Abstract
As RNA Polymerase II (Pol II) transcribes a gene, it encounters an array of well-ordered nucleosomes. How it traverses through this array in vivo remains unresolved. One model proposes that torsional stress generated during transcription destabilizes nucleosomes ahead of Pol II. Here, we describe a method for high resolution mapping of underwound DNA using next-generation sequencing, and show that torsion is correlated with gene expression in Drosophila melanogaster cells. Accumulation of torsional stress, through topoisomerase inhibition, results in increased. Pol II at transcription start sites. Whereas Topo I inhibition results in increased nascent RNA transcripts, Topo II inhibition shows little change. Despite the different effects on Pol II elongation, topoisomerase inhibition results in increased nucleosome turnover and salt solubility within gene bodies, suggesting that the elongation-independent effects of torsional stress on nucleosome dynamics contributes to the destabilization of nucleosomes.
Keywords: torsional stress, nucleosome turnover, nascent RNA, topoisomerase
Introduction
The wrapping of DNA around octameric histone proteins to form nucleosomes accomplishes the feat of packaging DNA within the nucleus with the consequence of restricting DNA access. This is problematic for DNA-templated processes including replication and transcription1. For transcription, cells have evolved to maintain promoter regions relatively depleted of nucleosomes, allowing DNA access to transcription factors and the RNA Polymerase II (Pol II) machinery2,3. However, as Pol II leaves the promoter region, it encounters a well-ordered nucleosomal array2,3. In vitro, a single nucleosome effectively blocks Pol II elongation4–6, but within the cell, Pol II transcribes chromatin templates with high efficiency. How does Pol II transcribe through the nucleosome? One model proposes that transcription generates torsional stress that destabilizes nucleosomes ahead of Pol II, allowing for efficient progression.
During transcription, the double helix is melted to form the transcription bubble, and resistance to Pol II rotation relative to DNA creates torsional stress that manifests as positive and negative supercoils downstream and upstream of Pol II, respectively7,8. Torsional stress has been detected in vivo, primarily through binding of psoralen and its derivatives, which preferentially intercalates into DNA that is underwound from negative supercoiling9. Psoralen binding has been used to visualize global unconstrained supercoiling in Drosophila polytene chromosomes10, and to map genome-wide supercoiling in yeast11 and human cells12,13. Whereas one study showed that supercoiling remodels large-scale chromosomal domains13, how torsion affects nucleosomes at the gene level remains unclear. In vitro, negative supercoiling promotes nucleosome assembly, whereas positive supercoiling inhibits it14 but in vivo evidence is lacking. Understanding this interplay has potential clinical implications, as widely used cancer chemotherapeutic anthracycline drugs that intercalate into DNA and induce positive torsion have recently been shown to increase nucleosome turnover and eviction around active promoters15,16.
To test the effect of transcription-generated torsional stress on nucleosome dynamics and Pol II kinetics in Drosophila cells, we measured torsional states at the gene level, and have perturbed torsion by inhibiting topoisomerases, enzymes that relieve supercoils. The resulting changes in torsion, Pol II levels, nascent RNA and nucleosome occupancy, solubility and turnover reveal an intricate balance between efficient Pol II progression and maintenance of the nucleosomal template.
Results
High-resolution genome-wide assay to detect torsion states
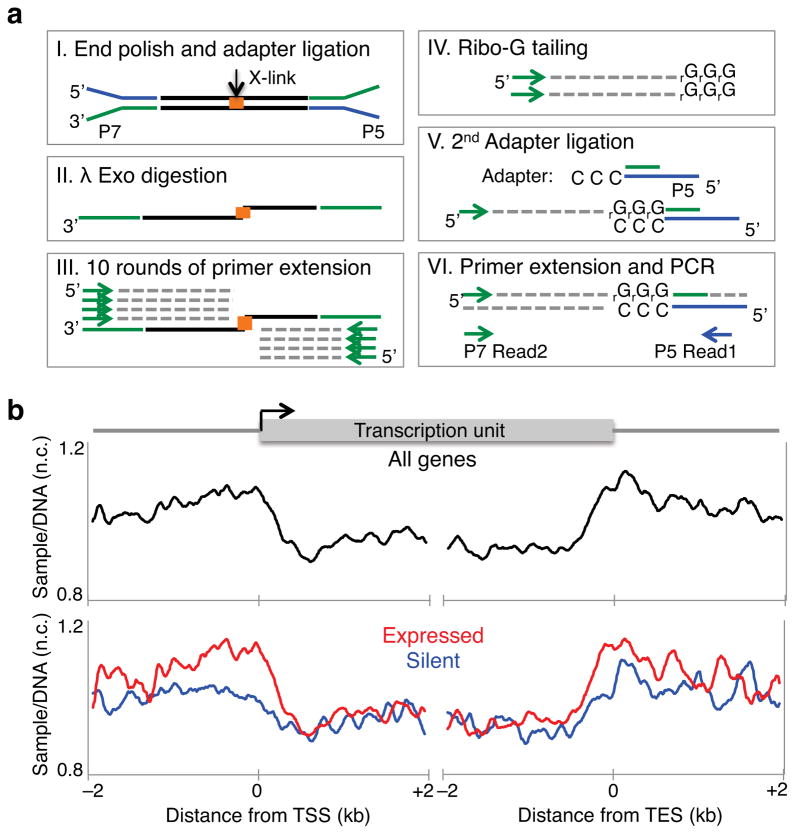
Several methods have recently been developed for large-scale detection of supercoils in yeast11 and in human cell lines12,13, but resolution has been insufficient to delineate torsional states at the nucleosome level. Therefore, we adapted a micro-array-based method11 to next-generation sequencing. We exposed Drosophila S2 cells to Trimethyl-psoralen (TMP) and covalently cross-linked both strands upon exposure to 365 nM UV light. Following DNA extraction and shearing to an average size of 250 bp, we enriched for cross-linked DNA fragments by multiple rounds of denaturation followed by Exonuclease I (Exo I) digestion, which preferentially digests single stranded DNA (Supplementary Fig. 1a)11. After end ligation of Illumina barcoded adapters17, we digested the 5′ strand with λ exonuclease until the cross-linked nucleotide inhibited further digestion (Fig. 1a). Using a primer complementary to the paired-end adapter, we performed 10 rounds of primer extension that end at the cross-linked site. When the λ exonuclease digestion was omitted, no single-stranded extension products were observed (Supplementary Fig. 1b). We then extended the ssDNA products with ribo-Gs using Terminal Transferase, and ligated a double stranded adapter that has a 5′ CCC overhang. After a round of primer extension followed by cycles of library amplification, we sequenced from the CCC overhang end to map the cross-linked site (Fig. 1a). We refer to this method as TMP-seq. As a control for sequence bias, we added TMP to purified genomic DNA, crosslinked by UV light exposure, and processed in parallel to TMP-treated S2 cells. We then mapped the nucleotide position of the cross-links from samples and genomic DNA onto the genome and fit a kernel density distribution around each site18. A representative region in chromosome 3R is shown (Supplementary Fig. 1c, top). The Hsp70C locus shows TMP binding upstream of the transcription start site (TSS) (Supplementary Fig. 1c, bottom) as confirmed by qPCR analysis of the Hsp70 gene (Supplementary Fig. 1d), consistent with previous studies19,20. To normalize for TMP sequence biases, we calculated the log-ratio of each sample to the genomic DNA control. We then averaged the normalized TMP-seq signals around the TSS and transcription end sites (TES) for all genes for each replicate, with a representative data set shown (Fig 1b). This shows that TMP signals are high upstream of genes and low within gene bodies (Fig. 1b, top). Furthermore, actively transcribed genes showed higher TMP levels at promoter regions than silent genes (Fig. 1b, bottom), consistent with previously published psoralen mapping studies11–13.
