Abstract
Genomic, transcriptional, and proteomic analyses of brain tumors reveal subtypes that differ in pathway activity, progression, and response to therapy. However, a number of small molecule inhibitors under development vary in strength of subset and pathway-specificity, with molecularly targeted experimental agents tending toward stronger specificity. The Notch signaling pathway is an evolutionarily conserved pathway that plays an important role in multiple cellular and developmental processes. We investigated the effects of Notch pathway inhibition in glioma tumor initiating cell (GIC, hereafter GIC) populations using γ secretase inhibitors. Drug cytotoxicity testing of 16 GICs showed differential growth responses to the inhibitors, stratifying GICs into responders and non-responders. Responder GICs had an enriched proneural gene signature in comparison to non-responders. Also gene set enrichment analysis revealed 17 genes set representing active Notch signaling components NOTCH1, NOTCH3, HES1, MAML1, DLL-3, JAG2 etc., enriched in responder group. Analysis of TCGA expression data set identified a group (43.9%) of tumors with proneural signature showing high Notch pathway activation suggesting γ-secretase inhibitors might be of potential value to treat that particular group of proneural GBM. Inhibition of Notch pathway by γ-secretase inhibitor treatment attenuated proliferation and self-renewal of responder GICs and induces both neuronal and astrocytic differentiation. In vivo evaluation demonstrated prolongation of median survival in an intracranial mouse model. Our results suggest that proneural GBM characterized by high Notch pathway activation may exhibit greater sensitivity to γ-secretase inhibitor treatment, holding a promise to improve the efficiency of current glioma therapy.
Keywords: Notch activation, proneural genes, γ secretase inhibitors, Glioma
Introduction
Glioblastoma (GBM) is the most intractable cerebral tumor; it progresses rapidly, demonstrating high resistance to both radiotherapy and most chemotherapeutic agents (1–3). Traditional genomic studies of GBMs have revealed common alterations in canonical central signaling pathways (4–8). Initial attempts at gene expression profiling have demonstrated the heterogeneous nature of glial tumors that were previously thought to represent a single tumor type. Analysis of large-scale genomic data sets has started to yield results for discriminating subgroups of patients with GBM who seem to respond differently to chemotherapeutic agents (9–11). As new-generation drugs are developed that target specific protein targets, personalized treatment for brain cancer may ultimately move beyond a gene expression-based molecular classification model towards the identification of small subsets of patients who are likely to respond to a particular agent.
Recent studies suggest that glioma tumor-initiating cell (GIC) have the capacity to repopulate tumors (12–14) and mediate radio- and chemo resistance (15, 16). However, the mechanism by which GICs maintain their immature stem-like state or, alternatively, become committed to differentiation is poorly understood (13, 14). The Cancer Genome Atlas (TCGA) Network described genomic abnormalities and gene expression-based molecular subtypes of GBM that showed a strong relationship between subtypes, genomic alterations, and different neural lineages (17). Future studies are required to elucidate the intricate relationship between tumor subtypes and treatment response. We have thus embarked on a comprehensive effort to detect expression signatures that are associated with response to therapy, and these signatures may allow prospective selection of patients with a high likelihood of response to therapy.
The Notch signaling pathway is an evolutionarily conserved pathway that plays an important role in multiple cellular and developmental processes, including cell fate decision, differentiation, proliferation, survival, angiogenesis, and migration (18–20). The Notch protein comprises both extracellular and intracellular regions. The extracellular region of the Notch receptor contains epidermal growth factor-like repeats essential for ligand binding and three copies of the juxta membrane repeats motif, known as the Lin-12 Notch repeats. Four Notch receptors (Notch1-4) and five ligands (Jagged1, Jagged2, and Delta-like 1, 3, and 4) have been identified in mammals. When the Notch receptor is triggered by the ligand, it promotes two proteolytic cleavage events at the Notch receptor. The first proteolytic step is carried out by ADAM17 at site S2 in the extracellular region. The cleaved extracellular subunit of the receptor is trans-endocytosed by the neighboring ligand-expressed cells. Binding of extracellular ligand to Notch also induces the second proteolytic cleavage event at the transmembrane region by a γ-secretase at site S3. This cleavage can release the Notch intracellular domain (NICD) (18, 19).
Recent studies indicate that the Notch signaling pathway regulates neural stem cells (21, 22)and several cancer cells with high Notch activity (23–25). Among central nervous system tumors, Notch signaling components have been found to be deregulated in meningiomas (26), and Notch activity has been observed to be critical in medulloblastomas (27). In glioma cells and GICs, the Notch signaling pathway plays an important role in proliferation, stem cell maintenance, cell differentiation and tumorigenesis (28–34). γ-Secretase inhibitor repressed the growth of several cancer cell types, such as breast cancer (25), leukemia (24), and glioma (33, 34). Therefore, the pharmacological inhibition of Notch signaling pathway has potential as a therapy for cancer.
In this study, we investigated the effects of Notch pathway inhibition on GIC growth using commercially available inhibitors of γ-secretase (DAPT, BMS-708163 and RO4929097). The cells responses to the γ-secretase inhibitor treatment divided GICs into groups of responders and non-responders based on their 50% growth inhibitory concentration(IC50) profiles. Furthermore, the GICs’ response to γ-secretase inhibitors was associated with their Notch signaling activity. γ-Secretase inhibitors impaired stem cell maintenance and induced neuronal and astrocytic differentiation. Notch-1 inhibition accounts for part of the γ-secretase inhibitors activity. Notch inhibition by short hairpin RNA (shRNA) knockdown induced only neuronal differentiation, whereas γ-secretase inhibitors induced both neuronal and astrocytic differentiation. Thus, we have here identified a responder signature for a group of GICs that can be targeted by Notch inhibitors, and this may allow the selection of patients with a high likelihood of responding to this therapy.
Materials and Methods
Cell lines and reagents
The GIC cell lines were established by isolating neurosphere-forming cells from surgical specimens of human GBM using a method described previously (35). The study was approved by the Institutional Review Board of MD Anderson Cancer Center, and informed consent was obtained from all subjects. These GICs were cultured as GBM neurospheres in DMEM/F12 medium containing B27 supplement (Invitrogen) and basic fibroblast growth factor and epidermal growth factor (20 ng/ml each). The γ secretase inhibitors N-[N-(3,5-difluorophenacetyl)-1-alanyl]-S-phenylglycine t-butyl ester (DAPT), BMS-708163 and RO4929097 were purchased from Selleck Chemicals (Houston, TX). For in vitro use, all the inhibitors were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO) to a concentration of 10 mmol/L, stored at −20°C, and further diluted to an appropriate final concentration in DMEM/F12 medium at the time of use. For in vivo studies, RO4929097 was formulated as a suspension in 1.0% carboxymethyl cellulose with 0.2% Tween 80 for oral administration.
