Abstract
ATP-dependent chromatin remodeling is an essential process required for the dynamic organization of chromatin structure. Here we describe the genome-wide location and activity of three remodeler proteins with diverse physiological functions in the mouse genome: Brg1, Chd4, and Snf2h. The localization patterns of all three proteins significantly overlap with one another and with regions of accessible chromatin. Furthermore, using inducible mutant variants, we demonstrate that the catalytic activity of these proteins contributes to the remodeling of chromatin genome-wide, and that each of these remodelers can independently regulate chromatin reorganization at distinct sites. Many regions require the activity of more than one remodeler to regulate accessibility. These findings provide a dynamic view of chromatin organization, and highlight the differential contributions of remodelers to chromatin maintenance in higher eukaryotes.
Organization of the eukaryotic genome into chromatin is essential for all DNA-templated processes. Packaging of DNA into nucleosomal arrays not only acts to condense the genome, allowing for efficient organization within the cell’s nucleus, but as an important mechanism to regulate access to DNA encoded information. Maintaining a balance between efficient packaging and accessibility is achieved through the combined activities of multiple specialized proteins that are critical for the dynamic alteration of chromatin structure. ATP-dependent chromatin remodeler enzymes play a key role in this process. Each member of this large family of enzymes is characterized by a highly conserved helicase-like ATPase domain, utilized to generate energy from ATP hydrolysis to reposition, evict, or otherwise modify nucleosomes 1. In terms of function, the outcome of remodeling is well understood to result in the regulation of chromatin accessibility and the exposure of DNA regulatory elements. Regions of accessible chromatin, often characterized as DNase I hypersensitive sites, have been mapped genome-wide in different cell types and shown to demarcate regulatory elements such as promoters, enhancers, silencers, and locus control regions 2–5. However, remodeler studies have focused predominantly on understanding the mechanism of ATP-mediated catalysis of nucleosome movement in vitro 6–9. Less is known concerning their distribution throughout the genome, and their individual roles in specific chromatin reorganization processes.
In vitro activity analysis demonstrates a common reaction mechanism is shared by remodeling complexes, suggesting functional differences seen between individual complexes in vivo may be due to regulatory differences 10. Indeed, the interaction of complexes with cofactors and the targeting of remodelers to specific modified regions of chromatin have been linked to distinct, and, in some cases, opposing functions 11–14. In particular, the recruitment of complexes by either repressors or activators to areas of accessible and inaccessible chromatin, respectively, would contribute to region specific activities of different complexes within the cell. The importance of these systems in cell selective gene expression has attracted increasing attention 15–17.
To gain an understanding of the potential interplay between multiple remodeling systems and their functions in cells, we have begun to build a comprehensive map of remodeler localization and genome-wide function in mouse cells. Using mutant variants of Brg1, Chd4, and Snf2h, we directly assigned remodeling activity at individual sites, demonstrating that each remodeler contributes to chromatin accessibility. Unexpectedly, many regions of accessibility require the concerted actions of all three proteins. Thus, we propose a general mechanism wherein the genome-wide organization of nucleosomes is a dynamic process requiring the activity of multiple remodeling systems.
RESULTS
Localization of three chromatin remodeler proteins genome-wide
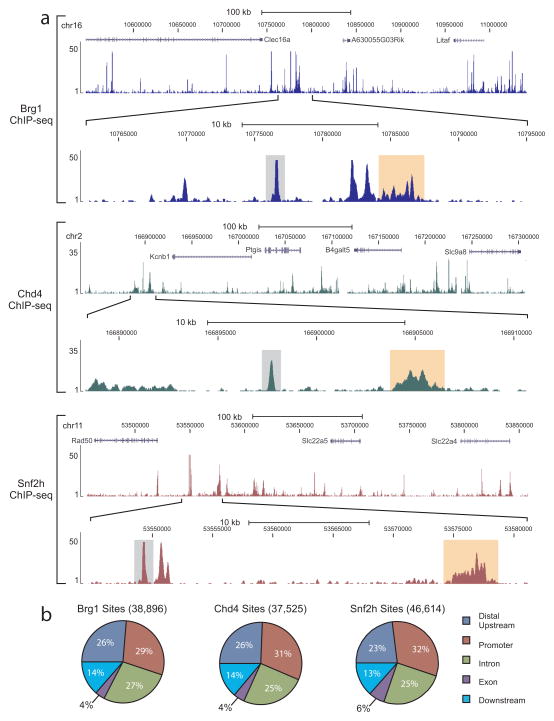
To expand our understanding of the interplay between remodeler proteins, we focused on the remodelers Brg1, Snf2h and Chd4, which are from the SWI-SNF, ISWI, and CHD families, respectively. Recent reports suggest that each of these remodelers perform unique roles in the regulation of chromatin structure making them ideal candidates for use in our studies 18. To begin our analysis of how these proteins function in vivo, we mapped their genome-wide locations in mouse mammary epithelial cells by ChIP-seq using both specific monoclonal and polyclonal antibodies (Supplementary Fig. 1a–b). Binding profiles of individual genomic regions demonstrate that each binds to defined locations within the genome (Fig. 1a, Supplementary Fig. 1c) and are characterized by a mix of binding events. In particular, we found two major site types composed of locally distributed regions characterized by single, defined peaks, very near the size of a transcription factor footprint (~150 bp) 19, 20, as well as regions more broadly distributed. Consistent with these two types of binding, we found the average size of a remodeler site to be 638 bp (589 bp for Brg1, 719 for Chd4, and 605 bp for Snf2h). In total, we identified 38,896, 37,525, and 46,614 sites occupied by Brg1, Chd4, and Snf2h, respectively.
Figure 1. Remodeler proteins bind to distinct regions of chromatin.
(a) Example ChIP-seq genome browser views of Brg1 (top, blue tracks), Chd4 (middle, green tracks), and Snf2h (bottom, dark red tracks) occupancy. Images represent tag densities (mapped sequence tags) relative to genome coordinates. For each remodeler, the lower browser image displays an expanded view of the selected region where examples of localized distributions (single peak, <500 bp) are highlighted by grey shading and broad distributions (>500 bp) are highlighted by light orange shading. (b) Distributions of remodeler occupancy at annotated genic regions. Sites are classified as promoter (−/+ 2.5 kb from TSS), exon (> 2.5 kb downstream from TSS, to the last intron, not intron), distal upstream (> 2.5 kb upstream from TSS), downstream (> 2.5 kb downstream from TSS, not exon or intron), or intron.
When we characterized the distribution of these sites relative to annotated genes, we found similar localization patterns for each remodeler in which ~60% of these sites were located in the promoter and body of genes, while ~40% of sites were found in intergenic regions (Fig. 1b). However, analysis of binding tag density values revealed differences between these remodelers at the level of enrichment (Supplementary Fig. 1d). Specifically, we found Brg1 enrichment to be higher at regions distal to promoters (P < 10−7), while we determined Chd4 to be distributed evenly between the different genomic regions. Unlike either of these remodelers, we found Snf2h enrichment to be higher at promoters than at any of the other regions (P < 10−7), with a slight enrichment at exons. Similar findings were observed for the Snf2h homolog ISWI in Drosophila indicating enrichment of Snf2h at promoter regions may be a conserved occurrence 17.
