Abstract
Amelogenin, the major extracellular matrix protein of developing tooth enamel is intrinsically disordered. Through its interaction with other proteins and mineral, amelogenin assists enamel biomineralization by controlling the formation of highly organized enamel crystal arrays. We used circular dichroism (CD), dynamic light scattering (DLS), fluorescence and NMR spectroscopy to investigate the folding propensity of recombinant porcine amelogenin rP172 following its interaction with SDS, at levels above critical micelle concentration. The rP172-SDS complex formation was confirmed by DLS, while an increase in the structure moiety of rP172 was noted through CD and fluorescence experiments. Fluorescence quenching analyses performed on several rP172 mutants where all but one Trp was replaced by Tyr at different sequence regions confirmed that the interaction of amelogenin with SDS micelles occurs via the N-terminal region close to Trp25 where helical segments can be detected by NMR. NMR spectroscopy and structural refinement calculations using CS-Rosetta modelling confirm that the highly conserved N-terminal domain is prone to form helical structure when bound to SDS micelles. Our findings reported here reveal interactions leading to significant changes in the secondary structure of rP172 upon treatment with SDS. These interactions may reflect the physiological relevance of the flexible nature of amelogenin and its sequence specific helical propensity that might enable it to structurally adapt with charged and potential targets such as cell surface, mineral, and other proteins during enamel biomineralization.
Keywords: Amelogenin, intrinsically disordered protein, enamel, SDS, CS-Rosetta
Introduction
Amelogenin, the major extracellular matrix protein present in the developing tooth enamel of all mammals, has been shown to be intrinsically disordered1,2. This protein, along with several other dental proteins, is produced by ameloblasts and carried by secretory vesicles to the mineralization site to assist enamel biomineralization via controlling the formation of highly organized enamel crystal arrays3,4. The expression and secretion of amelogenin has been proven to be essential for the formation of normal enamel5. Due to the presence of several proline and other disorder-promoting residues (E, K, R, G, Q, S, and A) in its sequence, amelogenin has a highly flexible backbone6,7. It is noteworthy that the amino acid sequences of the N and C terminal regions of amelogenins from various mammals are highly conserved, being unchanged for several million years8. Studies have further shown that either mutation or deletion of these conserved sequences lead to the formation of ill-defined enamel crystals9. Investigators have postulated that the supramolecular structure formed by the self assembly of amelogenin is important for the biomineralization and formation of enamel10–12. Its flexible nature and tendency to aggregate have made determination of the structure of amelogenin highly challenging12–16. The flexible structure is believed, however, to be important in the ability of amelogenin to adapt itself to various potential targets it interacts with during enamel formation1,2.
Structural analysis through solution NMR has revealed that in aqueous conditions a recombinant form of porcine amelogenin named rP172 is devoid of any global secondary structure but also confirmed the presence of several short structured regions2 (PPII, β-strand, turn/loop and helix). It also revealed that the N-terminal domain (the tyrosine-rich amelogenin polypeptide, or TRAP) is unfolded and the C-terminal domain is randomly coiled. Through CD and NMR measurements the PPII content in rP172 has been estimated to be between 20–37%2,6 and the presence of equilibrium between the disordered and PPII conformations was also identified6. The structure-function relationships of amelogenin have been addressed by investigators through the characterization of its different stable or meta-stable conformations under different experimental conditions17–19. Characterization of rP172 in the helix promoting organic solvent TFE suggested that the conformations at N- and C-termini are likely to interact with potential targets as they showed high helical propensity. In contrast, the Pro- and Gly-rich middle segment was resistant to induced folding, suggesting a different function20. We have further postulated the possibility that the transition of amelogenin's structure from disordered to order might facilitate its binding to enamel matrix targets1,21–24.
Our goal in the present study was to provide more insight into the folding propensities and behaviour of amelogenin by analyzing its intrinsically disordered behaviour in strong detergents like sodium dodecyl sulfate (SDS). SDS molecules have the ability to mimic biological cell membranes by forming amphiphilic micelles when present above critical micelle concentration (CMC)25–28. SDS has been used for denaturing folded proteins and peptides for gel electrophoresis analysis. In case of naturally unfolded or intrinsically disordered proteins (amelogenin in this case), ordered structures can be promoted by the presence of SDS, further highlighting regions that have high potential to undergo structural changes following their interactions with the target25,26,28.
In this study, we used circular dichroism (CD), dynamic light scattering (DLS), fluorescence and NMR spectroscopy to investigate the folding propensity of recombinant porcine amelogenin rP172 following its interaction with SDS, at levels above critical micelle concentration. We have recently reported that significant structural changes occur in the structure-stabilizing solvent TFE17. Here, rP172-SDS complex formation was first observed through DLS, while an increase in the structure moiety of rP172 was clearly noted through CD and fluorescence experiments. Further the fluorescence quenching analyses performed on several rP172 mutants confirmed that the N-terminal region has increased interactions with SDS micelles. Finally, NMR spectroscopy and structural refinement calculations and CS-Rosetta modelling confirm that the highly conserved N-terminal domain is prone to form helical structure when bound to SDS micelles. Our findings reported here reveal interactions leading to significant changes in the secondary structure of rP172 upon treatment with SDS that may reflect the physiological relevance of the amelogenin sequence.
Results and discussion
Formation of rP172-SDS complexes
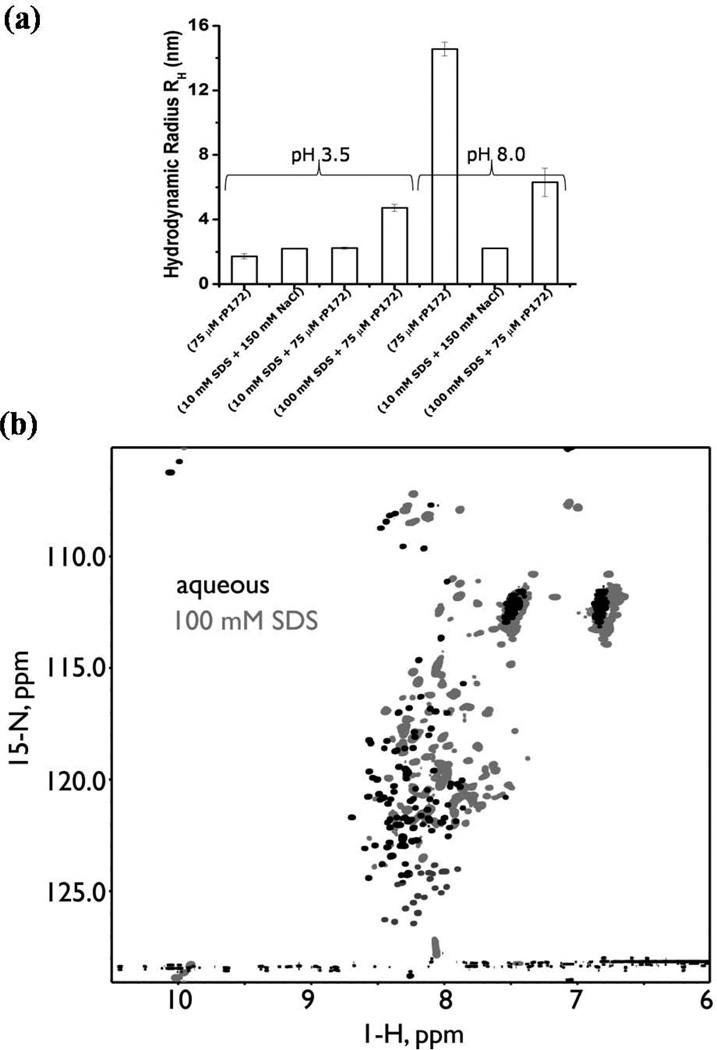
The interactions between rP172 and SDS micelles were analyzed by comparing the hydrodynamic radii (RH) of rP172 monomers and micelles with those of their complexes by Dynamic Light Scattering. Under both pH conditions (3.5 and 8), SDS micelles [above critical micellar concentration (CMC)] have a uniform radius averaging 2.2 nm. The monomers of rP172 formed at pH 3.5 possess an average RH of 1.72±0.16 nm (Fig. 1a). The increase in the hydrodynamic radii (from 2.19±0.33 to 4.72±0.22 nm) when 75 µM rP172 was mixed with 100 mM SDS micelles provides direct evidence for their close interaction of amelogenin and SDS although we are unable to accurately predict the shape and the stoichiometry of SDS-rP172 complexes. Under basic pH (8.0), due to the formation of amelogenin nanospheres, the average hydrodynamic radius of the particles was 14.56 nm11,14 while following the addition of SDS the nanospheres disassembled to particles with an average radius of 6.3±0.87 nm (Fig. 1a). It should be noted that this homogenous radius for the micelles could be unambiguously measured only when the solution had a specific ionic strength that stabilized the negatively charged head group of SDS molecules. Fig. 1a shows the average radius of SDS micelles measured in the presence of 150 mM NaCl. The similar charge-charge stabilization seen when 75 µM rP172 was added to 10mM SDS confirms the interaction between the protein and the detergent (Fig. 1a).