Figure 1. High resolution detection of supercoiling states.
(a) Strategy for paired-end sequencing of TMP cross-linked DNA fragments. I. Illumina barcoded adapters are ligated to cross-linked fragments. II. The 5′ strand is digested with λ exonuclease. III. Using a primer complementary to the paired-end adapter, 10 rounds of primer extension were performed. IV. Ribo-Gs were added at the 3′ end using Terminal Transferase, V. A double stranded adapter with a 5′ CCC overhang was ligated. VI. One round of primer extension followed by cycles of library amplification were performed. Libraries were sequenced from the CCC overhang end. (b) TMP-seq was performed on control samples with two replicates. Reads were normalized for DNA sequence bias. The average normalized TMP-seq (y-axis Ave. sample/DNA) signal for every 10 bp in a 4 kb region surrounding the transcription start site (TSS) and transcription end site (TES) of all genes is plotted (top), and for expressed and silent genes separately (bottom). n.c. Normalized counts.
Topoisomerase inhibition alters torsion genome-wide
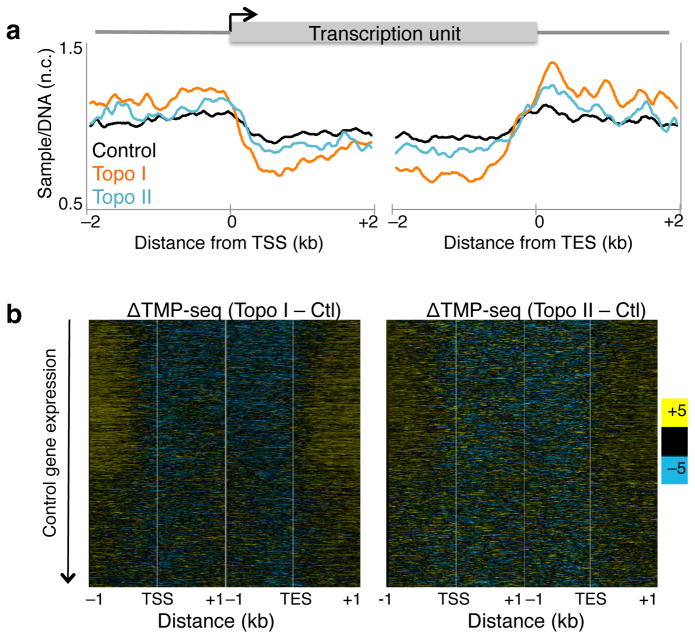
Toposiomerases relieve the torsional strain generated during transcription. We individually inhibited Topo I and Topo II in S2 cells using Camptothecin and ICRF-193, respectively, and measured changes in torsion levels using TMP-seq. We averaged the normalized TMP-seq signals genome-wide and found altered levels relative to control (Fig. 2a), suggestive of accumulated torsional strain in the absence of topoisomerase activity. The increased signal upstream of the TSS is consistent with increased negative torsion whereas decreased TMP binding within gene bodies suggests a relative increase in positive torsion compared to control. We then displayed the change in TMP-seq relative to control as heat maps with genes ordered by decreasing expression in control samples21, and found that the gene body effect is larger in transcribed genes than in silent genes (Fig. 2b, Supplementary Fig. 2a), consistent with the known role of both enzymes in transcription. Notably, Topo I inhibition resulted in a larger effect on accumulated torsional strain compared to Topo II inhibition, particularly for expressed genes. This is consistent with previous studies suggesting that Topo I is the major relaxer of transcription-generated torsional strain22,23.
Figure 2. Topoisomerase inhibition increases torsional stress genome-wide.
(a) TMP-seq was performed on control cells and cells treated with Topo I or II inhibitor with two replicates and samples were normalized for sequence bias. The average normalized signals surrounding the transcription start site (TSS) and transcription end site (TES) were determined. Data for control samples is as represented in Fig. 1b. n.c. Normalized counts. (b) The changes in normalized TMP-seq values were determined by subtracting the control from Topo I- (left) and Topo II- (right) inhibited samples, and were displayed as heat maps with genes ordered by decreasing gene expression in control samples. Contrast = 5. Ctl. control sample
Previous studies have shown that, under saturating TMP conditions, nucleosomes inhibit TMP crosslinking11,24. Although we used limiting amounts of TMP, it is possible that topoisomerase inhibition results in nucleosome occupancy changes, which in turn are responsible for the observed changes in the TMP-seq profile. We determined nucleosome occupancy by sequencing the total micrococcal nuclease (MNase) digested chromatin for control, Topo I-, and II-inhibited samples, and calculated the Pearson’s correlation for each gene between nucleosome occupancy and TMP-seq data in the 1 kb region surrounding the TSS. We then plotted the histogram of Pearson’s correlations (Supplementary Fig. 2b), and found that there is an overall slight anti-correlation (36% of genes have r ≤ −0.2) between nucleosome occupancy and TMP intercalation in untreated control cells, as expected from previous studies11,24. However, after Topo I or II inhibition, the correlation distributions did not shift. Furthermore, when we calculated the difference in nucleosome occupancy before and after topoisomerase inhibition, we saw very little change (Supplementary Fig. 2c), suggesting that the change in TMP-seq profile after topoisomerase inhibition is not a consequence of changes in nucleosome occupancy.
To quantify the accumulation of torsional stress for each gene, we calculated a torsion index for each gene by averaging the TMP-seq values for 1 kb upstream of the TSS and subtracting the average TMP-seq value for 1 kb downstream of the TSS. We then calculated the torsion difference (TD) by subtracting the torsion index of control samples from the Topo I- or Topo II-inhibited samples (see Methods) and displayed the TMP-seq data as heat maps with genes rank-ordered by the absolute value of TD for either Topo I or Topo II inhibition (Supplementary Fig. 2d). This analysis shows that most genes experience altered torsional stress, with varying degrees, from topoisomerase inhibition.
