Abstract
BACKGROUND
Rare variants in candidate atrial fibrillation (AF) genes have been associated with AF in small kindreds. The extent to which such polymorphisms contribute to AF is unknown.
OBJECTIVE
The purpose of this study was to determine the spectrum and prevalence of rare amino acid coding (AAC) variants in candidate AF genes in a large cohort of unrelated lone AF probands.
METHODS
We resequenced 45 candidate genes in 303 European American (EA) lone AF probands (186 lone AF probands screened for each gene on average [range 89–303], 63 screened for all) identified in the Vanderbilt AF Registry (2002–2012). Variants detected were screened against 4300 EAs from the Exome Sequencing Project (ESP) to identify very rare (minor allele frequency ≤ 0.04%) AAC variants and these were tested for AF co-segregation in affected family members where possible.
RESULTS
Median age at AF onset was 46.0 years [interquartile range 33.0–54.0], and 35.6% had a family history of AF. Overall, 63 very rare AAC variants were identified in 60 of 303 lone AF probands, and 10 of 19 (52.6%) had evidence of co-segregation with AF. Among the 63 lone AF probands who had 45 genes screened, the very rare variant burden was 22%. Compared with the 4300 EA ESP, the proportion of lone AF probands with a very rare AAC variant in CASQ2 and NKX2-5 was increased 3–5-fold (P < .05).
CONCLUSION
No very rare AAC variants were identified in ~80% of lone AF probands. Potential reasons for the lack of very rare AAC variants include a complex pattern of inheritance, variants in as yet unidentified AF genes or in noncoding regions, and environmental factors.
Keywords: Atrial fibrillation arrhythmia, Candidate genes, Family study, Genetic variation, Genetic epidemiology, Proarrhythmia, Rare variants
Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia in clinical practice and is associated with considerable morbidity and mortality.1 Whereas traditional risk factors for AF such as advancing age, coronary artery disease, and congestive heart disease are well described, there is accumulating evidence that genetic factors also play a role in the pathogenesis of AF. Epidemiologic studies have shown that offspring with one parent affected with AF have a three- to five-fold increase in the risk of developing AF.2
Multiple genes and genetic loci affecting AF susceptibility have been identified using positional cloning and candidate gene approaches.3–5 More recently, genome-wide association studies (GWAS) have revealed multiple novel AF susceptibility loci, although these findings only explain a small fraction of the interindividual risk for AF and the majority of common single nucleotide polymorphisms (SNPs) identified by GWAS reside in noncoding regions in which functional characterization remains a major challenge.6–9
Rare variants in candidate genes encoding cardiac ion channels, gap junction proteins, and signaling molecules have previously been reported in isolated AF cases and small kind-reds.6–11 Rare variants that co-segregate with AF in extended families also demonstrate abnormal electrophysiologic properties in vitro.12 Although previous studies have identified common loci in unrelated AF individuals, we still do not have a good estimate of the overall prevalence of rare variants in candidate AF genes among lone AF patients.6–9 Here, we determined the spectrum and prevalence of rare variants in AF candidate genes in a large lone AF cohort and tested for disease co-segregation.
Methods
Vanderbilt AF Registry
Since November 2002, subjects with AF have been prospectively enrolled in the Vanderbilt AF Registry, a clinical and genetic database. Patients were recruited from the Vanderbilt Cardiology and Arrhythmia Clinics, the emergency department, and in-patient services. Individuals older than 18 years with a confirmed diagnosis of AF on an electrocardiogram (ECG), rhythm strip, event recorder, or Holter monitor were included in the Vanderbilt AF Registry. The proband in each family was defined by having lone AF (i.e., early AF onset [≤ 65 years old]) and no predisposing risk factors for AF including hypertension) and being the first family member encountered in the clinic. Familial AF was defined as the presence of AF in one or more first-degree relatives of the proband.
We defined paroxysmal AF as self-terminating AF lasting > 30 seconds and persistent AF as AF lasting > 7 days that was terminated by either pharmacologic intervention or electrical cardioversion. Permanent AF was defined as AF resistant to any type of cardioversion or allowed to continue.
Study cohort
All probands in the Vanderbilt AF Registry of European American (EA) ancestry who presented with lone AF (2002–2012) were identified. Lone AF was defined as AF occurring at age ≤ 65 years with no evidence of structural heart disease, hypertension, or thyroid dysfunction as determined by clinical examination, ECG, echocardiography, and thyroid function tests. For the present study we also gathered echocardiographic information and body mass index (BMI, kg/m2) at the time of enrollment. Information on left atrial (LA; mm) size measurements was available in 254 lone AF probands, left ventricular ejection fraction (LVEF; %) in 270, left ventricular end-diastolic diameter (LVEDD; mm) in 241, left ventricular end-systolic diameter (LVESD; mm) in 228, and BMI in 291.