Figure 1.
Amelogenin-SDS interactions, (a)The mean hydrodynamic radius (RH) and standard deviation as determined from Dynamic Light Scattering (DLS) studies showing differences in the hydrodynamic radii of amelogenin-SDS complexes formed at pH’s 3.5 and 8.0 when compared with their free forms. (b) 1H, 15N HSQC spectra of DCN-rP172 recorded at 800 MHz in the monomeric aqueous state (red) and in the presence of 100 mM SDS (green).
Using NMR spectroscopy, we confirm that SDS micelles associate with rP172 and introduce changes in protein backbone dynamics. Fig. 1b, an overlay comparison of 1H, 15N HSQC spectra obtained for aqueous (AQ-) and 100 mM SDS (100-) DCN-rP172 samples at pH 3.8, reveals that significant changes occur to the amelogenin protein above the CMC of SDS as evidenced by significant shift and attenuation in the 1H, 15N resonances seen in the spectrum of 100-DCN-rP172. Relative to the AQ-DCN-rP172 sample where only the E40, Q126, and H132 resonances are unassigned, the 100-DCN-rP172 sample possesses significantly fewer 1H, 15N HSQC peaks (Fig. 1b). We were unable to assign 24 residues (i.e., P4 - H6; T21; I30 - H32; P41; L46 - Q56; P74 - Q77; P81 - Q83; P107 - Q115; P119 - Q120; P133 - Q135; and P141; representing 14% of the total sequence) due to either signal overlap or peaks attenuation (Fig. 1b, 2 and Fig. S1 in the supporting material). Selective loss or broadening of NMR signals was also observed in aggregation studies involving various proteins in buffer, salt environments, or alcohols16,17. Thus, the attenuation of cross peaks reflects that these residues are undergoing conformational exchange in the intermediate time scale. Furthermore, we also observed 17 additional cross peaks (shown in red) in the 1H, 15N HSQC spectra of 100-DCN-rP172 sample that we could not directly assign (Fig.S1in the supporting material). These additional peaks could arise from either the very slow cis-trans isomerization of the proline residues or from conformational exchange in the slow time scale in the presence of SDS micelles. In contrast we observed only four conformational exchange peaks (V54, A63, W161, T164) in the AQ-DCN-rP172 sample. We believe that this alteration in protein conformational exchange occurs in response to a number of factors, including SDS micelle-peptide backbone interactions and dynamics.
Figure 2.
Primary amino acid sequence of recombinant porcine amelogenin (rP172). Residue M1 is missing from the recombinant version. Regions denoted in red represent undetected NMR resonances in the 100 DCN-rP172 1H, 15N HSQC spectra. Underlined regions denote the highly conserved N-terminal or self-assembly A domain (purple), the PPII containing central domain (black), and the charged hydrophillic C-terminal domain (orange). Arrows indicate Trp (W) residues altered in the mutant proteins used, as described under methods and listed in Table 1.
SDS promotes conformational changes in rP172
After learning that rP172 binds to SDS our focus turned towards finding out how strongly or deeply the protein embeds within the micelle and the potential influence of SDS in promoting organized structure in rP172 under different pH conditions. It is well known that rP172 exists as monomers at pH 3.5, oligomers at pH 5.0 and as nanospheres at pH 8.014. The experiments presented in this study were carried out at pHs 3.5 and 8.0 to analyse what happens when monomers or nanospheres of rP172 interact with negatively charged SDS molecules.
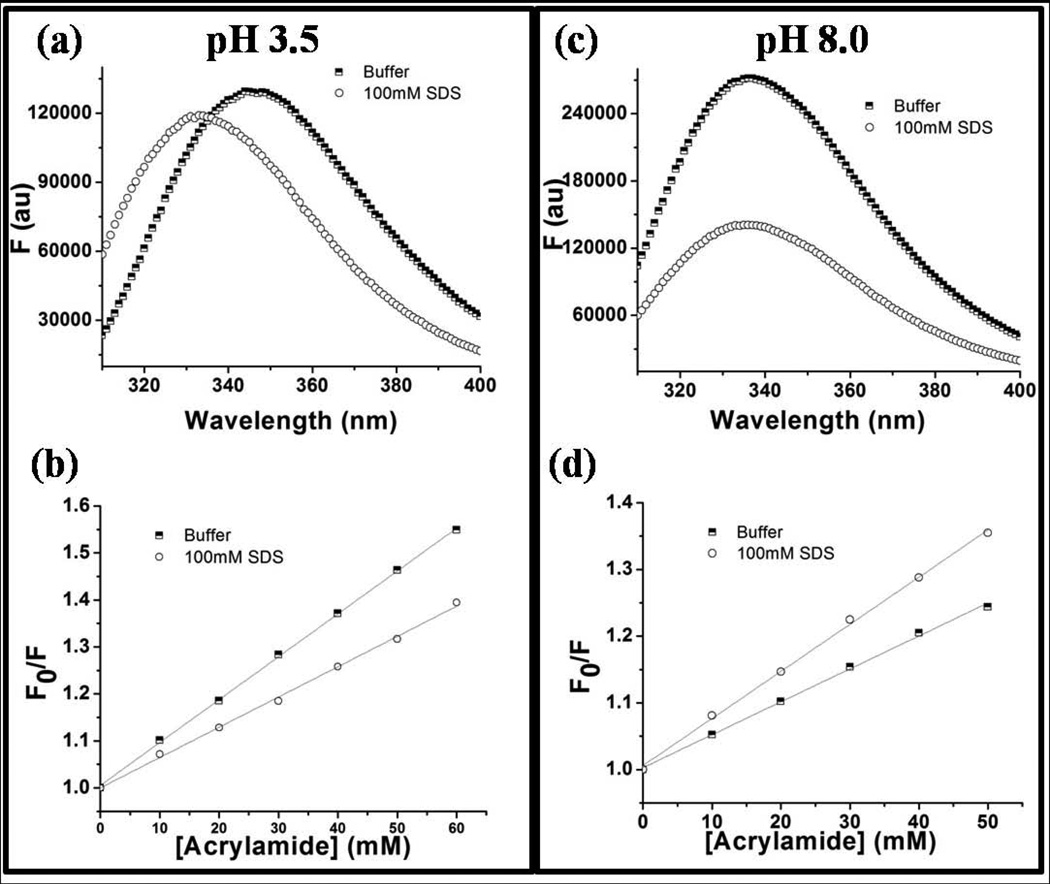