Cell proliferation assay
Briefly, cells were treated in triplicate for 72 hours with seven doses each of DAPT, BMS-708163 and RO4929097. Cell proliferation was estimated using the CellTiter-Blue (Promega, Madison, WI) viability assay. The IC50 value was calculated as the mean drug concentration required for inhibiting cell proliferation by 50% compared with vehicle-treated controls.
Gene arrays, bioinformatics
GICs were divided into responder and non-responder groups by their responses to γ secretase inhibitor treatments. To identify differentially expressed pathways between responder and non-responder cell lines, we performed gene set enrichment analysis (GSEA) (36) using canonical pathways from the Kyoto Encyclopedia of Genes and Genomes (37) (downloaded from MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp). Core enrichment genes were used as 17 gene signatures to represent the Notch pathway.
Affymetrix HGU133A CEL files of 533 TCGA GBM samples were downloaded from TCGA data portal and preprocessed using the Aroma (http://www.aroma-project.org/package version v2.8.0)(38). Applying single sample GSEA (ssGSEA)(39), we classified the GBMs into proneural, neural, classical and mesenchymal subtypes (17). To illustrate the expression pattern of the Notch pathway in cell lines and clinical samples, we projected the 17-gene signature to the cell line gene expression data set and TCGA data set.
Western blot analysis
Cells were harvested in lysis solution as previously described (40) and subjected to Western blotting. Membranes were probed with the following primary antibodies: Anti-Notch-1, anti-Cleaved Notch-1 (all from Cell Signaling, Boston, MA), anti-Hes-1, anti-Hes-5 (all from Millipore), and anti-Hes-3 (Sigma-Aldrich). Anti-β-actin antibody was purchased from Sigma and used as loading control.
Neurosphere formation assay
For the primary neurosphere formation assay, GICs were seeded in 96-well plates (100 cells/well) and were incubated with various concentrations of γ-secretase inhibitors (DAPT 10 μM, BMS-708163 2 μM and RO4929097 2 μM) for 7 days. For the secondary neurosphere formation assay, primary neurospheres were dissociated into single cells and 50 such cells were in each well of 96-well plates in the absence of inhibitors. After 7 days, total neurosphere numbers were counted under a dissection microscope using 4×4 objective magnification.
Cell Growth Assay
GICs were seeded in 6-well plates (1×105 cells/well). Cell numbers were counted using a hemocytometer and cell were counted every day for 7 days. Experiment was repeated three times and the average value was reported.
Indirect immunofluorescence
Immunofluorescence staining was performed as described previously (41). Cells were seeded at a concentration of 2 × 105 cells/well in 6-well plates with polylysine-coated coverslips inside and left for 24 hours in the incubator. The following day, cells were treated with the indicated dose of γ-secretase inhibitors (DAPT, BMS-708163 and RO4929097) for 5 days, media were aspirated, and the cells were washed with phosphate-buffered saline (PBS) once, before being fixed with 4% formaldehyde in PBS for 20 minutes. After another PBS wash, the cells were permeabilized with 0.1% Triton X-100 in PBS for 5 minutes and then blocked with 5% goat serum in PBS (containing 0.1% Tween 20) for 1 hour. Cells were then incubated with primary antibodies (Anti-Nestin and anti-GFAP from Cell Signaling, anti-CNPase from Millipore, and anti-TuJ-1 from Sigma-Aldrich) overnight at 4°C. After 2 washes with PBS (containing 0.1% Tween 20), the cells were incubated with the indicated fluorescently labeled secondary antibody in the dark at room temperature for 1 hour. The cells were counterstained with Vecta shield sealant containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, California, USA) and examined under a confocal laser-scanning microscope (Carl Zeiss Microscopy). Pictures were scanned with a DFC340FX camera. The percentage of positively stained cells in each section was determined at a magnification of 400 by counting 500 cells in a randomly selected field.
Knockdown of Notch-1 by lentiviral shRNA
Lentiviral vector encoding shRNA for Notch-1 was purchased from Genecopoiea Inc. (Rockville, MD). The lentiviral vector pLKO.1-mediated expression of shRNA for targeting human Notch-1 was performed according to the manufacturer’s instructions. Briefly, lentiviral particles expressing targeting or control scramble shRNA were produced in HEK293FT cells with the mixed set of packing plasmids, and the viruses were concentrated and titered as previously described (42). GSC35 cells were infected with the Notch-1 shRNA. Produced lentiviruses were concentrated using the Centricon Plus-20 centrifugal filter device (Millipore). To ensure the same number of lentiviral particles were used in each experiment, produced lentiviral stock was titered and stored at −80°C. For in vitro infection of GICs with the lentivirus, we disaggregated cultured tumorspheres prior to infection in order to increase the infection efficiency and uniformity. To infect target cells by lentiviruses, we exposed GSC35 24 hours. Cells were washed and then cultured with regular complete medium for 2 additional days in the presence of 2.5 μg/μl puromycin. Finally, the cells were washed and analyzed for protein expression by Western blotting.
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from GICs with the RNeasy Mini kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. RT-PCR was performed with SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen, Grand Island, NY), according to the manufacturer’s instructions.
Animal studies
Mice were maintained in a pathogen-free environment and used in accordance with the MD Anderson Animal Care and Use Guidelines. We examined the antitumor efficacy of RO4929097 in intracranial xenografts derived from GSC35. Nude (nu/nu) 6- to 8-week-old mice (n = 18) were obtained from the MD Anderson breeding facility. In this study, we implanted 5 × 105 GSC35 cells in DMEM/F12 serum-free media (SFM; 5 μL) intracranially into the caudate nucleus of each mouse using a guide-screw system, as described previously (43) and then randomly divided the mice into 3 groups of 6 mice each. Starting on day 4 after the tumor cells were implanted, mice were treated once per day by oral gavage with 10 or 20 mg/kg RO4929097 in 1.0% carboxymethyl cellulose with 0.2% Tween 80 or 1.0% carboxymethyl cellulose with 0.2% Tween 80 alone (control) on a 5-days-on, 2-days-off schedule for 4 weeks. Mice were monitored daily and euthanized when they became moribund. Then, the whole brain was extracted for rapid freezing in liquid nitrogen and storage at −80°C. At necropsy, all organs were analyzed grossly and microscopically to assess tolerability at the tested doses.
Immunohistochemical staining
Sections (5 μm thick) of formalin-fixed, paraffin-embedded whole brain from control vehicle- and RO4929097–treated animal specimens were stained with anti-Notch-1, anti-NICD, anti-Nestin, anti-Ki-67 (all from Cell Signaling), anti-Hes-1 (Millipore), and anti-TuJ1 (Sigma-Aldrich). The sections were visualized by using a diaminobenzidine substrate kit. The slides were examined under a bright-field microscope.
Statistical analysis
Statistical analysis was performed using the Student’s unpaired t–test. Results are presented as the mean of at least three independent experiments. Survival analysis was performed using the log-rank analysis module in SPSS 10.0 (SPSS Inc.). Differences were considered significant at p<0.05 for all comparisons.