Topo inhibition alters Pol II kinetics
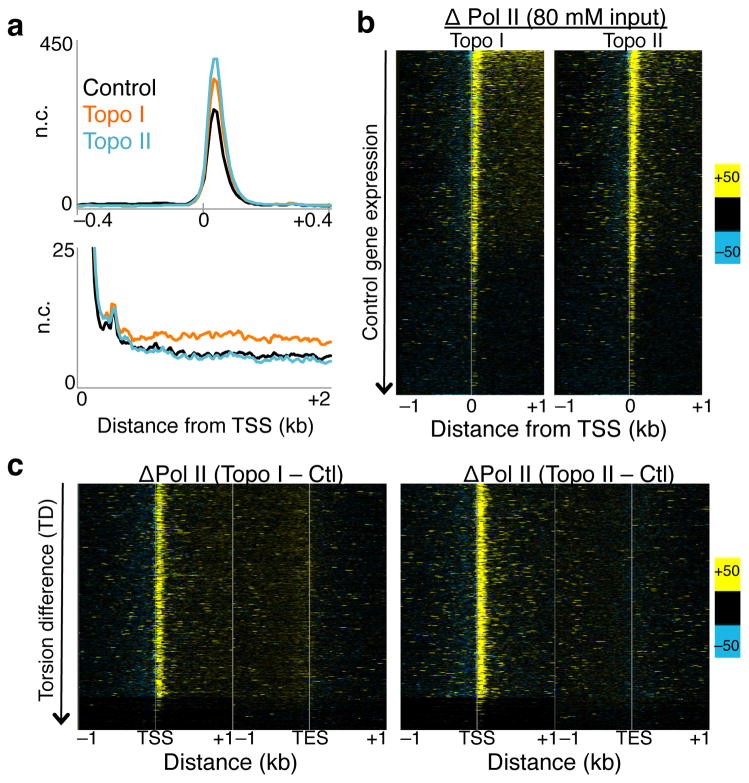
Torsional stress is believed to inhibit Pol II25. To test the effect of torsional stress on Pol II kinetics, we profiled the low-salt soluble Pol II, which we have shown previously to be highly enriched for the stalled species21,26. The stalled Pol II is a subset of Pol II that has initiated but has paused about 50 bp downstream of the TSS27–29. Notably, inhibition of either Topo I or Topo II led to increases in stalled Pol II just downstream of TSSs (Fig. 3a, top, Supplementary Fig. 3a–b, blue signal tracks), suggesting that topoisomerase inhibition affects Pol II pausing kinetics. When Pol II levels are displayed as heat maps, increases in stalled Pol II are seen at the TSS of most genes (Fig. 3b). Furthermore, Topo I inhibition resulted in increased Pol II levels within gene bodies compared to control, in contrast to Topo II inhibition (Fig. 3a, bottom), suggesting altered Pol II elongation kinetics upon Topo I inhibition.
Figure 3. Altered Pol II initiation kinetics upon topoisomerase inhibition.
(a) Pol II ChIP-seq, using input from 80 mM salt soluble fraction, was performed on control and treated cells with two replicates. Reads of length 25 – 75 bp were parsed out and mapped onto the genome, and the average signal 800 bp surrounding the transcription start site (TSS) of all genes were determined (top). The same data are shown for the region 2 kb downstream of the TSS (bottom). n.c. Normalized counts. (b) Changes in Pol II signals were determined by subtracting control signals from Topo I- (left), and Topo II-inhibited (right) Pol II ChIP, and values are presented in heat map format with genes arranged by decreasing gene expression in control samples. (c) Heat maps as in (b) arranged by decreasing TD. Contrast = 50. Ctl. control samples.
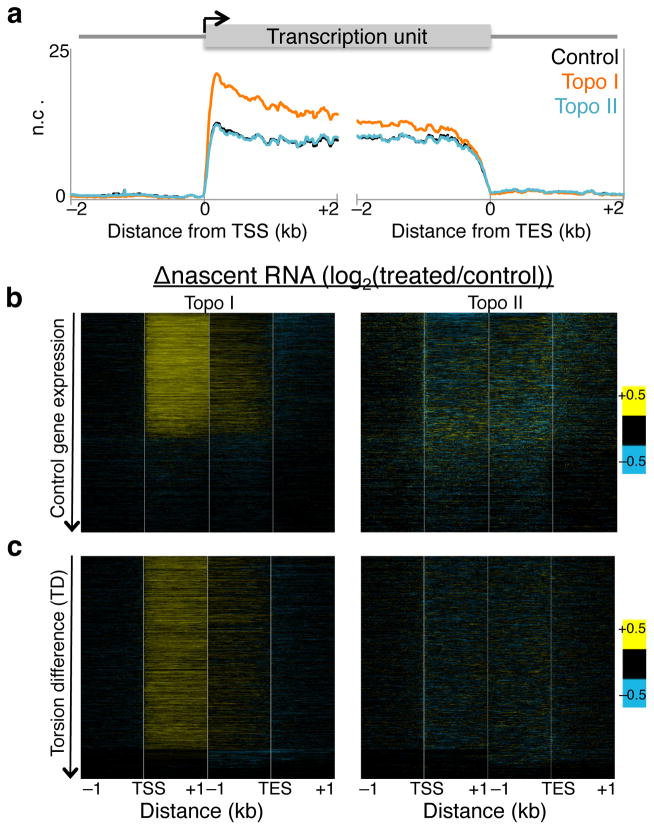
We then determined the kinetics of the elongating Pol II using Pol II-bound nascent RNA as proxy because nascent RNA is the most recent product of an elongating Pol II. Using a biochemical fractionation assay that enriches for chromatin-bound nascent RNA30,31 followed by high-throughput sequencing, we mapped nascent RNA levels before and after topoisomerase inhibition (Supplementary Fig. 3a–b, brown signal tracks). In parallel, we performed nascent RNA analysis for S2 cells heat shocked at 37°C for 15 minutes to confirm the validity of the assay, as heat shock is well known to rapidly decrease Pol II levels genome-wide32,33. When we averaged the normalized counts for all genes surrounding the TSS and TES for control, Topo I, and Topo II inhibited samples, we found that Topo I inhibition resulted in overall increased nascent RNA production near the 5′ end (Fig. 4a). In contrast, Topo II inhibition showed no overall change relative to control (Fig. 4a). When we displayed these changes as heat maps surrounding the TSS and TES, we found that Topo I inhibition affects primarily the transcribed genes, whereas Topo II inhibition results in heterogeneous changes in nascent RNA levels (Fig. 4b). Notably, the gain in nascent RNA for Topo I inhibition is most prominent within 1 kb of the TSS, whereas the TES shows little change, suggesting that less Pol II reaches the end of the gene than has initiated. Indeed long genes show decreased nascent RNA at the 3′ end compared to short genes (Supplementary Fig. 4a). Such increased nascent RNA after Topo I inhibition has been previously described for the DHFR gene in CHO cells34 and the Hif-1α in human cells35. Topo II inhibition results in decreased nascent RNA of only the most highly expressed genes (Fig. 4b), consistent with a secondary role for Topo II in transcription12. As an assay control, we measured the change in nascent RNA levels after heat shock and found decreased levels genome-wide (Supplementary Fig. 4b, top), as previously shown32, as well as increased expression of the highly induced Hsp genes (Supplementary Fig. 4b, bottom). We further repeated the nascent RNA isolation and performed qPCR on select genes that showed increased, decreased, and no change in nascent RNA levels upon Topo I or II inhibition. We found reproducible effects using qPCR (Supplementary Fig. 4c).
Figure 4. Altered Pol II elongation kinetics upon topoisomerase inhibition.