Genomic co-occupancy by remodeler proteins
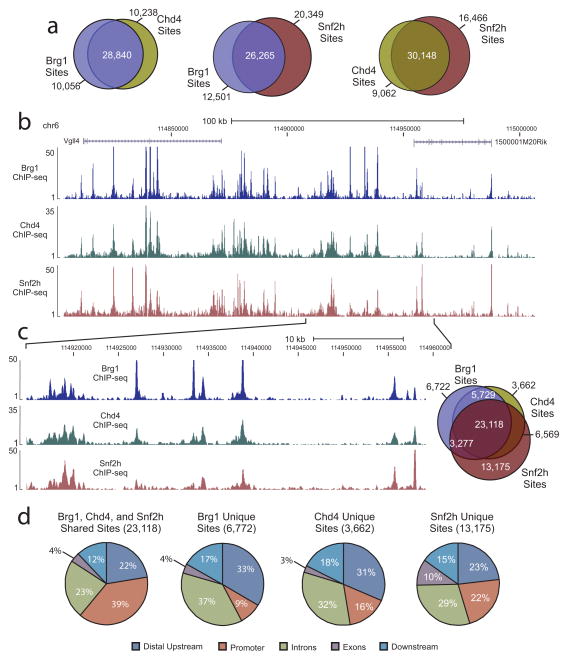
Given the similar distribution patterns, we wondered if common binding sites were shared by these proteins. To determine if they co-localized to the same genomic regions, pair-wise comparisons were performed of the genomic sites occupied by each remodeler. When Brg1 and Chd4 sites were compared, the majority of sites for each protein were shared with 74% of Brg1 sites overlapping with 76% of Chd4 sites. Comparisons between Brg1 and Snf2h revealed 68% of Brg1 sites were also bound by Snf2h, while 56% of Snf2h sites were occupied by Brg1. Similarly, the majority of Chd4 sites were co-occupied by Snf2h (76%), while 65% of Snf2h sites were bound by Chd4 (Fig. 2a). In comparisons of all three remodelers’ binding sites, a large proportion of each were found to be shared by the other two remodelers (59%, 62%, and 50% of Brg1, Chd4, and Snf2h sites, respectively) with binding profile comparisons further supporting these findings (Fig. 2b–c, Supplementary Fig. 2).
Figure 2. Brg1, Snf2h, and Chd4 tend to co-occupy the same genomic regions.
(a) Venn diagrams displaying overlaps of binding site occupancy between pairs of remodelers. (b) ChIP-seq genome browser view of Brg1 (blue track), Chd4 (green track), and Snf2h (dark red track) occupancy at the same genomic coordinates on chromosome 6. Mapped sequence tags represented as tag density are indicated on the y-axis. (c) An expanded view of the selected region in panel [(b)]. Displayed on the right-side is a three-way Venn diagram demonstrating the overlap between the binding sites of Brg1(blue), Chd4 (dark yellow), and Snf2h (red). (d) Distribution at annotated genic regions of shared and unique remodeler binding sites. Promoter represents region ± 2.5 kb from TSS.
The co-localization patterns could result from transient occupancy by separate remodelers in sub-sets of the cell populations, or could be due to a direct interaction between these proteins. We performed co-immunoprecipiation experiments and found that although each remodeler was capable of interacting with previously identified complex members, an association between the remodelers as soluble proteins was not detectable (Supplementary Fig. 3a). We also examined potential interactions at template sites by re-ChIP analysis Supplementary Fig. 3e–g). Very weak re-ChIP signals were detected, but these signals are so low (0.00003% range), they likely result from non-specific trapping of two remodelers in the same crosslinked chromatin complex. This lack of direct interaction suggests binding of remodelers to sites of co-localization occurs through transient, sequential binding events (see discussion).
To further characterize remodeler localization, we examined the tag density and distribution of both unique and shared sites to determine if there were distinct features associated with each type of region. In analyzing average tag density values, we found regions occupied by multiple remodelers and, in most cases, sites co-occupied by all three to display higher enrichment levels than sites occupied by a single remodeler (Supplementary Fig. 3b–d). Further, we found the distribution of these co-occupied sites to be very similar to the distribution patterns observed initially for individual remodeler-bound regions with a slight increase in the number of sites at promoters (39% vs. 29–32%). In contrast, the location of unique remodeler binding (sites bound by a single remodeler) tended to occur at regions distal to promoters (Fig. 2d). Taken together, our initial findings suggest these remodelers may act in coordination at co-occupied regions. Additionally, these results indicate the co-localization of these proteins is not exclusively a consequence of promoter occupancy, and hence activity at promoters, but may be a feature associated with various DNA regulatory elements.
DNA binding factor motifs at remodeler binding sites
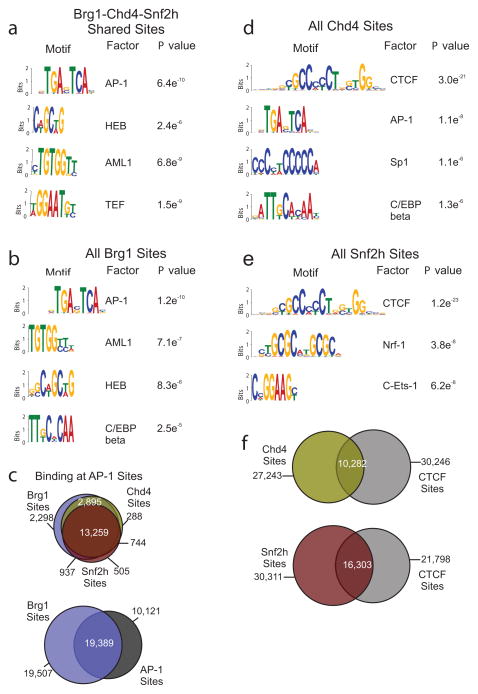
A likely quality of these bound regions is their content of DNA sequence-specific protein binding sites. Several reports indicate chromatin remodeler complexes interact with regulatory factors as a mechanism of targeting to chromatin 21–23. To investigate this possibility, we used the de novo DNA motif discovery program MEME 24 to identify consensus sequence motifs associated with the most enriched 2,000 overlapping and unique remodeler-bound regions. This analysis revealed significant enrichment of several motifs associated with each of these regions (P < 10−4). In sites co-occupied by Brg1, Chd4, and Snf2h, these included HEB, AML1, TEF and most significantly, AP-1 (Fig. 3a). We found AP-1 to be the most enriched motif at Brg1-occupied sites, including regions uniquely bound by Brg1 (Fig. 3b, Supplementary Fig. 4a). These results are in line with previous reports of Brg1 binding to AP-1 sites, and AP-1 maintenance of baseline chromatin accessibility 25, 26. Further, when we compared the binding sites of Brg1, Snf2h, and Chd4 to available AP-1 ChIP-seq data 27, we found 57% of the sites shared by all three remodelers overlap with 45% of AP-1 sites (Fig. 3c, Supplementary Fig. 4b). Unlike Brg1, the most significantly enriched motif associated with both Snf2h and Chd4 sites, including unique sites, was CTCF (Fig. 3d–e, Supplementary Fig. 4c–d). Notably, the remodeler Chd8 has been shown to functionally interact with CTCF and is required for enhancer blocking activity, but neither Snf2h nor Chd4 has been directly linked to CTCF association 28. When we analyzed and compared the localization of CTCF with these remodelers we found 27% of Chd4 sites overlap with 25% of CTCF sites, while 35% of Snf2h sites overlap with 40% of CTCF sites (Fig. 3f, Supplementary Fig. 4e). Thus, the association of multiple DNA sequence-specific motifs with remodeler-bound sites may reflect the range of factors that can either interact, recruit, or be targeted by these remodelers allowing for varied downstream effects on chromatin. Moreover, the predominance of the same factor at both highly enriched unique and shared regions emphasize the importance of not only binding, but enrichment levels, which differ between the sites found for Brg1, Chd4, and Snf2h (refer to Supplementary Fig. 1d).
