HDM2, and DAPI images together (MERGE), the data showed that overexpression of AURKA leads to a significant increase in HDM2 protein level. The percentage of cells that presented expression of HDM2 was averaged from six different microscopic fields for more than 100 cells. The IF data are represented as the mean of three different experiments.
P53 suppression by AURKA is dependent on regulation of HDM2 in gastric cancer
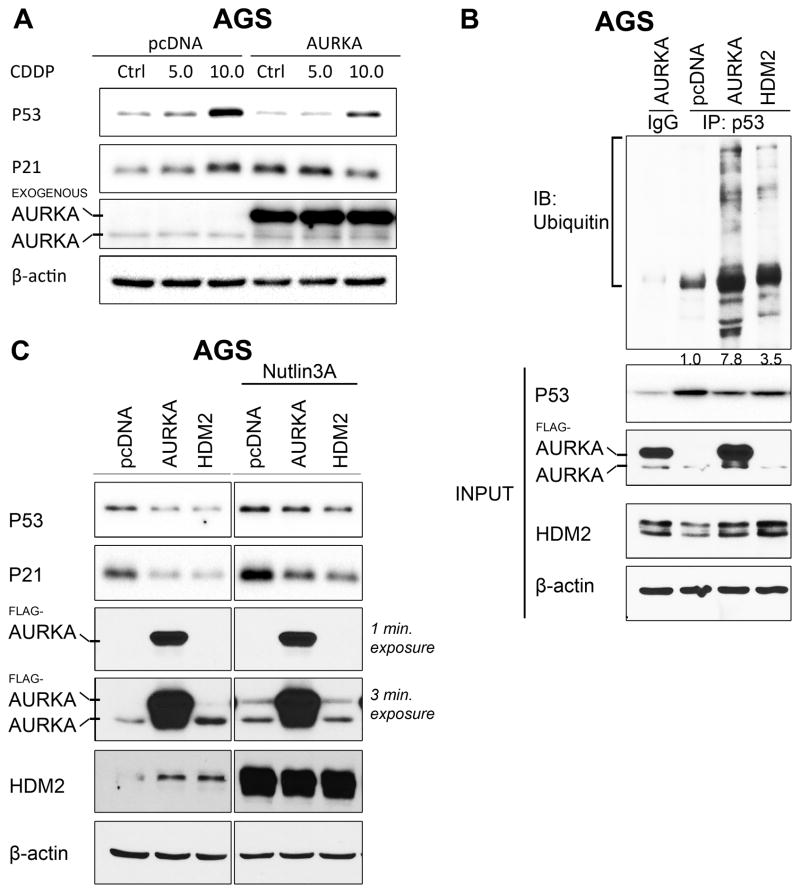
On the basis of our results showing that AURKA up-regulates HDM2 expression, we postulated that AURKA can regulate P53 protein expression and function through modulation of HDM2 in response to DNA damage in GC cells. To test this hypothesis, we transiently overexpressed AURKA in AGS and SNU-1 cells, and treated them with vehicle or cisplatin (CDDP). The Western blot data showed that AURKA overexpression attenuated CDDP-induced upregulation of P53 and P21 expression in AGS and SNU-1 cells (Figure 3A & Supplemental Figure 3A). Since HDM2 regulates P53 through ubiquitination and proteasomal degradation, we hypothesized that AURKA can enhance P53 ubiquitination. To examine this hypothesis, we assessed the effect of AURKA and/or HDM2 expression on P53 ubiquitination in AGS cells. The IP data indicated that transient overexpression of AURKA or HDM2 significantly enhanced P53 ubiquitination (Figure 3B). To confirm that inhibition of P53 by AURKA is dependent on HDM2, we transiently overexpressed AURKA or HDM2 and treated with vehicle or Nutlin3A in AGS and SNU-1 cells. Nutlin3A is a HDM2 inhibitor that prevents HDM2 from binding to P53, thereby preventing ubiquitination and degradation of P53 protein. Western blot data indicated that AURKA-mediated down regulation of P53 and its downstream target P21 was attenuated by Nutlin3A in AGS cells (Figure 3C). As a confirmation of the effect of Nutlin3A, the data showed that exogenous HDM2-mediated decrease in P53 and P21 expression was partially blocked by Nutlin3A (Figure 3C). Similar results were obtained in SNU-1 cells (Supplemental Figure 3B). Genetic knockdown of HDM2 by siRNA abrogated AURKA-induced suppression of P53 and P21 in AGS cells, confirming the results obtained by Nutlin3A (Supplemental Figure 3C). Together, these results indicated that AURKA-induced down-regulation of P53 is dependent on HDM2. In addition, we evaluated resistance to cytotoxic effects of CDDP in AGS cells overexpressing AURKA. The survival data indicate that transient overexpression of AURKA promotes resistance to CDDP in AGS cells (Supplemental Figure 4).