(a) Nascent RNA-seq was performed on control, Topo I-, and Topo II-inhibited cells, and average profiles were determined for 4 kb surrounding the transcription start site (TSS) and transcription end site (TES) of all genes. A second replicate was performed and analyzed by qPCR (Supplementary Fig. 4c) n.c. Normalized counts. (b) The log-ratio of nascent RNA for Topo I- (left) or II-inhibited (right) samples over control was determined and shown in heat map format with genes ordered by decreasing expression in control samples. (c) Heat maps as in (b) arranged by decreasing TD. Contrast = 0.5.
We then analyzed the change in stalled Pol II and nascent RNA by TD and displayed the heat maps with genes ordered by decreasing TD. With the exception of the genes that have the lowest TD, most genes experience increased stalled Pol II after Topo I or II inhibition (Fig. 3c), suggesting that increased torsional stress triggers increased Pol II stalling near the TSS. When arranged by TD, the increase in nascent RNA near the 5′ end but not at the 3′ end after Topo I inhibition becomes evident (Fig. 4c), further suggesting that Topo I is important for both Pol II initiation and elongation. The lack of change in nascent RNA after Topo II inhibition suggests that Topo II acts mainly at the 5′ end on initiated Pol II.
We also used our nascent chain mapping data to identify genes that expression due to topoisomerase inhibition by measuring changes in complete nascent RNA transcripts. About 90% of genes showed little or no change in expression with topoisomerase inhibition based on nascent RNA level. However, of those that changed more than 30%, the most prominent changes were just upstream of the 3′ end for both Topo I (Supplementary Fig. 5a) and Topo II (Supplementary Fig. 5b). It is possible that torsional differences that we observed around the TES (Fig. 2) modulate pausing and transcript release at these genes, and that both topoisomerases facilitate this process. Furthermore, when the nascent RNA data are displayed with genes arranged by change in stalled Pol II, genes that increase in stalled Pol II after Topo I inhibition show the highest increase in nascent RNA near the TSS, whereas Topo II inhibited samples show a mild increase in nascent RNA only for genes that have gained the most stalled Pol II (Supplementary Fig. 5c). Taken together, these analyses support a general role for both Topo I and II in Pol II initiation kinetics and for Topo I in Pol II elongation kinetics.
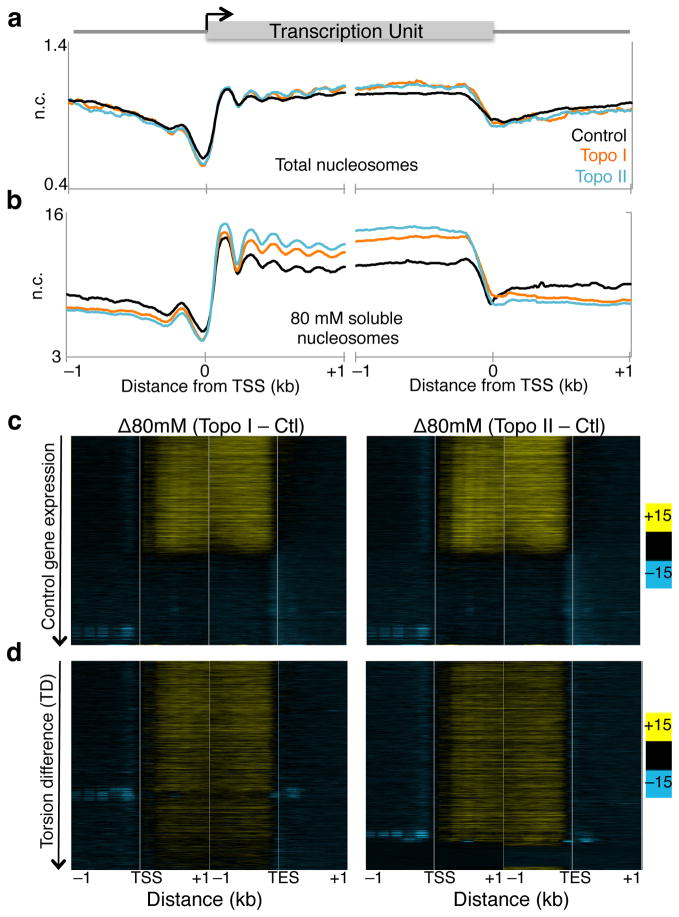
Topo inhibition increases nucleosome low-salt solubility
A previous study has shown that Pol II-generated supercoils affect large-scale chromosome organization, forming underwound and overwound domains13. However, how torsion affects organization of individual nucleosomes within genic regions is unclear. Chromatin of genic regions is characterized by reduced inter-nucleosomal interactions such that, after MNase-digestion, these nucleosomes become readily soluble in low salt, whereas condensed chromatin requires high salt for solubilization36,37. We therefore asked whether inhibition of topoisomerases affects nucleosome organization by extracting low-salt soluble chromatin followed by sequencing21,36. After mapping reads >120 bp onto the genome (Supplementary Fig. 3a–b, green signal tracks), we averaged the normalized counts for 2 kb surrounding the TSS and TES of all genes for control and Topo I- or Topo II-inhibited cells. We also performed the same analysis for total nucleosomes to determine changes in occupancy (Supplementary Fig. 3a–b, black signal tracks). For total nucleosomes, topoisomerase inhibition led to only a slight increase in overall occupancy near 3′ ends of genes (Fig. 5a, Supplementary Fig. 2c). However, low-salt solubility of nucleosomes dramatically increased within gene bodies while promoter regions experienced decreased solubility (Fig. 5b). This suggests that, although nucleosome occupancy is subtly affected, topoisomerase inhibition markedly alters the inter-nucleosomal interactions that determine salt solubility. When displayed as heat maps by control expression, the low-salt solubility of transcribed genes is seen to increase, unlike that of silent genes (Fig. 5c). However, when genes are ordered by TD, low-salt solubility of nucleosomes increased with increasing TD after Topo I inhibition, whereas Topo II inhibition resulted in a uniform increase in low-salt solubility (Fig. 5d). These analyses suggest that torsional stress affects the organization of genic nucleosomes. In contrast, nucleosome occupancy is not affected by torsional stress (Supplementary Fig. 6).
Figure 5. Topoisomerase inhibition affects low-salt chromatin fractionation.
(a) Two replicates of total MNase digested chromatin were sequenced from treated and untreated cells, and the average signal surrounding the transcription start site (TSS) and transcription end site (TES) in all genes for >120 bp reads were plotted. n.c. Normalized counts. (b) Sequenced reads from 80 mM salt soluble fraction for two replicates were mapped onto the genome and the average signals surrounding the TSS and TES were determined. (c) The difference in low-salt soluble signals between control and Topo I- (left), and II-inhibited (right) samples were determined and displayed as heat maps with genes ordered in decreasing expression in control samples for reads of length >120 bp. (c) Heat maps as in (b) arranged by decreasing TD. Contrast = 15.
To determine whether changes in transcriptional competence affect low-salt solubility of nucleosomes after topoisomerase inhibition we displayed changes in low salt soluble nucleosomes as heat maps with genes ordered by decreasing change in stalled Pol II (Supplementary Fig. 7a). For both Topo I and Topo II, we observed increases in low-salt solubility for genes with stalled Pol II, suggesting that altered Pol II kinetics also affect chromatin organization.
