Abstract
Chronic inflammation has been implicated in the etiology of colorectal adenoma and cancer; however, few key inflammatory genes mediating this relationship have been identified. In this study, we investigated the association of germline variation in innate immunity genes in relation to the risk of colorectal neoplasia. Our study was based on the analysis of samples collected from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. We investigated the association between 196 tag single nucleotide polymorphisms (SNPs) in 20 key innate immunity genes with risk of advanced colorectal adenoma and cancer in 719 adenoma cases, 481 cancer cases and 719 controls. Logistic regression was used to estimate odds ratios and 95% confidence intervals. After Bonferroni correction, the AG/GG genotype of rs5995355, which is upstream of NCF4, was associated with an increased risk of colorectal cancer (odds ratio [OR] 2.43, 95% confidence interval [95% CI] 1.73 – 3.39; P<0.0001). NCF4 is part of the NAPDH complex, a key factor in biochemical pathways and the innate immune response. While not definitive, our analyses suggest that the variant allele does not affect expression of NCF4, but rather modulates activity of the NADPH complex. Additional studies on the functional consequences of rs5995355 in NCF4 may help to clarify the mechanistic link between inflammation and colorectal cancer.
Keywords: colorectal cancer, single nucleotide polymorphism, innate immunity, NCF4
Introduction
Despite declining trends in recent years, colorectal cancer remains the third leading cause of cancer-related deaths and is a significant public health burden. In the period from 1990 to 2007, there was an approximate 30% reduction in colorectal cancer mortality, most of which has been attributed to advances in screening 1. Projected incidences for 2013 indicate that 142,820 men and women will be diagnosed with colorectal cancer in the United States 1.
Similar to most other complex disease traits, risk factors for colorectal cancer include a combination of genetic (e.g., family history of colorectal cancer, familial adenomatous polyposis, hereditary non-polyposis colorectal cancer) and lifestyle (e.g., infrequent physical activity, obesity, low fruit and vegetable intake, alcohol and tobacco) factors 2, 3. Chronic inflammation, arising from either immune deregulation or autoimmunity, is also a risk factor for colorectal cancer. The association is exemplified by the relationship between inflammatory bowel disease (IBD) and colorectal neoplasia. Crohn's disease (CD) and ulcerative colitis (UC) represent the two most common forms of IBD 4, 5. Individuals with these conditions are up to five times more likely to develop colorectal cancer than the general population and risk increases with the duration of disease 6, 7. In addition, precursor lesions such as adenomas frequently exhibit signs of inflammation 8 and studies have demonstrated that use of non-steroidal anti-inflammatory drugs, such as aspirin, confer a 40-50% reduction in colorectal cancer risk 9, 10.
The natural history of colorectal neoplasia is characterized by a loss of homeostatic regulatory controls and the acquisition of mutations. Chronic inflammation is linked to 20% of all cancers and may mediate an increase in mutation rates 11. One possible contributing factor to immune deregulation is carriage of single nucleotide polymorphisms (SNPs) in innate immunity genes. In support of this hypothesis, results from several candidate SNP and genome-wide association studies (GWAS) implicate SNPs in innate immunity genes with increased risk of developing CD and UC 12-17. Moreover, recent reports found that SNPs in innate immunity genes are associated with colon cancer risk and survival 18 and that expression of inflammatory protein coding genes and a microRNA, miR-21, are associated with prognosis of colon cancer 19. As the body's initial defense mechanism, the innate immune system plays an important role in the acute response to, and modulation of, infection. We examined 196 SNPs in 20 innate immunity genes, selected for biological plausibility, with risk of both adenoma formation and colorectal cancer. For this purpose we performed a nested case-control study based within the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial.
Materials and methods
Participants and recruitment
The PLCO Cancer Screening Trial is a large randomized controlled trial designed to assess the efficacy of various screening methods on the early detection and mortality of prostate, lung, colorectal and ovarian cancer in men and women aged 55-74. Participants were recruited from 10 screening centers throughout the US; (Birmingham, AL; Denver, CO; Detroit, MI; Honolulu, HI; Marshfield, WI; Minneapolis, MN; Pittsburgh, PA; Salt Lake City, UT; St. Louis, MO; and Washington, DC). The trial recruited ∼ 155,000 individuals between 1993 and 2001. For the colorectal cancer portion of PLCO, the objective was to determine the efficacy of 60 cm flexible sigmoidoscopy screening exams of the distal colon (including the descending colon, sigmoid colon, and the rectum) to reduce mortality, as has been described in further detail elsewhere 20. In the intervention arm of the trial, exams were conducted at study entry and then repeated at 3 or 5 years post-enrollment 21. The control arm received usual care. Suspicious lesions detected by the sigmoidoscopy exam were referred for follow up by the participant's primary care physician. Patient data were subsequently tracked by expert medical personnel to identify patients who developed pathologically verified colorectal adenoma or cancer. Cancers that were reported on the annual questionnaire sent to all participants, or reported on any death certificates, were also verified by a review of the medical records. Information regarding demographics, lifestyle habits, personal and family history of cancer, was collated through a questionnaire that was administered at study entry in both arms of the trial. Data regarding diet and alcohol were collected from a food frequency questionnaire administered at baseline in the intervention arm of the trial. The study was approved by the Institutional Review Boards of the National Cancer Institute and all 10 centers. Written informed consent was obtained from each participant.
For this case-control study, participants had to provide a blood or buccal specimen and consent to participate in etiologic studies of cancer and other diseases. Individuals were excluded if they had a self-reported history of colorectal polyps, UC, CD, familial polyposis, Gardner's syndrome, or cancer (except basal cell or squamous cell skin cancer). Although the PLCO trial recruited participants from many ethnic groups, 92% of the population in this study was Caucasian, and as such, we restricted our analyses to this group. Our study therefore included 481 Caucasian colorectal cancer cases and 719 adenoma cases. All cancers and adenomas were confirmed by a review of medical records. All adenoma cases were left-sided and considered advanced (defined as ≥ 1cm or having villous characteristics or with high-grade dysplasia or carcinoma in situ). Seven hundred and twenty controls that underwent a successful sigmoidoscopy exam and were found to be negative for polyps on the left-side at the baseline screen were selected from the screening arm. One control patient, that later developed colorectal cancer, was dropped from the analysis. Seven adenoma cases subsequently developed colorectal cancer. The controls were matched to the adenoma cases on gender. The advanced adenomas and controls were selected from the screening arm of the trial; colorectal cancers were selected from both arms.