At pH 3.5, rP172 monomers showed a maximum fluorescence emission (λmax) at 347 nm, whereas in the presence of 100 mM SDS micelles the λmax blue-shifted to 332 nm (Fig. 3a). This result confirmed that there is a change in the local hydrophobic environment around any or all of the three tryptophan residues present in rP172 in the presence of SDS. At pH 8.0 the λmax of rP172 nanospheres, originally at 336 nm, only shifted slightly, to 335 nm, in the presence of SDS (Fig. 3c). Due to the very similar emission maxima at pH 8.0 we initially thought the tryptophan residues were in similar hydrophobic environments in the presence or absence of SDS micelles. This view changed, however, when we did fluorescence quenching studies. Through collision mechanisms, acrylamide (quencher) can quench the fluorescence signal of tryptophan (fluorophore), and the extent of this quenching depends on the exposure/accessibility of the tryptophan residues. Fig. S2 shows the results of fluorescence quenching studies performed to compare the solvent accessibility of tryptophan residues in the absence and presence of SDS micelles. The slopes of the Stern-Volmer plots obtained from equation 1 are directly proportional to the accessibility of the tryptophan to acrylamide. The results show that at pH 3.5 (Fig. 3b) the tryptophan residues were less accessible in presence of SDS micelles than in their absence, and this is due to the interaction of tryptophan with the hydrophobic core of the micelles. However, the slopes obtained from Stern-Volmer plots29 (Fig. 3d) confirm that tryptophan residues are more accessible in the presence of SDS micelles at pH 8.0. The low accessibility of rP172 tryptophan residues at pH 8.0 in the absence of SDS is attributable to the fact that the protein is assembled into nanospheres at pH 8.0. The increase in accessibility that occurs under these conditions when SDS is added thus indicates that the SDS micelles can disassemble the nanospheres and reorganize them.
Figure 3.
Tryptophan Fluorescence emissions of 10 mM wild-type rP172 (a and c) and the Stern-Volmer plots of acrylamide quenching Trp fluorescence (b and d) in the presence (circles) and in absence (squares) of SDS micelles when performed at pH 3.5 (left panel) and at pH 8.0 (right panel).
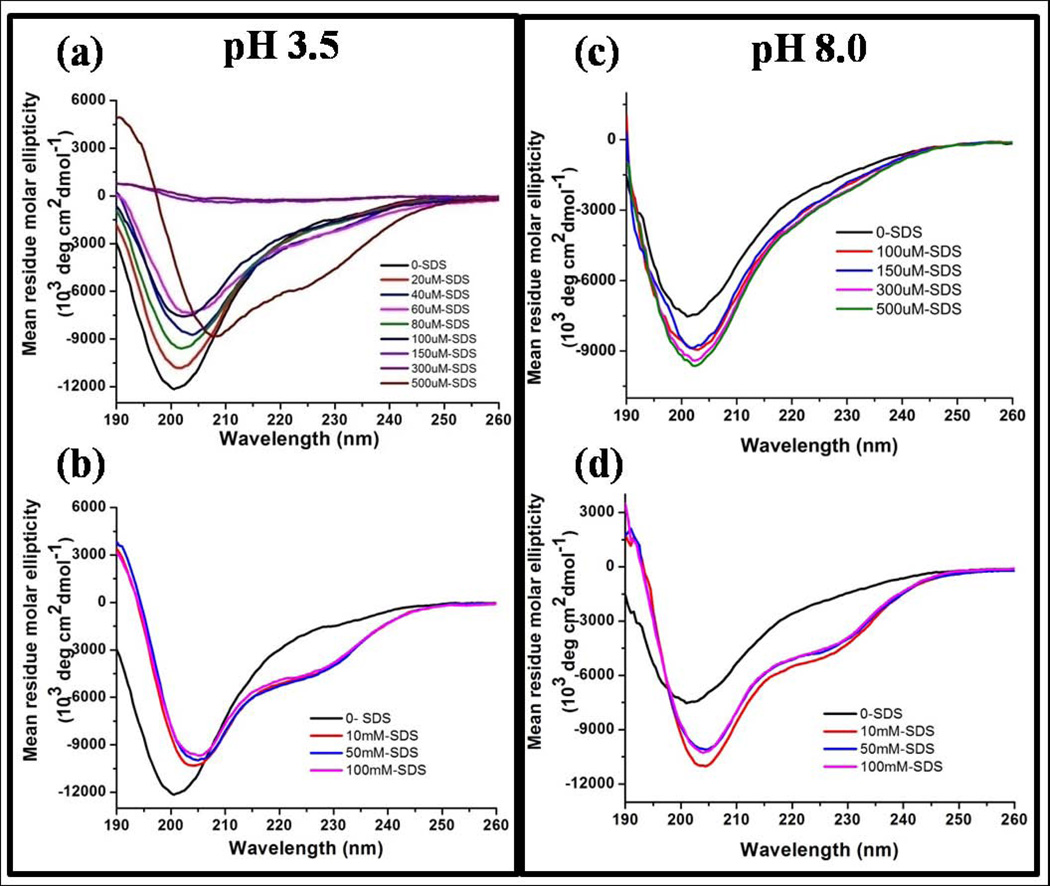
The secondary structure of rP172 in presence of SDS micelles can be more clearly revealed by studying the influence of micelles on the backbone structure of rP172. We find that SDS micelles increased the helicity of rP172, which is otherwise identified as an intrinsically disordered protein containing regions of PPII and short helices2,6. The CD spectrum of free rP172 when recorded at pH 3.5 had a negative minimum at 201 nm with no strong signal above 205 nm (Fig. 4a). We titrated a 10 µM solution of rP172 at pH 3.5 with SDS well below its CMC30 As the concentration of the titrate increase from 10 to 100 µM SDS, there was a steady decrease in the intensity of negative signal value at 201 (owing to minor structural change) until complete disappearance of the entire signal was seen at SDS concentrations of 150 µM and 300 µM (Fig. 4a). At a concentration of 500 µM the monomeric SDS molecules are able to induce and stabilize helical conformation in rP172 proving that the interaction is merely initiated through electrostatic interactions as the same phenomenon is not seen at pH 8.0 where rP172 is already assembled and has low overall positive charge. On the other hand similar stabilization is seen on 10mM of SDS when 75 µM of rP172 was added (Fig. 1a). In the presence of SDS (at any concentration above its CMC) the protein adopted helical-like structure as shown by the appearance of two negative peaks at 208 and 220 nm, and one positive peak at 190 nm (Fig. 4b and 4d). At pH 3.5 all the histidine residues of rP172 are protonated and attract the negative head groups of SDS molecules by a charge-charge interaction leading to charge neutralization-induced precipitation of rP172. This phenomenon did not happen at pH 8.0 since rP172 already exists as nanospheres at that pH, resulting in a considerable decrease in the protein’s overall positive charge (Fig. 4c). In contrast, when SDS concentrations were increased above the CMC, the CD profiles at both pH 3.5 and 8 were very similar showing the strong interaction of micellar SDS with the amphipathic and highly flexible rP172 (Fig. 4b and 4d). Thus above the CMC (from 10 mM – 100 mM), SDS micelles induce similar helical folding in rP172 under both pH conditions.