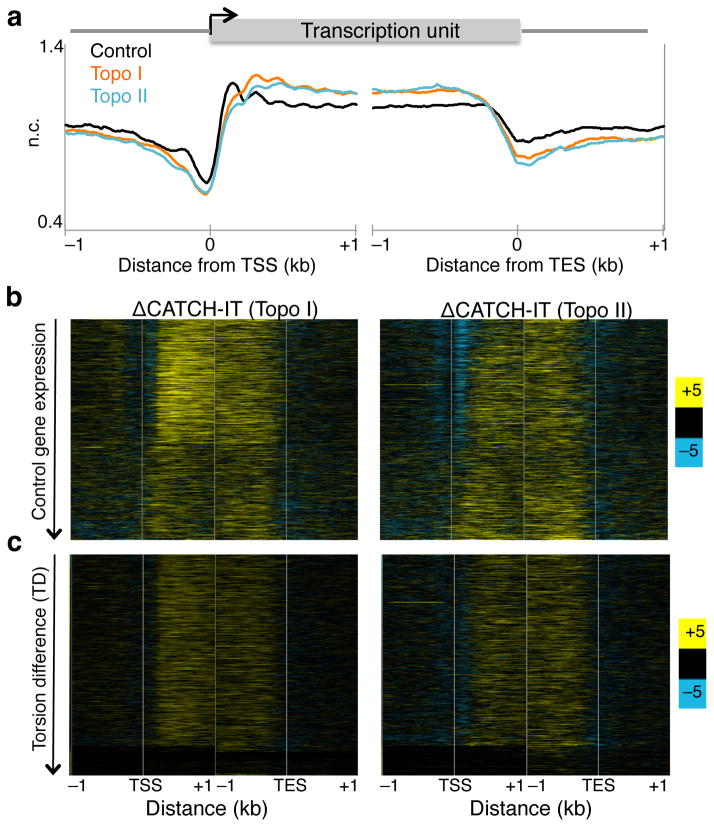
Topo inhibition increases nucleosome turnover within genes
Previous studies have shown that positive torsion inhibits nucleosome assembly in vitro14 and alter chromatin accessibility to nucleases ex vivo38, leading the authors to propose that positive torsion disrupts the nucleosomal template within gene bodies. To test this in vivo, we inhibited topoisomerases individually and measured nucleosome turnover by CATCH-IT, which allows for profiling of newly synthesized H3–H4 particles after a re-assembly process occurs during turnover39. After methionine depletion, S2 cells were labeled with the analog Azidohomoalanine in the presence of either Camptothecin or ICRF-193. Both labeling and topoisomerase inhibition were performed for 15 minutes at room temperature, and CATCH-IT was performed as previously described21,39. Representative regions of mapped reads are shown (Supplementary Fig. 3a–b, red signal tracks). Inhibition of Topo I or Topo II led to a decrease in turnover of the +1 and promoter nucleosomes but to an increase within gene bodies (Fig. 6a). To view the change in nucleosome turnover on a gene-by-gene basis, we displayed the difference in CATCH-IT between topo-inhibited and control samples as a heat map with genes arranged by decreasing expression in control. Topo I inhibition increased the overall nucleosome turnover within gene bodies but the effect for transcribed genes was greater than for non-transcribed genes (Fig. 6b, left). In contrast, Topo II inhibition led to increased nucleosome turnover within gene bodies regardless of expression levels (Fig. 6b, right). When genes were ordered by TD, nucleosome turnover increased similarly, with the exception of genes with the lowest TD values (Fig 6c). These results suggest that once a threshold of torsional stress is reached, nucleosomes become destabilized.
Figure 6. Altered nucleosome turnover under torsion.
(a) Control and treated samples were subjected to CATCH-IT followed by paired end sequencing for two replicates. Mapped reads were averaged surrounding the transcription start site (TSS) and transcription end site (TES) were averaged for all genes. (b) The changes in CATCH-IT values were determined by subtracting the control from Topo I- (left) and Topo II-inhibited (right) signals, and were displayed as heat maps with genes ordered by decreasing expression in control samples. (c) Heat maps as in (b) arranged by decreasing TD. Contrast = 5.
We have previously shown that increased expression results in increased nucleosome turnover21. That nucleosome turnover increased after Topo I and II inhibition, despite Topo II inhibition showing no changes in Pol II elongation, suggests that Pol II elongation and the torsion it generates have separable contributions on nucleosome turnover. To further examine these contributions, we analyzed the nucleosome turnover data relative to changes in stalled Pol II (Supplementary Fig 7b). We find that changes in nucleosome turnover caused by topoisomerase inhibition are associated with both increased and decreased expression, which implies that changes in turnover are not simply consequences of torsion-induced Pol II stalling. Rather, we propose that Pol II transit drives torsion, which in turn alters nucleosome stability.
Discussion
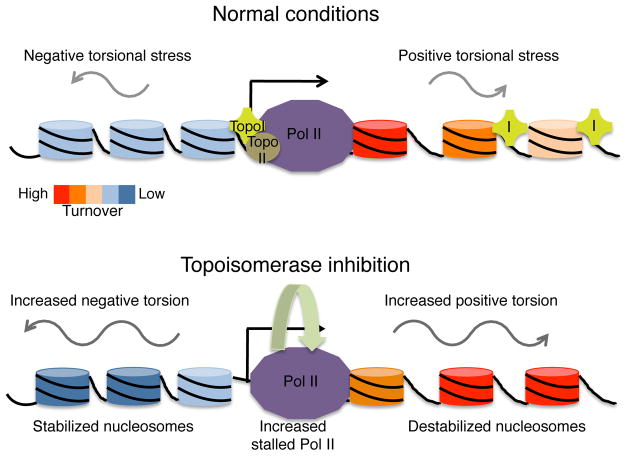
In this study, we have examined the effect of accumulated torsional stress on nucleosomes and Pol II in vivo. We have developed a genome-wide sequencing based assay to determine DNA torsional states genome-wide at nucleosome resolution in Drosophila cells. Using this assay, we have shown that inhibition of topoisomerases leads to rapid accumulation of torsional strain, which is accompanied by changes Pol II kinetics and chromatin properties. The stalled Pol II accumulates immediately downstream of the TSS after inhibition of either Topo I or Topo II, but the elongating Pol II, as measured by nascent RNA production, is affected differently. Topo I inhibition results in increased nascent RNA levels near the 5′ end of genes. In contrast, Topo II inhibition only affects nascent RNA levels of highly expressed genes, while most genes remain unaltered, suggesting that only Topo I acts within gene bodies to regulate Pol II elongation kinetics. We have shown that, despite the changes in expression and Pol II kinetics after topoisomerase inhibition, accumulation of torsional stress results in increased nucleosome turnover within gene bodies genome-wide, providing direct evidence for an in vivo influence of DNA torsion on nucleosome dynamics that is separable from Pol II elongation-associated dynamics. Our data support a model whereby the transient wave of positive torsion downstream of Pol II destabilizes genic nucleosomes to allow progression, and the transient negative torsion stabilizes nucleosome formation behind Pol II to maintain chromatin structure38. In this way, a delicate balance between nucleosomal destabilization, maintenance, and Pol II progression is achieved (Fig. 7).
