Abstract
The assembly of nanomaterials using DNA can produce complex nanostructures, but the biological applications of these structures remain unexplored. Here we describe the use of DNA to control the biological delivery and elimination of inorganic nanoparticles by organizing them into colloidal superstructures. The individual nanoparticles serve as building blocks, whose size, surface chemistry, and assembly architecture dictate overall superstructure design. These superstructures interact with cells and tissues as a function of their design, but subsequently degrade into building blocks that can escape biological sequestration. We demonstrate that this strategy reduces nanoparticle retention by macrophages and improves their in vivo tumour accumulation and whole-body elimination. Superstructures can be further functionalized to carry and protect imaging or therapeutic agents against enzymatic degradation. These results suggest a new strategy to engineer nanostructure interactions with biological systems and highlight new directions in the design of biodegradable and multifunctional nanomedicine.
Inorganic nanoparticles can be synthesized in the 1–100nm size range with precise shapes, surface chemistries, and physical properties. This engineering flexibility has enabled their design as novel therapeutics, contrast agents, and integrated systems for the diagnosis and treatment of diseases1–4. To optimally deliver these nanoparticles to their biological targets with low toxicity, recent studies have focused on understanding the effects of nanoparticle size, shape, and surface chemistry – known as the physicochemical properties – on interactions with cells and tissues5–8. While several formulations have been shown to effectively target diseased tissues (e.g. tumours)9–11, these designs diverge from those required for mitigating toxicity. Tumour targeting nanoparticles require sufficiently large sizes to reduce clearance and improve retention within tumours12,13, yet such inorganic nanoparticles will remain in the body for a long time because they do not biodegrade14. This in vivo persistence has raised concerns of chronic toxicity due to the possibility for inorganic nanoparticles to aggregate15,16, generate harmful metabolites17,18, and redistribute to vital organs within the body19–21. Few studies have demonstrated how the physicochemical properties of inorganic nanoparticles can be engineered to mediate both delivery and elimination22. This design bottleneck will stall the clinical translation of these nanotechnologies. Here we explore the use of DNA to organize sub-6nm inorganic nanoparticles, a size that can be cleared through the kidneys, into larger superstructures to mediate their biological delivery and elimination. This strategy combines the engineering flexibility of inorganic nanoparticles with the biodegradability of organic molecules, which should open new avenues to rationally engineer the interactions of inorganic naonparticles with complex biological systems.
Assembly of nanoparticle superstructures using DNA
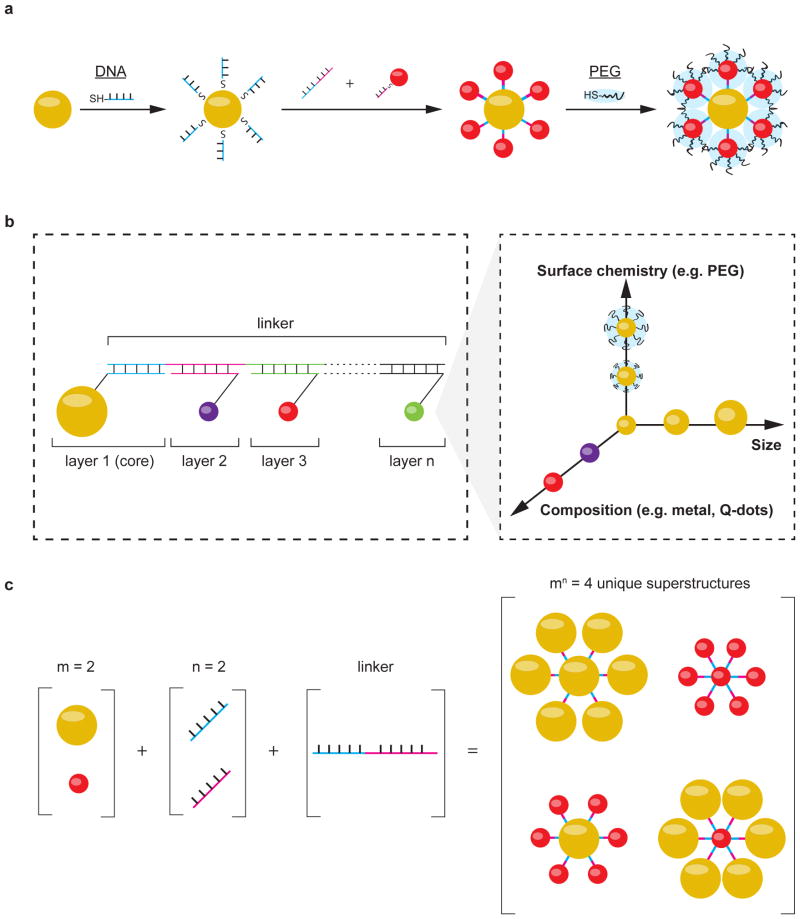
Figure 1a illustrates the principles of using DNA-nanoparticle assembly to engineer colloidal superstructures with different physicochemical properties. First, we used metal-thiol or streptavidin-biotin chemistry to functionalize inorganic nanoparticles with single stranded DNA. We then mixed DNA-functionalized nanoparticles together with linker DNA strands containing complementary sequences to initiate their assembly into colloidal superstructures. The architecture of the assembled superstructure was controlled by using both nanoparticle geometry and DNA grafting density, the latter determined the number of connections each nanoparticle makes with other building blocks. Finally, the outer surface of the resulting superstructure was coated with additional ligands to present the appropriate surface chemistries for interfacing with biological systems. This was achieved by assembling nanoparticles with low DNA grafting densities on the outer layer of the superstructure, such that their unsaturated surfaces provide binding sites for ligand attachment.