Figure 4.
Circular dichroism spectra showing structural transitions in rP172 when titrated with SDS at pH 3.5 (left panel) and pH 8.0 (right panel) below (a and c) and above (b and d) the critical micelle concentrations of SDS. SDS, at a concentration of 500 M and at pH3.5, stabilizes the alpha helical structure in rP172 likely through charge-charge interaction (a).
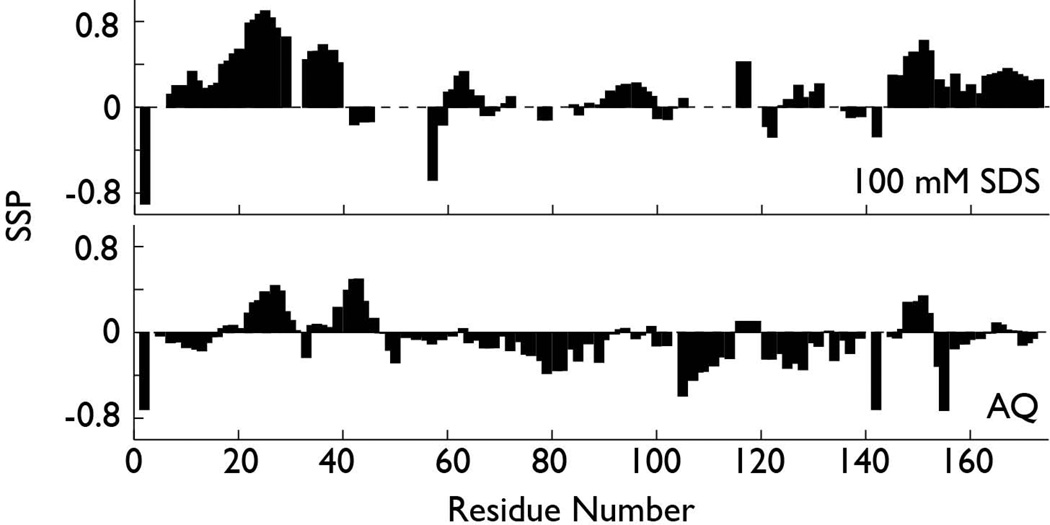
The regions within rP172 having SDS-induced helical folding propensity were identified using NMR spectroscopy (Fig. 5; Fig. S1). We compared the secondary structure probabilities for observable 100-DCN-rP172 sequence regions (Fig. S1) alongside AQ-DCN-rP172, using the SSP scoring system for IDP proteins (Fig. 5). A SSP score of +1 or −1 at a given residue reflects fully formed alpha-helix or beta-strand, respectively, while a score of 0.5 indicates that 50% of the conformers in the disordered state ensemble are helical at that position31. As shown in Fig. 5, AQ-DCN-rP172 does not feature any fully formed helical or beta strand as evidenced by SSP scores. However, if we examine the (+) or (−) SSP trend, we observe that a greater probability of forming α-helix exists within the N-terminus (Y17 – R31, Y34 – L46), in limited regions of the central domain (F147 – L152), and in the C-terminus (D165 – R169). Conversely, there is some propensity to form β-strand is the following regions: P10 – I13 (N-terminus), Q49 – P114, P121 – P142 (central domain) and P154 – E159 (C-terminus). When we compare these structural probabilities with those obtained for 100-DCN-rP172, we observe that SDS micelles induce overall higher SSP scoring (0.5 to 1.0), indicating that more stable secondary structures are forming. Moreover, it is clear that SDS micelles induce a beta structure-to-alpha helical conversion throughout the rP172 sequence, with the highest alpha helical probabilities observed for the P7 – Y39 (N-terminal) and P145 – V172 (C-terminal) regions. There are some other regions in the central domain that also show some helical propensity: Q60 – Q65 and Q91 – H99. These results are similar to those reported for rP172 in 70% v/v 2,2,2-trifluoroethanol (TFE) at pH 3.817. Hence, our SSP calculations indicate that the rP172 molecule possesses residual beta strand structures under monomeric conditions, and it is the terminal regions which transform into a predominantly helical structure in the presence of SDS micelles. We note that recent studies with the Leucine-Rich Amelogenin Polypeptide (LRAP) also document conformational folding from a disordered state into a helical state when this polypeptide adsorbs onto hydroxyapatite32. This suggests that the terminal regions of amelogenin exhibit similar folding propensities to a wide range of targets.
Figure 5.
Secondary structure propensity scoring (SSP) for AQ-DCN-rP172 (bottom) and 100-DCN- rP172 (100 mM SDS) (top). These plots document occurrence of beta strand (negative SSP) and helix (positive SSP) structures in monomeric and SDS-induced rP172.
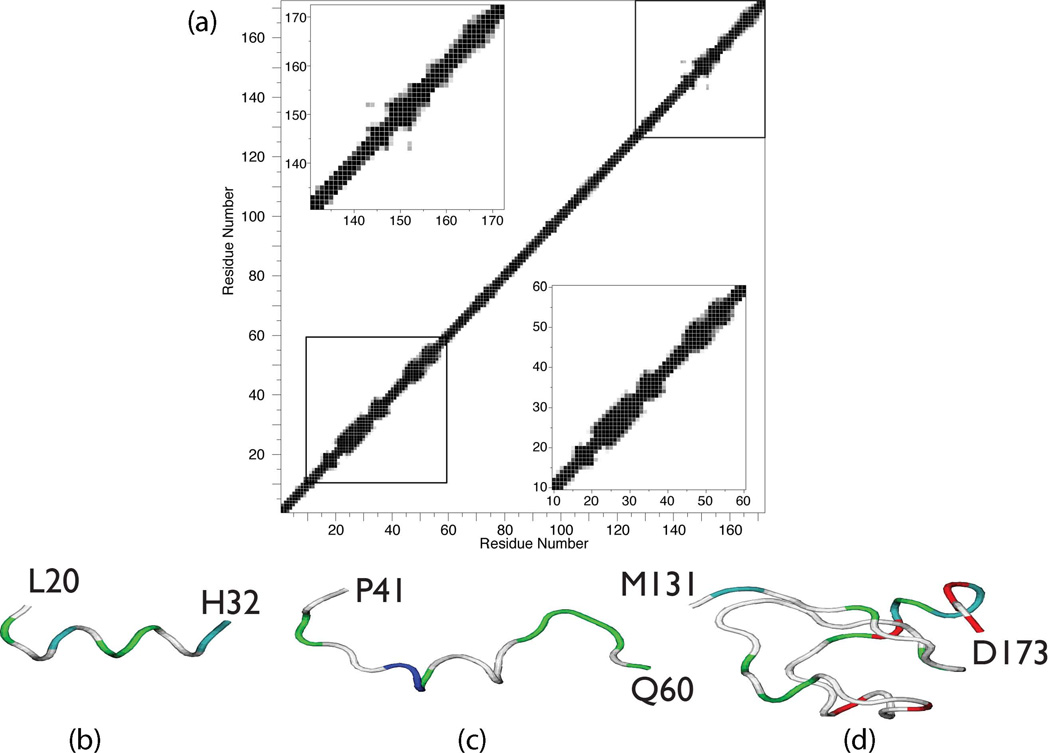
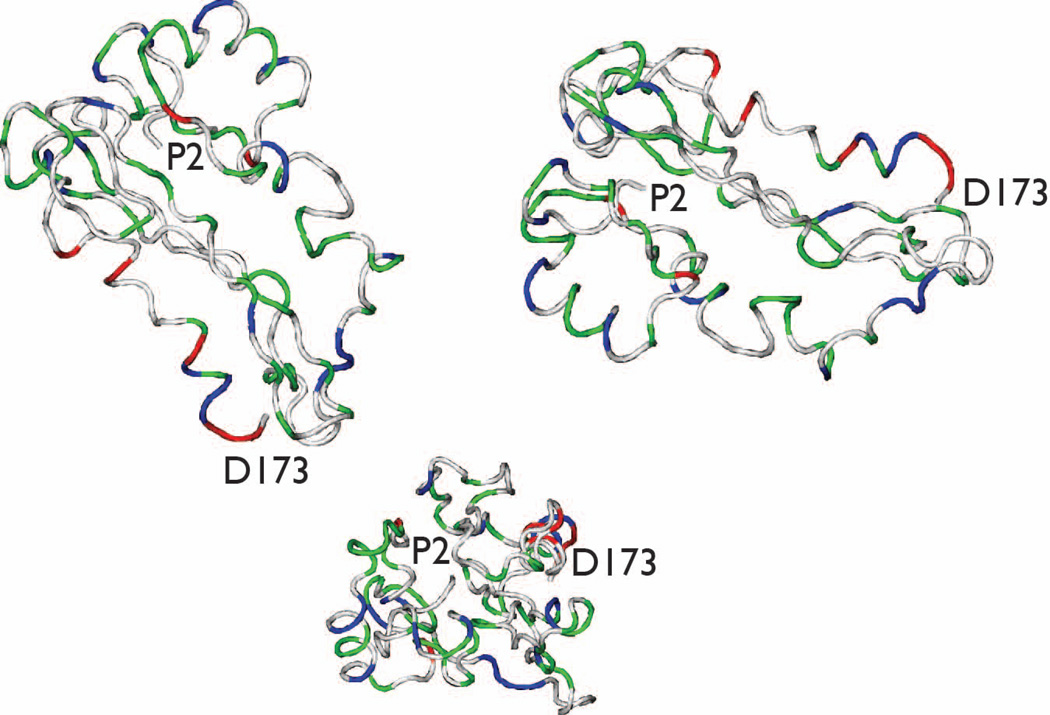