Results
GICs exhibit differential sensitivities to γ secretase inhibitors
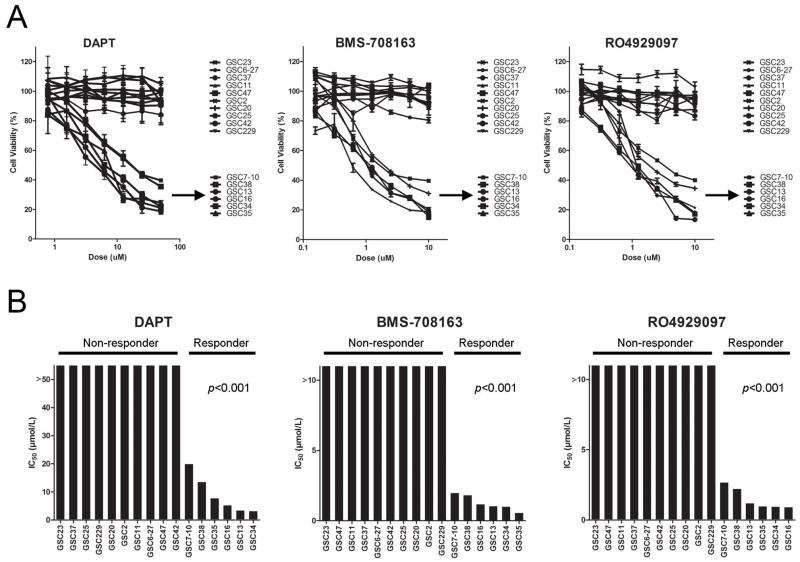
We quantified the sensitivity in a panel of 16 cell-lines to γ secretase inhibitors by measuring the concentration needed for each compound to inhibit proliferation by 50% (designated the IC50, the half maximal inhibitory concentration) after 72 hours of continuous exposure. Γ secretase inhibitors effectively showed dose-dependent growth inhibition of GICs (Fig. 1A). Representative waterfall plots showing responses to the γ secretase inhibitors are shown in Figure 1B; they reveal groups of cells showing differential responses to γ secretase inhibitors, dividing the GICs into a responder group and a non-responder group. Responder group was defined by the cell lines showing an IC50 values ranging between 5–15 μmol/L for DAPT and 0.5–2 μmol/L for BMS-708163 and RO4929097. GICs showing IC50 values above 5–15 μmol/L for DAPT and 0.5–2 μmol/L for BMS-708163 and RO4929097 were considered as non-responders.
Figure 1. γ Secretase inhibitors effectively showed dose-dependent growth inhibition of GICs.
A) A panel of GICS lines was treated with various concentrations of γ secretase inhibitors DAPT, BMS-708163 and RO4929097. Cells were treated with increasing concentrations of γ secretase inhibitors in triplicate wells for 72 hours, and cell viability was assessed by CellTiter-Blue assay as described in Materials and Methods. The results shown are of a single experiment with 3 independent replicates cell viability was measured by CellTiter-Blue assay. The graph depicts cell viability at 72 hours. Cell viability in the vehicle control was considered as to be 100%. B). Waterfall diagram of IC50 of 16 GICs. *p<0.001 responder GICs vs non-responder GICs.
We studied the proliferative potential and in-vitro tumorogenicity of responder (GSC13 and GSC35) and non-responder (GSC2 and GSC20) GICs. All the responder cell lines and non-responder cell lines showed similar exponential growth pattern during 7 days of growth curve with GSC35 a responder cell line with slightly slower growth rate (Supplemental Fig. S1). In an in-vitro tumorigenicity assay as assessed by the ability to form primary neurospheres, all 4 GICS (responder: GSC 35 and GSC13 and non-responder: GSC2 and GSC20) showed similar neurosphere formation ability showing that the GICs had tumorigenic potential in-vitro(Supplemental Fig. S2). The in-vivo biological behaviors of two groups were studied by injecting cells orthotopically into mouse brain and GICs from two groups (responder: GSC 35 and GSC13 and non-responder GSC2 and GSC20) formed tumors in mice clearly showing that both responders and non-responders are tumorigenic. We also show that in-vivo tumors from responder GICs exhibit proneural characterstics as shown by OLIG2 and Nestin positive staining where as non-responder GICs tumors show mesenchymal marker YKL-40 (Supplementary figure S2).
γ Secretase inhibitor responder GICs are enriched in proneural signature
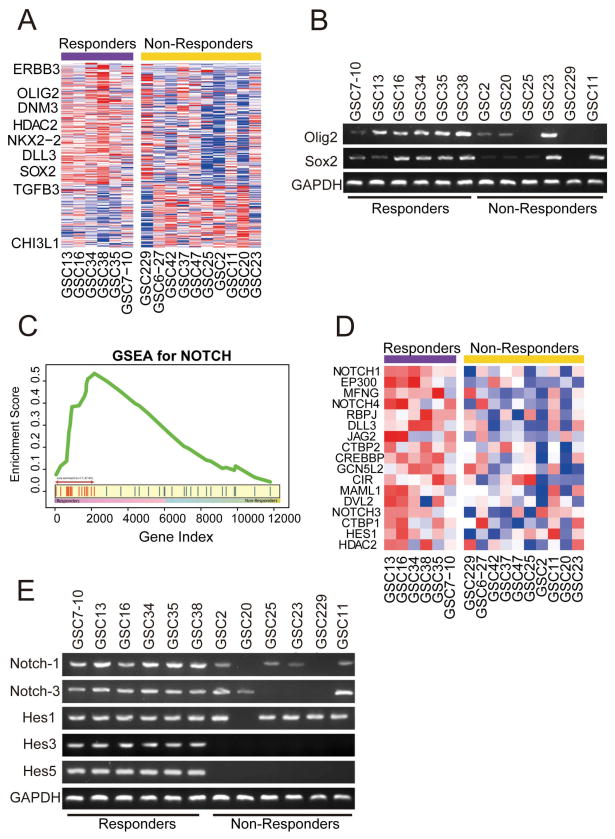
We compared the expression profile of responders and non-responders GICs and applied TCGA subtype gene cluster on gene expression data (Affymetrix U133A2) from 14 GIC cell lines (Fig. 2A). Expression data analysis identified several genes highly expressed in the responder group and divided 14 GICs into two major groups TCGA gene signature. The responder cell lines strongly associated with response to γ secretase inhibitors included the subtype with a proneural background, showing enrichment of proneural TCGA signature, including OLIG2, SOX2, and ERB3 (Fig. 2A). Rest of the cell lines showed low expression of proneural gene signature and were designated as non-responders. It is important to note that some of the non-responder cell lines (GSC23) although showing proneural gene expression of Olig2 and Sox2 (Fig 2B) but did not show Notch pathway activation and response to γ-secretase inhibitors were classified as non-responders. The non-responder group in contrast shows expression of CD44, TGFβ1 and FGF13 factors essential for maintenance of non-responders (Supplemental Fig. S3). RT-PCR data validated some of the proneural genes present in responder GICs (Fig. 2B).