Figure 3. AURKA-mediated regulation of HDM2 promotes P53 ubiquitination and attenuates CDDP-induced P53 expression in GC cells.
A, AGS cells transiently expressing AURKA or pcDNA3.1 were treated with CDDP (5.0–10.0 μmol/L) for 12 hours and analyzed by immunoblotting. The data indicated that CDDP treatment induced P53 and P21 expression in control cells. In contrast, AURKA expression abrogated these effects in response to CDDP. B, Immunoprecipitation of endogenous P53 was done in AGS cells transiently expressing pcDNA3.1, AURKA, or HDM2. Western blot analysis showed that overexpression of AURKA significantly induced P53 ubiquitination. C, AGS cells transiently expressing pcDNA3.1, AURKA, or HDM2 were treated with Nutlin3A (4.0 μmol/L) for 12 hours. Western blot data suggested that AURKA-induced suppression of P53 and P21 expression is mediated by HDM2. IgG, immunoglobin G; IB, immunoblotting.
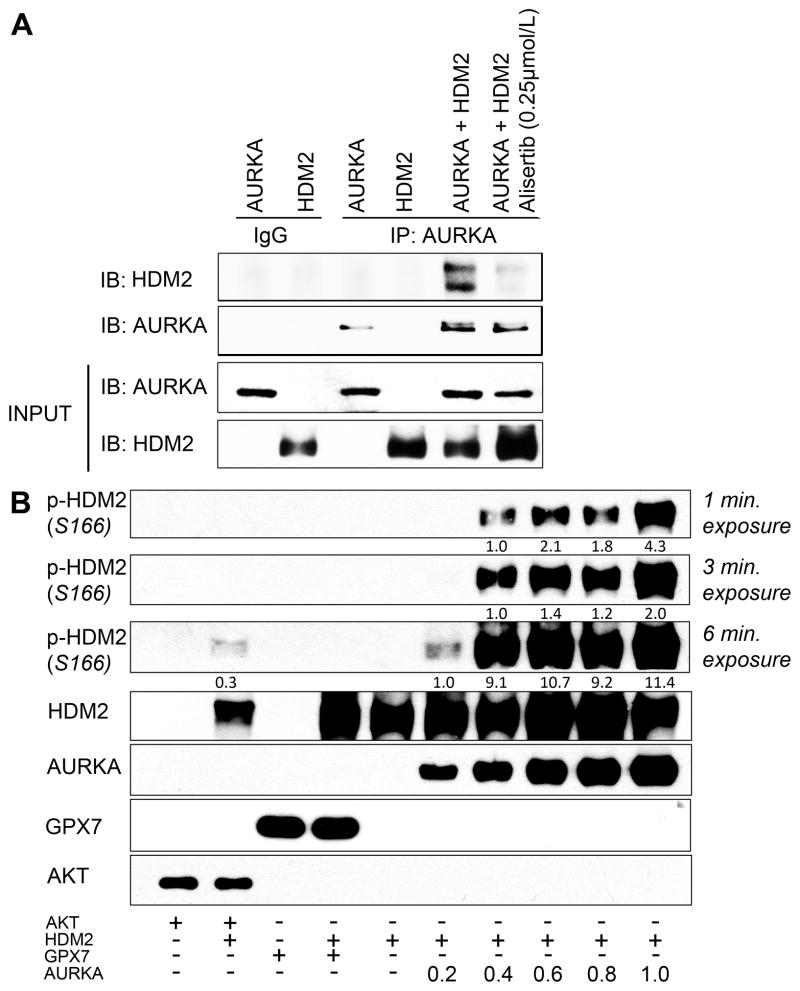
To ascertain if AURKA interacts with HDM2, we performed dual co-immunoprecipitation (Co-IP) following overexpression of AURKA in AGS cells. The dual Co-IP data indicated the presence of endogenous HDM2 protein in an immunocomplex with exogenous AURKA protein (Supplemental Figure 5). To investigate if AURKA directly interacted and phosphorylated HDM2 in the presence or absence of alisertib, we carried out an in vitro kinase assay using recombinant AURKA and HDM2 proteins followed by Co-IP assay. The data indicated that AURKA directly interacts and phosphorylates HDM2; these effects were abrogated upon inhibition of AURKA with alisertib (Figure 4A). To further confirm that AURKA directly phosphorylates HDM2, we performed the in vitro kinase assay with HDM2 and increasing concentrations of AURKA. The data indicated that AURKA protein can directly phosphorylate HDM2 protein at the Ser166 residue in a concentration-dependent manner (Figure 4B). Collectively, our data clearly indicated that AURKA can directly interact and phosphorylate HDM2.