SNP Selection and Genotyping
DNA was extracted from blood using QIAamp DNA Blood Midi or Maxi kits (Qiagen, Valencia, CA), and from buccal cells using phenol chloroform extraction. Briefly, the adenoma, cancer and control samples were genotyped at the NCI Core Genotyping Facility (Advanced Technology Corporation, Gaithersburg, MD) using a custom Illumina GoldenGate Oligo Pool Assay (OPA) panel of SNPs tagging candidate innate immunity genes and their surrounding regions. To reduce the potential for redundancy in the information, tag SNPs were chosen for each gene (including the region 20 kb upstream and 10 kb downstream of the genes) using the Caucasian 23 population genotyped as part of the International HapMap Project 22 and the Carlson algorithm as implemented in TagZilla 23, with the following conditions: 1) minor allele frequency >5% in HapMap Caucasian samples; 2) an r2 cut-off of >0.8 (assigned for inferring linkage); 3) design scores > 0.4, with preference given to SNPs with a design score > 1.1.
For quality control, 79 replicate samples from 26 subjects were interspersed among cases and controls. All SNPs showed >96% concordance among replicate samples. Assays with concordance rates <90% were excluded. Assays with a Hardy-Weinberg proportion P-value <1×10−5 among controls, or failed assay validation, were also excluded from the analysis.
Additional genotyping for rs5995355 was carried out on 40 tissue samples for which we also carried out gene expression profiling. DNA was isolated from buffy coat using the Qiagen FlexiGene DNA Kit (Qiagen, Tokyo, Japan). Genotyping was performed using a Taqman assay (Applied Biosystems, Foster City, CA) according to the supplier's instructions.
Data analysis
Tests for deviations from Hardy-Weinberg equilibrium were evaluated by calculating the expected genotype frequencies based on known allele frequencies and then comparing them to observed genotype frequencies using χ2 tests. Linkage disequilibrium (LD) was assessed using Haploview 24 among controls. Differences in the characteristics of colorectal cancer cases and controls were compared using the χ2 test for categorical values or by Student's t test for continuous measures, as indicated. Analyses were performed using STATA version 11 software (STATA Corp, College Station, TX). All statistical tests were two-sided.
Associations between genotypes and colorectal cancer or adenoma susceptibility were estimated by odds ratios (OR) using an unconditional logistic regression genotypic model. These models were adjusted for gender (male/female) and age at diagnosis (continuous). The test for trend analysis was computed using the linear contrast coefficients -1, 0, 1. An additional model adjusted for factors known to be associated with cancer/adenoma risk was also tested, i.e., smoking, BMI, alcohol, non-steroid anti-inflammatory drug (NSAID) use, folate intake and family history. If there were zero individuals with a particular genotype, the exact logistic model, which provides a better approximation of coefficients with small samples sizes, was used. The model was adjusted for age. Although the rate of transition from adenoma to colon cancer is similar between men and women, the prevalence of adenomas and the incidence of colorectal cancer are greater in men than in women 25, 26. Given these gender disparities, we also performed a series of analyses stratified by gender for each SNP. For rs5995355, additional subgroup analyses were performed with stratification by smoking status (current, former, never), pack-years of smoking (quartiles), aspirin intake (irregular [< 4 per month]/regular [> 1 per week]), ibuprofen intake (irregular [< 4 per month]/regular [> 1 per week]), aspirin or ibuprofen intake (irregular [< 4 per month]/regular [> 1 per week]), alcohol intake (quartiles, grams/day), total folate intake (quartiles, micrograms/day) and BMI (18-25/25-30/>30 kg/m2). A single model was fit for each of these factors and was adjusted for age, gender, and interaction terms for rs5995355 with each level of the factor. For example, for the analysis stratified by smoking, an interaction term was generated for rs5995355Xnever_smokers, rs5995355Xfromer_smokers and rs5995355Xcurrent_smokers was included in the model. rs5995355 was entered as a dominant variable. Stratum-specific odds ratios were then estimated based on linear combinations of the model parameters using the lincom code in STATA. To test for potential interactions between these variables and rs5995355 in colorectal cancer, we computed P-values from a one degree of freedom likelihood ratio test comparing nested models with and without the interaction term using the variables outlined above. Studies with moderate sample size and a large number of comparisons can increase the probability of generating false positive results. To limit this type 1 error, we used the Bonferroni correction to calculate α=2.9×10−4 (α=0.05/171 SNPs). This is a fairly conservative calculation given that all of the SNPs analyzed may not be independent of each other due to linkage disequilibrium.
mRNA array analysis
RNA was extracted from colorectal cancer tissues using Trizol, according to the manufacturer's instructions and analysed on the Affymetrix U133A arrays. Data was imported and RMA normalized using Partek Genomics Suite 6.5. Gene expression profiles were compared between samples with the AA and AG/GG genotypes combined using Partek. Data has been submitted and uploaded into GEOprofiles (GSE44861).