Figure 7. Model for transcription-generated torsional strain and nucleosome turnover.
Under normal conditions, RNA Polymerase II (Pol II) generates positive supercoils ahead, and negative supercoils behind, as it elongates a transcript along a gene. Topoisomerases I and II (purple cross labeled I and brown circle labeled II, respectively) mitigate the accumulation of torsional strain. Transcription results in nucleosome turnover and decondensed organization of transcribed nucleosomes. Upon topoisomerase inhibition, torsional strain, both positive and negative, accumulates, resulting in increased Pol II stalling and nucleosome destabilization, as measured by nucleosome turnover and low-salt solubility, within gene bodies. This supports a balance between destabilization of nucleosomes for Pol II passage and maintenance of chromatin structure for chromosomal integrity.
Recent studies have used TMP to profile supercoiling states genome-wide in yeast11, and in a portion of the human genome12,13. By adapting the method for high resolution sequencing analysis, we have confirmed that promoters and regions upstream of the TSS have high levels of TMP crosslinking, indicative of high negative torsion, while gene bodies have low levels of TMP crosslinking, as expected when positive torsion inhibits TMP intercalation. Whereas our TMP-seq profiles for Drosophila cells generally agree with those seen in human cells12,13, there are some interesting differences. First, in human cells, TMP-crosslinking peaked at the TSS and diminished beyond 1 kb upstream, leading the authors to conclude that supercoiling is a short-range force12. In Drosophila cells, we observed high levels of negative torsion beyond 2 kb upstream of the TSS. The difference might be due to the prevalent bi-directional transcription seen in mammalian genomes27,40 that is absent in Drosophila, which would concentrate negative torsion behind the Pol IIs at the TSS, whereas the uni-directional transcription of Drosophila promoters would allow for dissipation further upstream. Second, the 3′ end of Drosophila genes experienced high levels of TMP-crosslinking (Fig. 1c), which was further exacerbated by topoisomerase inhibition (Fig. 2). In yeast, topoisomerases are required for both transcription termination and the formation of the nucleosome-depleted region at the transcription end sites22,41. Similarly, Drosophila Hsp70 genes have Topo II both at the 5′ and 3′ end42, suggesting that at least with the Hsp70 gene, events at the 3′ end generate sufficient torsional strain to require topoisomerase activity.
It has been generally accepted that inhibition of topoisomerases results in an immediate halt in transcription due to accumulation of torsional stress. Early studies have shown that transcription of rDNA requires topoisomerase activity43,44, and in vitro transcription assays using viral RNA Polymerases are greatly inhibited when topoisomerase I activity is absent45. We have shown that topoisomerase inhibition alters Pol II kinetics, which is a potential mechanism for decreased expression. Topo I inhibition alters both the initiating and elongating form of Pol II,, and after a short inhibition period, results in little change in expression as measured by changes in complete nascent RNA transcripts. This result seems contradictory to the dogma that topoisomerase inhibition leads to transcription inhibition. However, various other studies inhibiting only Topo I have yielded mixed effects on transcription46–48. Topo I inhibition has been shown to increase Pol II and nascent RNA near the TSS but not at the 3′ end of the Dhfr gene in cultured CHO cells34, and to increase the release of paused Pol II and anti-sense transcription of HIF-1 gene in human cells35. These studies suggest that the highly conserved Topo I may also have a conserved effect on Pol II kinetics. In contrast to Topo I, Topo II inhibition resulted in altered Pol II elongation kinetics of only the most highly expressed genes, confirming that Topo I is the main relaxer of transcription-generated torsional strain and Topo II acts together with Topo I in highly expressed genes12,22,23.
It is noteworthy that the torsion-induced increase in nucleosome turnover is observed only beyond the first nucleosome downstream of the TSS (+1 nucleosome). In fact, relative to the genome-wide average, the +1 nucleosome, and promoter nucleosomes, showed decreased turnover upon topoisomerase inhibition. The +1 nucleosome has the highest density of stalled Pol II49, suggesting a specialized interaction between Pol II and the +1 nucleosome. Indeed, MNase-protected fragments spanning the +1 nucleosome and the footprint of Pol II are enriched immediately downstream of the TSS21. It is possible, therefore, that under topoisomerase inhibition, the interaction between Pol II and the +1 nucleosome is further stabilized, resulting in increased stalled Pol II at the TSS and the decreased turnover at the +1 nucleosome following topoisomerase inhibition. Furthermore, the decreased turnover of promoter nucleosomes may reflect the altered Pol II initiation kinetics after topoisomerase inhibition. But how are the effects of torsional stress generated at the TSS propagated beyond the +1 nucleosome? Mathematical modeling of transcription-generated torsion suggests that transcription of 5 bp is sufficient to propagate a wave of positive supercoils at a rate of 2 orders of magnitude faster than Pol II elongation50. Single molecule experiments on supercoil dynamics have shown that this propagation has two modes: the slower mechanism of diffusion that occurs in short range distances, and the much faster mode of “hopping” where supercoils are propagated in long distances under millisecond time frames51. Therefore, it is possible that the effect of increased turnover downstream of the +1 nucleosome is a result of torsional stress being propagated in long-range distances through the “hopping” mechanism. In this way, topological changes to DNA during transcription can act as the medium that connects events at the TSS to ones far downstream within gene bodies.
Online Methods
Cell culture and treatments
Drosophila S2 cells grown to log phase in HYQ-SFX Insect medium (Invitrogen) supplemented with 18 mM L-Glu and harvested as previously described. For heat shocked samples, cells were incubated at 37°C for 15 minutes and immediately harvested. Camptothecin (Sigma Aldrich C9911) and ICRF-193 (Sigma Aldrich I4659) were resuspended to 10 mM in DMSO and frozen in aliquots. For cell treatments, final concentration of 10 μM of either drug was added to cell medium for 15 minutes, and cells were harvested immediately. Trimethyl-psoralen (TMP) (Sigma Aldrich T6137) was resuspended in 0.5 mg/mL in ethanol.