Figure 3. Remodeler binding sites are associated with DNA sequence-specific regulatory elements.
(a) Results of de novo motif discovery using the top 2,000 binding sites (based on tag density) co-occupied by Brg1, Chd4, and Snf2h. Shown are the most significantly enriched motifs identified by MEME analysis (P < 10−4). The AP-1 motif is the most highly enriched motif (MEME E value = 1.9e−2110). (b) Results of de novo motif discovery (top 2,000 sites) of all Brg1 sites. AP-1 is the most highly enriched motif for these sites (MEME E value = 5.5e−2277). (c) Venn diagrams of sites shared between remodelers and AP-1. Top, three-way Venn diagram representing the overlap between remodeler sites that specifically co-localize with AP-1 sites. Bottom, Venn diagram of the overlap between Brg1 and AP-1 sites. (d–e) Similar de novo motif analysis as described above was performed for Chd4 [(d)] and Snf2h [(e)]. For both remodelers at each site type, the motif identified as CTCF was found to be the most highly enriched motif (Chd4 MEME E value = 2.0e−490; Snf2h MEME E value = 2.7e−983). (f) Venn diagrams representing the overlap of binding sites for Chd4 or Snf2h with CTCF sites.
Association of remodelers with accessible chromatin regions
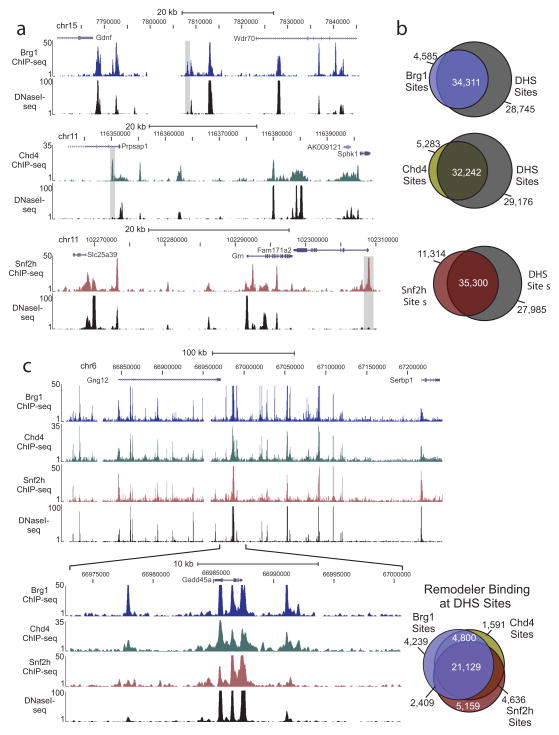
Since we know remodelers regulate the packaging of chromatin, it seems likely we would find these proteins at accessible chromatin. To determine if these proteins specifically localized to remodeled regions of chromatin, we identified genome-wide DHS sites by DNase I-seq and compared the location of these sites to those bound by remodelers. Binding profile comparisons between remodeler and DHS sites revealed strikingly similar patterns (Fig. 4a). Global analysis of site overlap indicated 88% of Brg1 sites, 85% of Chd4 sites, and 75% of Snf2h sites correspond to sites of chromatin accessibility (Fig. 4b). Yet, for each, there are clear examples of sites not associated with DHS (Fig. 4a–b). When we compared DHS sites bound by each remodeler, we discovered 21,129 sites to be co-occupied by all three proteins indicating the majority (91%) of the regions shared by Brg1, Chd4, and Snf2h occur at open chromatin (Fig. 4c, Supplementary Fig. 5a). In an examination of the average tag density values for remodelers bound at DHS sites, we found enrichment levels to be significantly (P < 2.2e−16) higher at these regions than for remodeler binding away from DHS sites (Supplementary Fig. 5b–d). Of the sites away from DHS, between 23% (Snf2H) and 27% (Brg1 and Chd4) were within 500 bp of a DHS site, which may indicate a small number are not truly bound to inaccessible regions, but may, in fact, be the tail end of a larger remodeler site. Nonetheless, this does not undermine the finding that there are regions of remodeler binding not associated with DHS. Similar findings were found for DHS sites occupied by remodelers indicating a strong correlation between the level of accessibility at these sites and binding by these specific remodelers (Supplementary Fig. 5e–f).
Figure 4. Remodeler protein binding highly overlaps with accessible chromatin regions.
(a) Genome browser view examples of remodeler ChIP-seq occupancy and DNase I hypersensitivity (measure of chromatin accessibility, DNaseI-seq) patterns. Images represent tag densities (mapped sequence tags) relative to genome coordinates. Examples of binding sites that do not overlap with accessible chromatin are highlighted by grey shading. (b) Venn diagrams representing the overlap of binding sites for each remodeler with DNase I hypersensitive (DHS) sites. (c) Genome browser view of Brg1 (blue track), Chd4 (green track), and Snf2h (dark red track) ChIP-seq occupancy and DNaseI-seq patterns at a region on chromosome 6 are displayed. An expanded view of the selected region is shown below this image. Displayed on the right-side is a three-way Venn diagram representing the overlap between remodeler sites that specifically co-localize with DHS sites.
Analysis of genome-wide remodeling activity
Given the strong association of each remodeler with accessible chromatin and their known functions in remodeling, we next sought to determine the role of each in the regulation of chromatin accessibility. To analyze remodeling function, we took advantage of the fact that the catalytic ATPase domain is highly conserved and, for each protein, created a dominant-negative variant by mutating a conserved lysine in this region (Supplementary Fig. 6a). Similar mutations have been successfully used for these and other remodelers in yeast and mammalian cells 29–34. Using these mutant constructs, we then created three stable congenic cell lines under control of the tetracycline (Tet)-off conditional system, which enabled expression of each dominant-negative variant in the absence of Tet (Supplementary Fig. 6b–d). A fourth cell line expressing the tetracycline transactivator (tTA) protein system alone acted as a control for genomic effects mediated by expression of the tTA regulatory protein in our cells. Induction of a given dn-remodeler had no effect on expression levels for the other remodelers under consideration (Supplementary Fig. 6e–g).