Figure 4. AURKA directly associates and phosphorylates HDM2 protein.
A, In vitro kinase assay followed by a subsequent IP with AURKA antibody was done on recombinant AURKA and HDM2 proteins. Western blot analysis of the immunoprecipitates showed that AURKA-mediated interaction of HDM2 is dependent on AURKA kinase activity. B, Western blot analysis of in vitro kinase assay with recombinant AURKA, HDM2, AKT and GPX7 proteins indicated that AURKA directly phosphorylates HDM2 in a concentration-dependent manner. AKT and GPX7 recombinant proteins were used as positive and negative controls, respectively. GPX7, glutathione peroxidase enzyme 7.
Pharmacological inhibition of AURKA induces P53 through down-regulation of HDM2 in vivo
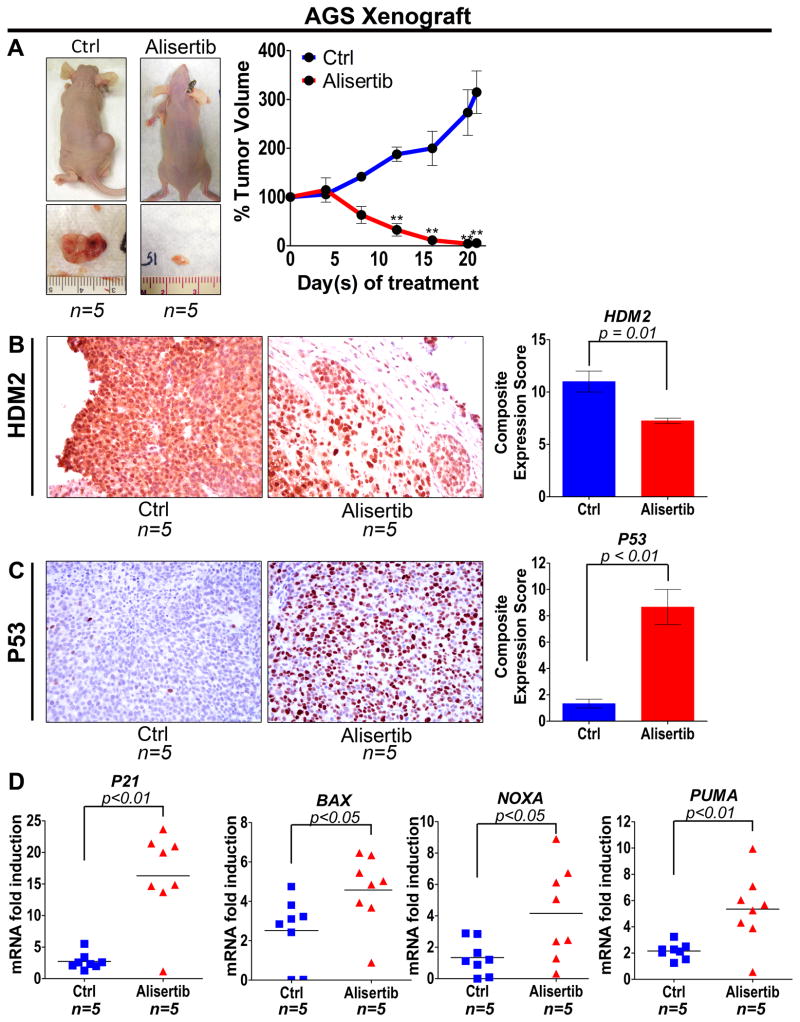
To establish the role of AURKA in regulating HDM2 and thereby modulating P53 expression and function in vivo, we treated AGS mouse xenografts with alisertib and examined the tumor growth and expression levels of HDM2 and P53. Additionally, we assessed P53 transcriptional activity by measuring the mRNA levels of P53 downstream targets, PUMA, NOXA, P21 and BAX. The treatments were initiated after the tumor xenografts reached 200 mm3 in size. The endpoint data for day 21 indicated that treatment with alisertib significantly reduced tumor volume by approximately 55-fold relative to control (P<0.01; Figure 5A). The IHC data revealed that alisertib-mediated inhibition of AURKA significantly suppressed HDM2 expression (P=0.01) and induced P53 expression (P<0.01) in the AGS xenografts (Figure 5B & 5C). The qRT-PCR data showed a significantly higher induction of P53 downstream targets, P21 (P<0.01), BAX (P<0.05), NOXA (P<0.05), and PUMA (P=0.01) in alisertib-treated AGS xenografts than control (Figure 5D). In accordance with the in vitro data, the xenograft results clearly show that AURKA inhibition induces P53 through down-regulation of HDM2 in vivo.
Figure 5. Alisertib exhibits anti-tumor activity, suppresses HDM2, and enhances P53 function in vivo.