Screening of candidate genes
Resequencing of 45 candidate genes was performed in lone AF probands (2002–2012) (Online Supplemental Table 1) and in family members who provided consent. Lone AF probands enrolled in the Vanderbilt AF Registry were screened for genes associated with AF at the time of enrollment. Hence, genes that have only recently been implicated with AF (e.g., MYH6) will tend to have a greater proportion of lone AF proband individuals screened compared to genes that were screened earlier in development of the resource (e.g., KCNQ1).8,13
In brief, coding and flanking regions were amplified by polymerase chain reaction using primers designed to obtain fragments of appropriate size for each gene. Polymerase chain reaction–amplified DNA fragments were analyzed using the Reveal Discovery System (based on temperature gradient capillary electrophoresis) to identify aberrant conformers, which were then directly sequenced.
Rare variants and segregation analysis
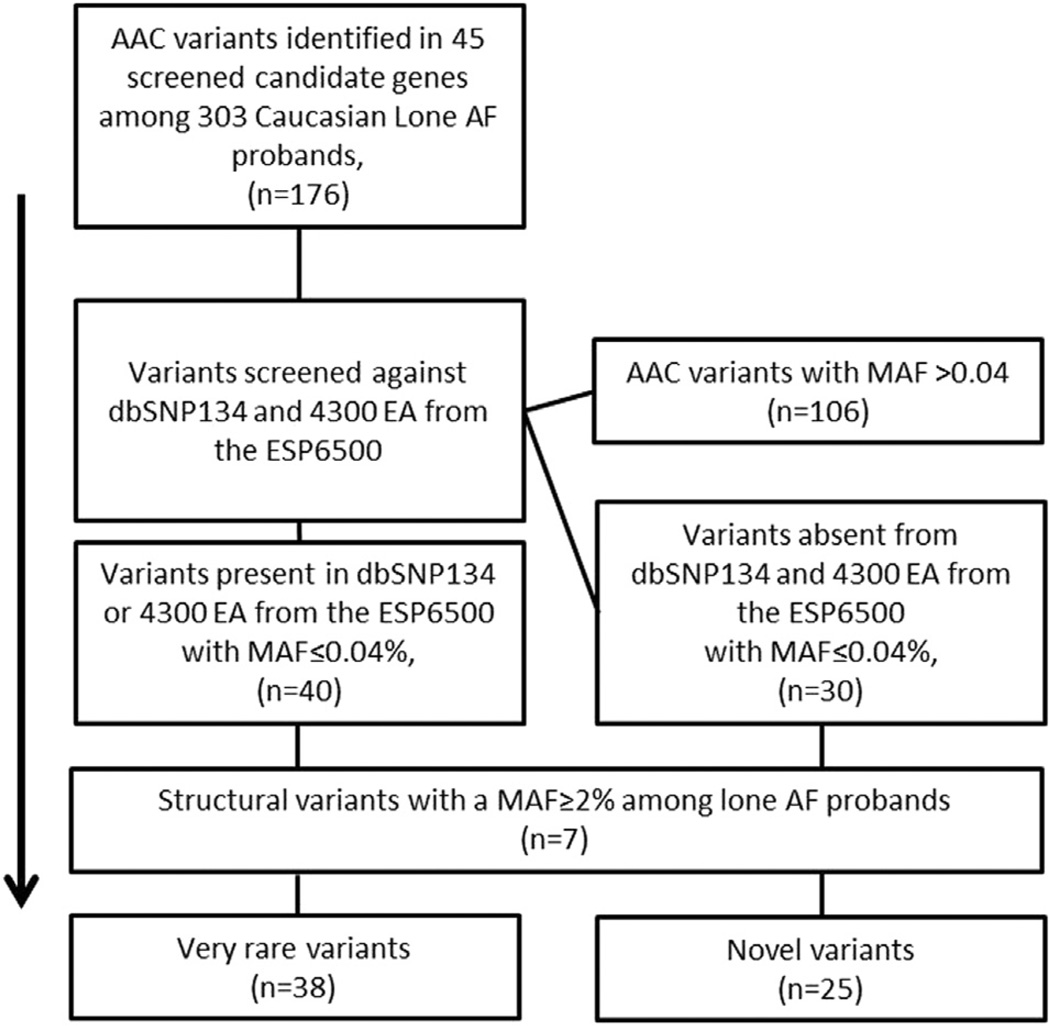
Very rare amino acid coding (AAC) variants (i.e., missense, nonsynonymous, splice site, insertions, and deletions) in screened candidate genes among the lone AF probands were analyzed using the filtering steps shown in Figure 1. First, all AAC variants in screened candidate genes were identified. Second, we screened all of the identified AAC variants against dbSNP134 and the 4300 EAs in the National Heart, Lung, Blood Institute (NHLBI) Exome Sequencing Project (ESP) of 6500 individuals, excluding variants with a reported minor allele frequency (MAF) > 0.04%.14 Third, because structural variants may be less well reported in publically available databases compared to singletons, we calculated the MAF among the lone AF probands excluding insertions and deletions with an internal MAF > 2%. Thus, we assumed that if an insertion or deletion is common among the lone AF probands, it is also likely to be common in the general population even though it may be absent from publically available databases (Figure 1). We assessed conservation and predicted functional effects for all variants with SIFT and PhastCons, GERP, Grantham scores, and PolyPhen2 using the SeattleSeq Genomic Variation Server (http://snp.gs.washington.edu/SeattleSeqAnnotation134/)
Figure 1.