In order to understand SDS-induced structural perturbations, we generated ensembles of structures for rP172 N- and C-terminal fragments (Fig. 6) and for the full length protein molecule (residues 2–173, Fig. 7) in the presence of 100 mM SDS using CS-ROSETTA, the chemical shift-based structure calculation program33. For each set, 10–40 lowest energy structures were selected whose Φ/Ψ angles were present in the most favoured region of the Ramachandran plot, and the lowest energy structure was then extracted for visual comparison. Fig. 6A shows the contact plot averaged over 20 lowest energy structures generated using CS-ROSETTA. This plot represents the average fraction of structures that have a pair of residues that are in contact. A pair of residues is in contact when their ‘all atoms’ are within 0.6 Å of each other. The residues that have contact distance below 0.3 Å are shown in black, above 0.6 Å as white and between 0.3 – 0.6 Å are shown in various shades of gray. It is clear from the plot that full length rP172 structures lacks tertiary fold but show regions that may have secondary structures as evident by strong contact seen within residues 22 – 32, 45 – 56 and 142 – 160. Further inspection of the full-length rP172 structures and fragments revealed that the N-terminal L20 – H32 region forms a helix (Fig.6b and Fig. 7), with a backbone rmsd of 0.42 Å for the 18 member ensemble. Helical regions were also noted for the N-terminal 45 – 56 sequence (Fig. 6c); however, note that this region lacked chemical shift data due to intermediate time-scale conformational exchange broadening and thus the structural elements seen in these models are solely based upon homology. Surprisingly, the C-terminal region, which had nearly complete assignments, showed no consistent structural features except for helical structure in the 141–151 sequence regions (Fig.6d and Fig. 7). Note that the full-length rP172- SDS model represents L20 – H32, P41 – Q60, and M131 – D173 within the context of the other unstructured regions of the protein. Due to the lack of quantitative NMR data (i.e., NOE's), we cannot ascertain the global folding state (i.e., condensed or extended conformation) of the full-length rP172 at this time. Thus, we caution that the full-length rP172-SDS model (Fig. 7) is highly qualitative, lacks tertiary information, and thus will be subject to reinterpretation at a later date pending additional NMR studies. As a side note, we also generated a full-length CS- aqueous rP172 model using CS-ROSETTA and reported chemical shift data (data not shown)2. Here, residues 22 – 30 also exhibited some tendency to form a helix but was less consistent (~ 40% of the structures, backbone rmsd ~ 0.35 Å). This suggests that these residues are in equilibrium between the disordered and structured conformation in aqueous solution and the equilibrium is shifted towards the formation of more structured conformation (i.e., helix) in presence of SDS micelles.
Figure 6.
(a) Contact plot representing the average fraction of structures that have a pair of residues that are in contact, averaged over 20 lowest energy structures of rP172 calculated using CS-ROSETTA. The contact distance between ‘all’ atoms within a pair of residues is below 0.3Å then it is shaded black and if above 0.6Å then shaded white. The contact distance between 0.3 to 0.6Å is shown in different shades of gray. Lower and upper insets show the expanded view of the contact plot for residues between 10 – 60 and 130 – 173, respectively. CS-ROSETTA – generated lowest energy backbone structure for sequence regions of P172 in 100 mM SDS. (b) L20 – H32; (c) P41 – Q60; (d) M131 – D173. Color scheme denotes side chain classification: White = non-polar, Blue = cationic; Green = polar; Red = anionic.
Figure 7.
Three views of the lowest energy CS-ROSETTA hypothetical model of rP172 in 100 mM SDS. Color scheme denotes side chain classification: White = non-polar, Blue = cationic; Green = polar; Red = anionic.