Figure 2. Enrichment of Notch pathway components and proneural signature in responder GICs.
A) Hierarchical cluster analysis on gene expression data from 16 GICs identified several genes highly expressed in the responder group and classified 16 GICs into two major groups with unique gene signatures. The proneural gene signature defined by Verhaak et al was projected to responder and non-responder GICs. (B) RT-PCR validating expression of proneural marker genes Olig2 and Sox2 in responder GICs. (C) Up regulation of Notch pathway in responder cell lines by Gene Set Enrichment Analysis. X-axis represents gene ordered by expression changes between responders and non-responders and Y-axis represents cumulative enrichment score. (D) Expression pattern of NOTCH pathway genes in the GIC microarray data set showing enrichment of Notch pathway components mainly NOTCH1, NOTCH3, HES1, MAML1, JAG2, and DLL3 in responder GICs. (E) RT-PCR data validated expression of the Notch pathway genes Notch1, Notch3, Hes1, Hes3 and Hes5 in the responder GICs.
Identification of subtype pathway markers in cell-line clustering
To identify differentially expressed pathways between responder and non-responder cell lines, we performed GSEA using canonical pathways from Kyoto Encyclopedia of Genes and Genomes (Kanehisa et al., 2012). Notch pathway was significantly up regulated in responder (p<0.05) group(Fig. 2C). Of the 38 genes in the Notch pathway, 17 were “core enrichment” genes that were adopted as a gene signature to represent this pathway (Fig. 2D). Core genes were the most deregulated genes and the major contributors to the enrichment score. Here, these genes included NOTCH1, NOTCH3, HES1, MAML1, DLL-3 and JAG2, among others (44, 45). RT-PCR data validated expression of the Notch pathway genes Notch-1, Notch-3, Hes1, Hes3 and Hes5 in the responder GICs (Fig. 2E).
Analysis of human tumor gene expression profiles identifies proneural subtype as having high Notch pathway activity
To investigate the Notch pathway in clinical samples, we projected the 17 Notch gene signatures onto GBM cohort collected by TCGA (Cancer Genome Atlas Research Network, 2008). Affymetrix HGU133A CEL files of 533 TCGA GBM samples were downloaded from the data portal and preprocessed using the aroma package (38). Using ssGSEA(39), these samples were classified into proneural, neural, classical and mesenchymal subtypes (17). As anticipated, the proneural subtype highly expresses the proneural gene signature (Supplemental Fig. S3). We compared expression of Notch signatures between the proneural group and the other groups and applied 17 gene signatures identified above as core enrichment genes. The results show that 43.9% of the proneural GBMs had higher Notch pathway gene expression, suggesting activation of the pathway in a subset of patients (Supplemental Fig. S4).
Notch blockade is pronounced in GICs with high Notch activity
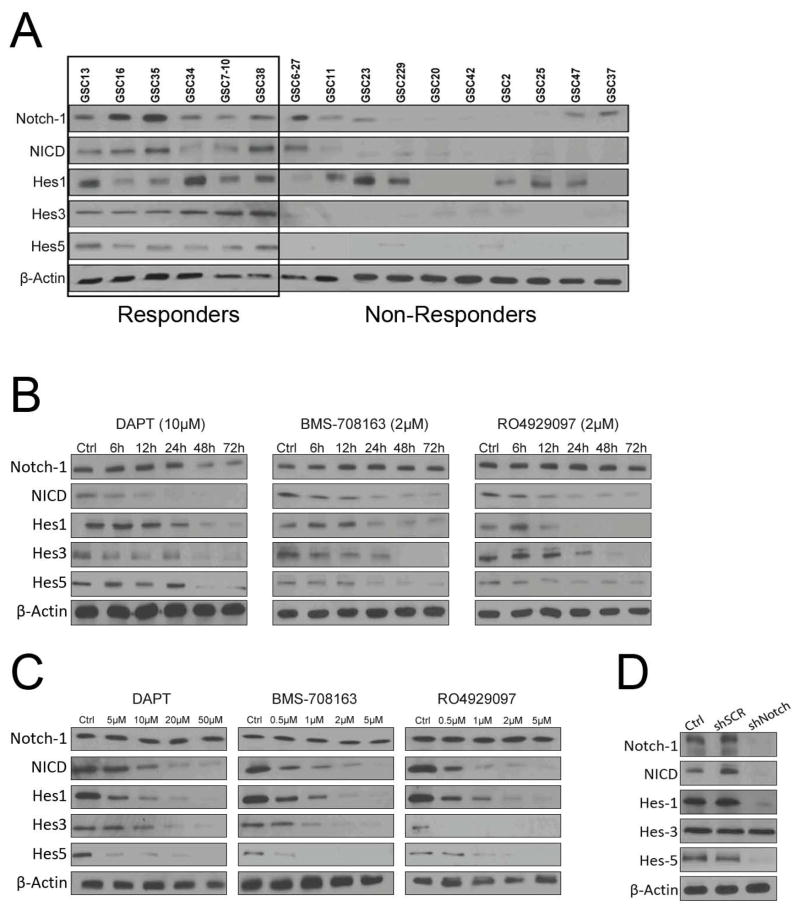
In order to identify any relationship between pathway activation of Notch signaling and the sensitivity of γ secretase inhibitors, we tested expression of genes in the Notch signaling pathway in 16 GICs by Western blot analysis (Fig. 3A). Activated Notch-1 NICD and Notch-1 pathway components, including Hes1, Hes3 and Hes5, were all expressed in the responder group and either absent or present at very low levels in non-responder GICs, indicating that response may be related to activation of the Notch signaling pathway. Hes1 was expressed in 12 GICs, and Hes3 and Hes5 were expressed only in the responder group. It has been reported that Hes1 expression is regulated by genes other than Notch-1, but it is unclear what genes were regulating Hes1 in GICs.
Figure 3. γ Secretase inhibitors suppresses Notch signaling pathway in a dose- or time-dependent manner.
A) Western blotting was performed to analyze the cellular protein levels of Notch signaling pathway in 16 GICs. Activated Notch-1 (NICD) and Notch-1 pathway components Hes1, Hes3 and Hes5, were all expressed in the responder GICs. β-Actin was used as loading control. B) A panel of GICs was treated with the indicated doses of DAPT, BMS-708163 and RO4929097 for indicated time intervals. All the γ secretase inhibitors inhibited expression of NICD, Hes1, Hes3 and Hes5 in a time-dependent manner. The decrease in NICD was followed by a decrease in Hes1, Hes3 and Hes5 expression, whereas the expression of Notch-1 did not change. C) A panel of GICs was treated with the indicated doses of DAPT, BMS-708163 and RO4929097 for 48 hours. γ Secretase inhibitors inhibited expression of NICD, Hes1, Hes3 and Hes5 in a dose-dependent manner. D) Western blot analysis confirmed the knockdown effect of Notch shRNA lentivirus on GSC35. shRNA knockdown of Notch-1 in GICs decreased Hes1 and Hes5 protein expression.