Flow chart identifying rare variants in screened candidate genes. AAC = amino acid coding; AF = atrial fibrillation; ESP6500 = Exome Sequencing Project of 6500 individuals using the reported minor allele frequency (MAF) for 4300 European Americans (EA).
We tested each variant for co-segregation with AF if possible. A variant was designated to have evidence of co-segregation with AF if ≥ 1 first-degree relative was affected with AF and also carried the same very rare variant as the proband.
Variant burden in the ESP6500
Using the reported MAF among 4300 EA in the ESP, we identified all very rare AAC (MAF ≤ 0.04%). Because individual level data are not available in the ESP, we estimated the rare AAC variant burden (i.e., the proportion) for each of the 45 candidate genes screened among the lone AF probands assuming that every individual carried only one very rare AAC variant across the 45 genes.
Statistical analysis
Statistical comparison of variant counts was performed with the Fisher exact test. All statistical analyses were performed using SAS (version 9.2, SAS institute, Cary, NC).
Ethics
The study protocol was approved by the Vanderbilt University Institutional Review Ethics Board, and informed written consent was obtained prior to enrollment.
Results
Lone AF cohort
The study cohort consisted of 303 lone AF probands of EA ancestry (72.6% male), with paroxysmal AF being the most predominant type of AF (73.3%). Median age at diagnosis was 46.0 years (interquartile range [IQR] 33.0–54.0 years), and 35.6% of the lone AF probands had a positive family history of AF. The clinical characteristics for the lone AF study cohort are listed in Table 1.
Table 1.
Clinical characteristics of 303 European American probands with lone atrial fibrillation
| Current age (years) | 58.2 (46.8–67.6) |
| Age of onset (years) | 44.0 (29.0–52.0) |
| Age of diagnosis (years) | 46.0 (33.0–54.0) |
| Male sex (%) | 220 (72.6) |
| Family history of AF (%) | 108 (35.6) |
| AF type (%) | |
| Paroxysmal | 222 (73.3) |
| Chronic | 10 (3.3) |
| Persistent | 71 (23.4) |
| LA size (mm) | 39 (35–44) |
| LVEF (%) | 55 (55–60) |
| LVEDD (mm) | 48 (44–52) |
| LVESD (mm) | 31 (27–35) |
| BMI (kg/m2) | 27.6 (25.1–31.7) |
Continuous variables are presented as median and interquartile range.
AF = atrial fibrillation; BMI = body mass index; LA = left atrium; LVEDD = left ventricular end-diastolic diameter; LVEF = left ventricular ejection fraction, LVESD = left ventricular end-systolic diameter.
Coding variants
We screened for coding variants in 45 AF-associated genes. An average of 186 lone AF probands (range 89–303) were screened for each gene (Online Supplemental Table 1). A subset of 63 of 303 lone AF probands (20.8%) were screened for all 45 genes. Among the 303 lone AF probands, we identified 63 very rare AAC variants, for a minimum frequency of 20.0%. Similarly, among the 63 of 303 lone AF probands who were screened for all 45 candidate genes, the very rare AAC frequency was 22.2%. All very rare AAC variants (n = 63) are listed in Online Supplemental Table 2 along with predicted conservation, functional scores, MAF among the lone AF probands, and reported MAF from the 4300 EAs in the ESP6500. The variant selection process is shown in Figure 1.
Five very rare variants were identified in > 1 lone AF proband, and 26 variants were predicted to be functionally deleterious by SIFT and 9 by PolyPhen2. Insertions and deletions comprised 15.9% (10/63) of all very rare AAC variants (Online Supplemental Table 2). The genes that harbored the most very rare or novel variants were ZFHX3 (n = 15), SCN5A (n = 7), and SCN10A (n = 7) (Online Supplemental Table 2). Variants in genes encoding sodium channels and their subunits (SCN5A, SCN10A, and SCNβ1–3) and potassium channels or their subunits (KCNAB1, KCNIP2, KCNQ1, KCNJ5, KCNJ11, KCNJ12, KCNE1, KCNN3, and KCNA5) accounted for 38.1% (n = 15 and n = 9, respectively) of all very rare AAC variants.
