Abstract
Phage-encoded serine-integrases are large serine-recombinases that mediate integrative and excisive site-specific recombination of temperate phage genomes. They are well suited for use in heterologous systems and for synthetic genetic circuits as the attP and attB attachment sites are small (<50 bp), there are no host factor or DNA supercoiling requirements, and they are strongly directional, doing only excisive recombination in the presence of a recombination directionality factor. Combining different recombinases that function independently and without cross-talk to construct complex synthetic circuits is desirable, and several different serine-integrases are available. However, we show here that these functions are not reliably predictable, and we describe a pair of serine-integrases encoded by mycobacteriophages Bxz2 and Peaches with unusual and unpredictable specificities. The Integrases share only 59% amino acid sequence identity and the attP sites have fewer than 50% shared bases, but they use the same attB site and there is non-reciprocal cross-talk between the two systems. The DNA binding specificities do not result from differences in specific DNA contacts, but from the constraints imposed by the configuration of the component half-sites within each of the attachment site DNAs.
Introduction
An unusual feature of the serine-integrases (Int) is their ability to recognize two distinctly different attachment sites, attP and attB, that share little sequence resemblance to each other 1; 2; 3, and select only these for site-specific integrative recombination. Most serine-integrases do so with impressive specificity and discrimination between att and non-att sites, such that targeted recombination can be accomplished in large eukaryotic genomes 4. Single base substitutions in either ϕC31 attB 5 or Bxb1 attP 6 are sufficient to interrupt recombination, and DNA binding alone is insufficient for recombination. Recombination is mediated by binding of integrase dimers to attP and attB DNA 7, protein-mediated synapsis 3, cleavage of both strands about a shared central dinucleotide 2; 3; 8, rotation of one set of covalently-linked half site-Int complexes about a common axis 9; 10, and religation. Integrative recombination is specific for attP x attB, is efficient, and is strongly directional 11; 12. Only permissive combinations of attachment sites (e.g. attP, attB) support synapsis, which presumably requires specific and compatible integrase conformations that are DNA-directed 1; 2; 13. Integrase-DNA complexes are symmetric, synapse in either orientation, and alignment of the asymmetric central dinucleotide determines the polarity of integration 2; 3; 14. Excisive recombination (i.e. attL x attR) is also catalyzed by Int, but requires a recombination directionality factor (RDF) that associates with Int-DNA complexes and converts non-compatible conformations with attL or attR into those permissive for synapsis 15; 16; 17.
Several serine-integrases have a proteolytically-sensitive site that defines a catalytic N-terminal domain (~150 residues) and a large C-terminal DNA binding domain 2; 7; 18. Bxb1, ϕC31 and A118 Ints bind as dimers to DNA 2; 7; 19 with one protomer bound to each half site (B, B′, P or P′; see Fig. 1). The C-terminal domain is a monomer in solution and has DNA binding activity 2; 7; 18, although it’s binding pattern is system specific. For example, ϕC31 CTD binds with cooperativity at least to some att sites, but cooperativity has not been observed with Bxb1 or A118 CTD2; 7. The ϕC31 CTD also forms synaptic interactions mediated by a coiled-coil (CC) motif 18, which are not seen in Bxb1 or A118 on the DNA substrates tested 2; 7. The structure of the Listeria innocua (LI) Int CTD of the integrase – a close relative of A118 Int – bound to attP half-site DNA has recently been described 20; 21. This supports a model for site selection in which the coiled-coil domain plays a key role, promoting synaptic interactions between attB and attP complexes, and interfering with other synaptic interactions either because of lack of CC interactions, stearic interference, or competitive intra-complex CC interactions.
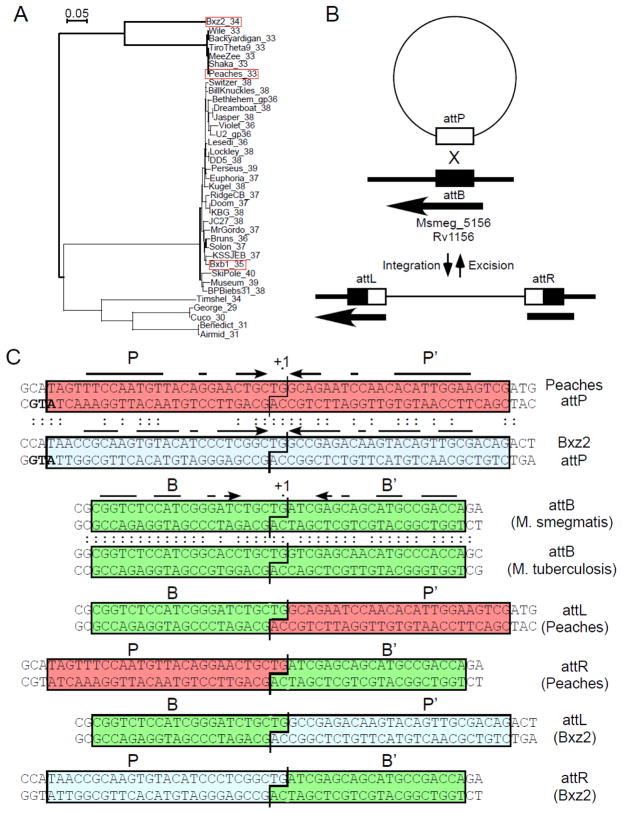
Figure 1. Attachment sites for Peaches and Bxz2 Integrases.
(A) Phylogeny of mycobacteriophage serine-integrases with the three integrases discussed here shown in red boxes; labels show the phage name and the gene number. (B) Schematic representation of Peaches and Bxz2 integration, showing site-specific recombination between the phage attP site and chromosomal attB site. Thin and thicker lines correspond to phage and bacterial DNA respectively. The arrow indicates the position and direction of Msmeg_5156 into which these phages integrate. (C) Attachment site sequences with shared bases between the Peaches and Bxz2 attP sites, and between the M. smegmatis and M. tuberculosis attB site, shown by colons. Required sequences for Bxz2 attP, attB (blue, green boxes respectively; see Fig. 2), and those inferred for Peaches-attP (red box) are shown; interrupted arrows indicate symmetrically conserved positions. The int translation initiation codons overlapping attP sites are shown in bold.