γ Secretase inhibitors show dose- and time-dependent response in GICs
Notch receptor triggered by the ligand promotes its cleavage by γ secretase. This cleavage can release the NICD, and released NICD translocates into the nucleus and binds CBF-1 to activate Notch targeting genes, such as the Hes genes, to antagonize the activity of proneural genes like Mash1, Math, NeuroD, and Neurogenins (18, 19). To examine the effect of γ secretase inhibitors on the fate of GICs, we used GSC35 to study the time- and dose-dependent effects of γ secretase inhibitors on Notch signaling in vitro. As shown in Fig. 3B, DAPT, BMS-708163 and RO4929097 inhibited expression of NICD, Hes1, Hes3 and Hes5 in a time- and dose-dependent manner (Fig. 3B and 3C). The decrease in NICD was followed by a decrease in Hes1, Hes3, and Hes5 expression, whereas the expression of Notch-1 did not change. This result indicates that γ secretase inhibitors inhibit the cleavage event at the Notch receptor and suppress downstream targets of Notch. Since γ secretase inhibitors have several sites of action and are not specific inhibitors of Notch, the effect of Notch inhibition by γ secretase inhibitors does not reflect specific Notch inhibition.
To gain a better understanding of whether Notch receptors are involved in response to γ secretase inhibitors, we determined the effects of knockdown of Notch-1 through lentivirus-mediated expression of shRNA targeting Notch-1. The efficiency of downregulation of the corresponding Notch receptors and Notch target genes were determined by Western blot analysis (Fig. 3B). To examine the function of Notch in GICs, we compared inhibition by © secretase inhibitors with that of shRNA knockdown of Notch-1 (Fig. 3D). shRNA knockdown of Notch-1 in GICs decreased Hes1 and Hes5 protein expression, unlike treatment with γ secretase inhibitors, which decreased Hes1, Hes3 and Hes5 protein expression. Several reports have suggested that Hes genes are functionally classified into two groups and that not all are regulated by Notch (46). Hes3 is not a downstream target of Notch-1 in GICs, but it inhibited by © secretase inhibitors.
Notch inhibition attenuates proliferation and decreases self-renewal of GICs
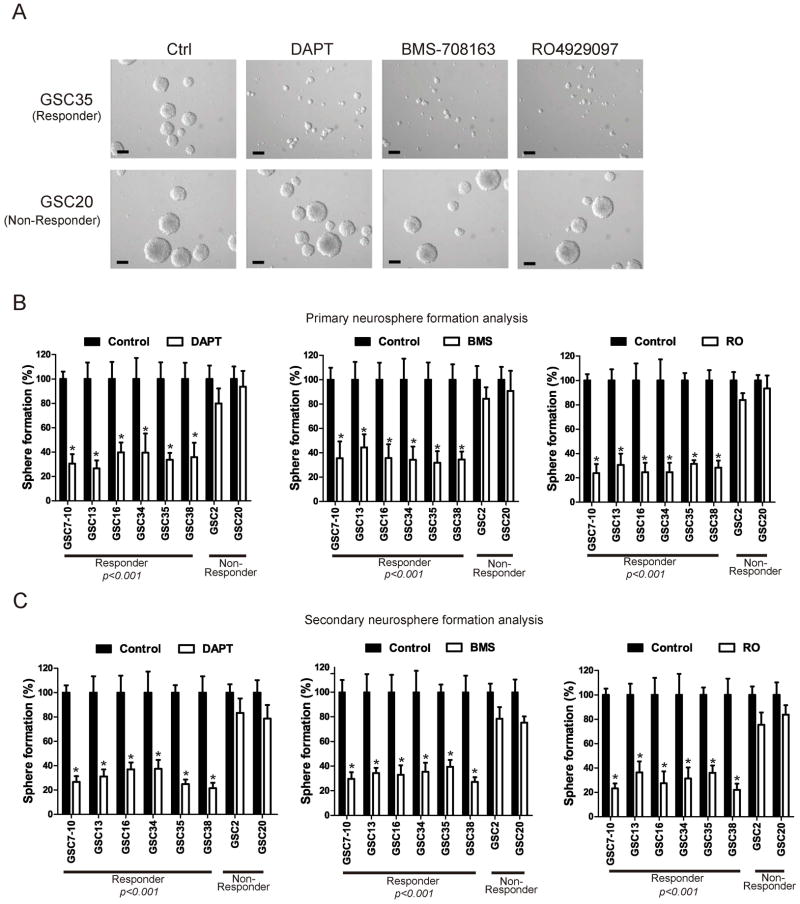
Recent data suggest the role of Notch signaling in the maintenance of normal neural stem cells (21, 22), but whether Notch signaling is involved in tumor stem cells is not fully clear. We analyzed the impact of Notch inhibition on GICs by exposing cells to the γ secretase inhibitors DAPT, BMS-708163 and RO4929097 for 7 days in a neurosphere formation assay. We selected 6 GICs in the responder group (GSC7-10, GSC13, GSC16, GSC34, GSC35 and GSC38), and 2 cell lines in the non-responder group (GSC2 and GSC20). Fig. 4A shows that only responder GICs (GSC35) showed decreased neurosphere formation ability in the presence of the three γ secretase inhibitors. The size of responder GSC35 neurospheres decreased in the presence of the γ secretase inhibitors, whereas inhibitor treatment had no effect on the size of non-responder (GSC20) neurospheres (Fig. 4A). To study the effect of γ secretase inhibitors on GIC proliferation, the primary neurosphere analysis showed that the number of neurospheres decreased significantly in responder GICs (Fig. 4B). To analyze the impact of Notch inactivation on self-renewal, we performed secondary neurosphere formation assays. 7 days after continuous DAPT, BMS-708163 and RO4929097 treatment, cells in the neurospheres were dissociated and placed under neurosphere growth conditions. A significant decrease in secondary neurosphere formation (Fig. 4C), by an average of 84%, was observed in responders after DAPT, BMS-708163 and RO4929097 treatment, compared with non-responder controls (n = 3, p<0.001). These findings suggest that inhibiting Notch results in a decrease in the self-renewal potential of tumor cells, as well as their proliferation.
Figure 4. γ Secretase inhibitors decrease self-renewal capacity of responder GICs.
A) GICs were treated with of γ secretase inhibitors DAPT, BMS-708163 and RO4929097 at doses described in Materials and Methods for 7 days in a neurosphere formation assay and numbers of neurospheres were counted in 10 separate fields using a microscope. All the tested γ secretase inhibitors reduce the number and size of neurosphere after 7 days of treatment in responder GICs. Micrographs showed the representative neurosphere results of for GSC35 (Responder) and GSC20 (Non-Responder). Bars, 250 μm. B) We performed primary neurosphere assay to show the effect on proliferation. All the tested γ secretase inhibitors significantly reduce the number of neurosphere after 7 days of treatment in responder GICs (p<0.001). Neurosphere formation in the vehicle control was considered to be 100%.C) For secondary neurosphere assay to show the effect on self-renewal, neurospheres after 7 days of treatment as indicated above were dissociated in to single cells and grown as single cells into a 96-well plate. Cells were grown for another 7 days after which cells as spheres were counted for self-renewal capacity. The bar graph depicts secondary neurosphere formation after 7 days of replating. Neurosphere formation in the vehicle control was considered as to be 100%.