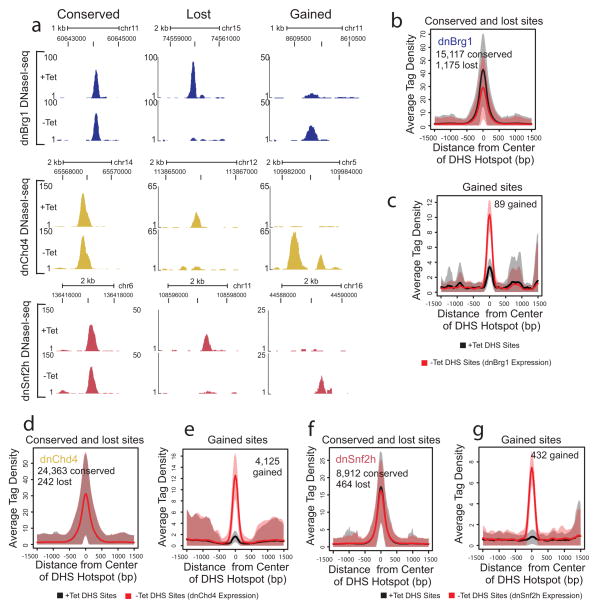
To examine the genome-wide roles of these remodelers in the regulation of chromatin structure, we analyzed changes in DNase I hypersensitivity by performing DNase I-seq in the absence or presence of each dominant-negative (dn) remodeler protein. Following expression of each of these variants, we found examples of chromatin where regions were unchanged (conserved), rendered inaccessible (lost), or newly opened, indicating each remodeler is capable of opening and closing sites (Fig. 5, Supplementary Fig. 7a–c). However, when we globally analyzed the effects of remodeler inhibition, we noted remodeler-specific trends in chromatin structure regulation (Fig. 6, Supplementary Fig. 7d–f, Supplementary Fig. 8, Supplementary Fig. 9, Supplementary Table 1). In particular, we noted predominant trends in the effect on accessibility following the expression of dnBrg1 and dnChd4 (Fig. 5b–g, Supplementary Fig. 7d–f)). Following inhibition of Brg1 activity, 1,175 DHS sites were lost, while approximately 3,247 DHS sites were reduced at least two-fold in size indicating a principal requirement for Brg1 maintenance of a subset of remodeled chromatin. In contrast, inhibition of Chd4 activity led to little change in the size of conserved DHS sites, but did lead to the opening of chromatin at a subset of regions (4,688 sites) indicating a role for Chd4 in the maintenance of chromatin in a closed state. Expression of dnSnf2h also produced changes in chromatin accessibility, with both the loss and opening of a small number of sites, consistent with a previous report of this remodeler’s involvement in controlling subtle changes in Drosophila nucleosome positioning 17. The examination of sites lost and gained in the presence of dnBrg1 and dnChd4, respectively, for DNA binding factor-specific motifs, indicates these sites may represent potential functional regulatory elements. Specifically, the remodeled regions that are newly opened following expression of dnChd4 contain motifs which would otherwise be inaccessible (Supplementary Fig. 4f–g).
Figure 5. Remodeler protein distribution at conserved and lost sites.
(a) Browser view examples of DHS sites (DNaseI-seq) in the absence (+Tet) or presence (−Tet) of each dnRemodeler. Tag densities (y-axis) are indicated for sites located at the displayed genomic coordinates. (b–g) Aggregate plot of average DHS tag density values over conserved and lost sites for dnBrg1 (b), gained sites for dnBrg1 (c), conserved and lost sites for dnChd4 (d), gained sites for dnChd4 (e), conserved and lost sites for dnSnf2h (f), and gained sites for dnSnf2h (g). The shaded areas are up to +/− standard deviation from the average profile.
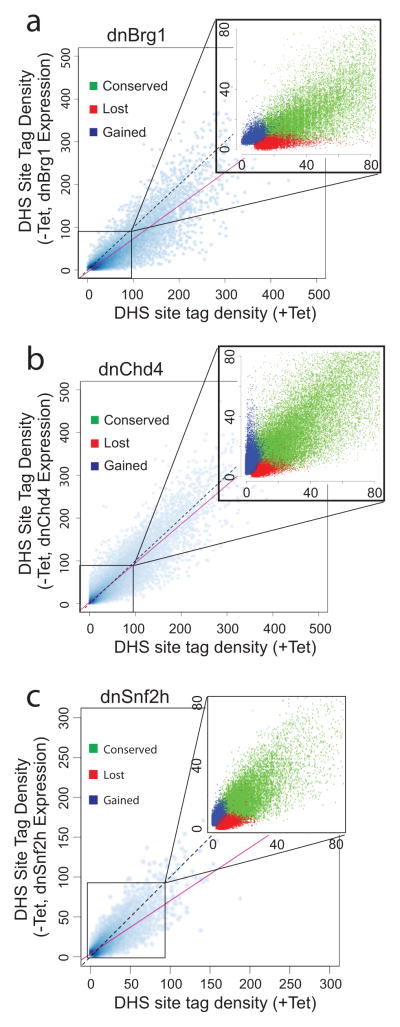
Figure 6. Trends in remodeler protein regulation of chromatin accessibility.
DNase I hypersensitivity (DHS site tag density) of DHS sites following expression (−Tet, y-axis) of dnBrg1 (a), dnChd4 (b), or dnSnf2h (c) compared to DHS sites in the absence (+Tet, x-axis) of the indicated dnRemodeler. Insets, expanded views of selected scatter plot regions. Conserved (green) are sites that existed prior to and after the expression of dnRemodeler; lost (red) are sites lost following dnRemodeler expression; and gained (blue) are newly opened sites. Solid red line; trend line used to indicate direction of DHS tag density change following dnRemodeler expression. Dotted black line, diagonal line indicating position of trend line if there were no changes in hypersensitivity. Control −Tet/+Tet distributions for parental cells with no dnRemodelers are shown in Supplementary Fig. 8.