Identification of interacting segments of amelogenin using Trp mutants
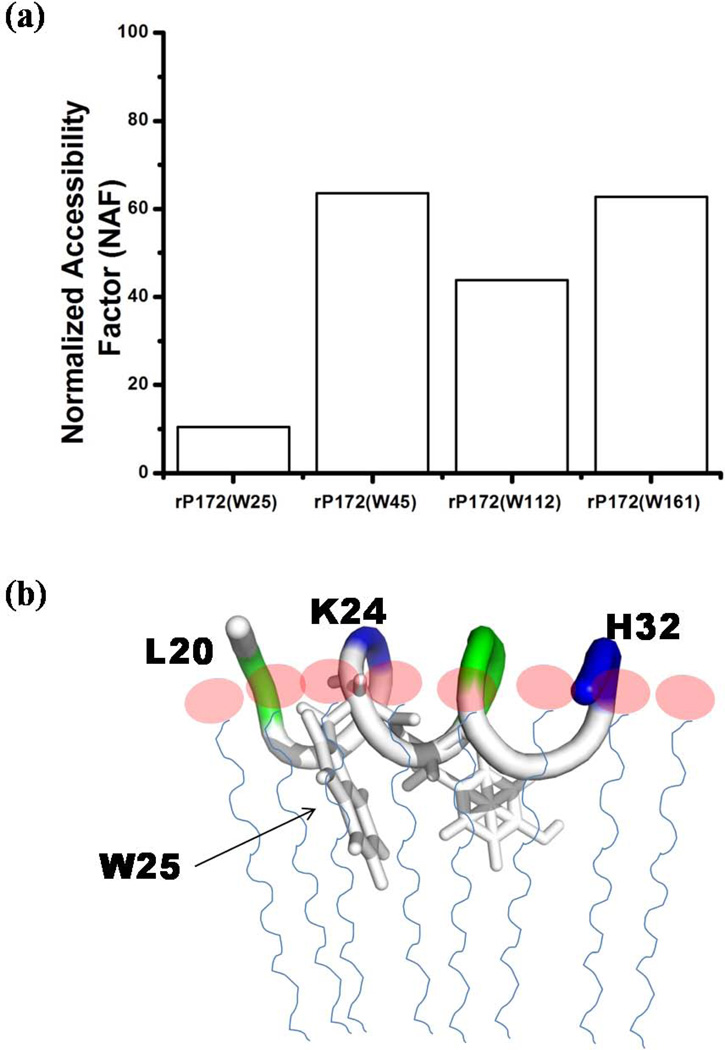
After careful analyses of overall binding and the secondary structural changes in SDS-bound rP172 we further sought to identify the motifs within the amelogenin protein that interact with SDS micelles. This will uncover the location of the sequence which has the propensity to coil into helix and thus be the primary region for binding potential targets. To achieve this goal, we studied four different rP172 mutants each of which had only a single remaining tryptophan, with its other tryptophan residues mutated to Tyrosine (Fig. 2, refer to Table 1). The CD spectra of all four mutants were compared with the wild type rP172 in order to ensure that there was no major structural change in their free forms (Fig. S3). The blue shift in λmax (Fig. S4) shown by every mutant when treated with the SDS micelles revealed a noticeable change in the local hydrophobic environments of the remaining tryptophans. When bound to SDS, the mutants rP172(W25) and rP172(W45) showed either considerable decrease or increase in their respective emission intensities compared to rP172(W112) and rP172(W161), suggesting that interactions with the N-terminus of rP172 could be more significant than interactions with the middle segment or the C-terminus (Fig. S4). The quantitative measurement of the extent of binding of each mutant to the micelle was analyzed by measuring the accessibility using acrylamide as the fluorescence quencher. Based on the Net or Normalized Accessibility Factor (NAF) (Fig.8a), the residue W25 was found to be most deeply buried in the hydrophobic core of SDS micelle. From the structural models we know that residue W25 is located within the helical L20-H32 segment (Fig. 6b). Upon closer examination it is quite clear that there are distinct polar and non-polar surfaces featured on this helical segment. Hypothetically, it is likely that the hydrophobic surface of this helix interacts with the hydrophobic core of the SDS micelle through hydrophobic-hydrophobic interactions and makes the residue W25 less accessible. Thus through these results we could certainly conclude that the N-terminal region’s W25 is buried in the hydrophobic core of micelles and we propose a schematic representation (Fig. 8b) of the rP172-SDS complex. This model explains how the hydrophobic side chains of Y24 and W25 would interact with the interior of SDS aliphatic chains with the positively charged K20, R31 and H32 residues interacting with the negative head groups outside the micelle. As the quenching studies did not show significant interaction between the C-terminus with SDS micelles, the helix formation between P145 to V172 could be a due to the allosteric influence of N-terminal interaction.
Table 1.
List of recombinant proteins and the nomenclature used to refer to them in the text.
| S. No. | Protein or its Mutant | Referred in text as |
|---|---|---|
| 1. | Recombinant Porcine Amelogenin | rP172 |
| 2. | Triply labelled rP172[U-2H,13C,15N] | DCN-rP172 |
| 3. | rP172(W45Y, W161Y) | rP172(W25) |
| 4. | rP172(W25Y, W161Y) | rP172(W45) |
| 5. | rP172{(W25, 45, 161Y), (F112W)} | rP172(W112) |
| 6. | rP172(W25Y, W45Y) | rP172(W161) |
Figure 8.
(a) Net or Normalized Accessibility Factor of the tryptophans present in the rP172 mutants calculated from their respective Stern-Volmer constants. (b) Schematic representation of rP172’s N-terminal segment showing the protein backbone interacting with the SDS head groups (pink circles). The protein hydrophobic side chains buried in the aliphatic tail of the SDS micellemodel (thin blue lines).
During the process of biomineralization amelogenin interacts with several potential charged substrates, including the ameloblasts plasma membrane phospholipids, proteins, and hydroxyapatite. Through our previous in-situ AFM studies34 we showed that, at pH 8.0, amelogenin nanospheres disassembled to form a uniform layer of monomers on the negatively charged surface of mica (may be by interacting with the positively charged surface of the helix). Such interactions may also be relevant while in contact with liposomes of secretory vesicles during the secretion of amelogenin or while interacting with the charged faces of HAP during biomineralization. Under the same pH condition, amelogenin oligomerized on the positively charged surface of 3-aminopropyltriethoxysilane (APS) mica, which may well be relevant to amelogenin's potential interaction with positively charged surfaces of HAP34. Amelogenin nanosphere disassembly following their adsorption onto fluoroapatite and other organic surfaces has been also demonstrated35,36. These observations all together show that the dynamics of amelogenin’s assembly and oligomerization can be influenced by the surface charge of its substrates. Studies have further shown that either mutation or deletion of these conserved sequences lead to misfolding37,38 and the formation of ill-defined enamel crystals9. These point mutations that lead to amelogenesis imperfecta (a series of inherited diseases of enamel malformation) are actually located at the N-terminal region very close to W25 and W45 (T21I and P41T in the case of human amelogenin) emphasizing the significance of our current finding. In this manuscript we have shown that the influence of negatively charged detergents can promote ordered structures in specific regions that are highly conserved in amelogenin and that can bind to specific targets. Thus this finding supports a possible functional role for the flexibility of amelogenin's structure and the importance of N-terminal region in the in vivo context.
Conclusions
We demonstrate that amelogenin forms complexes with SDS at two different pH conditions (3.5 and 8.0), and at above its CMC (SDS=100mM) exhibiting similar secondary structural behaviour. We used four different independent spectroscopy techniques to show that amelogenin binds to SDS via the N-terminal domain close to W25 where a helical segment can be formed following amelogenin-SDS interaction. Our study supports the notion that intrinsically disordered proteins are functionally important in biomineralization in vivo and they attain stable structures (e.g., α helix) upon binding with prospective targets, such as amphipathic assemblies (SDS micelles) or mineral phases19,32. Our findings further suggest that the amelogenin N terminal segment may be important in interacting with the membranous regions of secretory ameloblast cells as well as secretory vesicles.
Materials and methods
Protein expression and purification
All recombinant proteins including wild-type rP172, isotopically labelled, and mutant amelogenins (Table 1) used in this study were produced in E. coli DE3 codon plus RP cells as described elsewhere39. Briefly, the expressed protein was specifically precipitated by 20% ammonium sulfate solution and dissolved in 0.1% 2,2,2-trifluroacetic acid. This acidic solution was loaded on a reverse phase C4 column (10 × 250 mm, 5 µm) mounted on a Varian (Palo Alto, CA) prostar high performance liquid chromatography system and eluted using a linear gradient of 60% acetonitrile at a flow rate of 1.5 mL/min−1.