AGS xenograft tumors were treated with alisertib (30mg/kg) for 21 days and tumor size was measured every four days. A, The data indicated that alisertib has significant anti-tumor activity against AGS xenografts. B & C, IHC of AGS xenograft tumors showed that inhibition of AURKA suppressed HDM2 expression and induced P53 in vivo. D, qRT-PCR analysis of AGS xenograft tumors revealed that blocking AURKA with alisertib enhanced P53 transcriptional activity as indicated by elevated mRNA levels of P21, BAX, NOXA and PUMA downstream target genes. ** P<0.01.
Frequent overexpression of AURKA and HDM2 are directly correlated in human gastric cancer
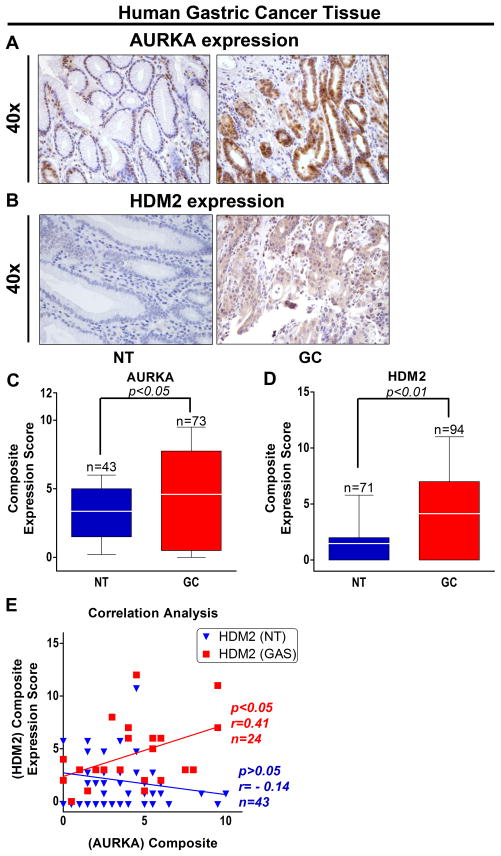
We examined AURKA and HDM2 protein expression in human gastric tissue microarray containing 94 gastric adenocarcinoma (GC) and 71 normal gastric (NT) tissue samples by IHC. The IHC data showed that both AURKA and HDM2 expression levels were highly overexpressed in gastric adenocarcinoma tissue samples as compared to normal tissue samples (Figure 6A & B). The data indicated that AURKA was localized in both nucleus and cytoplasm, whereas HDM2 was mainly localized in the nucleus (Figure 6A & B). The IHC analyses revealed that HDM2 was overexpressed in approximately 56% (53/94) of gastric adenocarcinoma tissue samples (Figure 6D). In addition, AURKA was similarly overexpressed in 70.4% (50/73) of gastric adenocarcinoma tissues (Figure 6C). The data also indicated a significant direct correlation between AURKA and HDM2 protein expression in gastric adenocarcinoma tissues (r=0.41, P=0.04; Figure 6E). Similar results with regard to AURKA and HDM2 overexpression and correlation were obtained in human esophageal tissue microarray containing 113 esophageal adenocarcinoma (EAC) and 26 normal esophageal (NT) tissue samples by IHC analysis (Supplemental Figure 6). Together, these results demonstrate that frequent overexpression AURKA and HDM2 is directly correlated in human gastric and esophageal adenocarcinoma.