Serine-integrases have been shown to function in a variety of heterologous hosts including bacteria, flies, plants, parasites, mouse and human cells 4; 22; 23; 24; 25. Bxb1 and TP901-1 Ints have been deployed for microbial computation by mediating regulated and well-maintained conversion between two alternate states 26; 27. Greater computational complexity requires the use of multiple systems that operate independently and without cross-talk, i.e. restricting usage to cognate attachment sites 26; 27. Many different serine-integrases have been identified, although the lack of extensive symmetry can make attP sites difficult to recognize, and attB sites cannot be predicted at all. Although sequence divergence between integrases and their attP sites is expected to reflect independent functions, we describe here an unusual pair of Ints that show non-reciprocal cross-talk and unexpectedly use the same attB site.
Results
Bxz2 and Peaches integrases using the same attB site
The 37 serine-integrases coded by mycobacteriophages span considerable sequence diversity (Fig. 1A). Many are close relatives of the previously studied Bxb1 Int 3; 7; 12 and contain identical or closely related attP sites. Others are more distantly related and share no more than 32% amino acid identity. In a search for additional serine-integrase systems we investigated Mycobacteriophages Peaches, a distant relative of Bxb1 (26% identity). To identify Peaches attP and chromosomal attB sites, we constructed a non-replicating integration-proficient plasmid vector (pKR04; coordinates 25,077 – 26,730 including Peaches int and flanking DNA) that efficiently transforms both M. smegmatis and M. tuberculosis (105 transformants/μg DNA) by chromosomal integration. Comparison of the recombinant junctions at attL and attR (Fig. S1) surprisingly showed that Peaches uses the same attB site within Msmeg_5156 (Fig. 1B) as previously described for Bxz2 Int 28, although the Peaches and Bxz2 Ints share only 59% amino acid identity (Fig. S2). Alignment of the Bxz2 and Peaches attP sites shows they have the same central nucleotide (5′-TG) about which strand exchange occurs, but little overall sequence similarity (24/52 bp shared; Fig. 1C). Both attP sites have imperfect inverted symmetry, but only seven positions (−2/+2, −3/+3, −5/+5, −14/+14, −15/+15, −16/+16, −20/+20) are both shared between the two attP sites and symmetrically conserved in both sites (Fig. 1C).
In vitro integration and attachment site size
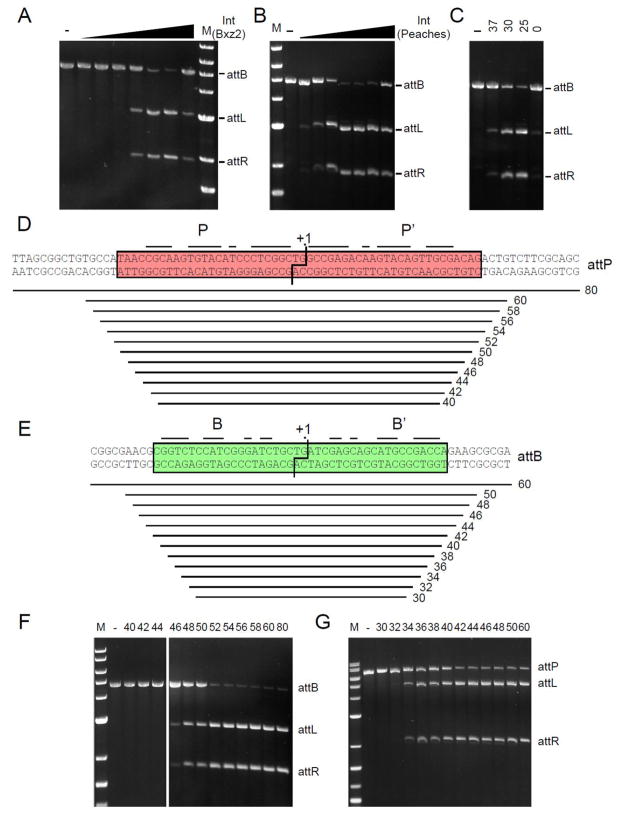
To further characterize the Bxz2 and Peaches systems, we overexpressed and purified the Ints and established in vitro integration systems (Fig. 2A–C). The recombination requirements are simple and include only Int, attP and attB DNAs, and a simple buffer, although Peaches Int is somewhat temperature sensitive (Fig. 2C); unlike what is reported for ϕC31 Int 29 but similar to Bxb1 and A118 neither reaction is influenced by zinc. Neither requires protein or high-energy cofactors nor DNA supercoiling, like other serine-integrases 2; 11; 30; 31; 32; 33. Mutations at the center of the Bxz2 site confirm the 5′-TG as the central dinucleotide (Fig. S3). To determine the site requirements for Bxz2 Int we constructed nested sets of DNA substrates and tested recombination with linear partner DNAs (Fig. 2D–G). For attP, efficient recombination requires 52 bp (Fig. 2D, F), although substrates as small as 46 bp show some activity; the outer three bp at each end may involve DNA backbone interactions rather than base-specific contacts. A similar pattern is seen with attB (Fig. 2E, G), where 42 bp are required for fully efficient recombination, but a 34 bp substrate has some activity.
Figure 2. Requirements for Peaches and Bxz2 integration.
(A) Bxz2 Int-mediated recombination in vitro between linear attB (pSS1 linearized with BamH1) and 60 bp attP DNA substrates; Int concentrations are: 0, 16, 33, 66, 125, 250, 500, 1000 nM. (B) Peaches-Int-mediated recombination in vitro. Peaches-Int concentrations: 0, 125, 250, 500, 600, 700, 1000, 2000 nM. (C) In vitro integration varying temperature (°C); Peaches-Int concentration, 700 nM. (D, E) Sizes of synthetic Bxz2 attP and M. smegmatis attB substrates used to determine site requirements, and (F, G) determination of their recombination activities with Bxz2 Int. The sizes of the attP (F) and attB (G) substrates are shown above each lane. Positions of attL and attR products are indicated.