Multiple chromatin remodelers at individual DHS sites
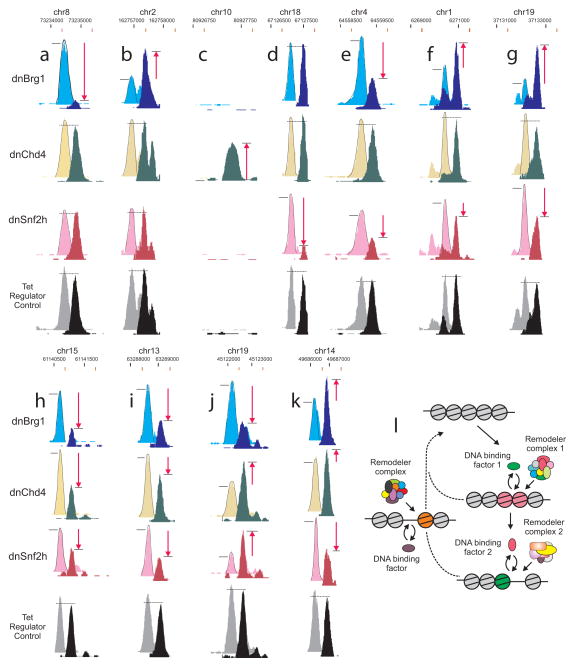
Assessment of remodeler binding at affected regions revealed a major proportion to be occupied by multiple remodelers (Supplementary Fig. 7). The finding of redundant remodelers at affected sites suggests an additional layer of complexity, where activity may be dependent on the sequence of recruitment. To determine the potential contribution of multiple remodeling activities at individual DHS sites, we examined the DNase I-seq data sets from the dominant-negative variant cell lines for examples of DHS sites affected by either a single remodeler or multiple remodelers. Surprisingly, the extent of remodeling at a substantial fraction of DHS elements is affected by multiple systems (Fig. 7). Panels a–d (Fig. 7) show examples of sites primarily controlled by one remodeling system, while panels e–g present examples of elements where two systems are functioning, either collaboratively (e) or in opposition (f, g). There are also examples where all three remodelers function at a single element (h–k). Occurrence frequencies for each of the 27 possible interaction classes are presented in Supplementary Table 1 and Supplementary Fig. 9 for over 95,000 individual sites pooled from the three dn cell lines. These values represent underestimates because of the following considerations. (1) There are likely to be cases where the equilibrium result of multiple functional systems may lead to compensating effects. If one system is abrogated by activation of the dominant negative, the contribution of a second system may increase, masking the effect. (2) Many members of the large class of sites that manifest no change with any of the remodelers could originate from other members of the large family of remodeling systems in mammalian cells.
Figure 7. Multiple remodelers contribute to the regulation of an individual DHS site.
(a–k) Examples of DHS sites affected by expression of each of the indicated dominant-negative remodelers. (a–d) Single remodeler effects. (e–g) Double synergism. (h–k) Triple synergism. For comparison, the −Tet induction of dominant negative tracks are displaced down and to the right (black coordinates +Tet; red coordinates −Tet). −/+ dnBrg1 [(Cyan (−), Blue (+)]; −/+ dnChd4 [(Yellow (−), Green (+)]; −/+ dnSnf2h [(Pink (−), Red (+)]; −/+ Tet regulator control [(Gray (−), Black (+)]. Tet Regulator Control; cell line expressing only the tetracycline transactivator protein demonstrating effects at these DHS sites are due exclusively to the expression of the dominant-negative variant. Red arrows denote increases or decreases in accessibility, while dotted horizontal line indicates no change. (l) Mechanism of dynamic transitions in chromatin structure mediated by transient recruitment of remodelers and their associated activity. Remodeler complexes are targeted to a nucleosomal region by specific DNA-bound factors. Both events, remodeler recruitment and factor binding, are transient. Transitions may involve a unique remodeler, or multiple complexes acting sequentially (right side). Furthermore, some reactions may lead to chromatin closing, rather than opening (left side). Thus, localized chromatin states monitored by current methodologies represent population averages of complex processes that sometimes involve multiple remodeling systems.
We also examined effects of the dn-remodelers on selective gene expression by global analysis of RNA expression patterns with activation of each dn-remodeler (Supplementary Fig. 10a–e). Two examples are shown in panels a–e for genes that are strongly de-repressed with dnChd4. In each case, a DHS element immediately upstream of the promoter is dramatically induced by activation of dnChd4. Importantly, induction of w.t. Chd4 has no effect on expression (panels a, c, e). As summarized in panel f, more than 800 genes are de-regulated by the three dn-remodelers.
DISCUSSION
Many multiprotein remodeling complexes have been characterized, but how these complexes interact with each other, and with site-specific DNA binding proteins, to generate specific chromatin structures remains a conceptual puzzle, especially in higher eukaryotes. Through ChIP-seq analysis with specific antisera, coupled with the controlled expression of inducible dominant-negative remodeling system variants, we address this problem by assigning site specific remodeling activities, genome-wide, for three remodeling systems in mouse cells. We report that each system is capable of both opening and closing chromatin. While remodeling systems are generally considered to be involved in the repositioning or disassembly of localized nucleosome structures, there is also precedence 35 for reversing access, or “closing,” localized structures, presumably by reassembly of nucleosome structures. Alternatively, repositioning of nucleosome(s) could modulate factor binding in a way that would lead to return of localized chromatin structure to a closed state. Our data sets reveal the widespread, and unexpected, extent of this chromatin closing phenomenon (Fig. 5). While Chd4 is the most frequent participant in chromatin closing events, the other two remodelers are also capable of catalyzing this process.
Current models regarding the mechanism of remodeling action suggest that a unique remodeling system is recruited to a given DHS element 14, 18. However, of the approximately 90,000 DHS elements we have characterized in this study, a substantial fraction are associated with multiple remodeling systems. Indeed, we identify many DHS sites bound by all three remodelers. Our studies also show these redundant systems not only co-occupy the DHS elements, but function in many combinatorial modes involving both synergistic and antagonistic action (Fig. 7a–k). Verrijzer and colleagues recently reported little overlap in remodeler distributions genome-wide in Drosophila 36. Yet, further support for the potential contributions of redundant remodelers comes from studies in yeast wherein deletions of multiple remodelers resulted in numerous alterations in chromatin structure, but individual deletions produced relatively minor effects 37, 38. GIven the much higher complexity of mammalian genomes, It seems possible that more complex mechanisms have evolved to provide control of chromatin remodeling in these systems.
Additionally, studies are emerging that support antagonistic remodeler complex functions at individual sites 39–42. Given these findings, we propose a general mechanism of cyclical, targeted remodeler activity to maintain a fluid chromatin state (Fig. 7l). Under this model, chromatin remodelers and transcription factors both cycle dynamically through a complex series of states. The nature and sequence of these states would be dependent both on the chromatin context (histone-DNA marks) and the specific transcription factors that recruit remodelers to the site. An important consideration is the timescale limitations inherent to ChIP and DHS assays, which give the appearance of relatively static states because the signals are averaged across heterogeneous cell populations. However, many lines of evidence support highly transient interactions of remodelers and DNA-binding proteins during remodeling, consistent with the model discussed here 43–50.
How remodelers are targeted to specific sites in chromatin remains a central question. Motifs for transcription factors cluster at regulatory elements such as enhancers and promoters, providing binding sites for multiple factors that could, in turn, recruit multiple remodeling systems 51. It is also well established that a specific remodeling system can be required for function at selected elements, as is observed for neural development 52. In our analysis of recognition elements present at remodeler-bound sites, we identified several distinct motifs, pointing to a subset of factors involved in selective recruitment. Regulatory protein motifs most frequently associated with each remodeler (AP-1 for Brg1 and CTCF for Chd4 and Snf2h) represent constitutive nuclear proteins that have previously been linked to regulation of chromatin accessibility and genomic organization. Specifically, we observed that blocking of AP-1 binding to chromatin reduced chromatin accessibility at GR response elements in mammary cells 26. Presumably, this occurs through AP-1 targeting of a remodeling complex. While transcription factors may recruit selected remodeling proteins by direct protein-protein contacts 53, 54, it is also likely that specific histone modifications at a given site contribute to remodeler selectivity. Many of the remodeling proteins contain bromo-chromo domains, which recognize specific histone marks 55, 56. Thus, selective recruitment is likely achieved through the combined action of site-specific DNA binding proteins and histone modifications; however, the enzymes directing these marks are also recruited in turn by transcription factors.