Figure 1. Design of nanoparticle superstructures using DNA assembly.
a, Individual nanoparticles (yellow and red spheres) were coated with thiolated, single stranded DNA, and then assembled using linker DNAs containing complementary sequence regions. Nanoparticles located on the surface of superstructures were coated with additional ligands (e.g. PEG, illustrated as blue clouds) to control superstructure interactions with cells and tissues. b, This study focused on the design of “core-satellite” superstructures, in which a central nanoparticle (i.e. core) is surrounded by one or multiple layers of satellite nanoparticles (i.e. layers 2 to n). Each layer is encoded by a unique DNA sequence and can contain nanoparticles of different size, surface chemistry, or composition (conceptually shown in the inset). The combination of these building blocks determines the overall dimension and functionality of the superstructure. c, Large numbers of unique superstructures can be generated and screened by combining different building blocks. An example is shown here, where combining 2 different nanoparticle designs (m) with 2 unique DNA sequences (n) gives 4 possible unique superstructures.
Here, we used a “core-satellite” architecture to build DNA-assembled superstructures where one or multiple layers of satellite nanoparticles surround a central core nanoparticle23,24 (Fig. 1b). Each layer of the core-satellite was encoded by a unique DNA sequence, such that nanoparticles grafted with the specific DNA sequence inserted into the corresponding layer. A linker DNA containing complementary regions to every layer joined the nanoparticles together. Each layer of nanoparticles can be designed with a different composition, size, or surface chemistry (Fig. 1b). This modularity allowed us to construct superstructures with controlled dimensions and multiple functionalities from relatively simple building blocks. The permutations amongst nanoparticle designs and DNA sequences can also quickly generate superstructures with distinct physiochemical properties. Figure 1c shows the use of 2 unique nanoparticle building blocks and 2 DNA sequences (e.g. 2 layers) to give 22=4 unique core-satellite superstructures. The total number of unique superstructures increases exponentially with increasing number of core-satellite layers and nanoparticle designs. For example, the combination of 10 nanoparticle designs in a 3-layer (e.g. 3 DNA sequences) core-satellite would give 310= 59049 unique superstructures; the use of “n-layer” core-satellites with m nanoparticle designs gives nm unique superstructures, each may interact differently with cells and tissues. This diversity of superstructure candidates will allow us to identify designs with high biological stability, low non-specific biological interactions, and favourable pharmacokinetics for disease targeting.
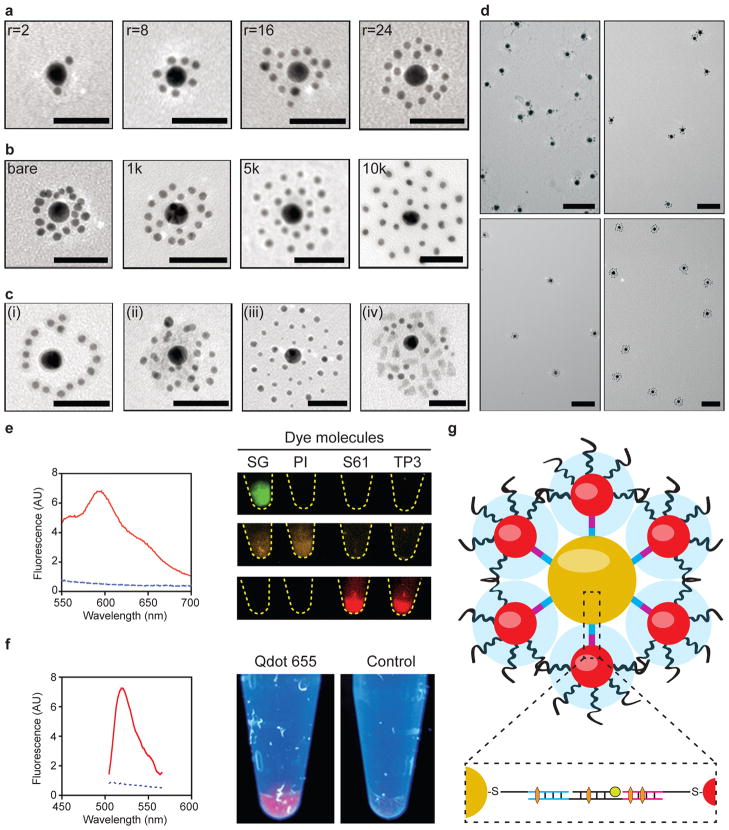
Based on these principles, we generated a sub-library of colloidal superstructures with different hydrodynamic sizes and surface chemistries to study the impact of their design on molecular and cellular interactions. Figure 2a–c shows the simplest 2-layer core-satellite structures that were synthesized for these experiments. First, we synthesized 13nm gold nanoparticles and used them as the core by grafting them with thiolated core oligonucleotides at a density of ~0.12DNA/nm2. This density corresponded to a valency of 80 to 90 DNA strands per particle, allowing them to make a large number of connections with the satellites. DNA grafting density was controlled by varying the DNA-to-nanoparticle grafting stoichiometry and quantified by using a fluorescence depletion assay (Supplementary Fig. 1). We then synthesized 3 and 5nm gold nanoparticles as the satellites by coating them with the satellite oligonucleotide sequence at a density of ~0.05 DNA/nm2. This density corresponded to 2 to 3 DNA strands per particle, which was sufficient to stabilize the satellites against aggregation but minimized their probability of cross-linking superstructures into macroscopic aggregates. We note that this low DNA coverage also left the rest of the satellite nanoparticle surface available for further ligand conjugation. A linker DNA containing complementary regions to both the core and satellite sequences was used to join these nanoparticles together. To assemble core-satellites, we first annealed a stoichiometric amount of linker DNAs with the core nanoparticles in a hybridization buffer that was first heated to 60°C for 10 minutes and then kept at 37°C for 2 hours. Linker-hybridized core nanoparticles were then purified by centrifugation