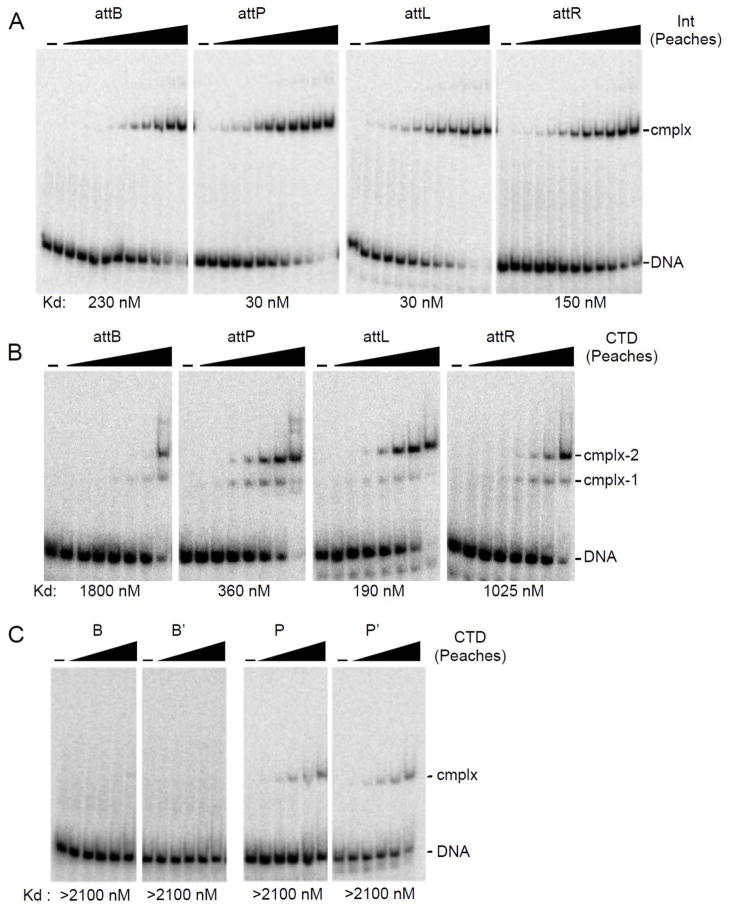
Binding of Bxz2 Int and CTD to att site DNA
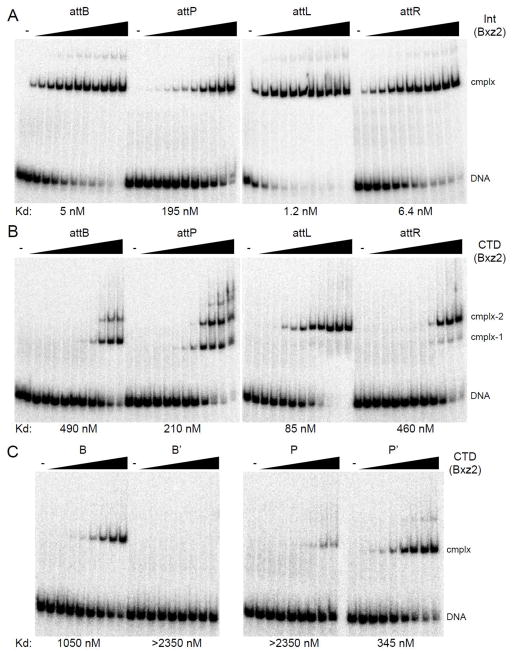
To explore site recognition and specificity, we analyzed the binding of Bxz2 Int and its DNA-binding C-terminal domain (CTD) to att site DNA (Fig. 3). Bxz2 Int binds to attB, attL and attR with affinities that are about 10-fold tighter than observed for Bxb1 Int 7, but binding to attP is substantially weaker (Fig. 3A). Int forms one major complex with each substrate – although weakly formed slower migrating complexes are also visible – and this major complex is a dimer of Int bound to DNA (Fig. 4A). To explore the protein interactions with each of the half-att sites we overexpressed and purified the 352-residue Bxz2 CTD (171–522; based on alignment with other serine integrases and secondary structure predictions; Fig. S2). Bxz2 CTD binds to all four att sites but generates different types of protein-DNA complexes (Fig. 3B). With attB DNA, two complexes are observed (complex-1, complex-2); complex-1 is a monomer of CTD bound to one of the half sites, and complex-2 contains two CTD monomers, as confirmed by formation of CTD and MBP-CTD hybrids (Fig. 4B). Binding to attP is similar, although at high CTD concentrations slower migrating complexes are seen (Fig. 3B) that might involve synaptic interactions as described for ϕC31 Int 18. The binding to attL and attR is somewhat different. First, the predominant complex formed migrates similarly to attP and attB complex-2, suggesting substantial cooperativity of CTD binding. Secondly, the affinity is substantially weaker for attR than for attL (Fig. 3B). Surprisingly, Bxz2 CTD shows little or no binding to the B′ and P individual half-sites, even though it binds reasonably well to both the B and P′ half sites (Fig. 3C). These patterns illustrate an unusual and interesting feature of Bxz2 Int, in that the affinity of Int for its att sites is not simply defined by the affinity of CTD for the component half sites. We note that the A118 Int binds better to B and P half sites than to B′ and P′ half sites, but that an attB substrate containing two B-type half sites is a poor recombination substrate 2, indicating a functional asymmetry of these half sites. In addition, the LI Int CTD-attP half-site DNA complex structure 20 shows DNA contacts by the C-terminal domain of the E helix at the center of attP (equivalent to Bxz2/Peaches positions −3 – +3), and exclusion from our CTD constructs may affect the affinity of CTD binding, although we note that the central six base pairs are identical in the Bxz2 and Peaches attP sites (Fig. 1).
Figure 3. DNA binding of Bxz2 Int and CTD.
(A) Binding of Bxz2 Int to attB, attP, attL and attR. Int concentrations are as follows: 0, 1, 2, 4, 8, 16, 31, 62, 125, 250, 500 and 1000 nM. DNA substrates used are 60 bp for attB, attP, attL and attR. (B) Binding of Bxz2 CTD to attB, attP, attL and attR DNA. CTD concentrations are as follows 0, 2, 4, 9, 18, 37, 73, 147, 294, 588, 1175 and 2350 nM. DNA substrates are same as in panel A. (C) CTD binding to half site substrates. CTD concentrations are as follows: 0, 18, 37, 73, 147, 294, 588, 1175 and 2350 nM. DNA substrates used are 50 bp, for B, B′, P and P′. Apparent binding affinities (Kd) are indicated.