Rare variants segregating with AF
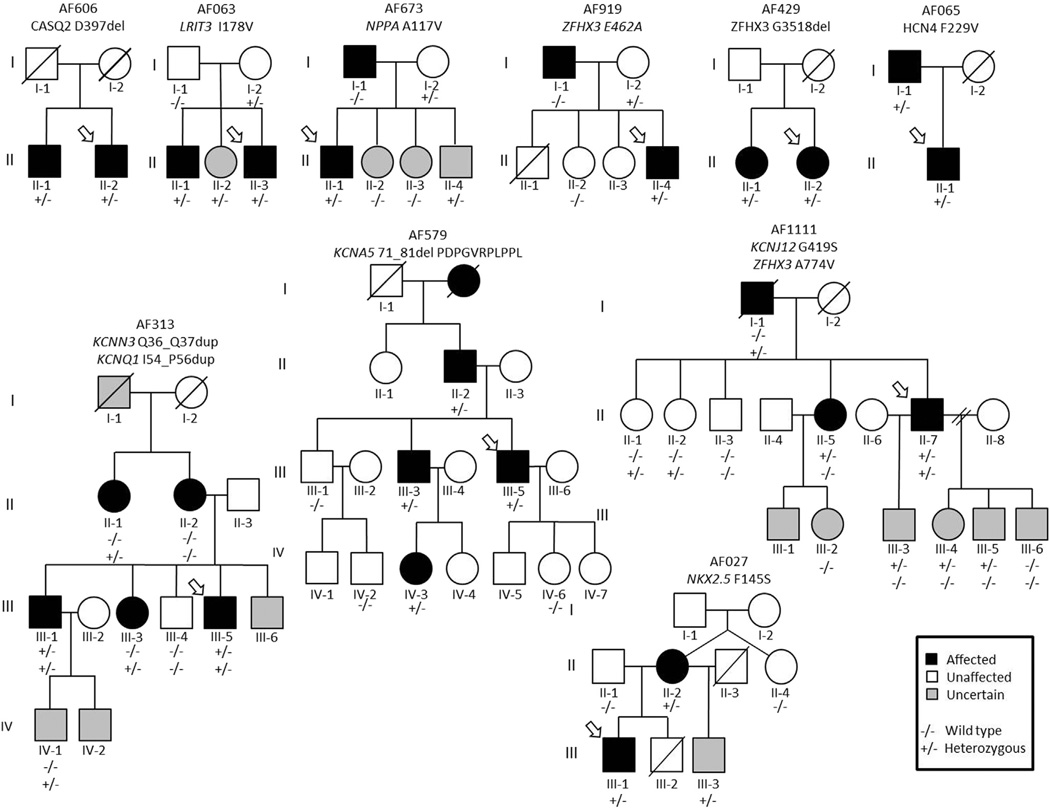
We were able to test 19 of the very rare AAC variants for disease co-segregation within a total of 18 families. Ten of nineteen very rare AAC variants (52.6%) had evidence of co-segregating with AF (i.e., present in ≥ 1 first-degree relative). Figure 2 shows the pedigrees of the families that we were able to test for AF co-segregation. Evidence of a very rare AAC variant co-segregating with AF was found in the following genes: CASQ2, KCNN3, KCNA5, KCNJ12, LRIT3, NKX2-5, KCNQ1, HCN4, and ZFHX3. Due to the lack of affected family members or available DNA in affected family members, we were unable to interrogate 45 very rare AAC variants for evidence of co-segregation with AF.
Figure 2.
Pedigrees of families with evidence of very rare amino acid coding (AAC) variants co-segregating with lone atrial fibrillation (AF). A variant was designated to have evidence of co-segregation with AF if ≥ 1 first-degree relative was affected with AF and also carried the same very rare variant as the proband.