and subsequently combined with satellite nanoparticles under similar hybridization conditions. We used a 100X molar excess of satellite nanoparticles per core nanoparticle to further eliminate the probability of superstructure cross-linking. Following core-satellite assembly, colloidal superstructures were back-filled with the polymer poly(ethylene glycol) (PEG) to improve their biological stability and reduce non-specific interactions with biomolecules and cells25. We used 4 different linker stoichiometries (2, 8, 16, 24 linkers per core, see characterization in Supplementary Fig. 2), which generated superstructures with different satellite-to-core ratios (Fig. 2A). We used 3 different lengths of PEG (1, 5, 10kDa) to control overall superstructure surface chemistry and morphology (Fig. 2b). We also generated 3-layer core-satellite structures in which a third DNA sequence (satellite2) hybridizes to an internal region of the linker (see schematic in Supplementary Fig. 3 and images in Fig. 2c–i). By grafting this DNA sequence onto other sets of nanoparticles, superstructures with additional satellite layers could be constructed (Fig. 2c ii–iv and Supplementary Fig. 4). Varying these parameters generated a diverse set of superstructures with hydrodynamic sizes ranging from 50–150nm (Supplementary Fig. 5). Transmission electron microscopy (TEM, Fig. 2d) and UV-Vis absorbance characterizations (Supplementary Fig. 6) demonstrated that these superstructures were monodisperse and colloidally stable in saline.
Figure 2. Characterization of core-satellite superstructures.
TEM characterization of 2-layer core-satellites as a function of a, satellite-to-core ratio (r=2, 8, 16 and 24) and b, satellite PEG length (MW=bare, 1, 5 and 10kDa). c, 3-layered core-satellites were synthesized by introducing a third DNA sequence (Satellite2) which inserts into the linker DNA. (i) Attaching this sequence to the linker increased core-satellite separation distance when viewed under TEM. Other nanomaterials grafted with this sequence were used to generate various 3-layer superstructures (ii = 5nm gold nanoparticles, iii = 3nm + 5nm gold nanoparticles, and iv = quantum dots + 3nm gold nanoparticles). Scale bars = 50nm. d, Low-magnification TEM images of the core-satellites shown in a, showing their colloidal stability and monodispersity in saline. Scale bars = 100nm. e, Left: fluorescence spectra of core-satellites with (red solid line) and without (blue dashed line) doxorubicin incorporation; right: fluorescence images of vials containing superstructures labelled with different colours of DNA-binding dyes (SG=Sybr Gold, PI=Propidium Iodide, S61=Syto61, TP3=TO-PRO-3) captured using 3 filter sets (top: 460/535nm, middle: 560/600nm, bottom: 650/700nm). f, Left: fluorescence spectra of core-satellite superstructures with (red solid line) and without (blue dashed line) FAM incorporation; right: vials of superstructures labelled with or without quantum dot incorporation under UV excitation. g, Cross-sectional view of a core-satellite. Inset shows the positioning of payloads encapsulated either via intercalating (orange hexagon) or hybridizing (green circle) to the DNA strands within the superstructure.
A key question regarding the biological application of colloidal superstructures is whether they can carry and protect pharmaceuticals against biological degradation. We found that therapeutic or imaging agents such as doxorubicin and several fluorescent molecules can be incorporated into superstructures through DNA intercalation or groove binding (Fig. 2e). Incorporation efficiency was dependent on linker sequence, improving with increasing number of TCG repeats which is a known binding site for doxorubicin26. Other agents such as quantum dots and fluorescein amidite (FAM), which do not intercalate or bind DNA directly, could be incorporated within superstructures as hybridized DNA conjugates (Fig. 2f). An advantage of using assembly to store these agents is that they are embedded within the superstructure and not exposed on the nanoparticle surface (Fig. 2g). By selecting the appropriate core and satellite building blocks, superstructures enhanced DNA resistance against nuclease and serum degradation by up to 5-fold relative to non-assembled nanoparticles (Supplementary Fig. 7). This improvement in DNA stability effectively protected the superstructures and its payloads from disintegrating in biological solutions. These results provide the first example of using assembly architecture to mediate payload stability, and highlight a novel strategy to build integrated platforms that carry multiple functionalities.