The finding of a functional involvement of multiple remodeling systems at many DHS sites implies a directed, or sequential order of recruitment. DHS sites completely lost or gained following the suppression of a single remodeler is the simplest case. For Brg1, several sites completely lost in the presence of the mutant variant were, in fact, bound by multiple remodelers. In this case, it is likely the activity of Brg1 is an initial event, while the function of other remodelers bound to the same site is unclear. Reports of Brg1-Chd4-NuRD complex co-bound sites demonstrated a NuRD complex-dependence on Brg1 binding and less of a dependence for Brg1 on the NuRD complex 39, 42. Thus, the activity of the initial recruited complex may affect the recruitment and activity of subsequent complexes. However, for the large majority of elements characterized in our study, multiple systems are shown to be not only present, but functional. It will be very difficult to determine the order of events in these complex examples through whole cell investigations. Biochemical reconstruction of these processes in vitro will eventually contribute to a detailed understanding of the sequence of events.
Given the large number of remodeler complexes in the mammalian genome, the potential number of remodeler interactions with chromatin is clearly extensive. The findings presented here demonstrate a further unanticipated complexity. A process that seems to move from one static state to another when examined by population averaged and time averaged methodologies is in fact highly dynamic. The localized reorganization of nucleosome structures will require intensive examination, both in vivo and in vitro, to develop a detailed understanding of these important processes.
METHODS
Cell lines and culture conditions
The 3134 mouse mammary epithelial cell line, originally derived from a subclone of 904.13 57, was maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 2 mM L-glutamine, and 5 mg/ml penicillin-streptomycin in a 37°C incubator with 5% C02. To generate dominant-negative variant cell lines, the coding sequence of human SNF2H (hSNF2H) fused to the FLAG tag sequence at the N-terminus in the pCI-neo vector was received as a gift from David Picketts (U. of Ottawa). Using BamHI and SalI restriction enzyme sites, FLAG-hSNF2H was cloned into a tetracycline-inducible retroviral vector (pRevTRELink, 58 followed by site-directed mutagenesis of lysine 211 to arginine using the QuikChange XL Site Directed Mutagenesis Kit according to the manufacturer’s instructions (Stratagene, La Jolla, CA). This dominant-negative variant of FLAG-hSNF2H (dnSnf2h) was then stably integrated into a cell line containing the tetracycline transactivator regulatory system (3134Tet, 7110) as described previously 50. Similarly, the coding sequence of mouse Chd4 (a gift from John Svaren (U. of Wisconsin)) was cloned into pRevTRELink containing a triple FLAG tag sequence using NotI/SalI restriction enzyme sites followed by site-directed mutagenesis of lysine 250 to cysteine and integration into the 3134Tet cell line. The tetracycline-inducible dnBrg1 cell line was described previously 59. Constructs were fully sequenced to confirm accuracy prior to cell line integrations. All tetracycline-regulated cell lines were maintained in DMEM and 5 μg/ml tetracycline to repress expression of the dominant-negative proteins. For experiments, cells were plated in DMEM supplemented with 10% charcoal-dextran treated FBS with or without tetracycline for 48 hrs.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as per standard protocols (Millipore, Billerica, MA) with minor modifications. Briefly, cells were crosslinked for 10 min with 1% formaldehyde at 37°C followed by quenching of this reaction with 150 mM glycine for 10 min. Each ChIP contained 400 μg of soluble, sonicated chromatin. DNA-protein complexes were immunoprecipitated using the following antibodies: anti-BRG1 (1 μg, EPNCIR11A, made in collaboration with Epitomics, Burlingame, CA), anti-Chd4 (7 μg, ab72418, Abcam, Cambridge, MA), anti-SNF2H (5 μg, ab72499, Abcam), and anti-CTCF (10 μL, 07-729, Millipore, Billerica, MA); validation of the antibodies is provided on the manufacturer’s website. DNA isolated from ChIPs were validated and-or confirmed by real-time quantitative PCR amplification using SyBr green mix (BioRad, Hercules, CA). Primer sequences are available upon request. Three samples from two biological replicates were pooled as a single sample before generating sequencing libraries. Two replicates per condition were sequenced.
Preparation of DNase I-digested DNA
DNase-I digested DNA were prepared for sequencing as previously described with minor modifications 5. Briefly, expression of dominant-negative variant proteins was induced by removal of tetracycline for 48 hrs prior to harvest by trypsinization. Nuclei from harvested cells were isolated and digested with 60–80 U/ml DNase I (Roche, Indianapolis, IN) for 3 min at 37°C. Digested DNA were incubated at 55°C with 10 μg/ml RNase A (Roche) for a few hours to overnight followed by addition of 25 μg/ml Proteinase K (Ambion, Austin, TX) and incubation at 55°C for at least 4 hrs. DNA fragments were purified by phenol-chloroform extraction and ultracentrifugation through a sucrose gradient. After purification, fragments (between 100 and 500 bp in size) were then pooled, precipitated and assembled into libraries for sequencing.
ChIP-seq and DNase I-seq data analysis
Sequence reads (36-mer) were generated for ChIP-seq and DNaseI-seq experiments on the Illumina Solexa genome analyzer platform and tags were uniquely aligned to the mouse reference genome (UCSC mm9 assembly). A description of the replicate datasets is presented in Supplementary Table 2. In total, 58.6, 69.3, 56.7, and 8.4 million uniquely aligned reads were obtained for Brg1, Chd4, Snf2h, and CTCF, respectively. For the DNase I-seq experiments, 39.7, 32.8 (39.3), 17.5 (40.2), 57.6 (52), and 54.8 (50.1) million uniquely aligned reads were obtained for 3134, 3134Tet +Tet (−Tet), dnBrg1 +Tet (−Tet), dnChd4 +Tet (−Tet), and dnSnf2h +Tet (−Tet), respectively. Regions of enriched tags known as ‘hotspots’ were called and determined significant using algorithms and methods previously described with minor modifications 60, 61. Briefly, ChIP-seq and DNaseI-seq data set tag density values were normalized to 10 million reads to adjust for differences in sequencing depth, and to allow for cross data set comparisons, before hotspots were called at 0% FDR. Replicate concordance was then calculated between replicates. ChIP-seq data sets were additionally normalized by subtracting tags found in the corresponding input data (sequenced sonicated genomic DNA after sequencing depth normalization). In all data, artifacts from sequencing (small regions of high density tags) were filtered out including satellites, long interspersed repetitive elements, and short single tandem repeats after extending these regions on either side to 150 bp. To increase the robustness of called ChIP-seq hotspots, tag density thresholds were determined and applied based on the calculated mode for each set. The final hotspots for each group were required to exceed this value and were: 16 for Brg1, 8 for Chd4, and 9 for Snf2h. In comparisons of data sets, regions were considered to overlap if at least 2 bp were shared. Changes in chromatin accessibility in the presence or absence of dominant-negative variants (changes in DNaseI-seq hotspot tag density values in the presence or absence (expression of dominant-negative variant) of tetracycline (Tet)) were determined by initially filtering each set of hotspots against hotspots generated in the control 3134Tet cell line. Specifically, hotspots with −Tet/+Tet ratios >2 in the 3134Tet data were removed in the corresponding dominant-negative sets. Hotspots in these cell lines were defined as ‘lost’ if the site is found only in the +Tet (dominant-negative variant not expressed) DNase data, ‘gained’ if found only in the −Tet (dominant-negative variant expressed) DNase data, and ‘conserved’ if found in the both +Tet and −Tet DNase data sets. To rule out differences created by comparisons of very small hotspots (e.g., the difference between 24/12 is the same as 4/2), tag density thresholds based on mode were applied to dominant-negative variant DNaseI-seq hotspots before classifications. These values were: 16 for dnBrg1 +Tet, 9 for dnBrg1 −Tet, 8 for dnChd4 +Tet, 5 for dnChd4 −Tet, 8 for dnSnf2h +Tet, and 8 for dnSnf2h −Tet.