Results
Characteristics of participants
Our study included an analysis of 196 SNPs in 20 inflammation-associated genes. Of these, 171 SNPs were successfully genotyped and analyzed; 12 failed assay validation, 3 had < 98% concordance among HapMap samples, 9 had a low call rate and 1 violated the Hardy-Weinberg assumption (Supplemental Table 1). The completion rate for rs5995355 was 91%. The 20 genes included in the study were ALOX12, CCL5, CRP, IL8, IL8RA/IL8RB, JAK3, ALOX15, ALOX5, ALOX5AP, CYBA, MBL2, MIF, NCF2, NCF4, NOS2A, NOS3, RAC2, STAT3/STAT5A, STAT5B and XOR (Supplemental Table 1). The characteristics of the cases and controls are detailed in Table 1. The cancer cases had a significantly greater proportion of females (41%) compared to the population controls (31.1%) (P<0.0001). In addition, they were more likely to be older (P <0.0001) and to have higher pack-years of smoking (P=0.028). The adenoma cases were also more likely to be older (p<0.0001), current smokers (P<0.0001) and have had a previous family history of cancer (12.8% vs. 8.8%) (P=0.018). In addition, adenoma cases had higher pack-years of smoking compared to controls (P<0.0001) and were more likely to have a lower total folate intake (cases vs. controls: 1st quartile [30.6% vs. 24%]; 4th quartile [20.2% vs. 25%]). These factors were included in an adjusted logistic regression model.
Table 1. Characteristics of adenoma and colorectal cancer cases and controls.
| Controls n=719 | Adenoma n=719 | Pa | Cancer n=481 | Pb | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| N | % | N | % | N | % | ||||
| Sex | Male | 495 | 68.9% | 504 | 70.1% | 284 | 59.0% | ||
| Female | 224 | 31.1% | 215 | 29.9% | 0.619 | 197 | 41.0% | <0.0001 | |
| Age | 59 or less | 338 | 46.9% | 244 | 33.9% | 97 | 20.2% | ||
| 60-64 | 185 | 25.7% | 220 | 30.6% | 160 | 33.3% | |||
| 65-69 | 134 | 18.8% | 160 | 22.3% | 127 | 26.4% | |||
| 70-74 | 62 | 8.6% | 95 | 13.2% | <0.0001 | 97 | 20.2% | <0.0001 | |
| Family History | No | 656 | 91.2% | 626 | 87.2% | 424 | 88.9% | ||
| Yes | 63 | 8.8% | 92 | 12.8% | 0.018 | 53 | 11.1% | 0.184 | |
| Aspirin | Irregular(<4/week) | 373 | 51.9% | 387 | 53.8% | 263 | 54.9% | ||
| Low (1-4/week) | 203 | 28.2% | 216 | 30.0% | 131 | 27.4% | |||
| Moderate (1/day) | 77 | 10.7% | 58 | 8.1% | 48 | 10.0% | |||
| High (2/day) | 66 | 9.2% | 58 | 8.1% | 0.878 | 37 | 7.7% | 0.702 | |
| Ibuprofen | Irregular(<4/week) | 450 | 62.6% | 312 | 65.0% | 394 | 82.3% | ||
| Low (1-4/week) | 85 | 11.8% | 49 | 10.2% | 50 | 10.4% | |||
| Moderate (1/day) | 152 | 21.1 | 96 | 20.0% | 7 | 1.5% | |||
| High (2/day) | 32 | 4.5% | 23 | 4.8% | 0.263 | 28 | 5.9% | 0.632 | |
| Aspirin/Ibuprofen | Irregular(<4/week) | 286 | 23.8% | 297 | 41.3% | 196 | 41.0% | ||
| Low (1-4/week) | 207 | 28.8% | 234 | 32.6% | 143 | 29.2% | |||
| Moderate (1/day) | 90 | 12.5% | 77 | 10.7% | 58 | 12.4% | |||
| High (2/day) | 136 | 136.0% | 111 | 15.4% | 0.145 | 81 | 18.1% | 0.830 | |
| BMI (Kg/m2) | <25 | 203 | 28.5% | 185 | 25.8% | 203 | 28.5% | ||
| >25-30 | 332 | 46.6% | 325 | 45.3% | 331 | 46.5% | |||
| >30 | 178 | 25.0% | 207 | 28.9% | 0.214 | 178 | 25.0% | 0.931 | |
| Smoking | Never | 298 | 43.9% | 246 | 36.0% | 190 | 41.0% | ||
| Former | 335 | 49.3% | 344 | 50.3% | 226 | 48.7% | |||
| Current | 46 | 6.8% | 94 | 13.7% | <0.0001 | 48 | 10.3% | 0.088 | |
| Pack-years | No use | 298 | 44.2% | 246 | 36.3% | 190 | 41.7% | ||
| 0-20 | 188 | 27.8% | 169 | 24.9% | 102 | 22.4% | |||
| 20-40 | 101 | 14.9% | 134 | 19.8% | 89 | 19.5% | |||
| 40+ | 88 | 13.0% | 129 | 19.0% | <0.0001 | 75 | 16.5% | 0.028 | |
| Alcohol (gm/day) | 0.5 or less | 173 | 24.7% | 178 | 25.9% | 74 | 26.3% | ||
| 0.5-3.0 | 140 | 19.9% | 109 | 15.9% | 63 | 22.4% | |||
| 3.0-17.0 | 187 | 26.6% | 181 | 26.4% | 62 | 22.1% | |||
| 17+ | 201 | 28.7% | 219 | 31.9% | 0.208 | 82 | 29.2% | 0.475 | |
| Folate (μg/day) | 1st Quartile | 168 | 24.0% | 210 | 30.6% | 81 | 28.8% | ||
| 2nd Quartile | 192 | 27.4% | 170 | 24.8% | 67 | 23.8% | |||
| 3rd Quartile | 166 | 23.7% | 168 | 24.5% | 77 | 27.4% | |||
| 4th Quartile | 175 | 25.0% | 139 | 20.2% | 0.018 | 56 | 19.9% | 0.106 | |
Pa compares the adenoma cases to controls.
Pb compares the colorectal cancer cases to controls.
Alcohol and folate intake classified based on quartile intake of controls.