Isotopically labelled rp172 for NMR spectroscopy
Uniformly triple labelled recombinant rP172 [U-2H, 13C, 15N] (hereafter referred to as DCN-rP172) was produced via recombinant bacterial over expression in the presence of 15NH4Cl, 13C-Glucose, and 100% D2O (Cambridge Isotope Laboratories, Andover, MA). The extent of triple labelling was verified to be 98.9% using ESI-ms TOF (Mass = 21777.0 Da). These labelled and unlabelled rP172 lack the N-terminal methionine and the phosphate in the Serine at the 16th position when compared to their wild type analogues.
Trp mutant rP172 for fluorescence spectroscopy
Three double mutants; rP172(W45Y,W161Y), rP172(W25Y,W161Y) and rP172(W25Y,W45Y); were produced using Quick Change® mutagenesis (Agilent Technologies, Inc.) as described elsewhere14. A new mutant, rP172 [(W25Y, W45Y, W161Y), (F112W)] was made in a similar manner in order to introduce a tryptophan residue at the middle region of rP172. These tryptophan mutants will be referred to hereafter in the text as rP172(W25), rP172(W45), rP172(W112) and rP172(W161) (Table 1), the parentheses showing the position of the single tryptophan present in the sequence. All mutants were expressed and purified employing a similar protocol to that used for rP172.
Protein sample preparations
For CD, DLS and Fluorescence experiments the protein stock solutions were prepared by dissolving 3 mg of lyophilized rP172 (or the mutant) in 1 mL of de-ionized water and left overnight at 4 °C on a rocker. The clear solution thus obtained was centrifuged at 10000 rpm for 10 min and only 95% of the supernatant at the top was withdrawn to avoid insoluble protein. The exact concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Products, Wilmington, DE). The extinction coefficients (ε) were determined experimentally using classical methods40 to be rP172: ε =17144, rP172(W25): ε =13947, rP172(W45): ε =12997, rP172(W112): ε =16830, rP172(W161): ε =10876.3 Liters.mol−1.cm−1. Two different buffer solutions, 25 mM Sodium acetate at pH 3.5, and 25 mM Tris pH 8.0, were prepared and used to dilute the protein stock to a final concentration of 10 µM. Stock solutions of 200 mM SDS (Sigma Aldrich) were made in the above mentioned two buffers and used as needed. All SDS titration experiments were performed by preparing individual samples with specific ratios in order to avoid any complicacies that might occur while increasing SDS concentration when bound to protein28.
For NMR experiments, lyophilized DCN-rP172 was used to create three different samples, each 75 µM in 300 µL volume. The first, designated as AQ-rP172, was created by dissolving an appropriate amount of lyophilized DCN-rP172 in Milli-Q pure un-buffered deionized distilled water [90% v/v UDDW / 10% v/v D2O (99.99 atom % D, Cambridge Isotope Laboratories, Lowell, MA)]. This sample is identical to the one utilized in earlier studies of the monomeric low pH form of rP172 and gives rise to a 15N - 1H HSQC spectrum that exactly matches the earlier sample2. The other two samples, designated as 50-DCN-rP172 and 100-DCN-rP172, utilized DCN-rP172, UDDW, and an appropriate amount of sodium dodecyl sulfate- d25 (SDS) (98% atom D, Cambridge Isotope Labs, Lowell, MA) to make a 50 mM and a 100 mM SDS / 75 µM DCN-rP172 sample. For all NMR samples, after mixing of components, the pH was adjusted to 3.8. Sample pH was monitored periodically between experiments and it was found that the sample pH remained stable near 4. Samples were placed in 5 mm symmetrical D2O-matched Shigemi NMR microtubes (Shigemi, Inc, Alison Park, PA). No visible aggregation/degradation was evident, as verified by periodic recording of 15N - 1H HSQC experiments2.
Dynamic light scattering
The hydrodynamic radius (RH) of rP172 in the presence or absence of SDS was measured using Wyatt Dyna pro Nanostar equipment and Dynamics 7 software. All experiments were recorded at 22 °C under both pH conditions. The hydrodynamic radii calculated were sphere-equivalent radii as the Rayleigh sphere model was used for the analyses41. For measuring the radii of SDS micelles alone, 150 mM NaCl was added to avoid the heavy background.
Circular dichroism spectroscopy
The far-UV CD spectra were acquired on a JASCO J-815 spectropolarimeter to obtain the secondary structural properties of rP172 in the presence and absence of SDS. The temperature was maintained at 22 °C for all the experiments and 200 µL samples were placed in a 0.1 cm path length cuvette to record each spectrum. The concentrations of all protein samples were maintained at 10 µM while SDS was taken at concentrations varying from 10 µM to 100 mM. The wavelength scan was performed between 190 and 260 nm with a resolution of 0.5 nm. An average of three independent scans of each sample, accumulated at a rate of 50 nm per minute, was analyzed using the in-built JASCO spectra-manager analysis software, and the data are presented in the form of Mean residue molar ellipticity.
Fluorescence spectroscopy
The intrinsic tryptophan fluorescence emissions of all proteins were obtained from a PTI QuantaMaster QM-4SE instrument (Photon Technology International, Birmingham, NJ) at 22 °C, with the excitation and emission slit widths set at 5 nm. The protein samples were excited at 295 nm and their characteristic emissions were monitored by recording emission spectra between 310 and 400 nm. An average of two scans was taken and analyzed using the built in Felix32 software. The scatter contributions from SDS micelles were removed by subtracting their blank spectrum from every measurement.
Tryptophan fluorescence quenching by acrylamide
The extent of tryptophan exposure/accessibility was quantitatively evaluated by quenching its fluorescence with known amounts of acrylamide. Increasing concentrations of acrylamide were added to protein samples in the presence of SDS, and the decrease in the λmax value was measured. The extent of decrease was compared between the free protein and SDS bound protein samples by plotting Stern-Volmer plots obtained with the following equation29.
| Eq. 1 |
Where F0 is the fluorescence intensity in the absence of quencher, F is the fluorescence intensity in the presence of quencher, [Q] is the concentration of the quencher, and KSV is the Stern–Volmer quenching constant. Further, the net or normalized accessibility factor (NAF) was calculated from the ratios of KSV obtained from quenching of tryptophan fluorescence in the presence and absence of SDS micelles.
NMR experiments
NMR data of rP172 in SDS was collected at 20°C using a 800 MHz spectrometer (Bruker) equipped with 4-channel cryoprobe. The operating Larmor frequencies were 800.234 MHz for proton, 201.255 MHz for carbon, and 81.096 MHz for nitrogen. The backbone assignment of rP172-SDS was done using TROSY and HNCO/HN(CA)CO (512, 50 and 32 complex points with sweep widths of 12, 8 and 24 ppm in the 1H, 13C and 15N dimensions respectively) and HN(CO)CACB/HNCACB experiments (512, 50 and 32 complex points with sweep widths of 12, 65 and 24 ppm in the 1H, 13C and 15N dimensions respectively). The HSQC (TROSY) (512 and 128 complex points with sweep widths of 12 and 24 ppm in the 1H and 15N dimensions respectively) spectra were used to monitor if there is any aggregation of the sample during the course of data collection by following the line width of the peaks. No aggregation was observed during data collection. NMRPipe42 software was used to process all data and NMRView43 was used for sequential assignments. The spectra were referenced with respect to the temperature-corrected water resonance and 13C and 15N chemical shifts were referenced on the basis of the 1H IUPAC guidelines using the unified chemical shift scale. A TALOS file was generated with the respective 13Cα, 13Cβ, 13CO, 15Nα, 1HN chemical shifts for further structure calculations.