Burden of rare variants
At least one very rare variant was identified in 60 of 303 lone AF probands. Because not all probands were screened for all genes, the minimal estimate is that 20% of lone AF subjects carry such variants. Notably, a subset analysis among the 63 lone AF probands who were screened for all 45 genes yielded a similar rare variant burden: 22.2% (14/63). Among the 303 lone AF probands, very rare AAC variants were identified in 25 of the 45 candidate genes screened and 2.9% of probands (9/303) had ≥ 2 rare AAC variants. We identified 1 of 303 lone AF proband (0.3%) with ≥ 2 very rare AAC variants in the same gene (ZFHX3). The burden of very rare AAC variants for the lone AF probands and the 4300 EA from the ESP6500 is listed in Table 2. Compared with the 4300 EA from the ESP6500, the lone AF probands had a three- to five-fold greater burden of very rare AAC variants in CASQ2 and NKX2-5 (P = .01 and P = .049, respectively). No other genes were associated with greater very rare AAC variant burden, although 15 genes trended toward a greater very rare AAC variant burden among the lone AF probands (Table 2). No very rare AAC variant was identified in 20 of 45 candidate genes among the lone AF probands. The rare AAC variant burden among the 4300 EA from the ESP6500 for these 20 genes is listed in Online Supplemental Table 3. The estimated overall burden of very rare AAC variants in the 45 candidate AF in the 4300 EA in the ESP6500 is 31.9% (1372/4300).
Table 2.
Burden of very rare AAC variants (MAF ≤ 0.04%) in 25 of 45 candidate genes in 303 probands with lone atrial fibrillation and 4300 European Americans from the Exome Sequencing Project
| Gene | Very rare AAC variants among lone AF probands |
% | Very rare AAC variants among 4300 EA in the ESP6500 |
% | Proportional enrichment (lone AF probands/EA ESP6500) |
P value |
|---|---|---|---|---|---|---|
| ANK2 | 1/148 | (0.7) | 159/4300 | (3.7) | 0.2 | .07 |
| CACNA1C | 1/218 | (0.5) | 56/4300 | (1.3) | 0.4 | .53 |
| CACNA2D2 | 2/217 | (0.9) | 47/4300 | (1.1) | 0.8 | 1 |
| CASQ2 | 4/148 | (2.7) | 22/4300 | (0.5) | 5.3 | .01 |
| GSK3B | 1/148 | (0.7) | 10/4300 | (0.2) | 2.9 | .31 |
| HCN4 | 3/303 | (1.0) | 36/4300 | (0.8) | 1.2 | .52 |
| KCNA5 | 1/130 | (0.8) | 32/4300 | (0.7) | 1.0 | 1 |
| KCNAB1 | 1/148 | (0.7) | 25/4300 | (0.6) | 1.2 | .59 |
| KCNE1 | 1/106 | (0.9) | 10/4300 | (0.2) | 4.1 | .24 |
| KCNIP2 | 1/148 | (0.7) | 10/4300 | (0.2) | 2.9 | .31 |
| KCNJ12 | 1/218 | (0.5) | 31/4300 | (0.7) | 0.6 | 1 |
| KCNJ4 | 1/148 | (0.7) | 7/4300 | (0.2) | 4.2 | .24 |
| KCNJ5 | 1/148 | (0.7) | 28/4300 | (0.7) | 1.0 | 1 |
| KCNN3 | 1/241 | (0.4) | 18/4300 | (0.4) | 1.0 | 1 |
| KCNQ1 | 1/89 | (1.1) | 31/4300 | (0.7) | 1.6 | .14 |
| LRIT3 | 3/241 | (1.2) | 30/4300 | (0.7) | 1.8 | .09 |
| MYH6 | 5/303 | (1.7) | 106/4300 | (2.5) | 0.7 | .55 |
| NKX2-5 | 1/241 | (0.4) | 6/4300 | (0.1) | 3.0 | .049 |
| NPPA | 1/172 | (0.6) | 13/4300 | (0.3) | 1.9 | .42 |
| NPPB | 1/218 | (0.5) | 10/4300 | (0.2) | 2.0 | .42 |
| SCN10A | 6/264 | (2.3) | 125/4300 | (2.9) | 0.8 | .71 |
| SCN2B | 1/202 | (0.5) | 21/4300 | (0.5) | 1.0 | 1 |
| SCN5A | 7/303 | (2.3) | 80/4300 | (1.9) | 1.2 | .38 |
| TTR | 2/264 | (0.8) | 6/4300 | (0.1) | 5.4 | .07 |
| ZFHX3 | 19/241 | (7.9) | 221/4300 | (5.1) | 1.5 | .15 |
In columns 2 and 4, x/y denotes very rare AAC variant carrier/number screened.
AAC = amino acid coding; AF = atrial fibrillation; EA = European American; ESP6500 = Exome Sequencing Project of 6500; MAF = minor allele frequency.
Discussion
In this study, we resequenced the coding regions of 45 candidate AF genes encoding cardiac ion channels, gap junction proteins, cytoskeletal proteins, and signaling molecules associated with AF in order to estimate the prevalence of very rare AAC variants in a large cohort of lone AF probands. At least 20% of the lone AF probands carried a very rare AAC variant in a candidate AF gene of which ~ 53% had evidence of co-segregating with AF. However, the majority of lone AF probands did not carry very rare AAC variant in the screened candidate AF genes, which underscores the complexity of lone AF and suggests that variants in important AF genes yet to be identified, epigenetic or environmental factors, or variants outside the screened regions contribute to the risk of lone AF.