Design-dependent uptake of nanoparticle superstructures
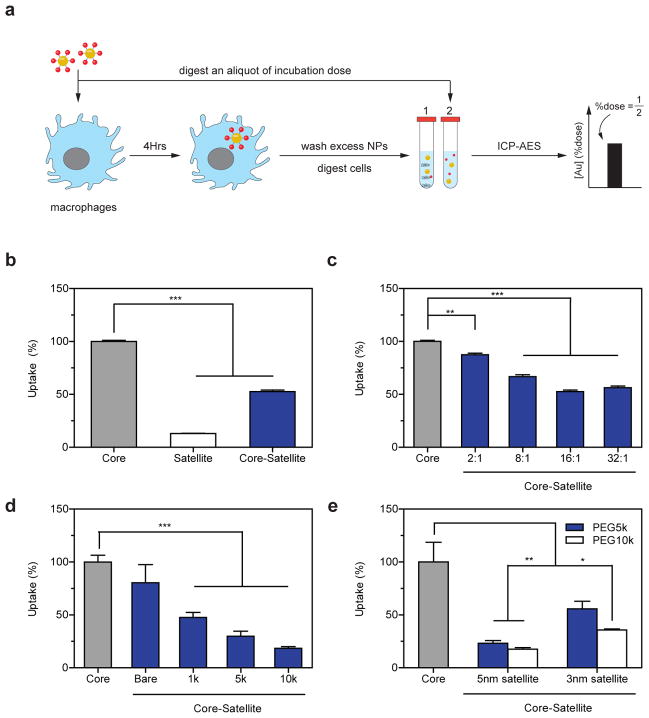
The potential application of colloidal superstructures as delivery platforms motivated us to further investigate their interactions with cells. We selected J774A.1 murine macrophages as a model cell system because macrophages sequester the majority of in vivo administered nanoparticles27. Sequestration of nanoparticles by macrophages not only limits the dose that is available to accumulate at diseased sites but is further associated with immune-toxicity28,29. The ability to control nanoparticle interactions with macrophages could improve disease-specific delivery and reduce toxicity. We measured macrophage uptake by incubating J774A.1 cells in culture media containing gold nanoparticles for 4 hours and then analysing the total cellular gold content using inductively coupled plasma atomic emission spectroscopy (ICP-AES, Fig. 3a). To assess the impact of nanoparticle design and assembly on uptake, we first exposed macrophages separately with 13nm core nanoparticles, 5nm satellite nanoparticles coated with 1kDa PEG, as well as superstructures assembled using these two components. Figure 3b shows that macrophages sequestered 13nm core nanoparticles 7 times more effectively than 5nm satellite nanoparticles coated with PEG 1kDa, consistent with previous findings that macrophage uptake correlates with nanomaterial size and surface charge30. Interestingly, the core-satellite superstructure was 2.5 times larger than its core component but resulted in 2-fold lower uptake into macrophages, suggesting that the superstructure displayed a different surface chemistry which inhibited its uptake. These results also motivated us to systematically characterize the impact of building block design and their assembly architecture on macrophage uptake. We used serum-free culture media for these experiments because our results (Supplementary Fig. 8) and a previous study31 have shown that DNA coated nanomaterials are taken up by cells through direct interactions with receptors (e.g., scavenger receptors) on the cell surface rather than through interactions with serum proteins adsorbed on the nanomaterial surface. Here we observed a monotonic decrease in superstructure uptake by macrophages as a function of satellite-to-core ratio (Fig. 3c), suggesting that satellites inhibited macrophages from interacting with the core. This hypothesis is further supported by the different dose-responses between these nanomaterials; macrophages sequestered DNA coated core nanoparticles in a dose-independent manner suggesting that cells take up such nanoparticles efficiently. In contrast, core-satellite structures exhibited a dose-dependent decrease in cell uptake similar to PEGylated nanomaterials (Supplementary Fig. 9). The length of PEG on the satellites also impacted macrophage uptake, where an increase from PEG 1 to 10kDa reduced macrophage uptake by an additional 30% (Fig. 3d). Interestingly, 5nm nanoparticles were ~2 times more effective than 3nm nanoparticles at mitigating core-satellite uptake by macrophages, implying they provide a denser PEG surface chemistry (Fig. 3e). Taken together, the optimal superstructure design reduced macrophage uptake by 80% relative to the core nanoparticle, despite being 3 times larger in size. This was achieved by using 5nm nanoparticles coated with PEG 10kDa as satellites at a saturating satellite-to-core ratio. Other parameters such as linker length (Supplementary Fig. 10) had relatively little effect on uptake. These results highlight the central role of satellite design and assembly stoichiometry in dictating superstructure interactions with cells. Nanoparticle assembly can reduce macrophage uptake by: 1) burying DNA within the superstructure to decrease their accessibility from cellular interactions, and 2) using nanoparticles as scaffolds to increase the density of PEG coverage above the DNAs.