Notch inhibition induces neuronal and astrocytic differentiation
To test the role of Notch signaling in stem cell maintenance, we performed the differentiation assay in vitro. GSC35 (in the responder group) and GSC20 (in the non-responder group) were plated on polylysine-coated coverslips overnight before exposing the cells to all the three γ secretase inhibitors individually for 5 days. Immunostaining was performed to detect the levels of CNPase (an oligodendrocytic marker), GFAP (an astrocytic marker), TuJ1 (a neuronal marker), and Nestin (a neural stem cell marker). Results show that γ secretase inhibitor treatment decreased the expression of Nestin and increased that of TuJ1 and GFAP in the responder group of GIC: GSC 35 (Fig. 5A), while as there was no effect on non-responder GIC: GSC20. Responder GICs have the potential to differentiate into multiple lineages, and γ secretase inhibitors act as triggers of neuronal and astrocytic differentiation, suggesting that inhibiting Notch signaling in GICs leads to an increase in the number of differentiated cells. However, knockdown of Notch-1 by shRNA decreased the expression of only Nestin and increased that of TuJ1 (Fig. 5B). Therefore, we conclude that Notch knockdown induces only neuronal differentiation, whereas γ secretase inhibitors induce both neuronal and astrocytic differentiation. The difference between γ secretase inhibitor treatment and shRNA knockdown of Notch-1 is in the expression level of Hes3. Notch inhibition by γ secretase inhibitors reduced Hes1, Hes3 and Hes5 expression and induced neuronal and astrocytic differentiation. On the other hand, Notch knockdown reduced Hes1 and Hes5 expression and induced only neuronal differentiation. These findings indicate Hes1, Hes5 and Hes3 expression might exhibit differential effects on neural differentiation in GICs.
Figure 5. Notch inhibition induces neuronal and astrocytic differentiation.
A)GICs were treated with indicated doses of DAPT, BMS-708163 and RO4929097 for 5 days. Cells were stained with CNPase (an oligodendrocytic marker), GFAP (an astrocytic marker), TuJ1 (a neuronal marker), and Nestin (a neural stem cell marker). Nuclei were stained with DAPI. Micrographs showed the representative staining results of for GSC35 (Responder) and GSC20 (Non-Responder) cells. Bars, 100 μm. DAPT, BMS-708163 and RO4929097 induce neuronal and astrocytic differentiation in responder GICs (GSC35). n=4, p<0.05, as compared to marker staining positivity in vehicle control. B) Knockdown of Notch-1 by shRNA decreased the expression of only Nestin and increased that of TuJ1 showing that Notch knockdown induces only neuronal differentiation. Bars, 100 μm.
RO4929097 improves survival in an orthotropic mouse model of GBM
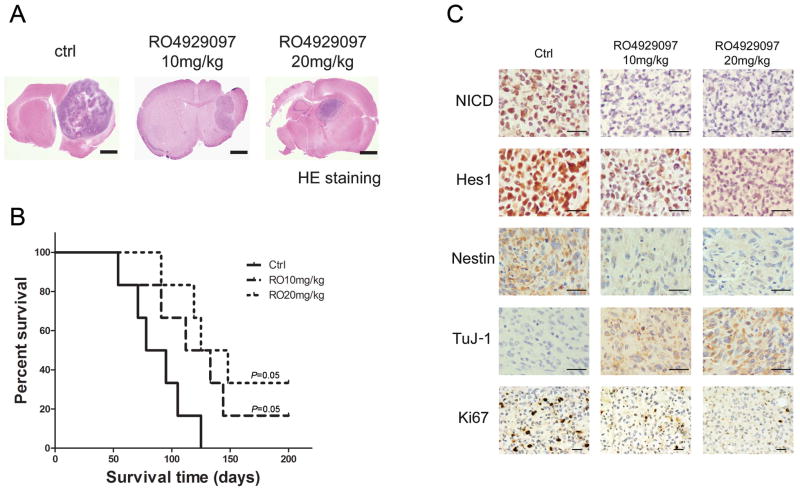
We assessed the anti-glioma efficacy of RO4929097 in vivo using a GSC35 orthotopic model of human gliomaxenografts in immunocompromised mice. The median survival duration for animals treated with methylcellulose [control] was 85 days (Fig. 6A and 6B). Treatment with 10 mg/kg RO4929097 and 20 mg/kg RO4929097 alone extended the survival duration to a median 132 and 185 days, respectively (p= 0.05, log-rank test for both experiments). The results show the potential therapeutic efficacy of RO4929097 for the treatment of human gliomas. The orthotopic tumors after RO4929097 treatment for 4 weeks were analyzed for biological effects. The levels of Notch pathway proteins NICD and Hes1 were lower in RO4929097-treated GSC35 tumors than in untreated tumors, suggesting RO4929097 suppressed Notch pathway activities in vivo. In addition, the treated tumors showed decreased Ki-67 positivity (Fig. 6C) indicating that RO4929097 suppressed proliferation in the responder GIC tumors. Furthermore, tumors showed increased TuJ1 and decreased stem cell marker Nestin positivity after RO4929097 treatment.
Figure 6. RO4929097 extends survival in an intracranial animal model.
GSC35 cells were implanted intracranially in nude mice (n=6 in each group), and treatment was commenced 4 days later. RO4929097 (10 and 20 mg/kg/day) was administered orally 5 times a week for 4 weeks. Control group mice were treated with 1.0% carboxymethyl cellulose with 0.2% Tween 80. Mice were sacrificed at morbidity, and survival curves were compared using Prism 5 software. A)Representative H&E-stained whole brain sections at 4 weeks post-treatment showing the tumor mass. Bars, 1 mm B). Kaplan-Meier survival probability plots of tumor-bearing mice in vehicle or RO4929097 treatment groups (n = 6), using the log-rank method to test for a difference between groups. Treatment of RO4929097 (10 and 20 mg/kg/day) showed a statistically significant improvement over control (p= 0.05, log-rank test for both experiments). C) Immunostaining of the brain sections of animals treated with RO4929097 for 4 weeks. The tissue section was incubated with antibodies against NICD, Hes1, Nestin, TuJ1 and Ki67. Bars, 100 μm.