The estimated burden of very rare AAC variants in the present study was found to be ~ 20% among 303 lone AF probands. However, because the lone AF probands were only screened for the genes associated with AF at the time of enrollment, not every lone AF proband was not screened for all candidate genes; this implies that the true proportion of lone AF probands with a very rare AAC variant is likely to exceed 20%. In agreement with the latter notion, the rare AAC variant burden among the 63 lone AF probands who had all 45 candidate genes screened was 22.2%. In contrast, the estimated burden from the ESP is likely to be considerably lower than the estimated 31.9% because some individuals will have >1 very rare AAC variant; indeed, 2.9% of the lone AF proband cohort had ≥ 2 very rare AAC variants in the 45 candidate genes, but only 0.3% lone AF probands had ≥ 2 variants in the same gene. Hence, a comparison of overall burden is not as meaningful as a gene-by-gene comparison of variant burden between the lone AF probands and the EA in the ESP6500. Although the ESP6500 is a very useful reference for assessing the MAF of specific variants in EA and African Americans, the lack of individual level data does limit its usability (e.g., in aggregate rare variant or burden analysis). Although the 1000 Genomes Project does provide individual level data, population stratification and the small sample sizes are of concern, especially in studies of rare variants that may be subpopulation specific; the 1000 Genomes Project comprises 14 distinct populations (range 14–100 individuals).15
The lone AF probands were significantly more burdened by very rare AAC variants in two genes: CASQ2 and NKX2-5. CASQ2 encodes a calcium buffering protein located in the sarcoplasmic reticulum, which forms a complex with triadin and junctin that is associated with the cardiac ryanodine receptor 2 (RyR2).16 Disruption of calcium homeostasis is thought to be a key a contributor to generating the AF-prone substrate.17 Of the three very rare AAC variants identified in CASQ2, we were able to test for and establish co-segregation with one (AF606 family; Figure 2). A three-fold increase in burden of very rare AAC variants in NKX2-5 was identified among the lone AF probands compared to the EAs in the ESP6500 (0.4% vs 0.1%, P = .049). NKX2-5 encodes a homeodomain-containing transcription factor that is essential for normal cardiovascular morphogenesis and has previously been linked with atrial septal defects and AV nodal conduction disturbances.18 Interestingly, the NKX2-5 F145S variant identified in the present study has recently been implicated in autosomal dominant familial AF, with functional studies demonstrating the F145S variant to be associated with significantly decreased transcriptional activity compared to wild type.19 In the AF027 family, three individuals (II-2, III-1, and III-3) carried F145S mutation, in which II-2 and III-1 were affected with AF. One brother (III-3) has an atrial septal defect but has not yet developed AF. Although the F145S carrier status of one deceased brother (III-2) is unknown, it is noteworthy that he was born with tetralogy of Fallot and underwent heart transplantation (Figure 2).
Very rare AAC variants that co-segregate with disease are likely to be of importance,3,12,13 although caution should be exercised before any variant is defined as being causative without supporting segregation and/or functional studies. In the present study, we tested 19 very rare AAC variants for disease co-segregation and found that ~ 53% had evidence of co-segregating with AF. Conversely, ~ 47% of the very rare AAC variants did not. These may be false positives, or it is possible that low-frequency variants still affect disease risk without demonstrating a clear mendelian inheritance pattern.20 For example, variants may not segregate with AF if a second “hit” is required before AF is manifest. This second hit may be a clinical risk factor such as hypertension or diabetes. Alternatively, other AF susceptibility alleles, common or rare, may modulate the penetrance of AF.21
Cardiac potassium channel genes have previously been linked to familial AF13,22 but rarely cause typical AF.23 Similar conclusions were drawn in another study that showed that variants in KCNQ1 are an uncommon cause of lone AF.24 In agreement with the latter study, we identified (and have previously reported) only one gain-of-function variant in KCNQ1, a nine-base-pair insertion that showed augmented potassium currents in IKs compared to wild type.12 However, very rare AAC variants in genes encoding potassium channels or their subunits (KCNAB1, KCNIP2, KCNQ1, KCNJ5, KCNJ11, KCNJ12, KCNE1, KCNN3, and KCNA5) accounted for 14.2% (9/63) of all very rare AAC variants, which suggests that variants in genes encoding potassium channels and their subunits could be important modulators of lone AF disease susceptibility (Table 2). Moreover, four of four very rare AAC variants in genes encoding potassium channels or their subunits had evidence of AF co-segregation (Figure 2). Thus, our findings support the idea that families with AF have an excess of rare functional potassium channel gene variants that may contribute to an atrial arrhythmogenic substrate.25