Figure 3. Macrophage uptake of superstructures as a function of design.
a, Workflow for assessing superstructure uptake into J774A.1 macrophages (blue cells) by ICP-AES. Relative cell uptake efficiency was determined by measuring the amount of gold in the cells (tube 1) relative to the total dose of gold administered (tube 2). b, Relative uptake efficiency of 13nm core nanoparticles, 5nm-PEG1kDa satellite nanoparticles, and their corresponding superstructures by macrophages. c, Macrophage uptake of superstructures as a function of satellite-to-core ratios. d, Macrophage uptake of superstructures with varying PEG lengths (satellite-to-core ratio kept constant at 24). e, Effect of satellite size on superstructure uptake by macrophages. Error bars represent S.E.M. from at least three independent replicates; *P<0.05, **P<0.01, ***P<0.001.
Cellular degradation and exocytosis
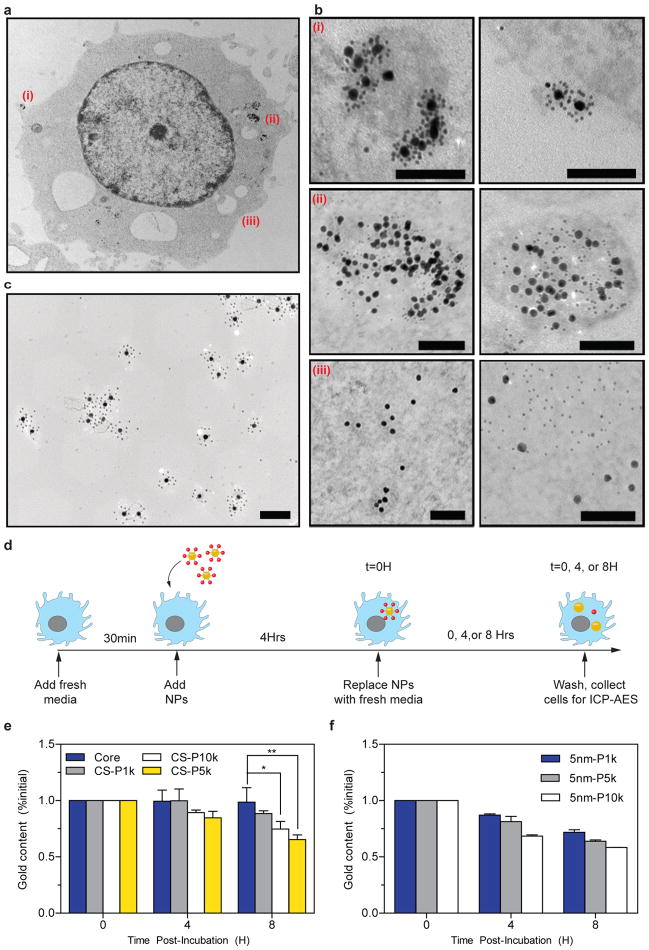
Nanomaterials internalized by macrophages are sequestered within the cells if they are not biodegraded. This contributes to the persistence of inorganic nanoparticles within the body14,21. To investigate how superstructures are processed within macrophages, we washed the cells following their incubation with superstructures and chemically fixed them for visualization under TEM (Fig. 4a). Electron micrographs reveal that superstructures associated with the extracellular membrane of macrophages both as single entities (Supplementary Fig. 11) and as clusters (Fig. 4b–i). Associated superstructures were eventually internalized by macrophages within vesicles, in which superstructures disassembled into their respective building blocks (Fig. 4b–ii). We did not observe intact superstructures within macrophages, even in cells fixed immediately following exposure to the superstructures (Supplementary Fig. 12), suggesting that the intracellular degradation of superstructures occurred rapidly. In contrast, superstructures incubated in culture medium alone (i.e. without cells) remained largely intact over 8 hours of incubation (Fig. 4c), indicating that superstructure degradation was intracellularly-mediated. Phagocytic vesicles are known to contain a complex mixture of 40 or more hydrolytic enzymes that are responsible for digesting foreign pathogens or endogenous debris. It is possible for this mixture to quickly hydrolyse the DNA linkages that connect the nanoparticles together32,33. While many nanoparticle formulations have been reported to aggregate under such environments6, superstructure components remained dispersed following breakdown. These building blocks eventually escaped from the vesicles and distributed throughout the cellular cytoplasm (Fig. 4b–iii). To test whether this intracellular behaviour is mediated by superstructure assembly, we incubated core and satellite nanoparticles separately with macrophages under identical conditions and then examined their subcellular localization over time under TEM. Here we observed that while both core and satellite nanoparticles were endocytosed within vesicles, some satellite nanoparticles may have also entered cells via vesicle-independent pathways (Supplementary Fig. 13–14). In cells incubated with core nanoparticles, endocytosis resulted in the appearance of nanoparticle clusters that were confined within vesicles and grew in size over time, suggesting core nanoparticles are actively sorted into phagosomes from which they fail to escape. Satellite nanoparticles, in contrast, could be identified within cells as both being confined to vesicles and as individual, discrete nanoparticles within the cytoplasm. Supplementary Figure 14 further shows several instances where satellite nanoparticles originally confined within vesicles were released into the cytoplasm or excreted across the plasma membrane. These results suggest that, when delivered to the cells alone, the intracellular behaviour of nanoparticles is determined by their design. However, the assembly of these nanoparticles into superstructures alters their intracellular behaviour.