Aspirin/Ibubrofen (irregular < 4/week; low 1-4/week; moderate 1/day; high 2/day)
Folate quartiles: 1st ≤ 337.4 μg/day, 2nd >337.4 μg/day/≤ 547.6 μg/day, 3rd > 547.6 μg/day/≤ 796.5 μg/day, 4th >796.5 μg/day
Alcohol quartiles: 1st ≤ 0.177 mg/day, 2nd >0.177 mg/day/≤ 2.354 mg/day, 3rd > 2.354 mg/day/≤ 15.086 mg/day, 4th > 15.086 mg/day
Association between SNPs in innate immunity genes and colorectal cancer risk
In the analysis of colorectal cancer risk, one SNP was deemed statistically significant at α < 2.9×10−4 (Table 2). The AG/GG genotype of rs5995355 (A > G upstream of promoter) in neutrophil cytosolic factor 4 (NCF4) conferred a 2.43 increased odds of colorectal cancer in a model adjusted for age and gender (95% C.I. 1.73 – 3.39; P=2.4−7) (Table 2). A fully adjusted model, corrected for additional co-variates including smoking status, pack-years of smoking, family history of colorectal cancer, folate intake, alcohol and NSAID and BMI use did not alter the risk estimate more than 10% (ORAG/GGvs. AA 2.56 [1.83 – 3.60]; P=4.0−7) (OR minimally adjusted 2.43 vs. OR fully adjusted 2.56); therefore the minimally adjusted model was used in subsequent analyses. Results for SNPs that were not significant following Bonferroni correction are shown in Supplementary Table 2. When we stratified our analyses by gender, we did not observe any significant differences between men and women (Supplementary Tables 3 and 4).
Table 2. Association between rs5995355 with colorectal cancer risk.
| Controls | Cases | Univariable | Multivariablea | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| N | % | OR | 95% C.I. | P | OR | 95% C.I. | P | |
| AA | 616 (85.8) | 212 (70.0) | Referent | Referent | Referent | Referent | ||
| AG | 99 (13.8) | 82 (27.0) | 2.31 | 1.68 – 3.17 | 1.6 × 10−5 | 2.23 | 1.58 – 3.06 | 1.7 × 10−4 |
| AG/GG | 102 (14.2) | 91 (30.0) | 2.59 | 1.88 – 3.58 | 7.5 × 10−9 | 2.43 | 1.73 – 3.39 | 2.4 × 10−7 |
| P trend | 1.2 × 10−10 | 2.6 × 10−9 | ||||||
Adjusted for age and gender
Characters in bold represent SNPs significant after Bonferroni correction
Association between SNPs in innate immunity genes and adenoma risk
rs206811 (G >A, intron 1) in xanthine oxoreductase (XOR) was the most significant SNP associated with risk of adenoma formation (ORAAvs. GG 1.57 [1.13 – 2.18]; p=0.007). However, this P-value was not significant following Bonferroni correction. Supplementary Table 5 shows analyses for SNPs and adenoma risk.
Stratified analyses of rs5995355
Previous data have highlighted the association between several lifestyle and demographic variables with both adenoma risk and colorectal cancer formation. Moreover, certain factors including BMI, smoking and NSAID use are known to modulate inflammation. Some of these variables were captured in the questionnaires administered during the PLCO Cancer Screening Trial. Therefore, to check for modification of the association between rs5995355 and colorectal cancer by these variables we performed a series of stratified analyses. Results for rs5995355 are presented as a dominant model.
We found that rs5995355 was associated with an increased risk of cancer in individuals who drank up to 17 g/day of alcohol; however, a dose-related increase in risk was not observed (Table 3). Similarly, this SNP was associated with increased risk of cancer in individuals with low to moderate BMI, but not in those with a BMI>30. While there were differences between some of the strata (for example alcohol, smoking and BMI), there were no significant interactions between rs5995355 and alcohol, smoking, folate, aspirin, BMI and family history of colorectal cancer (Table 3).
Table 3. Stratum-specific odds ratios for colorectal cancer risk with rs5995355 (NCF4).
| Stratum 1 OR (95% C.I.) | Stratum 2 OR (95% C.I.) | Stratum 3 OR (95% C.I.) | Stratum 4 OR (95% C.I.) | |
|---|---|---|---|---|
| Alcohol (gm/day) GA/GG vs. AA | 1st Quartile (n=234) 2.79 (1.47 – 5.32) P=0.002 | 2nd Quartile (n=193) 4.13 (1.89 – 9.04) P<0.0001 | 3rd Quartile (n=239) 2.82 (1.47 – 5.43) P=0.002 | 4th Quartile (n=273) 1.37 (0.74 – 2.56) P=0.318 |
| Folate (μg/day) GA/GG vs. AA | 1st Quartile (n=237) 2.81 (1.49 – 5.31) P=0.001 | 2nd Quartile (n=255) 1.37 (0.69 – 2.73) P=0.372 | 3rd Quartile (n=229) 1.47 (0.77 – 2.79) P=0.239 | 4th Quartile (n=218) 6.86 (3.33 – 14.16) P<0.0001 |
| Pack-years GA/GG vs. AA | No use (n=433) 3.01 (1.82 – 4.96) P<0.0001 | 0-20 (n=274) 3.16 (1.67 – 5.97) P<0.0001 | 20-40 (n=162) 2.43 (1.11 – 5.36) P=0.027 | 40+(n=139) 1.18 (0.47 – 2.93) P=0.722 |
| Smoking GA/GG vs. AA | Never (n=402) 2.99 (1.81 – 4.934) P<0.0001 | Former (n=454) 2.49 (1.56 – 3.97) P<0.0001 | Current (n=73) 0.77 (0.27 – 2.22) P=0.631 | |
| Aspirin GA/GG vs. AA | Irregular (n=613) 2.51 (1.69 – 3.73) P<0.0001 | Low (n=109) 1.74 (0.61 – 5.03) P=0.299 | Moderate (n=207) 2.95 (1.48 – 2.87) P=0.002 | High (n=47) 2.58 (0.59 – 11.24) P=0.207 |
| BMI (Kg/m2) GA/GG vs. AA | < 18.5 (n=4) 0.34 (1.51 – 5.09) P=0.594 | 18.5 – 25 (n=297) 2.76 (1.51 – 5.09) P<0.0001 | ≥25 -30 (n=499) 2.34 (1.49 – 3.67) P<0.0001 | > 30 (n=255) 2.43 (1.21 – 4.88) P=0.013 |
| Family History GA/GG vs. AA | No (n=884) 2.77 (1.98 – 3.86) P<0.0001 | Yes (n=93) 0.95 (0.34 – 2.68) P=0.926 |
All data adjusted for age and gender and interaction terms for each level of the factor with rs5995355
OR denotes odds ratio
Data in italics denotes statistically significant data
Alcohol and folate intake classified based on quartile intake of controls.