The gene harboring the most very rare AAC variants was ZFHX3 (zinc finger homeobox 3). Although the lone AF probands were 1.5 times more burdened by rare variants in ZFHX3 than the 4300 EA in the ESP6500, this finding was not statistically significant (Table 2). ZFHX3 encodes a transcription factor that was originally identified as being a regulator of α-fetoprotein expression.26 Common noncoding variants in ZFHX3 have previously been associated with typical AF7–9 but were not associated with early disease onset in a Danish cohort of lone AF patients.27 Given that common variants in ZFHX3 have been associated with AF and the largest number of very rare AAC variants was identified in this gene, it is possible that both common and rare variants in ZFHX3 modulate AF susceptibility.21
Eighty percent of lone AF probands did not carry a very rare AAC variant in any of the interrogated candidate AF genes. This finding lends support to the hypothesis that the genetic architecture of AF includes multiple variants, and these can be rare or common.28 In particular, the recent study by Ritchie et al21 found that common AF-associated 4q25 SNPs modified the clinical expression of latent cardiac ion channel and signaling molecule gene mutations associated with familial AF. There are other potential explanations for our finding that only a minority of subjects carried very rare AAC variants in the interrogated candidate AF genes. First, even though we resequenced high-priority candidate AF genes, it is likely that there are still many genes involved in AF pathophysiology that have not yet been identified. Second, given that the vast majority of common AF susceptibility polymorphisms identified by GWAS have been in intronic regions, it is possible that by e-sequencing coding regions we may have missed potentially important noncoding variants.6–9 Third, by focusing on very rare AAC variants and ignoring common and low-frequency variants with MAF > 0.04%, we may have missed variants, or combinations of variants, with small or intermediate effects.20 Fourth, identifying the underlying genetic basis of AF is complicated because AF can, in many patients, be asymptomatic. Furthermore, the high prevalence of AF in the general population and the likely modifying effects of environmental factors, as well as the late age of onset of the arrhythmia, make the assignment of the correct clinical phenotype a challenge.
Other limitations to the present study warrant consideration. We acknowledge that by not screening every lone AF proband for each gene, our estimation of very rare AAC variant burden likely underestimates the true variant burden among lone AF probands, although our subset analysis of the 63 lone AF probands who had all candidate genes screened (n = 63) yielded a similar estimate of the very rare AAC variant burden. Similarly, by assuming that each very rare AAC variant among the 4300 EA in the ESP6500 is unique to one person, our estimation of the variant burden in the EA from the ESP may be inflated. However, by using an MAF ≤ 0.04% cutoff, we reduced the likelihood of each individual contributing with multiple variants, particularly within genes (0.3% of the lone AF probands had ≥ 2 very rare AAC variants within the same gene). Given these limitations, it is important to note that we consider a gene-to-gene comparison of rare variant AAC burden to be more meaningful than overall comparison of all candidate genes. The latter is highlighted by the fact that evaluating very rare AAC variant burden in all ~ 25,000 genes in the human genome undoubtedly would increase the proportion to 100%, although it is important to note that the genes included in the present study are based on previous findings and known associations (i.e., the present study is hypothesis driven). We acknowledge that most of the AF kindreds were small, which limited our ability to perform formal segregation analysis and calculate LOD scores because clinical data and/or DNA were not available from some family members. However, studying complex phenotypes such as AF is challenging because families tend to be small due to variable age-dependent penetrance and the paroxysmal nature and variable symptoms associated with AF, which can make assignment of the clinical phenotype difficult given the high prevalence of AF in the general population. In addition, rare variants may not segregate with AF in the absence of a “second hit,” which may be a modifier common AF susceptibility SNP or a clinical risk factor.21 Although the segregation analysis performed in the present study may be indicative of a causative variant (e.g., the NKX2.5 F145S variant in the AF027 family demonstrated imperfect segregation, although a previous study demonstrated that this variant is functionally important in familial AF),19 it is important to note that the chance of first-degree relative sharing a given variant is 50%.
Conclusion
This study evaluated the prevalence of very rare AAC variants in candidate AF genes using a large lone AF cohort. At least one in five lone AF probands carried ≥ 1 very rare AAC variant in a candidate AF gene, of which 53% had evidence of AF co-segregation. The fact that very rare AAC variants in candidate genes were absent in ~ 80% of lone AF probands is suggestive of a complex AF inheritance pattern, unidentified candidate AF genes, or epigenetic or environmental factors. Collectively, our findings represent important progress toward molecular phenotyping of lone AF.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants U19 HL65962 and R01 HL092217, and an American Heart Association Established Investigator Award (0940116N). Dr. Weeke was funded by an unrestricted research grant from the Tryg Foundation (J.nr. 7343-09, TrygFonden, Denmark).