Figure 4. Intracellular processing of superstructures by J774A.1 macrophages.
a, Overview of subcellular localization of superstructures. Outlined areas are magnified in b: (i) Intact superstructures first interact with the plasma membrane, and (ii) are internalized by the macrophage within vesicles, where they undergo disassembly. (iii) Individual superstructure components eventually escape from vesicles and were seen distributed throughout the cytosol. The panel on the right depicts similar stages of processing occurring in another macrophage cell. c, Core-satellite structures show minimal structural disintegration after 8 hours of incubation in culture media containing 10% fetal bovine serum. All scale bars = 100nm. d, Workflow for assessing superstructure excretion by J774A.1 macrophages by ICP-AES. e, Time-course changes in intracellular gold content following exposure to superstructures containing 5nm satellite nanoparticles with varying PEG lengths (P1k=PEG 1kDa, P5k=PEG 5kDa, P10k=PPEG 10kDa). f, Time-course changes in intracellular gold content following exposure to 5nm satellite nanoparticles with varying PEG lengths; *P<0.05, **P<0.01, ***P<0.001. CS=core-satellite.
These results prompted us to carry out a parallel experiment, in which we measured changes in total intracellular gold content to assess whether dispersed building blocks could escape from these macrophages following uptake (Fig. 4d). In cells treated with superstructures, we found a 10 to 40% reduction in intracellular gold content over the course of 8 hours (Fig. 4e). The extent of this reduction was dependent on satellite design. In contrast, no change in gold content was measurable in cells incubated with core nanoparticles alone (Fig. 4e), suggesting that measured differences were attributable to the satellites. These differences occurred independently of changes in cell density (Supplementary Fig. 15) and plasma membrane permeability (Supplementary Fig. 16), and were apparent when cells harvested from different time points were cross-examined under TEM (Supplementary Fig. 17). Control experiments further verified that satellites alone escaped macrophage sequestration in a time- and PEG length-dependent manner (Fig. 4f and Supplementary Fig. 18). While the role of nanoparticle and PEG size in cellular uptake has been widely reported, results herein suggest that these design parameters also define the thresholds for cellular excretion, which has implications for the in vivo clearance and toxicity of nanomaterials. Additionally, molecular assembly techniques may offer a unique approach whereby PEGylated satellite nanoparticles can be used to facilitate therapeutic delivery without contributing significantly to overall in vivo persistence of nanomaterials.
Elimination and tumour targeting in mice
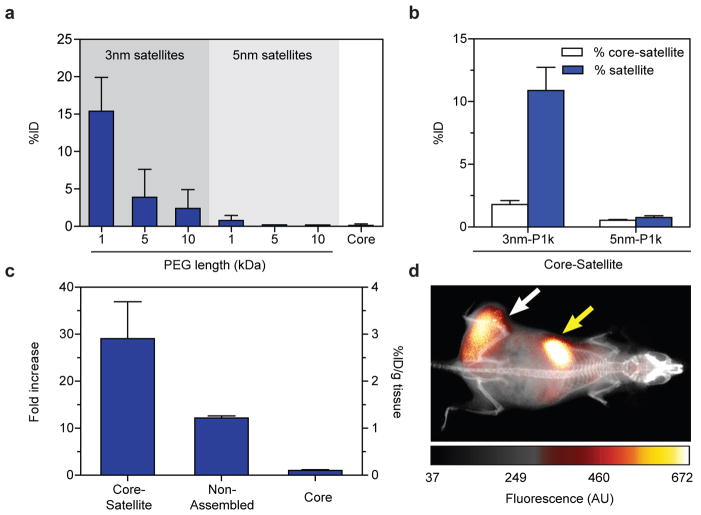
If small nanoparticles degraded from the superstructures can escape macrophage sequestration, we suspect that they can be designed small enough for renal elimination in vivo. This would decrease the biological persistence of nanoparticles injected into the body and eliminate their risks of chronic toxicity. To test this, we synthesized a panel of satellite building blocks and administered them intravenously into CD1 athymic nude mice. We housed the mice within metabolic cages, collected their urine for up to 48 hours post-injection, and analysed the urine for gold content. Urinary excretion was highest for the smallest satellites at 15% of the injected dose and diminished rapidly with nanoparticle size (Fig. 5a). Building on these results we assembled core-satellites with the smallest satellite nanoparticles to test the ability of superstructures to undergo renal clearance. Urinary excretion efficiency of superstructures mirrored closely with the clearance behaviour of their building blocks (Fig. 5b), suggesting they can be engineered to eliminate from the body unlike larger solid nanoparticles. More importantly, this result underscores an approach to tailor the size and surface chemistry of nanostructures for mediating their delivery while allowing them to clear from the body.