Western blots
Cells were grown with and without tetracycline (for dominant-negative variant expression) for 48 hrs followed by two washes in cold PBS and suspension in PBS containing protease inhibitors (EDTA-free complete protease inhibitor cocktail, Roche). Cell pellets isolated by centrifugation were then suspended in buffer (50 mM Tris (pH 8.0), 250 mM NaCl, 5 mM EDTA, 20 mM Na2HPO4, and 0.1% NP-40) containing protease inhibitors and lysed by freeze-thaw (3X). Protein concentrations of collected supernatants (whole cell lysates) were measured by Bradford assay and 50 to 75 μg of cell lysates were separated by electrophoresis on 3–8% NuPAGE Novex Tris-Acetate gels (Invitrogen) followed by transfer to PVDF membranes. After blocking in TBST containing 5% milk, membranes were probed with primary antibodies for anti-FLAG (1:1000, F1804, Sigma-Aldrich, St. Louis, MO), anti-tetracycline transactivator (tTA, 1:5,000, TET01, MoBiTec, Boca Raton, FL), and anti-Actin (1:500, sc-1615, Santa Cruz, Santa Cruz, CA) in TBST with 5% milk overnight at 4°C. Following several washes, membranes were probed with HRP-conjugated secondary antibodies, extensively washed, and visualized with Super Signal Pico detection reagent (Pierce (Thermo Scientific), Rockford, IL). Membranes were exposed to X-ray film to capture images.
De novo DNA sequence motif discovery analysis
Analysis of de novo DNA sequence motifs were performed on ChIP and DNase I hotspots using the MEME algorithm 62. For ChIPs, 150 bp peaks derived from the top 2,000 hotspots (by tag density) were analyzed for each of the indicated groups, while the top 1,000 hotspots from the selected DNase I data sets were analyzed using a width of 150 bp. The minimum and maximum motif size was 8 bp and 40 bp, respectively, with a maximum of 50 motifs used for the search. Following MEME analysis, motif comparisons and identification of enriched sequences in unknown motifs (MEME E values < 10−2) were performed using a TOMTOM search against the Transfac database of characterized transcription factor motifs. Matches were considered significant if the majority of sequence nucleotides were shared and P values were < 10−4.
Preparation of nuclear extracts
Nuclear extracts were prepared according to standard nuclear fractionation protocols (Abcam). For co-immunoprecipitation experiments, 200 μg of nuclear extract were incubated overnight in a total volume of 1 ml at 4°C with or without 1 μg of antibody (anti-BRG1 (Epitomics), anti-CHD4 (Abcam), or anti-SNF2H (Abcam) in IP buffer (10 mM Hepes (pH 8.0), 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Tween-20, and 300 mM NaCl). To isolate antibody-protein complexes, 20 ul of washed Protein A beads (Sigma-Aldrich) were added to each sample and incubated at 4°C for 1–2 hours followed by two washes in wash buffer (10 mM Hepes, pH (8.0), 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Tween-20, 100 mM NaCl and 5% glycerol). Antibody-protein complexes were eluted from beads by heating for 5 min at 100°C in 20 μl SDS-PAGE sample buffer and DTT. Purified complexes were separated by electrophoresis on 3–8% NuPAGE Novex Tris-Acetate gels (Invitrogen) followed by transfer to PVDF membranes and blocking in TBST, 5% milk. Blocked membranes were incubated overnight at 4°C with the following antibodies in TBST, 5% milk: anti-BRG1 (1:30,000), anti-CHD4 (1:2,000), anti-SNF2H (1:2,000), anti-WSTF (1:500, W3641, Sigma-Aldrich), anti-BAF155 (1:500, B5186, Sigma-Aldrich), and anti-HDAC1 (1:4,000, PA1-860, Pierce). Following several washes, membranes were probed with HRP-conjugated secondary antibodies, extensively washed, and visualized with Super Signal Pico detection reagent (Pierce). Membranes were exposed to X-ray film to capture images.
Preparation of RNA and quantitative real-time PCR (QPCR) analysis
RNA was extracted from cells grown in media with or without tetracycline for 48 hers according to standard methods using Trizol reagent (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA). All RNA samples were treated with RNase-free DNase (Qiagen). Following purification, reverse transcription of total RNA was performed using the BioRad cDNA Synthesis Kit via the manufacturer’s instructions and analyzed by real-time QPCR using SyBr green (BioRad). Primer sequences are available upon request.
Statistical analysis/calculation of P values
Significantly different P values between box plot data sets were determined by calculating means and analyzing these values using a Kolmogorov-Smirnov test (R function KS.test). For box plots with multiple comparisons, significant differences between means were determined for pair-wise comparisons by variance analysis (one-way ANOVA) combined with Tukey’s Honestly Significant Difference test.
For Supplementary Figure 1d, we performed the nonparametric Kruskal-Wallis rank sum test for the equality of tag density medians on peaks among the five groups. For all 3 chromatin remodelers (Brg1, Chd4, Snf2h), the test results correspond to p-values less than 2.2e-16. Therefore, the null hypotheses were rejected and we concluded that tag densities were not identically distributed among the groups. Next, Tukey’s HSD (Honestly Significant Difference) test was performed for the equality between all possible pairs of groups. For Brg1, the test showed that the density mean of promoter hotspots is significantly lower than those of the other groups (all 4 corresponding p-values < 2.2e-16). However, for Chd4, only two pair-wise comparison tests between (intron vs. downstream, promoter vs. downstream) have p-values less than 0.001 that is adjusted for multiple comparisons using Bonferroni correction.