Figure 6. Frequent overexpression and direct correlation between AURKA and HDM2 in human gastric cancer tissues.
A & B, Representative images of AURKA and HDM2 IHC staining in normal gastric (NT) and gastric cancer (GC) tissue samples are shown at x40 magnification. The data showed that AURKA and HDM2 protein expression levels are significantly higher in GC than NT. C & D, IHC analysis of human gastric tissue array exhibited significantly high expression scores for AURKA and HDM2 proteins in GC than NT (P<0.05). E, AURKA and HDM2 protein expression correlation analysis in human gastric tissue array indicated strong direct correlation between AURKA and HDM2 expression (P=0.04, r=0.41, n=24) in GC.
Discussion
Gastric cancer is the second most frequently diagnosed form of cancer accounting for 10% of total cancer related deaths worldwide (38). Despite improvement in chemotherapy, the 5-year survival rate for gastric cancer patients is a dismal 22% (39, 40). Development of resistance to chemotherapeutic drugs is a major cause for disease recurrence and low patient survival rates (41–43). Therefore, identification of molecular mechanism(s) mediating chemotherapeutic drug resistance is vital for developing more effective therapeutic strategies against GC. In this study, we examined the role of AURKA in regulating cell survival and apoptosis in GC. Herein, we report for the first time that HDM2 and AURKA are frequently overexpressed in gastric cancer cell lines and in approximately half of primary GC tumors. Similar results were obtained from EAC tumors and cell lines, suggesting that overexpression of AURKA and HDM2 is a common molecular event in upper gastrointestinal cancers.
We have previously reported that AURKA suppresses P53 transcriptional activity in gastric cancer (36). P53 is a vital transcriptional factor that induces expression of pro-apoptotic proteins in response to DNA damaging chemotherapeutic agents. AURKA has been reported to inhibit P53 function by directly phosphorylating P53 at Ser315 and thereby inducing its proteolytic degradation in H1299 human non-small cell lung carcinoma cells (44). In addition, AURKA-mediated phosphorylation of P53 at Ser215 inhibits P53 transcriptional activity and suppresses expression of its downstream targets (24). Although we have previously shown that overexpression of AURKA inhibits P53 expression and activity (36), we did not observe AURKA/P53 interaction in GC cell models (data not shown). These results indicate that this protein interaction may be cell line dependent, and suggest that AURKA-induced suppression of P53 is mediated by other mechanisms. HDM2, a critical negative regulator of P53, blocks P53 function by inhibiting its transcriptional activity through nuclear export into the cytoplasm and ubiquitin-mediated proteasomal degradation (26). Our results showed that overexpression of AURKA significantly increased the HDM2 protein level coupled with downregulation of P53 and P21 expression. Conversely, knockdown of endogenous AURKA or pharmacological inhibition with alisertib attenuated HDM2 protein levels, and upregulated P53 and P21 expression. Additionally, mutant AURKA failed to affect expression of HDM2, indicating that AURKA function is dependent on its kinase activity. These results clearly showed that AURKA is a positive regulator of HDM2 in GC cells.
Phosphorylation of HDM2 (S166) by AKT is known to enhance HDM2-mediated P53 ubiquitination and degradation (26). Of note, we have previously shown that AURKA can phosphorylate AKT at Ser473 (36). Using immunofluorescence, we confirmed the AURKA mediates an increase in HDM2 protein levels. This is in accordance with previous reports showing that both AURKA and HDM2 localize to the nucleus, thereby promoting mitosis and suppressing P53 function, respectively (45, 46). The co-IP data confirmed that AURKA and HDM2 were present in the same immuno-protein complex in GC cells. The co-IP and in vitro kinase assay data indicated, for the first time, that recombinant AURKA directly interacts with and phosphorylates HDM2 recombinant protein. We further validated this novel finding and demonstrated that inhibition of AURKA with alisertib blocks AURKA/HDM2 interaction in vitro.