Aspirin (irregular < 4/week; low 1-4/week; moderate 1/day; high 2/day)
Folate quartiles: 1st ≤ 337.4 μg/day, 2nd >337.4 μg/day/≤ 547.6 μg/day, 3rd > 547.6 μg/day/≤ 796.5 μg/day, 4th >796.5 μg/day
Alcohol quartiles: 1st ≤ 0.177 mg/day, 2nd >0.177 mg/day/≤ 2.354 mg/day, 3rd > 2.354 mg/day/≤ 15.086 mg/day, 4th > 15.086 mg/day
Association between rs5995355 and gene expression
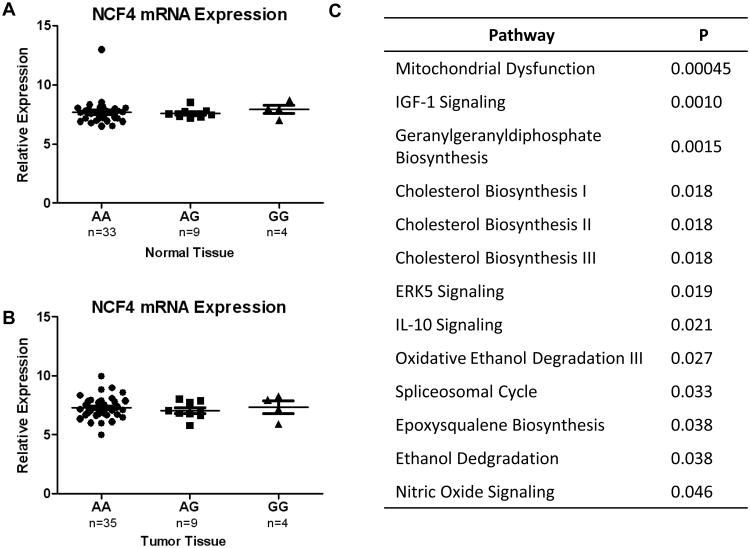
rs5995355 lies upstream of NCF4, potentially within a regulatory region that could alter NCF4 transcription. We therefore performed an expression quantitative trait (eQTL) analysis of NCF4 expression across rs5995355 genotypes. As we did not have access to tissue samples from the PLCO study, we performed the eQTL analysis on 49 tumor and 48 non-tumor in the NCI-MD case-control study. These samples were submitted for Affymetrix array analysis and genotyping for rs5995355 using Taqman assays. As shown in Figure 1, there was no difference in NCF4 expression across rs5995355 genotypes in either normal (Figure 1A) or tumor (Figure 1B) tissue. This suggests that rs5995355 does not modulate NCF4 expression.
Figure 1.
Relative mRNA expression of NCF4 in normal (A) and tumor (B) colon tissue, stratified by rs5995355 genotype. Data are not statistically significant. Ingenuity pathway analysis (IPA) of gene expression stratified by rs5995355 genotype. Genes differentially expressed at P<0.05 were submitted for the IPA analysis (C).
We then reasoned that the SNP could be associated with colorectal cancer risk through by modulating the function of NCF4 through its role as a scaffolding protein within the NADPH complex. Genes that were differentially expressed between AA and AG/GG tumor samples at p<0.05 were assessed for pathway enrichment using Ingenuity Pathway Analysis (IPA) software (Supplementary Table 6). As shown in Figure 1C, of the top pathways differentially regulated the majority were involved in biochemical pathways that involved the NADPH complex, such as mitochondria signaling, cholesterol, inositol and zymosterol biosynthesis. This suggests that the SNP may be related to colorectal carcinogenesis through a modulation of NADPH function.
Discussion
We investigated the association of innate immunity genes and their relationship to susceptibility of colorectal adenoma and cancer in a nested case-control study of the PLCO Cancer Screening Trial. We observed that a SNP, rs5995355, in NCF4 was significantly associated with risk of colorectal cancer after adjustment for both potential confounders and multiple comparisons.
The gene product of NCF4, also known as p40phox, is a partner in the nicotinamide dinucleotide phosphate (NAPDH) oxidase complex, a multi-component enzyme system whose main function is the elimination of invading microorganisms 27. Specifically, NCF4 acts as a scaffold for neutrophil cytosolic factor 2 (NCF2/p67phox), neutrophil cytosolic factor 1 (NCF1/p47phox) and RAC1. Upon infection, the complex is translocated to the cell membrane of phagocytic cells where it partners with gp91phox and p22phox to catalyze the production of the reactive oxygen species (ROS), both of which facilitate the eradication of invading bacteria 28, 29.
Despite the linkage of NCF4 to an increased predisposition to Crohn's disease in recent studies, including a GWAS (rs4821544) 30-33, a connection between this gene and colorectal cancer has not previously been reported. The SNP identified in our study, rs5995355, is weakly correlated in the HapMap CEU population with the CD GWAS SNP rs4821544 (D′=1.0, r2=0.14). We did not observe an association between rs4821544 and colorectal cancer risk in our study, though an association between this SNP and colorectal adenoma risk in females was found. rs5995355 lies upstream of NCF4, possibly within the promoter of NCF4 (Supplementary Tables 4 and 5). Thus, it is possible that the SNP affects transcription, and therefore expression, of the gene product. However, in an analysis of NCF4 mRNA expression across samples with known rs5995355 genotypes, we did not observe a change of expression in either tumor or normal tissue. Moreover, a recent analysis by Muise et al. did not find evidence for modulation of NCF4 expression by rs4821544 genotypes 34.