Discussion
The Notch signaling pathway involved in cell fate decisions during normal development (18–20)and in the genesis of several cancers (23–25) has been implicated both in the maintenance of neural progenitors and in the generation of glia during development of the brain (21, 22). In gliomas, Notch seems to confer radio-resistance to cancer stem cells(32), and inhibition of Notch signaling through γ secretase inhibitors(33) or Delta-4 monoclonal antibody (48) decreased the numbers of CSC and/or their tumorigenicity in some preclinical models, suggesting that therapeutic regimens including Notch inhibitors may be used in the clinic to target CSC and reverse or prevent chemo- or radio-resistance. We have shown that γ secretase inhibitor treatment, suppresses the oncogenic properties of a subgroup of GICs with high Notch pathway activation. Our results indicate that responder group had a gene signature enriched in proneural genes, suggesting γ secretase inhibitors might be of potential value to treat proneural glioma. Furthermore, we show that γ secretase inhibitor treatment decreases cell proliferation, self-renewal and induces differentiation accompanied with increases in animal survival in the γ secretase inhibitor sensitive responder group.
In recent years, a distinct cellular hierarchy has been identified in several hematopoietic and solid tumors, and efforts to personalize the treatment of solid tumors, including brain tumors, are aimed at identifying subsets of patients most likely to benefit from treatment and avoiding treatment-associated morbidity and mortality in patients who are unlikely to respond. Dysregulated Notch activity in human brain tumors was reported by several studies (28–31, 47). However, few studies have investigated the functional relevance of Notch in this tumor type and it is still not clear if and how Notch signaling affects glioma tumorigenesis and maintenance.
In the context of personalized medicine, for the best use of Notch inhibitors and other CSC-targeted agents, various factors need to be determined: 1) which cancers and specific cancer subtypes contain Notch-dependent CSC; (2) what role specific components of Notch signaling play in these CSC; and 3) how can one measure Notch activity in CSC from individual patients. The selection of cancer patients who would benefit from treatment with molecular inhibitors is one of the biggest challenges. TCGA Network described genomic abnormalities and gene expression-based molecular subtypes of GBM with strong relationships between subtypes and genomic alterations. Establishing the association between drug responses and GBM molecular subclasses may help to identify potential cohorts of patients for targeted therapy. The potential clinical utility of these findings is supported by concordance of in vitro-derived molecular predictors of response to therapeutic compounds and clinical results. In the current study, we used neurospheres derived from human GBM specimens (GICs) to examine the effects of Notch pathway blockade. Notch pathway blockade by three structurally distinct γ-secretase inhibitors slowed tumor growth in a subset of GICs with high Notch pathway activation. This subset of GICs was identified with gene set enrichment analysis matching that of the characteristic proneural GBM that predicted response to Notch inhibition. However the clinical application is hindered by complexities arising from intratumoral heterogeneity. Our study showed that only 43.9% of the TCGA proneural tumors were identified as strong Notch pathway expressers after applying the 17-gene Notch signature to the proneural TCGA data set (Supplemental Figure S4). It is therefore important to note that only about 50% of proneural GBMs were identified having the Notch pathway signature and most likely rest 50% of the proneural GBMs have other gene or pathway signatures. Further analysis defining the signatures should aid more specific therapeutic targeting. It is important to note that the frequency of functionally defined CSCs, can vary dramatically among different patients. Moreover, different subpopulations within the tumor possess stem cell properties, which may be isolated using a variety of cell surface markers. It is becoming evident that for the development of new therapeutic strategies it is essential to consider the heterogeneity of brain tumors like GMBs as well as different subtypes. Probably, a unique marker is not enough to identify and characterize tumor-initiating population within the tumor bulk. Moreover, during glioma progression stem cells may be generated via accumulation of more mutations in the cells initially manifesting glioma features. Tumor heterogeneity may represent inherent instability in gene expression markers that confers undifferentiated cell character as opposed to stage in the stem cell lineage defining patterns of gene expression.
Additionally, we showed that Notch signaling components are expressed in human GBM-derived neurospheres. These findings are consistent with those from other previous reports (28–31), showing Notch signaling components were expressed in different brain tumors and played important roles in their pathogenesis. Recently, Verhaak et al. reported that Notch signaling was highly expressed in the classic subtype of GBM (17). We further showed that γ secretase inhibitors inhibited Notch signaling and differentially inhibited GBM cell growth. The difference of growth inhibition could be due to the different cellular context and heterogeneous genetic background of these GBM cell lines. Blocking the Notch pathway suppressed the GICs’ tumorigenic features such as cell viability, self-renewal and differentiation.
These data suggest that Notch inhibition would be effective in targeting the bulk of the tumor as well as eventually minimizing recurrence by inhibiting the proliferation and differentiation of cancer stem cells. Results of this study are therefore anticipated to help identify potential GBM patient cohorts who would gain the most from Notch inhibition. Additional work remains before the signatures reported in this study can be used to select patients for clinical trials. Such future work would include the development of robust and reliable molecular assays that can be applied to clinical samples, establishment of predictive algorithms with decision-making thresholds optimized for clinical use, and validation of predictive power in multiple independent studies.
Supplementary Material
S1) GICs were seeded in 6-well plates with 1×105 cells/well. Cell number was counted by using a hemocytometer every day for 7 days. All the experiments were repeated three times and the average value was reported. All the responder cell lines and non-responder cell lines showed similar exponential growth pattern during 7 days of growth curve with GSC35 a responder cell line with slightly slower growth rate.
S2) In an in-vitro tumorigenicity assay as assessed by the ability to form primary neurospheres, all 4 GICS showed similar neurosphere formation ability showing that the GICs had tumorigenic potential in-vitro. Bars, 250 μm. To create the intracranial disease model, we engrafted GICs into the caudate nucleus of each mouse using a previously described guide-screw system (Lal et al., 2000) Briefly, 5×104 dissociated tumor cells in 5 μl neurobasal medium were injected stereotactically into 6- to 8-week-old NOD-SCID mouse. Animals were sacrificed when they developed weight loss greater than 10% or neurological symptoms. The tissue sections were incubated with antibodies against OLIG2, Nestin, and YKL-40. Bars, 100 μm.
S3) Immunostaining positivity of markers in responder and non-responder GIC lines. Cells were grown on chamber slides precoated with poly-lysine- coated coverslips. Cells were fixed with 4% paraformaldehyde, permeated with 0.25% Triton X-100, blocked with 3% normal goat serum. Immunostaining was performed using the appropriate primary and secondary antibodies, and images were acquired using a confocal laser scanning microscope (Carl Zeiss Microscopy). Immunostaining of various markers showed that all responder GSC lines exhibited high Olig2 and Sox-2 expression, whereas non-responder GICs were positive for CD44, TGFβ1 and FGF13 expression.
S4) Analysis of human tumor gene expression profiles identifies proneural subtype as having high Notch pathway activity. Expression of GBM proneural signature and NOTCH pathway in clinical GBM samples available through TCGA Expression of Proneural signature in 533 TCGA GBM samples.
S5) Expression of NOTCH pathway genes in the TCGA data set. 43.9% of Proneural GBMs have high expression of representative NOTCH pathway genes.
Acknowledgments
We thank Verlene Henry and Lindsay Holmes for performing the animal studies. We thank Kathryn Carnes (Department of Scientific Publications, The University of Texas MD Anderson Cancer Center) for editing the manuscript. This study was supported by grants from the National Cancer Institute (CA56041 and CA127001 to W.K.A.Y) and Cancer Center Support Grant (CA16672).