Since HDM2 is a major negative regulator of P53 (47), we examined the effects of AURKA on HDM2-mediated ubiquitination of P53. The IP data indicated that AURKA overexpression substantially enhanced HDM2-mediated ubiquitination of P53. To examine if AURKA-mediated suppression of P53 is dependent on HDM2/P53 interaction, we employed Nutlin3A, an investigational HDM2 specific inhibitor with antitumor activity (48), to disrupt this protein interaction. Indeed, the data showed that Nutlin3A treatment can abrogate AURKA-mediated inhibition of P53, confirming that HDM2/P53 interaction is critical for AURKA-dependent suppression of P53.
Our aforementioned results confirm that AURKA kinase activity is essential for enhancing HDM2-induced ubiquitination and degradation of P53. This could be an important mechanism that mediates resistance to chemotherapeutic drugs whereby activation of HDM2 prevents apoptosis by down-regulating P53 expression. Accordingly, our data indicated that AURKA overexpression significantly attenuated CDDP-mediated induction of P53 and P21 protein expression. This finding provides further evidence that AURKA can mediate resistance to CDDP through regulation of HDM2/P53 signaling. Conversely, the inhibition of AURKA with alisertib attenuated cell survival in a dose-dependent manner, and was associated with decreased expression of HDM2 coupled with increased P53 and P21 protein levels in GC cells. In line with the in vitro data, the tumor xenograft data and molecular analysis indicated that inhibition of AURKA with alisertib significantly reduced tumor growth coupled with down-regulation of HDM2 and induction of P53 and its downstream transcriptional targets. This underscores the importance of AURKA as an effective therapeutic target upstream of HDM2 in GC. Our data further support the ongoing clinical trials with AURKA-specific small molecule pharmacological inhibitors (clinicaltrials.gov). In addition, we have previously reported that AURKA regulates cell death in P53 deficient cancer cells by inhibiting P73 apoptotic protein expression and activity (8, 25). HDM2 has been shown to attenuate P73 protein function by suppressing P73 transcriptional activity (49). Therefore, in addition to AURKA-mediated direct regulation of p73 protein expression, AURKA-HDM2 axis could be an alternative mechanism that regulates cell death in P53 deficient cancer cells.
In summary, we demonstrated that AURKA and HDM2 are overexpressed in gastric cancer. Up-regulation of AURKA enhanced expression and phosphorylation of HDM2 coupled with suppression of P53 and its downstream targets. Therefore, targeting AURKA/HDM2 axis with AURKA inhibitors could be an effective therapeutic approach in GC.
Supplementary Material
Translational Significance.
Gastric cancer (GC) is characterized by poor patient survival and resistance to chemotherapy. Several chemotherapeutic agents used in the treatment of GC induce DNA damage and activate P53 pro-apoptotic functions. Conversely, AURKA and HDM2 suppress P53 protein expression and activity. Herein, we report for the first time that AURKA and HDM2 proteins are frequently co-overexpressed in GC tissues and cell lines. Our data indicate that AURKA regulates the expression and phosphorylation levels of HDM2 which could be a major mechanism attenuating P53 protein function in gastric cancer cells. The use of AURKA inhibitor, alisertib, reversed these effects leading to significant suppression of tumor cell growth in vitro and in vivo. The fact that AURKA activates HDM2 and is frequently overexpressed in GC strongly reasons for the use of AURKA inhibitors in the treatment of GC.
Acknowledgments
Grant support: This study was supported by grants from the National Institute of Health; R01CA131225 (WER), VICTR pilot project support (WER) from Vanderbilt CTSA grant UL1 RR024975; Vanderbilt SPORE in Gastrointestinal Cancer (P50 CA95103), Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or Vanderbilt University.
Footnotes
Conflict of interest: All the authors declared no conflict of interest for the purpose of this study.