Figure 5. Renal elimination and tumour accumulation of superstructures.
a, Total gold content in mice urine up to 48 hours following systemic injection of various superstructure components. b, Total gold content in mice urine up to 48 hours following systemic injection of superstructures assembled from the smallest satellite nanoparticles in a. Results show that urinary excretion of superstructures mirrors closely with their respective satellites building blocks. c, Tumour accumulation of core-satellite superstructures using 5nm-PEG10k satellite nanoparticles at 24 hours post-injection. Injection of core nanoparticles alone and mixtures of unassembled superstructure components were used as separate controls. Error bars represent S.E.M. from three to five independent replicates; *P<0.05, **P<0.01, ***P<0.001. d, Whole-animal imaging using fluorescently-labelled superstructures show tumour-specific accumulation (white arrow) as well as accumulation in the liver (yellow arrow). ID=injected dose.
Finally, we assessed the potential of using superstructures to target tumours via a passive mechanism. Preliminary results with xenograft tumour models demonstrate that one of our current superstructure formulations accumulated within tumours better than its controls (e.g. core nanoparticle and non-assembled mixture) following systemic administration (Fig. 5c). Using a previously published procedure for fluorescently-labelling gold nanoparticles34, we chemically conjugated this formulation with near-infrared dyes in order to monitor their distribution in tumour xenograft models in real time. Our characterization shows the structures were not altered during this modification (Supplementary Fig. 19). Whole-animal fluorescence imaging showed that this superstructure design increased tumour-specific fluorescence contrast steadily over time (Supplementary Fig. 20), achieving a final tumour-over-background ratio of 2.3±0.1 and a signal-over-noise ratio of 5.2±0.5 at 24 hours following administration (Fig. 5d and Supplementary Fig. 21a–b). Analysis of the fluorescence images estimated a blood circulation half-life of 5 hours (Supplementary Fig. 21c). To ensure that superstructures were non-toxic, we collected blood from these animals for biochemistry and haematology analysis, and harvested organs for biodistribution and histology analysis. Results show that, while a large proportion of these superstructures also accumulated in the liver and spleen (Supplementary Fig. 22), they did not cause acute toxicity and were well-tolerated by the animals at the given dose (Supplementary Fig. 23–24). Together, these results demonstrate promise of using molecularly assembled superstructures for in vivo biomedical applications. As a next step, we are currently preparing a library of nanoparticles to further investigate and understand the effect of design on the rate and efficiency of tumour accumulation and whole-body clearance, similar to our previous study9. Our current findings have now defined the required building block designs and assembly architectures to engineer superstructures that can accumulate in tumours and be eliminated from the body.
Conclusions
In summary, we demonstrated the use of molecular assembly to mediate the biological delivery and elimination of nanoparticles. We showed that colloidal superstructures assembled with the appropriate building blocks and architectures can reduce their uptake and sequestration by macrophages, improve their accumulation into tumours, and facilitate their elimination from the body. The use of DNA assembly to engineer nanodelivery vehicles offers five advantages: 1) accurate and programmable control over nanocarrier architecture, 2) modular construction of complex platforms from simple nanoparticle building blocks, 3) compartmentalization of imaging or therapeutic payloads against biological degradation, 4) enable the development of new strategies to control the release of therapeutics (e.g., DNAzymes), and 5) ability to control the design of multifunctional nanomedicines (e.g. delivery vehicle with PET, MRI, and optical imaging agents or therapeutics). The use of both DNA and nanoparticle technologies together can help translate fundamental nanomaterial design principles that are being discovered into clinically relevant nanomedicine solutions.
Supplementary Material
Acknowledgments
This research was funded by the Canadian Institute of Health Research (MOP-93532; COP-126588; RMF-111623), Natural Sciences and Engineering Research Council (RGPIN-288231), Collaborative Health Research Program (CPG-104290; CHRPJ385829), Canadian Foundation for Innovation, and Ontario Ministry of Research and Innovation. L.Y.T.C. acknowledges Canadian Breast Cancer Foundation for fellowship. L.Y.T.C. and K.Z. acknowledge NSERC for fellowship. We are also grateful to Chuen Lo and Bill Dai for assistance with animal blood collection; the Advanced Bioimaging Centre at Mt.Sinai Hospital, Toronto, Canada for the use of TEM, and the ANALEST facility in the Department of Chemistry, University of Toronto, for the use of ICP-AES.
Footnotes
Author Contributions
W.C.W.C, L.Y.T.C., and K.Z. conceived the idea. W.C.W.C and L.Y.T.C. wrote the paper. L.Y.T.C. and K.Z. designed and performed experiments. All authors analysed data.