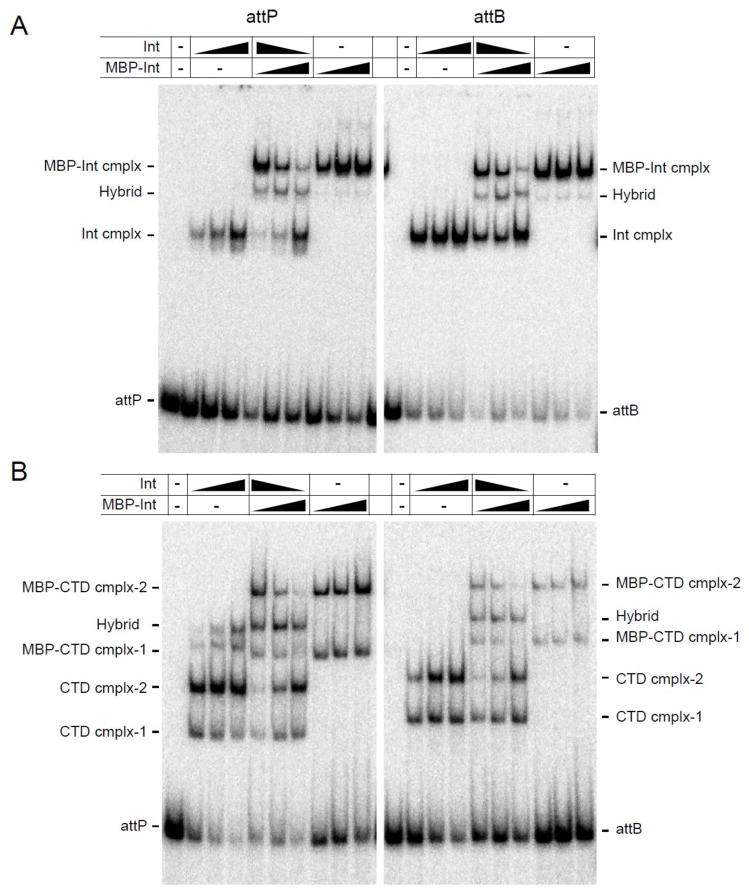
Figure 4.
Multimeric states of Int-DNA and CTD-DNA complexes. (A) Both Bxz2 Int and the larger MBP-Int fusion protein form a single complex with both attP and attB DNA, as indicated. Addition of both proteins forms a single hybrid complex consistent with both proteins binding as dimers to attP and attB DNA. The Int and MBP-Int concentrations are 125, 250 and 500 nM and the first lane is without protein. The ratio of Int and MBP-Int used in the central panels are as follows: 4:1 (lane 1), 1:1 (lane 2), 1:4 (lane 3). (B) As above but using Bxz2 CTD and Bxz2 MBP-CTD fusion protein. These data suggest that complex-1 contains a CTD monomer, and complex-2 contains two CTD protomers. CTD and MBP-CTD concentrations are 588, 1175 and 2350 nM and the first lane is without protein. Ratios of CTD and MBP-CTD in the central panel are as in panel A.
Binding of Peaches Int and CTD to att site DNA
Peaches Int has similar DNA binding properties to Bxz2 Int (Fig. 5A) but with three main differences. First, although the two Ints share the same attB site, the Peaches Int affinity for attB is substantially weaker than with Bxz2 Int (Figs. 3A, 5A). Secondly, Peaches Int binds tighter to its attP than Bxz2 Int does to its attP (Figs. 3A, 5A) such that the preferences for the two sites are inverted (Bxz2 Int, attB > attP; Peaches Int, attP > attB; Figs. 3A 5A). Thirdly, Peaches and Bxz2 Ints bind their respective attL sites somewhat tighter than their attR sites (5-fold), but the Peaches Int affinities are both 25-fold weaker than with Bxz2 Int. Peaches CTD binds its att sites although the affinity is weaker by ~5-fold for Peaches Int binding to each of the sites. Surprisingly, only very weakly formed complexes are observed with Peaches CTD and P and P′ half site DNA, and there are no observable complexes with B or B′ half site DNA (Fig. 5C). Thus, as with Bxz2 Int, DNA binding is not determined solely by the sum of the interactions with each of the component half sites.
Figure 5. DNA binding of Peaches Int and CTD.
(A) Protein-DNA complexes were separated from attB, attP, attL or attR DNA by native gel electrophoresis. Int concentrations are as in Fig. 3A. (B) Binding of Peaches-CTD to attB, attP, attL, and attR; CTD concentrations: 0, 2.9, 8.6, 26, 78, 234, 700 and 2100 nM. (C) Binding of Peaches-CTD to half-site substrates, B, B′, P, P′. CTD concentrations: 0, 26, 78, 234, 700 and 2100 nM. Apparent binding affinities (Kd) are indicated.
Cross-talk between the Bxz2 and Peaches Ints
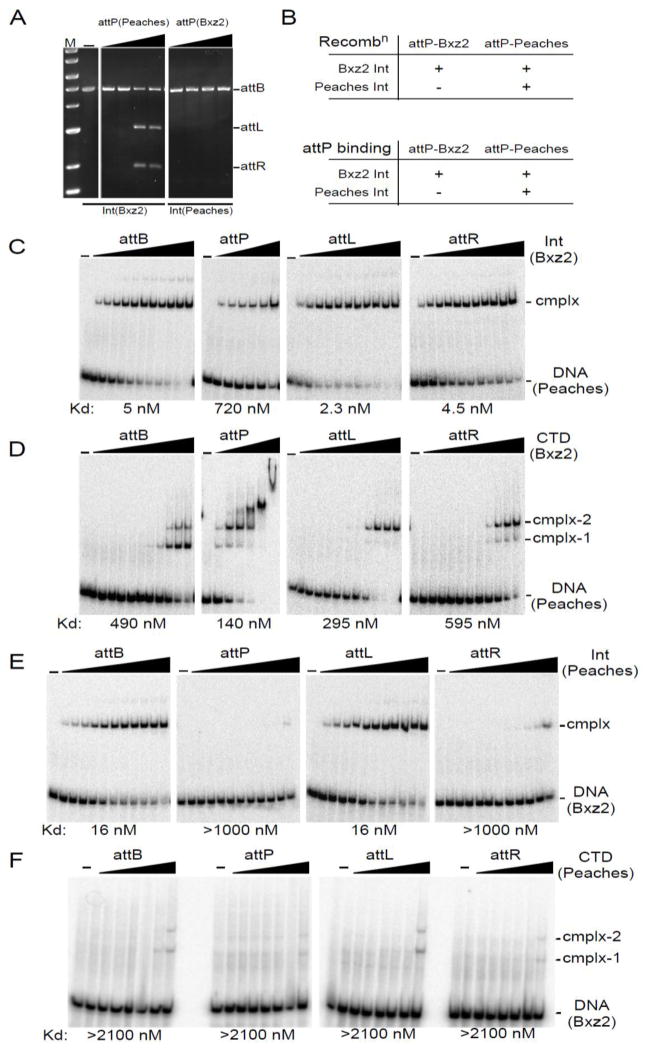
Because the Peaches and Bxz2 Ints use the same attB site, we tested whether they could also use each other’s attP sites, in spite of the evident lack of sequence similarity. Peaches Int shows strong specificity for its attP site and does not recombine Bxz2 attP (Fig. 6A, B). In contrast, Bxz2 Int efficiently catalyzes recombination with Peaches attP (Fig. 6A, B), demonstrating remarkable promiscuity given the differences between the attP sites (Fig. 1C). Bxz2 Int binds Peaches attP – albeit with slightly reduced affinity relative to its own attP (Figs. 3A, 6C) – and binds well to both Peaches attL and attR (Fig. 6C); the pattern of CTD binding to Peaches att sites is generally similar to the binding to its own (Fig. 3B, 6D). The failure of Peaches Int to recombine Bxz2 attP can be accounted for by its inability to bind Bxz2 attP DNA (Fig. 6E). Although this could simply be explained by failure of Peaches Int to recognize the Bxz2 P and P′ half sites, this is not supported. For example, Peaches Int binds well to Bxz2 attL showing that in that context, it recognizes the Bxz2 P′ site (Fig. 6E). Furthermore, the failure to bind well to Bxz2 attR (Fig. 6E) is not the result of failure to recognize the P half site, because in other substrate contexts it is able to do so (see below). We note that Peaches CTD binds poorly to Bxz2 attP, attL and attR and there are no cooperativity benefits as seen for its own att sites (Fig. 6F). Taken together, these data show not only that Int binding to its att sites does not equate to the sum of its half-site interactions but that this has important consequences for site specificity, which cannot be predicted from DNA sequence alone.