Sequential ChIP (re-ChIP) analysis
Re-ChIP experiments were performed as described previously 59, with changes as follows. The first IP was performed as described in John, et al with the exception of the elution step, which was performed in 10mM DTT at 37°C for 30 minutes. DTT elutions were diluted 1:60 in ChIP dilution buffer and antibodies for the second IP were then added for overnight incubation at 4°C. The Re-ChIP IPs were then processed as per 59.
Analysis of cross-regulation by dn-remodeler proteins
For Supplementary Figure 6 e–g, all cells were maintained in DMEM media (Invitrogen) supplemented with 10% Tet-approved FBS (Clontech cat no 631101) and 10 ug/ml tetracycline. Cells were washed twice with PBS, typsinized and 1 million cells plated in 10 cm dishes either in the presence or absence of Tet. Cells were collected 48 hours later by trypsinization and pelleted in growth media. The cells were washed twice with PBS and the pellet was lysed in 100 ul of RIPA Buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH8.0) supplemented with a protease inhibitor cocktail (Sigma cat no P2714). The protein concentrations of the resulting cell extracts were measured using the Pierce BCA Protein Assay Kit (Prod # 23227). 15 ug of each cell extract was loaded onto a 15 well 1 mm NuPAGE 4–12% Bis-Tris Gel (Invitrogen cat no. NP0323) along with 8 ul of Novex SeeBlue prestained marker (Invitrogen cat no. 100006636) and run at 200 volts in the Mops NuPage buffer system. The proteins were transferred to a nitrocellulose membrane in Tris Glycine transfer buffer supplemented with 0.037% SDS at 400 mA for 2 hours to facilitate transfer of high molecular weight proteins. The membranes were blocked with 5% non-fat dry milk (NFDM) and probed with the following antibodies diluted in NFDM: Rabbit anti-FLAG polyclonal 1:2000 (Sigma F7425, Lot No. 068K4800); Mouse anti-CHD4 monoclonal 3F214 1:2000 (Abcam ab70469, Lot No. GR104037-3); Rabbit anti-Snf2h/ISWI polyclonal 1:2000 (Bethyl A301-017A, Lot No A301-017A-1); Rabbit anti Brg1 custom monoclonal antibody (described in this manuscript) 1:20,000; Mouse anti-GAPDH 6C5 monoclonal antibody (Abcam ab 8245, Lot No. 917777). The blots were probed with the appropriate species anti-IgG antibodies conjugated to HRP at 1:5000 in NFDM (Jackson Labs Goat anti-Mouse cat no 115-035-003 and Goat anti-Rabbit cat no 111-035-144). Protein bands were detected using Pierce SuperSignal West Pico Chemiluminescent Substrate (Pierce Cat No. 34080, Lot No. MC154418) and imaged on a Biorad ChemiDoc MP imaging system.
Supplementary Material
Acknowledgments
The authors thank D. Picketts (U. Of Ottawa) and J. Svaren (U. Of Wisconsin) for the kind gift of remodeler cDNA constructs (hSNF2H and mChd4, respectively), A. Indrawan for technical assistance, the National Cancer Institute Advanced Technology Program Sequencing Facility for sequencing services, and Epitomics, Inc. for generation of the monoclonal rabbit BRG1 antibody. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and by a UNCF-Merck Postdoctoral Science Research Fellowship to S.A.M.
Footnotes
AUTHOR CONTRIBUTIONS
S.A.M. and G.L.H. conceived of and designed the study. S.A.M., M.W., T.J. and R.L.S. performed the experiments. S.J. provided technical advice. S.B. and M.S. conducted the bioinformatics analysis. S.A.M. performed the experimental analysis and data interpretation. S.A.M. and G.L.H. wrote the manuscript.
References
- 1.Bork P, Koonin EV. An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 1993;21:751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 7.Rippe K, et al. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci U S A. 2007;104:15635–15640. doi: 10.1073/pnas.0702430104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Vugt JJ, et al. Multiple aspects of ATP-dependent nucleosome translocation by RSC and Mi-2 are directed by the underlying DNA sequence. PLoS ONE. 2009;4:e6345. doi: 10.1371/journal.pone.0006345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer LA, et al. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes [In Process Citation] J Biol Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 11.Agalioti T, et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 12.Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5:464–469. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alenghat T, Yu J, Lazar MA. The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor. EMBO J. 2006;25:3966–3974. doi: 10.1038/sj.emboj.7601280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan C, Varga-Weisz P. The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res. 2007;618:41–51. doi: 10.1016/j.mrfmmm.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sala A, et al. Genome-wide characterization of chromatin binding and nucleosome spacing activity of the nucleosome remodelling ATPase ISWI. EMBO J. 2011;30:1766–1777. doi: 10.1038/emboj.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 19.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2010;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 22.Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p [In Process Citation] Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 23.Schultz DC, Friedman JR, Rauscher FJ., III Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao M, et al. Inhibition of cyclin D1 gene transcription by Brg-1. Cell Cycle. 2008;7:647–655. doi: 10.4161/cc.7.5.5446. [DOI] [PubMed] [Google Scholar]
- 26.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biddie SC, John S, Hager GL. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol Metab. 2010;21:3–9. doi: 10.1016/j.tem.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corona DF, et al. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 31.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 32.Hakimi MA, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 33.Dirscherl SS, Henry JJ, Krebs JE. Neural and eye-specific defects associated with loss of the imitation switch (ISWI) chromatin remodeler in Xenopus laevis. Mech Dev. 2005;122:1157–1170. doi: 10.1016/j.mod.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2006;281:15129–15137. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- 35.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 36.Moshkin YM, et al. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol. 2012;32:675–688. doi: 10.1128/MCB.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gkikopoulos T, et al. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao H, et al. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci U S A. 2009;106:11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol. 2012;32:1312–1320. doi: 10.1128/MCB.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagaich AK, Walker DA, Wolford RG, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- 45.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor regulated promoter. Mol Biol Cell. 2008;19:3308–3322. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA Binding Directs Nucleosome Sliding By Chd1. Mol Cell Biol. 2011;31:4746–4759. doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 50.Voss TC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Won KJ, et al. An integrated approach to identifying cis-regulatory modules in the human genome. PLoS ONE. 2009;4:e5501. doi: 10.1371/journal.pone.0005501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bultman S, et al. A Brg1 Null Mutation in the Mouse Reveals Functional Differences among Mammalian SWI/SNF Complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 53.Siatecka M, Xue L, Bieker JJ. Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol Cell Biol. 2007;27:8547–8560. doi: 10.1128/MCB.00589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sims RJ, III, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hassan AH, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 57.Fragoso G, Pennie WD, John S, Hager GL. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol. 1998;18:3633–3644. doi: 10.1128/mcb.18.6.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voss TC, et al. Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity. J Cell Sci. 2009;122:345–356. doi: 10.1242/jcs.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.John S, et al. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 60.Siersbaek R, et al. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baek S, Sung MH, Hager GL. Quantitative analysis of genome-wide chromatin remodeling. Methods Mol Biol. 2012;833:433–441. doi: 10.1007/978-1-61779-477-3_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.