Structure modelling of rP172 in the presence and absence of SDS with CS-Rosetta
CS-Rosetta calculations were done using the protocol given in the manual33,44. Briefly, chemical shifts obtained after backbone assignment were used for both fragment selection and re-scoring of the CS-Rosetta model. Fragment assembly was done using the regular Rosetta Monte Carlo fragment assembly method45, and this was used to generate all-atom models. A total of 10,000 structures were generated for full-length rP172 by using backbone chemical shifts obtained for rP172 under aqueous and 100 mM SDS conditions. The 1,000 lowest energy structures were selected and their all-atom energies were re-scored based upon the predicted backbone chemical shifts. For both samples subsequent shorter fragments, residues 1 – 60 and 131–173 were also used for the structure generation.
Supplementary Material
Acknowledgments
The study was funded by NIH-NIDCR grants DE-020099 and DE-013414 to JMO. We acknowledge Nanobiophysics Core Facility at the University of Southern California for circular dichroism spectropolarimetry and fluorescence spectroscopy and the Mass Spectrometry Facility, at department of pharmaceutical chemistry, San Fransisco, California (project 403). Portions of this work were supported by a grant from the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Award DE-FG02-03ER46099 to JSE. JSE is a member of the New York Structural Biology Center. The data collection at NYSBC was made possible by a grant from NYSTAR, ORIP/NIH facility improvement Grant CO6RR015495, NIH Grant P41GM066354, the Keck Foundation, New York State, and the NYC Economic Development Corporation. This paper represents contribution number 70 from the Laboratory for Chemical Physics, New York University.
References
- 1.Lakshminarayanan R, Moradian-Oldak J. In: Amelogenins: Multifaceted Proteins for Dental and Bone Formation and Repair. Goldberg M, editor. Oak Park, IL: Paris: Bentham Science Publishers Ltd.; 2010. pp. 106–132. [Google Scholar]
- 2.Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan YW, Moradian-Oldak J, Evans JS. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fincham AG, Moradian-Oldak J, Simmer JP. Journal of Structural Biology. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 4.Eastoe JE. Nature. 1960;187:411–412. doi: 10.1038/187411b0. [DOI] [PubMed] [Google Scholar]
- 5.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen EH, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Journal of Biological Chemistry. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 6.Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. Biophysical Journal. 2007;93:3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakshminarayanan R, Yoon I, Hegde BG, Fan DM, Du C, Moradian-Oldak J. Proteins-Structure Function and Bioinformatics. 2009;76:560–569. doi: 10.1002/prot.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sire JY, Delgado S, Fromentin D, Girondot M. Arch Oral Biol. 2005;50:205–212. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Paine ML, Luo W, Zhu DH, Bringas P, Snead ML. Journal of Bone and Mineral Research. 2003;18:466–472. doi: 10.1359/jbmr.2003.18.3.466. [DOI] [PubMed] [Google Scholar]
- 10.Fowler CE, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. European Journal of Oral Sciences. 2006;114:297–303. doi: 10.1111/j.1600-0722.2006.00288.x. [DOI] [PubMed] [Google Scholar]
- 11.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 12.Diekwisch T, David S, Bringas P, Santos V, Slavkin HC. Development. 1993;117:471–482. doi: 10.1242/dev.117.2.471. [DOI] [PubMed] [Google Scholar]
- 13.Katz EP, Seyer J, Levine PT, Glimcher MJ. Archives of Oral Biology. 1969;14 doi: 10.1016/0003-9969(69)90146-0. 533-&. [DOI] [PubMed] [Google Scholar]
- 14.Bromley KM, Kiss AS, Lokappa SB, Lakshminarayanan R, Fan D, Ndao M, Evans JS, Moradian-Oldak J. J Biol Chem. 2011;286:34643–34653. doi: 10.1074/jbc.M111.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Ramirez BE, Liao XB, Diekwisch TGH. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchko GW, Tarasevich BJ, Bekhazi J, Snead ML, Shaw WJ. Biochemistry. 2008;47:13215–13222. doi: 10.1021/bi8018288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ndao M, Dutta K, Bromley KM, Lakshminarayanan R, Sun Z, Rewari G, Moradian-Oldak J, Evans JS. Protein Science. 2011;20:724–734. doi: 10.1002/pro.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beniash E, Simmer JP, Margolis HC. Journal of Dental Research. 2012;91:967–972. doi: 10.1177/0022034512457371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarasevich BJ, Perez-Salas U, Masica DL, Philo J, Kienzle P, Krueger S, Majkrzak CF, Gray JL, Shaw WJ. J Phys Chem B. 2013;117:3098–3109. doi: 10.1021/jp311936j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bienkiewicz EA, Woody AYM, Woody RW. Journal of Molecular Biology. 2000;297:119–133. doi: 10.1006/jmbi.2000.3545. [DOI] [PubMed] [Google Scholar]
- 21.Ravindranath HH, Chen LS, Zeichner-David M, Ishima R, Ravindranath RMH. Biochemical and Biophysical Research Communications. 2004;323:1075–1083. doi: 10.1016/j.bbrc.2004.08.207. [DOI] [PubMed] [Google Scholar]
- 22.Fan DM, Du C, Sun Z, Lakshminarayanan R, Moradian-Oldak J. Journal of Structural Biology. 2009;166:88–94. doi: 10.1016/j.jsb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang XD, Fan DM, Mattew S, Moradian-Oldak J. European Journal of Oral Sciences. 2011;119:351–356. doi: 10.1111/j.1600-0722.2011.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallon V, Chen L, Yang X, Moradian-Oldak J. J Struct Biol. 2013 doi: 10.1016/j.jsb.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds JA, Herbert S, Polet H, Steinhardt J. Biochemistry. 1967;6:937–947. doi: 10.1021/bi00855a038. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds JA, Tanford C. J Biol Chem. 1970;245:5161–5165. [PubMed] [Google Scholar]
- 27.Garavito RM, Ferguson-Miller S. Journal of Biological Chemistry. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 28.Otzen D. Biochim Biophys Acta. 2011;1814:562–591. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Stern VO, Volmer M. Physikalische Zeitschrift. 1919;20:183–188. [Google Scholar]
- 30.Rahman A, Brown CW. Journal of Applied Polymer Science. 1983;28:1331–1334. [Google Scholar]
- 31.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu JX, Xu YS, Shaw WJ. Biochemistry. 2013;52:2196–2205. doi: 10.1021/bi400071a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CL, Bromley KM, Moradian-Oldak J, DeYoreo JJ. Journal of the American Chemical Society. 2011;133:17406–17413. doi: 10.1021/ja206849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarasevich BJ, Lea S, Bernt W, Engelhard M, Shaw WJ. Journal of Physical Chemistry B. 2009;113:1833–1842. doi: 10.1021/jp804548x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarasevich BJ, Lea S, Bernt W, Engelhard MH, Shaw WJ. Biopolymers. 2009;91:103–107. doi: 10.1002/bip.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bromley KM, Lakshminarayanan R, Lei YP, Snead ML, Moradian-Oldak J. Cells Tissues Organs. 2011;194:284–290. doi: 10.1159/000324342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakshminarayanan R, Bromley KM, Lei YP, Snead ML, Moradian-Oldak J. J Biol Chem. 2010;285:40593–40603. doi: 10.1074/jbc.M110.131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, Simmer JP. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 40.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Science. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.schartl w. Light Scattering from Polymer Solutions and Nanoparticle Dispersions. Germany: Heidelberg: Springer-Verlag; 2007. [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 44.Bowers PM, Strauss CEM, Baker D. Journal of Biomolecular Nmr. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- 45.Bradley P, Misura KM, Baker D. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.