Figure 6.
Crosstalk between Peaches and Bxz2 Integrases. (A) Recombination of Peaches-attP and Bxz2-attP by Bxz2-Int and Peaches-Int. Int concentrations: 125, 250, 500, 1000 nM. (B) Summary of att site usage. (C) Binding of Bxz2 Int to attB, attP-Peaches, attL-Peaches, and attR-Peaches. Int concentrations for attB, attL and attR are as in Fig. 3A. Int concentrations for attP: 0, 31, 62, 125, 250, 500 and 1000 nM. (D) Binding of Bxz2-CTD to attB, attP-Peaches, attL-Peaches and attR-Peaches. CTD concentrations are as in Fig. 3B, except for attP: 0, 73, 147, 294, 588, 1175 and 2350 nM. (E) Binding of Peaches Int to attB, attP-Bxz2, attL-Bxz2, and attR-Bxz2. Int concentrations are as in Fig. 3A. (F) Binding of Peaches CTD to attB, attP-Bxz2, attL-Bxz2, and attR-Bxz2. CTD concentrations are as in Fig. 5B. Apparent binding affinities (Kd) are indicated.
Activities of Bxz2-Peaches hybrid sites
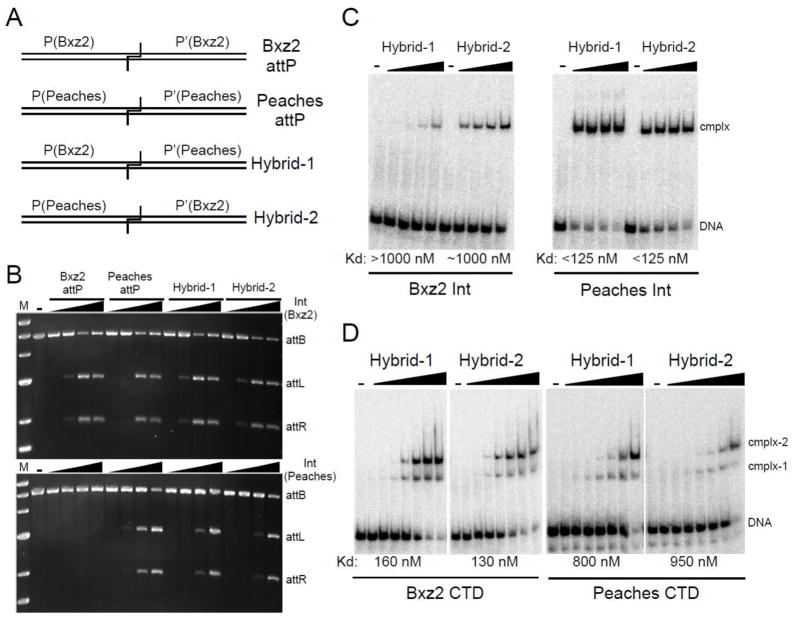
To further explore the specificity differences between these two Ints, we constructed hybrid sites each containing one half site from Bxz2 attP and one from Peaches attP (Fig. 7A). Bxz2 Int is able to recombine both hybrids efficiently, reflecting its greater promiscuity described above (Fig. 7B). Nonetheless, Bxz2 Int binding to both hybrids is relatively poor, especially to Hybrid-1 (Fig. 7C) suggesting that in this context, recognition of the Peaches P′ site is weak, even though it is not in the other contexts examined (Peaches attP, Peaches attL; Fig. 6D). In support of this, Bxz2 CTD binds better to both hybrids than full-length Int does, showing that although in some contexts Int dimerization via the N-terminal domain facilitates binding, in other contexts there is a significant binding penalty. Interestingly, Peaches Int also recombines both hybrid substrates (Fig. 7B) and binds well to them (Fig. 7C), showing no obvious difficulty in recognizing either the Bxz2 P or P′ site in these contexts; however, in this case Int binding is substantially stronger than CTD binding (Fig. 7C, D).
Figure 7. Peaches and Bxz2 Int usage of hybrid attP sites.
(A) Schematic representation of hybrids. (B) Recombination of hybrid attP sites by Peaches- and Bxz2-Integrases. Int concentrations: 0, 125, 250, 500, 1000 nM. (C) Binding of Ints to hybrid attP. Int concentrations: 0, 125, 250, 500, 1000 nM. (D) Binding of CTDs to hybrid sites. CTD concentrations are as in Fig. 5B. Apparent affinities (Kd) are indicated.
Discussion
The serine-integrases are a particularly interesting class of site-specific recombinases. They are able to recognize two small, distinctly different, quasi-symmetrical DNA sites, attP and attB, and are highly directional. As such, they must discriminate not only between att and non-att DNA, but also between the substrates for integrative recombination, attP and attB, and the products, attL and attR. This represents a conundrum for selectivity, in that the substrates and products differ only by their configurations of the component half-sites, B, B′, P and P′ (Fig. 1C), and this is accommodated in part because DNA binding (e.g. to attL or attR) is required but not sufficient for recombination. Here we show that selectivity is also determined at the level of DNA binding through the specific context of the two half-sites and that different Ints have different properties in interpreting this context-specific information.