References
- 1.Chang SM, Theodosopoulos P, Lamborn K, et al. Temozolomide in the treatment of recurrent malignant glioma. Cancer. 2004;100:605–611. doi: 10.1002/cncr.11949. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Purow B, Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass? Nat Rev Neurol. 2009;5:419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin CR, Laterra J. Neuro-oncology: unmasking the multiforme in glioblastoma. Nat Rev Neurol. 2010;6:304–305. doi: 10.1038/nrneurol.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald TJ, Brown KM, LaFleur B, et al. Expression profiling of medulloblastoma PDGFRA and the AS/MAPK pathway. Nat Genet. 2001;29:143–152. doi: 10.1038/ng731. [DOI] [PubMed] [Google Scholar]
- 7.Nagarajan RP, Costello JF. Molecular epigenetics and genetics in neuro-oncology. Neurotherapeutics. 2009;6:436–446. doi: 10.1016/j.nurt.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson JP, VanBrocklin MW, Guilbeault AR, et al. Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene. 2010;29:335–344. doi: 10.1038/onc.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mladkova N, Chakravarti A. Molecular profiling in glioblastoma: prelude to personalized treatment. Curr Oncol Rep. 2009;11:53–61. doi: 10.1007/s11912-009-0009-3. [DOI] [PubMed] [Google Scholar]
- 10.Colman H, Zhang L, Sulman EP, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 13.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikushima H, Todo T, Ino Y, et al. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2010;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- 21.Kageyama R, Ohtsuka T, Hatakeyama J, et al. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama J, Sakamoto S, Kageyama R. Hes1 and Hes5 regulate the development of the cranial and spinal nerve systems. Dev Neurosci. 2006;28:92–101. doi: 10.1159/000090756. [DOI] [PubMed] [Google Scholar]
- 23.Reedijk M, Odorcic S, Zhang H, et al. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33:1223–1229. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Breit S, Danckwardt S, et al. Downregulation of Notch signaling by gamma-secretase inhibition can abrogate chemotherapy-induced apoptosis in T-ALL cell lines. Ann Hematol. 2009;88:613–621. doi: 10.1007/s00277-008-0646-x. [DOI] [PubMed] [Google Scholar]
- 25.Watters JW, Cheng C, Majumder PK, et al. De novo discovery of a gamma-secretase inhibitor response signature using a novel in vivo breast tumor model. Cancer Res. 2009;69:8949–8957. doi: 10.1158/0008-5472.CAN-09-1544. [DOI] [PubMed] [Google Scholar]
- 26.Cuevas IC, Slocum AL, Jun P, et al. Meningioma transcript profiles reveal deregulated Notch signaling pathway. Cancer Res. 2005;65:5070–5075. doi: 10.1158/0008-5472.CAN-05-0240. [DOI] [PubMed] [Google Scholar]
- 27.Yokota N, Mainprize TG, Taylor MD, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 28.Kanamori M, Kawaguchi T, Nigro JM, et al. Contribution of Notch signaling activation to human glioblastoma multiforme. J Neurosurg. 2007;106:417–27. doi: 10.3171/jns.2007.106.3.417. [DOI] [PubMed] [Google Scholar]
- 29.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–63. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 30.Zhu TS, Costello MA, Talsma CE, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–6072. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Xing H, Kim TM, et al. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem Cells. 2012;30:1313–1326. doi: 10.1002/stem.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Wakeman TP, Lathia JD, et al. Notch Promotes Radioresistance of Glioma Stem Cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan X, Khaki L, Zhu TS, et al. Notch Pathway Blockade Depletes CD133-Positive Glioblastoma Cells and Inhibits Growth of Tumor Neurospheres and Xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert CA, Daou MC, Moser RP, et al. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010;70:6870–6879. doi: 10.1158/0008-5472.CAN-10-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhat KP, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bengtsson H, Jönsson G, Vallon-Christersson J. Calibration and assessment of channel-specific biases in microarray data with extended dynamical range. BMC Bioinformatics. 2004;5:177. doi: 10.1186/1471-2105-5-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koul D, Jasser SA, Lu Y, et al. Motif analysis of the tumor suppressor gene MMAC/PTEN identifies tyrosines critical for tumor suppression and lipid phosphatase activity. Oncogene. 2002;21:2357–2364. doi: 10.1038/sj.onc.1205296. [DOI] [PubMed] [Google Scholar]
- 41.Liu JL, Sheng X, Hortobagyi ZK, et al. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol Cell Biol. 2005;25:6211–6224. doi: 10.1128/MCB.25.14.6211-6224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lal S, Lacroix M, Tofilon P, et al. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92:326–333. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 44.Cooper LA, Gutman DA, Long Q, et al. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tso CL, Freije WA, Day A, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- 46.Katoh M, Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol. 2007;31:461–466. [PubMed] [Google Scholar]
- 47.Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–93. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 48.Hoey T, Yen WC, Axelrod F, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–77. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1) GICs were seeded in 6-well plates with 1×105 cells/well. Cell number was counted by using a hemocytometer every day for 7 days. All the experiments were repeated three times and the average value was reported. All the responder cell lines and non-responder cell lines showed similar exponential growth pattern during 7 days of growth curve with GSC35 a responder cell line with slightly slower growth rate.
S2) In an in-vitro tumorigenicity assay as assessed by the ability to form primary neurospheres, all 4 GICS showed similar neurosphere formation ability showing that the GICs had tumorigenic potential in-vitro. Bars, 250 μm. To create the intracranial disease model, we engrafted GICs into the caudate nucleus of each mouse using a previously described guide-screw system (Lal et al., 2000) Briefly, 5×104 dissociated tumor cells in 5 μl neurobasal medium were injected stereotactically into 6- to 8-week-old NOD-SCID mouse. Animals were sacrificed when they developed weight loss greater than 10% or neurological symptoms. The tissue sections were incubated with antibodies against OLIG2, Nestin, and YKL-40. Bars, 100 μm.
S3) Immunostaining positivity of markers in responder and non-responder GIC lines. Cells were grown on chamber slides precoated with poly-lysine- coated coverslips. Cells were fixed with 4% paraformaldehyde, permeated with 0.25% Triton X-100, blocked with 3% normal goat serum. Immunostaining was performed using the appropriate primary and secondary antibodies, and images were acquired using a confocal laser scanning microscope (Carl Zeiss Microscopy). Immunostaining of various markers showed that all responder GSC lines exhibited high Olig2 and Sox-2 expression, whereas non-responder GICs were positive for CD44, TGFβ1 and FGF13 expression.
S4) Analysis of human tumor gene expression profiles identifies proneural subtype as having high Notch pathway activity. Expression of GBM proneural signature and NOTCH pathway in clinical GBM samples available through TCGA Expression of Proneural signature in 533 TCGA GBM samples.
S5) Expression of NOTCH pathway genes in the TCGA data set. 43.9% of Proneural GBMs have high expression of representative NOTCH pathway genes.