Trimethyl-psoralen photobinding
Our protocol was adapted from a previously published assay11. S2 cells were diluted to 1 × 107 cells/mL in growth medium, and 2 mL aliquots in small plates were used. TMP was added to cells with and without drug treatments at a final concentration of 2 μg/mL for 10 minutes in the dark. Plates of cells were exposed to 3 kJ m−2 of 365 nm light (Fotodyne UV Transilluminator 3–3000 with 15 W bulbs). Cells were then collected and washed with cold 1x PBS and resuspended thoroughly in 0.5 mL of 1x PBS with 0.5% SDS. Proteins were digested using 4 μg of Proteinase K (Invitrogen) for 1 hour at 55°C. DNA was extracted by phenol/chloroform and precipitated with ethanol. After resuspension in 200 μL of H2O, RNA was digested using 1μg of RNase (Invitrogen) at 37°C for 30 minutes. DNA was extracted and precipitated as before and resuspended to 0.5 μg/μL. To achieve fragment sizes of 100 – 500 bp, 50 μg of DNA was sonicated in a Bioruptor 3× 15 minutes each at high setting with 30 seconds on/off in cold water. Cross-linked fragments were enriched by repeated rounds of denaturation and Exonuclease I (Exo I) digestion. Using 3 μg of sonicated DNA diluted to 250 μL, samples were boiled in water bath for 10 minutes and incubated in ice-water for 2 minutes. To each sample, 30 μL of 10X Exo I buffer and 10 μL of Exo I were added, and digestion was allowed to proceed for 1 hour at 37°C. Samples were boiled and cooled as before, and 10 μL of Exo I was added for a second round of 1 hour digestion. DNA was extracted and precipitated, and concentration was assayed by PicoGreen quantification (Invitrogen). Following λ exonuclease digestion, ssDNA products were used as template for qPCR analysis with primers spanning the Hsp70 genes, as previously described52. The primers used are listed in Supplementary Table 1.
TMP-seq
We devised a method for producing Solexa libraries to map the precise location of the irreversible TMP interstrand cross-link (Fig. 1a). Exo I digested DNA samples were subjected to enzymatic reactions for end polishing and ‘A’-tailing as previously described 17. After ligation of PE barcode adapters, the 5′ strand was digested using 25 U of λ exonuclease (NEB) for 30 minutes at 37°C. The DNA was purified using Ampure beads, and eluted in 35 μL of H2O. The resulting 3′ strand was used as template for 10 rounds of primer extension in 1x HiFi Phusion buffer, 0.8 mM dNTP, 2 U Phusion DNA Polymerase (NEB), and 40 nM of P7 extension primer (CAAGCAGAAGACGGCATACGA*G - *denotes phosphorothioate linkage) in the following cycling conditions: 95°C for 3 minutes, 10 rounds of linear amplification (95°C for 1 minute, 57°C for 1 minute, 72°C for 3 minutes), 95°C for 1 minute, hold at 8°C. The resulting single-stranded products were purified with Ampure beads, eluted at 35 μL of H2O, and concentrated down to 17 μL using a vacuum centrifuge. The purified products were then appended with ribo-G in 1x Terminal deoxynucleotidyl Transferase (TdT) buffer, 20 U TdT (NEB), and 4 mM of rGTP in 37°C for 15 minutes. The products were purified using Ampure beads, eluted in 35 μL of H2O, and concentrated to 18 μL volume as before. The single-stranded ribo-tailed products were ligated to a double stranded adapter with CCC-overhang (oligo 1: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCCC, oligo 2: [Phosphate]GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG) as described17. Prior to standard PCR for PE library17, we added an extension step to create double stranded templates (60°C for 3 minutes, 98°C for 30 seconds, 18 cycles of amplification (98°C for 10 seconds, 65°C for 30 seconds, and 60°C for 30 seconds), 60°C for 5 minutes and hold at 8°C). The final PCR products were purified with Ampure beads, eluted with 40 μL of H2O, and quantified by PicoGreen assay.
CATCH-IT
CATCH-IT was performed essentially as described 39 with the following modifications. For cells treated with either Topoisomerase I or II, drugs were added at final concentration of 10 μM immediately after Azidohomoalanine (Aha) addition and incubated for 15 minutes. Nuclei extraction, biotin coupling, MNase digestion and streptavidin pulldown were performed as described previously 39. The input and streptavidin ChIP DNA were prepared for paired end Solexa sequencing using the modified library preparation 17. Two replicates were performed and a representative data set is shown.
Salt fractionation and Pol II native ChIP
For cells treated with either Topoisomerase I or II inhibitors, drugs were added at a final concentration of 10 μM, and cells were incubated at room temperature for 15 minutes. 80 mM salt fractionation and stalled Pol II native ChIP was performed as described previously 21. For both 80mM salt fraction and stalled Pol II ChIP, two replicates were performed and representative data sets are shown.
Biochemical Fractionation for nascent RNA isolation
Biochemical fractionation of chromatin was performed as previously described 30 with the following changes. Briefly, ~1 × 108 cells were harvested and washed with PBS. Cells were resuspended in 800μL Buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, and protease inhibitor cocktail [Roche], 200 U Superase-in [Ambion]) supplemented with Triton X-100 (0.1% final) before use. After 8 minutes of incubation on ice, nuclei were pelleted (3 min, 3000 × g, 4°C), and lysed in 800μL of Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, and protease inhibitor cocktail [Roche], 200 U Superase-in [Ambion]) supplemented with Triton X-100 (0.1% final) before use. The samples were incubated on ice for 15 min and the pellet was collected by centrifugation as before. Buffer B washing was repeated twice. Pellets were collected and resuspended in Buffer B+ (20 mM EDTA, 0.2 mM EGTA, 2 mM spermine, 5 mM spermidine, 1 mM DTT, protease inhibitor cocktail [Roche], 200 U Superase-in [Ambion]), supplemented with Triton X-100 (0.1% final) before use. Pellets were incubated on ice for 10 minutes and collected by centrifugation as before. Buffer B+ wash was repeated 3 times and the final pellet was resuspended in 720 μL of Buffer B+ with 80 μL 10% SDS. Chromatin associated RNA was isolated by Trizol and quantified spectroscopically. Ribosomal RNA was depleted using Ribo-Zero Magnetic kit (Epicentre).
Nascent RNA-seq
Nascent RNA (200 ng) after rRNA depletion was fragmented to 50 – 300 nucleotides using the RNA Fragmentation Reagents (Ambion). After propanol precipitation, RNA was resuspended in 5 μL and used as input for reverse transcription using the random primers of the SuperScript III First Strand Synthesis System (Life Technologies). Following reverse transcription, sample volumes were increased to 100μL and cleared using Illustra Microspin S-200 HR columns (GE Healthcare). To create the second strand, 50 μL of master mix (0.4× First Strand buffer, 1 mM DTT, 0.8 mM dNTP [with U replacing T], 3× 2nd Strand Buffer, 40 U E.coli DNA Pol I [Invitrogen], 10 U E.coli DNA Ligase [Invitrogen], 2 U E.coli RNAse H [Invitrogen]) was added for a final volume of 150 μL per sample. Reaction proceeded for 2 hours at 16°C, and the resulting double stranded DNA was purified on Qiagen column, eluted in 40 μL, and subjected to modified Paired-End sequencing library protocol17. Prior to final PCR amplification, the second strand was digested using 1U UNG AMPerase (Invitrogen) in 40 μL TE 0.1× pH 7.5 for 15 minutes at 37°C. Libraries were amplified 12 to 18 cycles. qPCR analysis were performed using 10 ng of reverse-transcribed nascent cDNA in duplicate reactions using SYBR green PCR master mix (Applied Biosystems). Relative quantification was performed using Δ Ct method with the untreated sample as reference control. The primers used for each gene are listed in Supplementary Table 1.
Sequencing and data analysis
Solexa sequencing libraries were constructed as described17. Cluster generation and 25 rounds of paired-end sequencing were performed by the FHCRC Genomics Shared Resource using the Illumina Hi-Seq 2000. Base calling, data processing and analysis were performed as described17. Gene tracks from the sequencing data were visualized using Signalmap (Nimblegen, Inc.). Ends analysis, quintiles, and heat maps were performed as described. Data have been deposited with GEO (GSE47795).