Interestingly, the variant allele of rs4821544 has been linked to reduced ROS production in granulocytes 35, suggesting that SNPs in NCF4 can mediate a connection to the pathogenesis of CD through impaired NADPH function and subsequent bacterial clearance. Therefore, although rs5995355 is a synonymous SNP, we further tested whether expression profiles, and the pathways associated with them, were differentially affected in samples from patients with different genotypes. We found that the most significant pathway represented from this analysis was mitochondrial dysfunction. Moreover, the majority of top 20 pathways differentially regulated between samples with the G allele of rs5995355 was biochemical and involved NADPH. Notably, the genes involved in these pathways, including cholesterol and inositol biosynthesis, were downregulated – suggesting the NADPH function was somehow compromised in cells that carried the variant allele of the SNP. As noted, rs5995355 is located upstream of the NCF4 promoter. As such, it in itself is unlikely to affect either the three dimensional structure of NCF4 or its spatial ability to complex with NCF-1/NCF-2 and RAC1. It is more likely that the SNP is in linkage disequilibrium with an as yet unknown, causal allele.
In addition to various biochemical pathways, conservation of NADPH levels is also required to enable redox reactions that control the levels of reactive oxygen species, which are increased by metabolic stress. Deficiencies in assembly of the complex could leave individuals susceptible to recurrent infections that could increase their risk of cancer. A corollary of this argument is seen in patients with chronic granulomatous disease (CGD), a condition that is molecularly characterized by mutations in components of the NADPH oxidase complex that negate the ability of phagocytes to produce ROS and repel infections 36, 37. Decreased levels of NCF4 could also diminish toll-like receptor (TLR) activation and antigen presentation owing to inappropriate ROS generation 38, 39 and therefore prolong the infection period. Numerous studies suggest that persistent bacterial infections contribute to colorectal cancer risk 40, 41, thus presenting one mechanism by which modulation of NCF4 could contribute to colorectal carcinogenesis.
An analysis of publicly-available gene expression datasets shows that expression of NCF4 is reduced in colon cancer (Supplementary Figure 1). In addition, high expression is associated with a reduced risk of mortality, according to Prognoscan 42, an online database that compiles gene expression profiling and outcomes in cancer (Supplementary Figure 2) 42, 43. This may be mediated in part through sensitivity to targeted therapies. Our analysis of available public datasets also showed that NCF4 expression in at least 2 fold higher in samples sensitive to treatments such as Topecan, Irinotecan, Panobinostat and Vorinostat. Although the mechanisms that mediate disease initiation and progression are often distinct, these findings suggest that NCF4 downregulation, or attenuation of NCF4 function, are associated with colorectal carcinogenesis and those tumors with expression of NCF4 have a good outcome. rs5995355, or most likely a SNP in linkage disequilibrium with it, may diminish the activity of the NADPH complex, perturb normal biochemistry and thereby facilitate persistent infections. Moreover, given that NCF4 SNPs have not been identified as risk alleles in all GWAS of CD and that NCF4 specifically has been associated with perianal CD 33, it is possible that this gene is implicated in distinct CD phenotypes and forms of colorectal cancer.
Our analysis did not identify any SNPs associated with adenoma risk after correction for multiple comparisons. It is plausible that our sample size may have been a limiting factor in this analysis. However, 7/20 SNPs in XOR were associated with adenoma risk, prior to Bonferroni correction, indicating a potential association as a whole. At the protein level, the gene exists as two interconvertible forms: xanthine dehydrogenase (XDH), which primarily reduces NAD+, and xanthine oxidase (XO), which can only reduce O244. One of the enzymatic roles of XOR is the bioactivation of ethanol. Indeed, alcohol toxicity and DNA damage in the liver is mediated by XOR, and it is thought that the carcinogenic effects of alcohol in breast cancer are mediated by this gene 45-47. In addition, recent evidence shows that activation of alcohol by XOR is enhanced by purines 48. Thus, if a diet is rich in meat (a good source of purines) and has a high alcohol intake, is possible that the risk-associated effects of this SNP in both adenomas and cancer may be more pronounced. However, our study is limited in power to assess this possibility (n=126 for those participants with a high-meat and high alcohol diet). Therefore, the hypothesis that an association between XOR and colorectal neoplasia is modified by an interaction with meat and alcohol intake could theoretically be tested in a larger, appropriately powered, cohort.
Our study has both limitations and strengths. With reference to the latter, our study is based within a prospective cancer screening trial with limited potential for recall bias for dietary and lifestyle factors. However, as noted above, we had limited power to assess gene-environment interactions. In addition, all adenomas in this study were left-sided and advanced. If the left and right colon share different etiologies, the results from this section of the study may not be generalizable to right-sided polyps. In addition, the study was restricted to advanced polyps and thus our data may not reflect the etiology of less advanced disease. However, advanced adenomas are more likely to progress to colorectal cancer and therefore have greater clinical significance. Finally, although the PLCO Screening Trial, which is US-based, recruited from many ethnic groups, our study was restricted to Caucasians and therefore our findings may only be relevant to this population. The minor allele frequency of rs5995355 varies significantly across ethnic groups; it is 8% in Caucasian populations, but as high as 43% in some African populations (http://www.ncbi.nlm.nih.gov/SNP/), suggesting that carriage of the deleterious allele at a higher frequency in these populations could be associated with a higher risk of colorectal cancer. However, by limiting our population to those of European ancestry, we reduced the potential for population stratification. Another limitation of our study was the exclusion of SNPs we previously studied in colon cancer, i.e., ALOX5 49 and MBL2 18. However, in this study we did not see an association between the MBL2 SNPs rs920724, rs1838066 and rs930507 with colon cancer risk, which was in agreement with our previous result 18.