ABBREVIATIONS
- AAC
amino acid coding
- AF
atrial fibrillation
- BMI
body mass index
- ECG
electrocardiogram
- ESP
Exome Sequencing Project
- GWAS
genome-wide association study
- IQR
interquartile range
- LVEDD
left ventricular end-diastolic diameter
- LVEF
left ventricular ejection fraction
- LVESD
left ventricular end-systolic diameter
- MAF
minor allele frequency
- SNP
single nucleotide polymorphism
Footnotes
Appendix
Supplementary data
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.hrthm.2013.10.025.
References
- 1.Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98:946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 2.Arnar DO, Thorvaldsson S, Manolio TA, et al. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–712. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 3.Brugada R, Tapscott T, Czernuszewicz GZ, et al. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–911. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 4.Ellinor PT, Shin JT, Moore RK, Yoerger DM, MacRae CA. Locus for atrial fibrillation maps to chromosome 6q14-6. Circulation. 2003;107:2880–2883. doi: 10.1161/01.CIR.0000077910.80718.49. [DOI] [PubMed] [Google Scholar]
- 5.Volders PG, Zhu Q, Timmermans C, et al. Mapping a novel locus for familial atrial fibrillation on chromosome 10p11-q21. Heart Rhythm. 2007;4:469–475. doi: 10.1016/j.hrthm.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts JD, Gollob MH. Impact of genetic discoveries on the classification of lone atrial fibrillation. J Am Coll Cardiol. 2010;55:705–712. doi: 10.1016/j.jacc.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Darbar D. Genetics of atrial fibrillation: rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm. 2008;5:483–486. doi: 10.1016/j.hrthm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraham RL, Yang T, Blair M, Roden DM, Darbar D. Augmented potassium current is a shared phenotype for two genetic defects associated with familial atrial fibrillation. J Mol Cell Cardiol. 2010;48:181–190. doi: 10.1016/j.yjmcc.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 14.Norton N, Robertson PD, Rieder MJ, et al. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet. 2012;5:167–174. doi: 10.1161/CIRCGENETICS.111.961805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knollmann BC. New roles of calsequestrin and triadin in cardiac muscle. J Physiol. 2009;587:3081–3087. doi: 10.1113/jphysiol.2009.172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrev D, Nattel S. Calcium handling abnormalities in atrial fibrillation as a target for innovative therapeutics. J Cardiovasc Pharmacol. 2008;52:293–299. doi: 10.1097/FJC.0b013e318171924d. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez-Roelens I, De Roy L, Ovaert C, et al. A novel CSX/NKX2-5 mutation causes autosomal-dominant AV block: are atrial fibrillation and syncopes part of the phenotype? Eur J Hum Genet. 2006;14:1313–1316. doi: 10.1038/sj.ejhg.5201702. [DOI] [PubMed] [Google Scholar]
- 19.Huang RT, Xue S, Xu YJ, Zhou M, Yang YQ. A novel NKX2.5 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2013;31:1119–1126. doi: 10.3892/ijmm.2013.1316. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie MD, Rowan S, Kucera G, et al. Chromosome 4q25 variants are genetic modifiers of rare ion channel mutations associated with familial atrial fibrillation. J Am Coll Cardiol. 2012;60:1173–1181. doi: 10.1016/j.jacc.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson TM, Alekseev AE, Liu XK, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 23.Ellinor PT, Petrov-Kondratov VI, Zakharova E, Nam EG, MacRae CA. Potassium channel gene mutations rarely cause atrial fibrillation. BMC Med Genet. 2006;7:70. doi: 10.1186/1471-2350-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellinor PT, Moore RK, Patton KK, Ruskin JN, Pollak MR, Macrae CA. Mutations in the long QT gene, KCNQ1, are an uncommon cause of atrial fibrillation. Heart. 2004;90:1487–1488. doi: 10.1136/hrt.2003.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann SA, Otway R, Guo G, et al. Epistatic effects of potassium channel variation on cardiac repolarization and atrial fibrillation risk. J Am Coll Cardiol. 2012;59:1017–1025. doi: 10.1016/j.jacc.2011.11.039. [DOI] [PubMed] [Google Scholar]
- 26.Burgner D, Davila S, Breunis WB, et al. A genome-wide association study identifies novel and functionally related susceptibility Loci for Kawasaki disease. PLoS Genet. 2009;5:e1000319. doi: 10.1371/journal.pgen.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olesen MS, Holst AG, Jabbari J, et al. Genetic loci on chromosomes 4q25, 7p31, and 12p12 are associated with onset of lone atrial fibrillation before the age of 40 years. Can J Cardiol. 2012;28:191–195. doi: 10.1016/j.cjca.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.