For TMP-seq data, read 1 was trimmed of the CCC overhangs. The first sequenced nucleotide was mapped onto the genome and fitted with a kernel density function estimation with bandwidth of 20 bp18. The fact that there are clusters that don’t seem to exactly align between replicates is a consequence of the inevitable sparseness of data points subjected to kernel density smoothing. Ends analyses for TMP-seq data were performed as follows: For each 10-bp interval in a 4 kb region centered at either the TSS or TES, the average signal in that interval was normalized to the average signal in the whole 4 kb region. A running average of a 50 bp-window was used to smoothen the data. The torsion difference (TD) was calculated as follows: For each gene, we assigned a torsion index (TI) by averaging the TMP-seq values for 1 kb upstream of the TSS and subtracting the average TMP-seq value for 1 kb downstream. TDTopoI = TITopoI − TICtl and TDTopoII = TITopoII − TICtl. To determine changes in complete nascent RNA transcripts, log2(treated/Ctl) signals for the last 30 bp of each gene were averaged.
For nascent RNA-seq data, 98% of reads were stranded. Sense strands were mapped onto the genome, and normalized counts were calculated as previously described26. To determine changes in nascent RNA levels, the log2(treated/control) was determined for each Topo I- or II-inhibited sample. Changes in expression due to treatment were determined by averaging the log2 ratio of the last 30 bp of gene ends and a cut off of log2 ratio = 0.6 were used to determine increased and decreased changes. For Topo I-treated samples, 479 genes increased and 855 genes decreased in expression. For Topo II-treated samples, 419 genes increased and 660 genes decreased in expression.
Supplementary Material
Acknowledgments
We thank A. Marty (Fred Hutchinson Cancer Research Center Genomics Shared Resource) for Illumina sequencing, F. Steiner, F. Yang, C. Weber, and S. Ramachandran for critical comments, and J. Henikoff for help on sequencing data analysis. This work was supported by the US National Science Foundation Graduate Research Fellowship under Grant No. DGE-0718124 (S.T.), by the US National Institutes of Health grant U01 HG004274 (S.H.), and by the Howard Hughes Medical Institute (S.H.).
Footnotes
Author Contributions
S.S.T. and S.H. conceived of the project. S.S.T. designed and performed experiments. S.S.T. and S.H. analyzed data, and wrote the manuscript.
Accession Codes
All sequencing data have been deposited in the Gene Expression Omnibus database, under accession code GSE47795.
References
- 1.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 2.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 5.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 6.Shaw PA, Sahasrabuddhe CG, Hodo HG, Saunders GF. Transcription of nucleosomes from human chromatin. Nucleic Acids Res. 1978;5:2999–3012. doi: 10.1093/nar/5.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 8.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci. 1987;84:7024. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 11.Bermúdez I, García-Martínez J, Pérez-Ortín JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta P, Zlatanova J, Tomschik M. Nucleosome Assembly Depends on the Torsion in the DNA Molecule: A Magnetic Tweezers Study. Biophys J. 2009;97:3150–3157. doi: 10.1016/j.bpj.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang B, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F, Kemp CJ, Henikoff S. Doxorubicin Enhances Nucleosome Turnover around Promoters. Curr Biol. 2013;23:782–7. doi: 10.1016/j.cub.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jupe ER, Sinden RR, Cartwright IL. Specialized chromatin structure domain boundary elements flanking a Drosophila heat shock gene locus are under torsional strain in vivo. Biochemistry. 1995;34:2628–33. doi: 10.1021/bi00008a029. [DOI] [PubMed] [Google Scholar]
- 20.Jupe ER, Sinden RR, Cartwright IL. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. The EMBO journal. 1993;12:1067–75. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teves SS, Henikoff S. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev. 2011;25:2387–2397. doi: 10.1101/gad.177675.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand-Dubief Mel, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sperling AS, Jeong KS, Kitada T, Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc Natl Acad Sci. 2011;108:12693–8. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potter DA, Fostel JM, Berninger M, Pardue ML, Cech TR. DNA-protein interactions in the Drosophila melanogaster mitochondrial genome as deduced from trimethylpsoralen crosslinking patterns. Proc Natl Acad Sci. 1980;77:4118–4122. doi: 10.1073/pnas.77.7.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roca J. Transcriptional inhibition by DNA torsional stress. Transcription. 2011;2:82–85. doi: 10.4161/trns.2.2.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien T, Lis JT. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991;11:5285–90. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–8. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamrich M, Greenleaf AL, Bautz EK. Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster. Proc Natl Acad Sci. 1977;74:2079–2083. doi: 10.1073/pnas.74.5.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenney K, et al. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103:11970–4. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ljungman M. Effect of differential gene expression on the chromatin structure of the DHFR gene domain in vivo. Biochim Biophys Acta. 1996;1307:171–177. doi: 10.1016/0167-4781(96)00037-1. [DOI] [PubMed] [Google Scholar]
- 35.Baranello L, Bertozzi D, Fogli MV, Pommier Y, Capranico G. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1 gene locus. Nucleic Acids Res. 2009;38:159–171. doi: 10.1093/nar/gkp817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders MM. Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J Cell Biol. 1978;79:97–109. doi: 10.1083/jcb.79.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MS, Garrard WT. Positive DNA supercoiling generates a chromatin conformation characteristic of highly active genes. Proc Natl Acad Sci. 1991;88:9675–9679. doi: 10.1073/pnas.88.21.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durand-Dubief M, Svensson JP, Persson J, Ekwall K. Topoisomerases, chromatin and transcription termination. Transcription. 2011;2:66–70. doi: 10.4161/trns.2.2.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe TC, Wang JC, Liu LF. In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol Cell Biol. 1986;6:985–992. doi: 10.1128/mcb.6.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. DNA topoisomerase activity is required as a swivel for DNA replication and for ribosomal RNA transcription. NCI monographs. 1987:11–15. [PubMed] [Google Scholar]
- 44.Muller MT, Pfund WP, Mehta VB, Trask DK. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bendixen C, Thomsen B, Alsner J, Westergaard O. Camptothecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry. 1990;29:5613–5619. doi: 10.1021/bi00475a028. [DOI] [PubMed] [Google Scholar]
- 46.Collins I, Weber A, Levens D. Transcriptional consequences of topoisomerase inhibition. Mol Cell Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 49.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becavin C, Barbi M, Victor JM, Lesne A. Transcription within Condensed Chromatin: Steric Hindrance Facilitates Elongation. Biophys J. 2010;98:824–833. doi: 10.1016/j.bpj.2009.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Loenhout MTJ, de Grunt MV, Dekker C. Dynamics of DNA supercoils. Science. 2012;338:94–97. doi: 10.1126/science.1225810. [DOI] [PubMed] [Google Scholar]
- 52.Petesch SJ, Lis JT. Rapid, Transcription-Independent Loss of Nucleosomes over a Large Chromatin Domain at Hsp70 Loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.