The finding that phage Bxz2 and Peaches use the same attB site is surprising given the sequence divergence of both the Ints and their attP sites, and the non-reciprocal cross-talk between the two systems was not predictable. These relationships are interesting with regard to the specificity of site selection, but perplexing for predicting which integrase systems operate independently, especially when designing complex synthetic genetic circuits 26; 27. We note for example, that the Timshel and Benedict Ints share 52% sequence identity (Fig. 1A), and although plausible attP sites can be predicted with little or no discernable sequence similarity (Fig. S4), the data here suggest caution in interpreting the likelihood of cross-talk. In this case, assuming that the attP predictions are valid, they have different central dinucleotides (Fig. S4) and thus must use different attB sites.
Three aspects of DNA binding by Peaches and Bxz2 Int are particularly intriguing. First, there is good evidence for cooperative interactions between CTD protomers when bound to DNA – which has not been observed in Bxb1 or A118 2; 7 but is similar to ϕC31 18 – and is also att-site specific. Not only does the Bxz2 CTD predominantly form complex-2 containing two CTD protomers with both attL and attR DNA, but this occurs regardless of the nearly undetectable binding to individual B′ and P half sites. With attB, Bxz2 CTD predominantly forms complex-1 presumably by occupying the B half site (given the poor binding to B′; Fig. 3C), and there is only a minor cooperative benefit for binding of a second CTD protomer to form complex-2 (Fig. 3B). These observations are consistent with recent structural data suggesting that the Int CC domain promotes cooperativity of binding of Int (and CTD) at some sites (i.e. attL and attR) but not others (i.e. attP and attB) 20. Peaches CTD does not bind well to any of the half-site DNA substrates (Fig. 5C), but benefits from cooperative interactions in binding to full att sites, particularly attP and attL (Fig. 5B). Cooperative binding to attL may be facilitated by the CC interactions as suggested for Bxz2, but these are not predicted to promote attP cooperative binding 20.
A second striking aspect of DNA recognition by these Integrases is the critical nature of the context of the half-sites for Int recognition. Although there are several illustrations of this, the most striking is the ability of Peaches Int to bind to both hybrid sites (Fig. 7C) – each of which contains either a Bxz2 P or P′ half-site – even though it fails to bind those same half-sites in the context of Bxz2 attP (Fig. 6E). A plausible explanation is that critical bases for recognition may be positioned differently in different attP half sites – such that Int protomers have a different ‘register’ – and that similar registers must be adopted in both halves of an att site. The structure of the LI Int-DNA complex shows that two DNA binding motifs – the recombinase domain (RD, corresponding to Bxz2 Int residues 152 – 310) and the Zinc-nucleated domain (ZD, corresponding to Bxz2 residues ~324 – 383; Fig. S2) – are joined by an 8-residue linker, such that RD and ZD contact DNA at the inside and the outside of the site respectively 20; 21. Appropriate docking of these two domains may therefore correspond to different registers of Int-DNA interactions at different sites. Rutherford et al. 20 also noted that the different geometries of attP and attB sites could be accommodated by the LI Int ZD recognizing similar sequence motifs in the two sites, but displaced 5 bp towards the center in attB relative to their positions in attP, and that this is a common features of serine-integrases. Although the P′ site of Bxz2 attP and both B and B′ share a 5′-GTCNNNA motif (Fig. 1), this is poorly conserved in the Bxz2 P half-site, and absent in both P and P′ of the Peaches attP site. This is not inconsistent with the model, as the sequences common to attB and attP may not be easily recognizable (only one base is conserved in all four proposed ZD binding sites for Bxb1 Int 20), although it is unclear what common sequences in P, P′, B and B′ could be recognized by the Peaches ZD.
Third, the non-reciprocal cross-talk suggests that the two Ints have different conformational flexibility enabling or restricting protein configurations needed to bind different att sites. Such flexibility could arise from the connectivity of various domains, including the linkers between NTD and CTD (Bxz2 residues 165–170), between the D and E helices in the NTD (Fig. S2), between the RD and ZD domains, and between the ZD and CC domains 20; 21. The first is attractive, as substitution at residue 156 in Bxb1 Int (within the inter-domain linker) specifically interferes with binding to attB but not to attP 7, and altered specificity mutants of ϕC31 Int also have substitutions in this inter-domain region 34. The helix D-E linker is also implicated, as it is the site of activating mutations in Tn3 and γδ resolvases 35; 36. Flexibility of the CC domain is also anticipated to promote different subunit interactions. Although sequence differences are distributed across the spans of the Bxz2 and Peaches Integrases there are notable departures at the linker locations (Fig. S2). Thus, as the Peaches and Bxz2 Integrases diverged from a common ancestor, relatively subtle changes in overall protein configuration could have moved specificity in two opposing directions. Bxz2 Int has become promiscuous and able to bind and recombine distantly related att sites, whereas Peaches Int acquired a new specificity, and the inability to bind to Bxz2 attP site or use it for recombination, all while utilizing the same chromosomal attB site. As a consequence, serine-integrase specificities for attP and attB site should be predicted cautiously, and in each case tested experimentally.
Materials and Methods
Plasmids constructions
The Peaches integration region (coordinates 25,077 – 26,730) was amplified by PCR, digested with NdeI and cloned into similarly digested pMOS-Hyg, producing pKR04. Candidate integration-proficient vectors were verified by restriction digestion and sequencing. For transformation efficiencies, 100ng of pKR04 DNA was electroporated into either M. smegmatis mc2155 or M. tuberculosis mc27000 competent cells 37, recovered and plated on selective media. Confirmation of attB integration sites was completed with PCR using a vector specific oligonucleotide and a nonspecific host oligonucleotide. PCR products were sequenced and the site of integration into the host chromosome was confirmed.
The Bxz2 integrase gene (34) was PCR amplified from genomic DNA and cloned into the Nde1-HindIII site of bacterial expression vector pLC3 to obtain pSS4. The resulting plasmid, pSS4, expresses a 522-residue of full integrase and His6-MBP tag protein; the His6-MBP fusion tag can be removed by digesting with TEV protease. The Bxz2 C-terminal domain (CTD) was PCR amplified from plasmid pSS4 and cloned into the Nde1-HindIII site of bacterial expression vector pLC3 to obtain pSS3; it expresses residues 171–522 of Bxz2 Int.
The Peaches integrase gene (33) was PCR amplified from genomic DNA and cloned into the Nde1-HindIII site of bacterial expression vector pET-21a to obtain pSS25. The resulting plasmid, pSS25, expresses a 504-residue of full integrase and His6 tag protein. Peaches CTD was PCR amplified from plasmid pSS25 and cloned into the Nde1-HindIII site of bacterial expression vector pET-21a to obtain pSS26; it expresses residues 170–504 of Peaches Int.