In summary, we have provided evidence of an association between NCF4, an innate immunity gene, with colorectal carcinoma risk. Chronic inflammation, which precedes tumor development, is thought to play a distinct role in the carcinogenesis pathway compared to tumor-associated inflammation. Indeed, given the many chronic inflammatory disorders associated with cancer, it is plausible to think of inflammation-associated colorectal cancer as a distinct disease, perhaps similar to how we conceive the distinction between smoking and non-smoking associated lung cancer. When one considers that not all chronic inflammatory conditions are associated with an increased risk of cancer, identifying those genes and factors that do confer an increased risk of cancer will be important to both decipher the causal pathway between inflammation and carcinogenesis. In addition, it may highlight methods and points at which interventions will be effective. Thus, the identification of NCF4 in this study and its association with risk of cancer, combined with its known role in fighting off infections and association with increased risk of IBD, may highlight an underlying biological link in the causal pathway between inflammation and cancer. Our data suggests that the SNP, or one in which it is in linkage disequilibrium with, may alter functionality of the NADPH complex. Thus, additional functional studies investigating this SNP, and other SNPs in the region, may be warranted. These associations are promising; however additional epidemiological studies are needed to validate the findings and comparisons between African Americans and European Americans may also be informative given the higher frequency of the minor allele in African Americans, and their increased incidence of colorectal cancer1. Moreover, additional functional and translational work could help to dissect the mechanisms by which NCF4 contributes to the natural history of colorectal neoplasia.
Supplementary Material
Novelty and impact statement.
Colorectal cancer risk is modified by a SNP in NCF4, which is a member of the NADPH complex. While this gene was associated with risk of inflammatory bowel disease, this is the first study to link NCF4 with colon cancer. Our analysis suggests that the SNP disrupts functionality of the NADPH complex which could impede the ability of neutrophils to repel infectious agents. Future studies should examine the molecular functions of this gene in more detail.
Acknowledgments
We thank Karen Yarrick for administrative assistance. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute Center for Cancer Research and Division of Cancer Epidemiology and Genetics.
Financial Support: The work was supported by funding from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research and Division of Cancer Epidemiology and Genetics.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Terry MB, Neugut AI, Bostick RM, Sandler RS, Haile RW, Jacobson JS, Fenoglio-Preiser CM, Potter JD. Risk factors for advanced colorectal adenomas: a pooled analysis. Cancer Epidemiol Biomarkers Prev. 2002;11:622–9. [PubMed] [Google Scholar]
- 3.Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, Woodward M. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int J Cancer. 2009;125:171–80. doi: 10.1002/ijc.24343. [DOI] [PubMed] [Google Scholar]
- 4.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–40. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 5.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 6.Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526–35. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Roon AC, Reese G, Teare J, Constantinides V, Darzi AW, Tekkis PP. The risk of cancer in patients with Crohn's disease. Dis Colon Rectum. 2007;50:839–55. doi: 10.1007/s10350-006-0848-z. [DOI] [PubMed] [Google Scholar]
- 8.Higaki S, Akazawa A, Nakamura H, Yanai H, Yoshida T, Okita K. Metaplastic polyp of the colon develops in response to inflammation. J Gastroenterol Hepatol. 1999;14:709–14. doi: 10.1046/j.1440-1746.1999.01938.x. [DOI] [PubMed] [Google Scholar]
- 9.Cole BF, Logan RF, Halabi S, Benamouzig R, Sandler RS, Grainge MJ, Chaussade S, Baron JA. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst. 2009;101:256–66. doi: 10.1093/jnci/djn485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flossmann E, Rothwell PM. Effect of aspirin on long-term risk of colorectal cancer: consistent evidence from randomised and observational studies. Lancet. 2007;369:1603–13. doi: 10.1016/S0140-6736(07)60747-8. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal BB, Vijayalekshmi RV, Sung B. Targeting inflammatory pathways for prevention and therapy of cancer: short-term friend, long-term foe. Clin Cancer Res. 2009;15:425–30. doi: 10.1158/1078-0432.CCR-08-0149. [DOI] [PubMed] [Google Scholar]
- 12.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 13.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, Green T, Stevens CR, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–7. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slattery ML, Wolff RK, Herrick JS, Caan BJ, Potter JD. IL6 genotypes and colon and rectal cancer. Cancer Causes Control. 2007;18:1095–105. doi: 10.1007/s10552-007-9049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Polymorphisms in inflammatory pathway genes and their association with colorectal cancer risk. Int J Cancer. 2010;127:2822–30. doi: 10.1002/ijc.25299. [DOI] [PubMed] [Google Scholar]
- 18.Zanetti KA, Haznadar M, Welsh JA, Robles AI, Ryan BM, McClary AC, Bowman ED, Goodman JE, Bernig T, Chanock SJ, Harris CC. 3′ UTR and functional secretor haplotypes in mannose-binding lectin 2 are associated with increased colon cancer risk in African Americans. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, Hawkes JE, Croce CM, Leung SY, Harris CC. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–87. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, Fogel R, Gelmann EP, Gilbert F, Hasson MA, Hayes RB, Johnson CC, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 21.Weissfeld JL, Schoen RE, Pinsky PF, Bresalier RS, Church T, Yurgalevitch S, Austin JH, Prorok PC, Gohagan JK. Flexible sigmoidoscopy in the PLCO cancer screening trial: results from the baseline screening examination of a randomized trial. J Natl Cancer Inst. 2005;97:989–97. doi: 10.1093/jnci/dji175. [DOI] [PubMed] [Google Scholar]