References
- 1.Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–15. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- 2.Reim D, Gertler R, Novotny A, Becker K, Buschenfelde CM, Ebert M, et al. Adenocarcinomas of the Esophagogastric Junction Are More Likely to Respond to Preoperative Chemotherapy than Distal Gastric Cancer. Ann Surg Oncol. doi: 10.1245/s10434-011-2147-8. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 5.Cronin J, McAdam E, Danikas A, Tselepis C, Griffiths P, Baxter J, et al. Epidermal growth factor receptor (EGFR) is overexpressed in high-grade dysplasia and adenocarcinoma of the esophagus and may represent a biomarker of histological progression in Barrett’s esophagus (BE) Am J Gastroenterol. 106:46–56. doi: 10.1038/ajg.2010.433. [DOI] [PubMed] [Google Scholar]
- 6.Rugge M, Fassan M, Zaninotto G, Pizzi M, Giacomelli L, Battaglia G, et al. Aurora kinase A in Barrett’s carcinogenesis. Hum Pathol. 41:1380–6. doi: 10.1016/j.humpath.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sehdev V, Peng D, Soutto M, Washington MK, Revetta F, Ecsedy J, et al. The Aurora Kinase A Inhibitor MLN8237 Enhances Cisplatin-Induced Cell Death in Esophageal Adenocarcinoma Cells. Mol Cancer Ther. 11:763–74. doi: 10.1158/1535-7163.MCT-11-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lord RV, O’Grady R, Sheehan C, Field AF, Ward RL. K-ras codon 12 mutations in Barrett’s oesophagus and adenocarcinomas of the oesophagus and oesophagogastric junction. J Gastroenterol Hepatol. 2000;15:730–6. doi: 10.1046/j.1440-1746.2000.02163.x. [DOI] [PubMed] [Google Scholar]
- 10.Matuschek C, Bolke E, Peiper M, Knoefel WT, Budach W, Erhardt A, et al. The role of neoadjuvant and adjuvant treatment for adenocarcinoma of the upper gastrointestinal tract. Eur J Med Res. 16:265–74. doi: 10.1186/2047-783X-16-6-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res. 2000;60:3899–903. [PubMed] [Google Scholar]
- 12.Katayama H, Ota T, Jisaki F, Ueda Y, Tanaka T, Odashima S, et al. Mitotic kinase expression and colorectal cancer progression. J Natl Cancer Inst. 1999;91:1160–2. doi: 10.1093/jnci/91.13.1160. [DOI] [PubMed] [Google Scholar]
- 13.Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, et al. Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. Br J Cancer. 2001;84:824–31. doi: 10.1054/bjoc.2000.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–58. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa T, Sunamura M, Horii A. Molecular mechanisms of pancreatic carcinogenesis. Cancer Sci. 2006;97:1–7. doi: 10.1111/j.1349-7006.2005.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen CC, Yeh CN, Cheng CT, Jung SM, Huang SC, Chang TW, et al. Integrating bioinformatics and clinicopathological research of gastrointestinal stromal tumors: identification of aurora kinase A as a poor risk marker. Ann Surg Oncol. 2012;19:3491–9. doi: 10.1245/s10434-012-2389-0. [DOI] [PubMed] [Google Scholar]
- 18.Fang Z, Xiong Y, Li J, Liu L, Li M, Zhang C, et al. Copy-number increase of AURKA in gastric cancers in a Chinese population: a correlation with tumor progression. Med Oncol. 2011;28:1017–22. doi: 10.1007/s12032-010-9602-4. [DOI] [PubMed] [Google Scholar]
- 19.Dar AA, Goff LW, Majid S, Berlin J, El-Rifai W. Aurora kinase inhibitors--rising stars in cancer therapeutics? Mol Cancer Ther. 2010;9:268–78. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Cutsem E, Van de Velde C, Roth A, Lordick F, Kohne CH, Cascinu S, et al. Expert opinion on management of gastric and gastro-oesophageal junction adenocarcinoma on behalf of the European Organisation for Research and Treatment of Cancer (EORTC)-gastrointestinal cancer group. Eur J Cancer. 2008;44:182–94. doi: 10.1016/j.ejca.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Ilson DH. Docetaxel, cisplatin, and fluorouracil in gastric cancer: does the punishment fit the crime? J Clin Oncol. 2007;25:3188–90. doi: 10.1200/JCO.2006.10.2210. [DOI] [PubMed] [Google Scholar]
- 22.Sumi K, Tago K, Kasahara T, Funakoshi-Tago M. Aurora kinase A critically contributes to the resistance to anti-cancer drug cisplatin in JAK2 V617F mutant-induced transformed cells. FEBS Lett. 2011;585:1884–90. doi: 10.1016/j.febslet.2011.04.068. [DOI] [PubMed] [Google Scholar]