Over expression and purification of proteins
For overexpression and purification of Bxz2 Int, plasmid pSS4 was transformed into the E. Coli BL21 (DE3) pLysS cells and grown in Luria Bertani (LB) media at 37ºC to an OD600 of 0.5. His6-MBP-Int expression was induced by the addition of 0.8 mM IPTG at 18ºC for overnight growth. Cells were harvested and kept at −80ºC until used. The pelleted cells were resuspended by adding approximately 5 ml/g of lysis buffer to the cells and lysed by sonication. The lysis buffer composition was 10% glycerol, 500 mM NaCl, 25 mM Na2HPO4/NaH2PO4 (pH 8), 5 mM Imidazole, 1 mM β-mercaptoethanol. The homogenized cells were centrifuged and the cleared lysate was applied to a nickel column using Ni-NTA matrix, equilibrated with lysis buffer. The column was washed with lysis buffer followed by 1M NaCl then washed with lysis buffer + 10 mM and 20 mM imidazole, which removed most of the contaminant proteins. His6-MBP-Int was eluted with lysis buffer + 200 mM imidazole and then His6-MBP tag were cleaved via overnight digestion with TEV protease followed by chromatography through a heparin sepharose matrix to remove TEV and other contaminants. The heparin sepharose was equilibrated with buffer H (8% glycerol, 300 mM NaCl, 25 mM Na2HPO4/NaH2PO4 (pH 8), and 1 mM β-mercaptoethanol). The column was washed with buffer H + 300, 500, and 590 mM salt and integrase was eluted with buffer H + 700 mM salt. The resulting full-length integrase was dialyzed against storage buffer (pH 6.7) containing 50% glycerol, 300 mM NaCl, 25 mM Na2HPO4/NaH2PO4 (pH 8), and 1 mM β-mercaptoethanol. Bxz2 CTD expression and purification was similar to Bxz2 Int as described above.
For overexpression and purification of Peaches Int, Plasmid pSS25 was transformed into BL21 (DE3) Star cells and grown in LB broth to an optical density (OD) of 0.5 at 37ºC and induced for overnight at 17ºC with 0.7 mM IPTG. The cells were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 400 mM NaCl, 5% glycerol, and 10 mM imidazole, and the clarified supernatants were loaded onto Ni-NTA columns (Qiagen). The columns were washed with lysis buffer and 30 mM and 50 mM imidazole, and eluted with 180 mM imidazole. The purified integrase was dialyzed against storage buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 0.1 mM DTT and 50% glycerol. Peaches CTD expression and purification was similar to Peaches Int as described above.
Stocks of gpInt and CTD proteins were used as indicated and diluted as appropriate in 10 mM Tris (pH-7.5), 1 mg/ml Bovine serum albumin (BSA) and 1 mM Dithiothreitol (DTT).
DNA substrates and oligonucleotides
Oligonucleotides used in this study are shown in Table S1. DNA fragments of 508 bp (attB) and 2442 bp (attP) were amplified from genomic DNA of M. smegmatis and Bxz2, respectively. Plasmids pSS1 and pSS2 were obtained by cloning the attB (508 bp) and attP (2442 bp) DNA fragments into the HindIII site of pMOS Blue. Plasmid pSS2 also contains the full-length int gene along with attP. DNA fragments of 341 bp of Bxz2 attL and 340 bp of Bxz2 attR were amplified by PCR from the product of in vitro integration reactions (between pSS1 and pSS2) and cloned into the Nde1 and HindIII site of pMOS Blue to obtain pSS5 and pSS6, respectively. DNA fragments (402 bp) containing Peaches attP were amplified from genomic DNA and cloned into the Nde1 and HindIII site of pMOS Blue to obtain pSS27. DNA fragments (60 bp) containing wild type Bxz2 and Peaches attP, attB, attL and attR site were prepared by annealing complementary oligonucleotides (Table S1). Wild type attB and Bxz2 and Peaches attP DNAs containing a single CTD binding site were prepared by eliminating a half-site (attP-P half-site, attP-P′ half-site, attB-B half-site or attB-B′ half-site). These DNAs were obtained by annealing the appropriate pairs of nucleotides (Table S1).
DNA-binding assays
DNA substrates were prepared by 5′-end labeling of one oligonucleotide of each pair with T4 polynucleotide kinase (PNK) (Roche) prior to annealing to obtain single side labeled oligonucleotides. Approximately 5 ng of labeled Bxz2 DNA was incubated with the indicated amounts of gpInt and CTD in a buffer containing 20 mM Tris (pH-7.5), 10 mM NaCl, 6 mM spermidine, 1 mM DTT, 0.2 mg/ml BSA, 6% glycerol and 1 μg Calf Thymus DNA, in a total volume of 10 μl. Peaches DNA was incubated in a buffer containing 20 mM Tris (pH-7.5), 25 mM NaCl, 10 mM EDTA, 10 mM Spermidine, 1 mM DTT, and 1 μg Calf Thymus DNA, in a total volume of 10 μl. The Bxz2 and Peaches integration reactions were incubated at 37ºC and 25 ºC, respectively, for one and half hours and the protein-DNA complexes were separated from free DNA on a native 5% (unless otherwise stated) polyacrylamide gel at 4ºC. Gels were dried after electrophoresis and exposed to phosphorimager screen for overnight and then scanned (Fuji Phosphoimager). For each substrate the binding buffers are the same as used for recombination of that DNA substrate, and some affinities are somewhat buffer sensitive, such that Peaches Int binds to attB ~10-fold tighter in Bxz2 buffer (Fig. 6E) than in Peaches buffer (Fig. 5A).
In vitro recombination assays
In vitro integrative recombination assays for phage Bxz2 were performed in a recombination buffer containing 20 mM Tris (pH-7.5), 10 mM NaCl, 6 mM spermidine, 1 mM DTT, 0.2 mg/ml BSA, 6% glycerol and 1 mM DTT in final volume of 10 μl. Recombination buffer composition for the integrative assays for Peaches are as follows, 20 mM Tris (pH-7.5), 25 mM NaCl, 10 mM EDTA, 10 mM Spermidine, 1 mM DTT, in final volume of 10 μl. Integration reactions were performed with supercoiled DNA containing 0.03 pmol of pSS1 (pattB) or pSS2 (Bxz2 pattP), pSS27 (Peaches pattP) and synthesized oligonucleotides (60 bp) of wild type attP and attB, respectively. Amounts of DNA were added as indicated. Supercoiled DNAs were linearized with the appropriate restriction enzymes, as needed. The Bxz2 and Peaches integration reactions were incubated at 37ºC and 25 ºC respectively, for overnight and treated with 1 mg/ml Proteinase K and 0.5% SDS for 20 min at 55 ºC. The products were separated on 0.8% agarose gel in 1x TBE running buffer and visualized by ethidium bromide staining.