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Kim J, Piao Y, Hyeon T. Multifunctional nanostructured materials for multimodal imaging, and simultaneous imaging and therapy. Chem Soc Rev. 2009;38:372–90. doi: 10.1039/b709883a. [DOI] [PubMed] [Google Scholar]
- 2.Giljohann Da, et al. Gold nanoparticles for biology and medicine. Angew Chem Int Ed Engl. 2010;49:3280–94. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao J, Gu H, Xu B. Multifunctional magnetic nanoparticles: design, synthesis, and biomedical applications. Acc Chem Res. 2009;42:1097–107. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]
- 4.Smith AM, Duan H, Mohs AM, Nie S. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv Drug Deliv Rev. 2008;60:1226–40. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang W, Kim BYS, Rutka JT, Chan WCW. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–50. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 6.Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 7.Petros Ra, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615–27. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 8.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 9.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WCW. Mediating Tumor Targeting Efficiency of Nanoparticles Through Design. Nano Lett. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 10.Robinson JT, et al. In vivo fluorescence imaging in the second near-infrared window with long circulating carbon nanotubes capable of ultrahigh tumor uptake. J Am Chem Soc. 2012;134:10664–9. doi: 10.1021/ja303737a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, et al. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc Natl Acad Sci U S A. 2010;107:981–986. doi: 10.1073/pnas.0909565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi HS, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–70. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou C, Long M, Qin Y, Sun X, Zheng J. Luminescent gold nanoparticles with efficient renal clearance. Angew Chem Int Ed Engl. 2011;50:3168–72. doi: 10.1002/anie.201007321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballou B, et al. Sentinel lymph node imaging using quantum dots in mouse tumor models. Bioconjug Chem. 2007;18:389–96. doi: 10.1021/bc060261j. [DOI] [PubMed] [Google Scholar]
- 15.Albanese A, Chan WCW. Effect of gold nanoparticle aggregation on cell uptake and toxicity. ACS Nano. 2011;5:5478–89. doi: 10.1021/nn2007496. [DOI] [PubMed] [Google Scholar]
- 16.Díaz B, et al. Assessing methods for blood cell cytotoxic responses to inorganic nanoparticles and nanoparticle aggregates. Small. 2008;4:2025–34. doi: 10.1002/smll.200800199. [DOI] [PubMed] [Google Scholar]
- 17.Lovrić J, Cho SJ, Winnik FM, Maysinger D. Unmodified cadmium telluride quantum dots induce reactive oxygen species formation leading to multiple organelle damage and cell death. Chem Biol. 2005;12:1227–34. doi: 10.1016/j.chembiol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang RSH, et al. Persistent tissue kinetics and redistribution of nanoparticles, quantum dot 705, in mice: ICP-MS quantitative assessment. Environ Health Perspect. 2007;115:1339–43. doi: 10.1289/ehp.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JS, et al. Toxicity and tissue distribution of magnetic nanoparticles in mice. Toxicol Sci. 2006;89:338–47. doi: 10.1093/toxsci/kfj027. [DOI] [PubMed] [Google Scholar]
- 21.Ye L, et al. A pilot study in non-human primates shows no adverse response to intravenous injection of quantum dots. Nat Nanotechnol. 2012;7:453–458. doi: 10.1038/nnano.2012.74. [DOI] [PubMed] [Google Scholar]
- 22.Choi HS, et al. Design considerations for tumour-targeted nanoparticles. Nat Nanotechnol. 2010;5:42–7. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon J, Lim J, Yoon S. Controlled Assembly and Plasmonic Properties of Asymmetric Core-Satellite Nanoassemblies. ACS Nano. 2012:7199–7208. doi: 10.1021/nn302264f. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Rosi NL, Wang Y, Huo F, Mirkin Ca. Asymmetric functionalization of gold nanoparticles with oligonucleotides. J Am Chem Soc. 2006;128:9286–7. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knop K, Hoogenboom R, Fischer D, Schubert US. Poly(ethylene glycol) in drug delivery: pros and cons as well as potential alternatives. Angew Chem Int Ed Engl. 2010;49:6288–308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 26.Chaires J, Herrera J, Waring M. Preferential binding of daunomycin to 5′TACG and 5′TAGC sequences revealed by footprinting titration experiments. Biochemistry. 1990;29:6145–6153. doi: 10.1021/bi00478a006. [DOI] [PubMed] [Google Scholar]
- 27.Owens DE, Peppas Na. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Khlebtsov N, Dykman L. Biodistribution and toxicity of engineered gold nanoparticles: a review of in vitro and in vivo studies. Chem Soc Rev. 2011;40:1647–71. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 29.Clift MJD, et al. The impact of different nanoparticle surface chemistry and size on uptake and toxicity in a murine macrophage cell line. Toxicol Appl Pharmacol. 2008;232:418–27. doi: 10.1016/j.taap.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 30.He C, Hu Y, Yin L, Tang C, Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–66. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 31.Patel PC, et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 2010;21:2250–6. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claus V, et al. Lysosomal Enzyme Trafficking between Phagosomes, Endosomes, and Lysosomes in J774 Macrophages. Enrichment of Cathepsin H in Early Endosomes. J Biol Chem. 1998;273:9842–9851. doi: 10.1074/jbc.273.16.9842. [DOI] [PubMed] [Google Scholar]
- 33.Odaka C, Mizuochi T. Role of macrophage lysosomal enzymes in the degradation of nucleosomes of apoptotic cells. J Imunol. 1999;163:5346–52. [PubMed] [Google Scholar]
- 34.Chou LYT, Chan WCW. Fluorescence-tagged gold nanoparticles for rapidly characterizing the size-dependent biodistribution in tumor models. Adv Healthc Mater. 2012;1:714–21. doi: 10.1002/adhm.201200084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.