- 23.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, et al. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–82. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- 25.Dar AA, Belkhiri A, Ecsedy J, Zaika A, El-Rifai W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer Res. 2008;68:8998–9004. doi: 10.1158/0008-5472.CAN-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- 28.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–9. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soussi T. Handbook of p53 mutation in cell lines. 2007 [Google Scholar]
- 30.Dar AA, Belkhiri A, El-Rifai W. The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene. 2009;28:866–75. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, et al. Aurora Kinase A Promotes Inflammation and Tumorigenesis in Mice and Human Gastric Neoplasia. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soutto M, Belkhiri A, Piazuelo MB, Schneider BG, Peng D, Jiang A, et al. Loss of TFF1 is associated with activation of NF-kappaB-mediated inflammation and gastric neoplasia in mice and humans. J Clin Invest. 121:1753–67. doi: 10.1172/JCI43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu S, Hong J, Tripathi MK, Sehdev V, Belkhiri A, El-Rifai W. Regulation of CXCR4-mediated invasion by DARPP-32 in gastric cancer cells. Mol Cancer Res. 11:86–94. doi: 10.1158/1541-7786.MCR-12-0243-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorgun G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115:5202–13. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belkhiri A, Zaika A, Pidkovka N, Knuutila S, Moskaluk C, El-Rifai W. Darpp-32: a novel antiapoptotic gene in upper gastrointestinal carcinomas. Cancer Res. 2005;65:6583–92. doi: 10.1158/0008-5472.CAN-05-1433. [DOI] [PubMed] [Google Scholar]
- 36.Dar AA, Zaika A, Piazuelo MB, Correa P, Koyama T, Belkhiri A, et al. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer. 2008;112:1688–98. doi: 10.1002/cncr.23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 39.Howlader N, Ries LA, Stinchcomb DG, Edwards BK. The impact of underreported Veterans Affairs data on national cancer statistics: analysis using population-based SEER registries. J Natl Cancer Inst. 2009;101:533–6. doi: 10.1093/jnci/djn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth AD. Chemotherapy in gastric cancer: a never ending saga. Ann Oncol. 2003;14:175–7. doi: 10.1093/annonc/mdg081. [DOI] [PubMed] [Google Scholar]
- 41.Ohi S, Takahashi N, Ninomiya K, Nakajima M, Hashimoto H, Tachibana T, et al. Establishment and characterization of a cisplatin-resistant cell line (IGSK-1) from a poorly differentiated gastric adenocarcinoma. Hum Cell. 2007;20:15–22. doi: 10.1111/j.1749-0774.2007.00024.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L, Pan Y, Gang Y, Wang H, Jin H, Tie J, et al. Identification of GAS1 as an epirubicin resistance-related gene in human gastric cancer cells with a partially randomized small interfering RNA library. J Biol Chem. 2009;284:26273–85. doi: 10.1074/jbc.M109.028068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S, Chen M, Shen Y, Shen W, Guo H, Gao Q, et al. Inhibition of activated Stat3 reverses drug resistance to chemotherapeutic agents in gastric cancer cells. Cancer Lett. 315:198–205. doi: 10.1016/j.canlet.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 45.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 46.Schuster K, Fan L, Harris LC. MDM2 splice variants predominantly localize to the nucleoplasm mediated by a COOH-terminal nuclear localization signal. Mol Cancer Res. 2007;5:403–12. doi: 10.1158/1541-7786.MCR-06-0146. [DOI] [PubMed] [Google Scholar]
- 47.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–8. [PubMed] [Google Scholar]
- 48.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–41. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maisse C, Guerrieri P, Melino G. p73 and p63 protein stability: the way to regulate function? Biochem Pharmacol. 2003;66:1555–61. doi: 10.1016/s0006-2952(03)00511-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.