Supplementary Material
Highlights.
Serine-integrases of mycobacteriophages Bxz2 and Peaches are 59% identical
Bxz2 and Peaches integrases use the same attB site but different attP sites
Attachment site specificity cannot be simply predicted from half-site occupancies
Acknowledgments
We thank Carlos Guerrero for excellent technical assistance, Aubrey Lowen and Bethany Corbin for assistance with cloning, expression, and purification, and Pallavi Ghosh for comments on the manuscript. This work was supported by National Institutes of Health grant number AI059114.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith MC, Brown WR, McEwan AR, Rowley PA. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans. 2010;38:388–94. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- 2.Mandali S, Dhar G, Avliyakulov NK, Haykinson MJ, Johnson RC. The site-specific integration reaction of Listeria phage A118 integrase, a serine recombinase. Mob DNA. 2013;4:2. doi: 10.1186/1759-8753-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh P, Kim AI, Hatfull GF. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol Cell. 2003;12:1101–11. doi: 10.1016/s1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 4.Keravala A, Calos MP. Site-specific chromosomal integration mediated by phiC31 integrase. Methods Mol Biol. 2008;435:165–73. doi: 10.1007/978-1-59745-232-8_12. [DOI] [PubMed] [Google Scholar]
- 5.Gupta M, Till R, Smith MC. Sequences in attB that affect the ability of phiC31 integrase to synapse and to activate DNA cleavage. Nucleic Acids Res. 2007;35:3407–19. doi: 10.1093/nar/gkm206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S, Ghosh P, Hatfull GF. Attachment site selection and identity in bxb1 serine integrase-mediated site-specific recombination. PLoS Genet. 2013;9:e1003490. doi: 10.1371/journal.pgen.1003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh P, Pannunzio NR, Hatfull GF. Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites. J Mol Biol. 2005;349:331–48. doi: 10.1016/j.jmb.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 8.Smith MC, Thorpe HM. Diversity in the serine recombinases. Mol Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 9.Bai H, Sun M, Ghosh P, Hatfull GF, Grindley ND, Marko JF. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc Natl Acad Sci U S A. 2011;108:7419–24. doi: 10.1073/pnas.1018436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olorunniji FJ, Buck DE, Colloms SD, McEwan AR, Smith MC, Stark WM, Rosser SJ. Gated rotation mechanism of site-specific recombination by varphiC31 integrase. Proc Natl Acad Sci U S A. 2012;109:19661–6. doi: 10.1073/pnas.1210964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci U S A. 1998;95:5505–10. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S, Hatfull GF. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol. 2003;50:463–73. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 13.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 14.Smith MA, Till R, Smith MC. Switching the polarity of a bacteriophage integration system. Mol Microbiol. 2004;51:1719–28. doi: 10.1111/j.1365-2958.2003.03942.x. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh P, Bibb LA, Hatfull GF. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc Natl Acad Sci U S A. 2008;105:3238–43. doi: 10.1073/pnas.0711649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh P, Wasil LR, Hatfull GF. Control of Phage Bxb1 Excision by a Novel Recombination Directionality Factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khaleel T, Younger E, McEwan AR, Varghese AS, Smith MC. A phage protein that binds phiC31 integrase to switch its directionality. Mol Microbiol. 2011;80:1450–63. doi: 10.1111/j.1365-2958.2011.07696.x. [DOI] [PubMed] [Google Scholar]
- 18.McEwan AR, Rowley PA, Smith MC. DNA binding and synapsis by the large C-terminal domain of phiC31 integrase. Nucleic Acids Res. 2009;37:4764–73. doi: 10.1093/nar/gkp485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorpe HM, Wilson SE, Smith MC. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol Microbiol. 2000;38:232–41. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 20.Rutherford K, Yuan P, Perry K, Sharp R, Van Duyne GD. Attachment site recognition and regulation of directionality by the serine integrases. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Duyne GD, Rutherford K. Large serine recombinase domain structure and attachment site binding. Crit Rev Biochem Mol Biol. 2013 doi: 10.3109/10409238.2013.831807. [DOI] [PubMed] [Google Scholar]
- 22.Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006;3:615–21. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Ghosh P, Hatfull GF, Hong Y. Successive and targeted DNA integrations in the Drosophila genome by Bxb1 and phiC31 integrases. Genetics. 2011;189:391–5. doi: 10.1534/genetics.111.129247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–77. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson JG, Chan R, Smith J, Thilmony R, Yau YY, Wang Y, Ow DW. The Bxb1 recombination system demonstrates heritable transmission of site-specific excision in Arabidopsis. BMC Biotechnol. 2012;12:9. doi: 10.1186/1472-6750-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012;109:8884–9. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying Genetic Logic Gates. Science. 2013 doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 28.Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology. 2007;153:2711–23. doi: 10.1099/mic.0.2007/008904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McEwan AR, Raab A, Kelly SM, Feldmann J, Smith MC. Zinc is essential for high-affinity DNA binding and recombinase activity of PhiC31 integrase. Nucleic Acids Res. 2011;39:6137–47. doi: 10.1093/nar/gkr220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bibb LA, Hatfull GF. Integration and excision of the Mycobacterium tuberculosis prophage-like element, phiRv1. Mol Microbiol. 2002;45:1515–26. doi: 10.1046/j.1365-2958.2002.03130.x. [DOI] [PubMed] [Google Scholar]
- 31.Breuner A, Brondsted L, Hammer K. Resolvase-like recombination performed by the TP901-1 integrase. Microbiology. 2001;147:2051–63. doi: 10.1099/00221287-147-8-2051. [DOI] [PubMed] [Google Scholar]
- 32.Morita K, Yamamoto T, Fusada N, Komatsu M, Ikeda H, Hirano N, Takahashi H. In vitro characterization of the site-specific recombination system based on actinophage TG1 integrase. Mol Genet Genomics. 2009;282:607–16. doi: 10.1007/s00438-009-0490-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Ou X, Zhao G, Ding X. Highly efficient in vitro site-specific recombination system based on streptomyces phage phiBT1 integrase. J Bacteriol. 2008;190:6392–7. doi: 10.1128/JB.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–51. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold PH, Blake DG, Grindley ND, Boocock MR, Stark WM. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. Embo J. 1999;18:1407–14. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke ME, Arnold PH, He J, Wenwieser SV, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol Microbiol. 2004;51:937–48. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 37.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nature Methods. 2007;4:147–52. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.