- 22.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Brenner H, Hoffmeister M, Stegmaier C, Brenner G, Altenhofen L, Haug U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: estimates based on 840,149 screening colonoscopies. Gut. 2007;56:1585–9. doi: 10.1136/gut.2007.122739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferlitsch M, Reinhart K, Pramhas S, Wiener C, Gal O, Bannert C, Hassler M, Kozbial K, Dunkler D, Trauner M, Weiss W. Sex-specific prevalence of adenomas, advanced adenomas, and colorectal cancer in individuals undergoing screening colonoscopy. JAMA. 2011;306:1352–8. doi: 10.1001/jama.2011.1362. [DOI] [PubMed] [Google Scholar]
- 27.Zhan S, Vazquez N, Wientjes FB, Budarf ML, Schrock E, Ried T, Green ED, Chanock SJ. Genomic structure, chromosomal localization, start of transcription, and tissue expression of the human p40-phox, a new component of the nicotinamide adenine dinucleotide phosphate-oxidase complex. Blood. 1996;88:2714–21. [PubMed] [Google Scholar]
- 28.Ellson CD, Davidson K, Ferguson GJ, O'Connor R, Stephens LR, Hawkins PT. Neutrophils from p40phox-/- mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing. J Exp Med. 2006;203:1927–37. doi: 10.1084/jem.20052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh CI, Stull ND, Li XJ, Tian W, Price MO, Grinstein S, Yaffe MB, Atkinson S, Dinauer MC. The phosphoinositide-binding protein p40phox activates the NADPH oxidase during FcgammaIIA receptor-induced phagocytosis. J Exp Med. 2006;203:1915–25. doi: 10.1084/jem.20052085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561–5. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 32.Eglinton TW, Barclay ML, Gearry RB, Frizelle FA. The spectrum of perianal Crohn's disease in a population-based cohort. Dis Colon Rectum. 2012;55:773–7. doi: 10.1097/DCR.0b013e31825228b0. [DOI] [PubMed] [Google Scholar]
- 33.Eglinton TW, Roberts R, Pearson J, Barclay M, Merriman TR, Frizelle FA, Gearry RB. Clinical and genetic risk factors for perianal Crohn's disease in a population-based cohort. Am J Gastroenterol. 2012;107:589–96. doi: 10.1038/ajg.2011.437. [DOI] [PubMed] [Google Scholar]
- 34.Muise AM, Xu W, Guo CH, Walters TD, Wolters VM, Fattouh R, Lam GY, Hu PZ, Murchie R, Sherlock M, Gana JC, Russell RK, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–35. doi: 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somasundaram R, Deuring JJ, van der Woude CJ, Peppelenbosch MP, Fuhler GM. Linking risk conferring mutations in NCF4 to functional consequences in Crohn's disease. Gut. 2012;61:1097. doi: 10.1136/gutjnl-2011-301344. author reply -8. [DOI] [PubMed] [Google Scholar]
- 36.Volpp BD, Nauseef WM, Clark RA. Two cytosolic neutrophil oxidase components absent in autosomal chronic granulomatous disease. Science. 1988;242:1295–7. doi: 10.1126/science.2848318. [DOI] [PubMed] [Google Scholar]
- 37.de Oliveira-Junior EB, Bustamante J, Newburger PE, Condino-Neto A. The human NADPH oxidase: primary and secondary defects impairing the respiratory burst function and the microbicidal ability of phagocytes. Scand J Immunol. 2011;73:420–7. doi: 10.1111/j.1365-3083.2010.02501.x. [DOI] [PubMed] [Google Scholar]
- 38.Matsue H, Edelbaum D, Shalhevet D, Mizumoto N, Yang C, Mummert ME, Oeda J, Masayasu H, Takashima A. Generation and function of reactive oxygen species in dendritic cells during antigen presentation. J Immunol. 2003;171:3010–8. doi: 10.4049/jimmunol.171.6.3010. [DOI] [PubMed] [Google Scholar]
- 39.Nakahira K, Kim HP, Geng XH, Nakao A, Wang X, Murase N, Drain PF, Sasidhar M, Nabel EG, Takahashi T, Lukacs NW, Ryter SW, et al. Carbon monoxide differentially inhibits TLR signaling pathways by regulating ROS-induced trafficking of TLRs to lipid rafts. J Exp Med. 2006;203:2377–89. doi: 10.1084/jem.20060845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasan N, Pollack A, Cho I. Infectious causes of colorectal cancer. Infect Dis Clin North Am. 2010;24:1019–39. x. doi: 10.1016/j.idc.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 41.de Martel C, Franceschi S. Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol. 2009;70:183–94. doi: 10.1016/j.critrevonc.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 42.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, Eschrich S, Kis C, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–68. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stirpe F, Della Corte E. The regulation of rat liver xanthine oxidase. Conversion in vitro of the enzyme activity from dehydrogenase (type D) to oxidase (type O) J Biol Chem. 1969;244:3855–63. [PubMed] [Google Scholar]
- 45.Castro GD, de Layno AMAD, Costantini MH, Castro JA. Cytosolic xanthine oxidoreductase mediated bioactivation of ethanol to acetaldehyde and free radicals in rat breast tissue. Its potential role in alcohol-promoted mammary cancer. Toxicology. 2001;160:11–8. doi: 10.1016/s0300-483x(00)00433-9. [DOI] [PubMed] [Google Scholar]
- 46.Shaw S, Jayatilleke E. The role of cellular oxidases and catalytic iron in the pathogenesis of ethanol-induced liver injury. Life Sci. 1992;50:2045–52. doi: 10.1016/0024-3205(92)90570-f. [DOI] [PubMed] [Google Scholar]
- 47.Mira L, Maia L, Barreira L, Manso CF. Evidence for free radical generation due to NADH oxidation by aldehyde oxidase during ethanol metabolism. Arch Biochem Biophys. 1995;318:53–8. doi: 10.1006/abbi.1995.1203. [DOI] [PubMed] [Google Scholar]
- 48.Castro GD, Delgado de Layno AM, Costantini MH, Castro JA. Cytosolic xanthine oxidoreductase mediated bioactivation of ethanol to acetaldehyde and free radicals in rat breast tissue. Its potential role in alcohol-promoted mammary cancer. Toxicology. 2001;160:11–8. doi: 10.1016/s0300-483x(00)00433-9. [DOI] [PubMed] [Google Scholar]
- 49.Goodman JE, Bowman ED, Chanock SJ, Alberg AJ, Harris CC. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis. 2004;25:2467–72. doi: 10.1093/carcin/bgh260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.