Abstract
Quantitative susceptibility mapping (QSM) is a recently developed MRI technique that provides a quantitative measure of tissue magnetic susceptibility. To compute tissue magnetic susceptibilities based on gradient echoes, QSM requires reliable unwrapping of the measured phase images and removal of contributions due to background susceptibilities. Typically, the two steps are performed separately. Here we present a method that simultaneously performs phase unwrapping and HArmonic (background) PhasE REmovaL using the LAplacian operator (HARPERELLA). Both numerical simulations and in vivo human brain images showed that HARPERELLA effectively removes both phase wraps and background phase, while preserving all low spatial frequency components originating from brain tissues. When compared with other QSM phase preprocessing techniques, such as path-based phase unwrapping followed by background phase removal, HARPERELLA preserves the tissue phase signal in gray matter, white matter and cerebrospinal fluid with excellent robustness, providing a convenient and accurate solution for QSM. The proposed algorithm is provided together with QSM and susceptibility tensor imaging (STI) tools in a shared software package named “STI Suite”.
Keywords: Laplacian, phase unwrapping, background phase removal, phase contrast, quantitative susceptibility mapping
Introduction
Gradient echo (GRE) phase images provide better tissue contrast-to-noise ratio (CNR) compared to magnitude images, revealing many anatomical details of the brain that are not easily visible using other imaging contrasts (1,2). Furthermore, GRE signal phase shows excellent sensitivity to molecular and cellular components, such as iron and myelin (2–6), whose magnetic properties are different from those of bulk water. Alterations in iron content and myelination are hallmarks of many neurological diseases, such as Parkinson’s disease, Huntington’s disease, multiple sclerosis and others (3,7–10). However, the application of GRE phase images to studying these pathologies is significantly hampered by the nonlocal and orientation dependent dipolar interaction between tissue magnetic susceptibilities and the main magnetic field. To overcome this problem, quantitative susceptibility mapping (QSM) was developed to convert the nonlocal signal phase into magnetic susceptibility, which is directly related to local magnetic sources such as iron and myelin (6,11–18). Further, it is recognized that both tissue phase and magnetic susceptibility are dependent on orientation of the white matter fibers, which can be described by the magnetic susceptibility tensors of the molecular and cellular components of brain white matter (19–28). Both QSM and susceptibility tensor imaging (STI) require a series of processing steps, including 3D phase unwrapping, background phase removal, and dipole deconvolution to calculate magnetic susceptibility. Since the dipole deconvolution in QSM is an ill-posed problem, the quality of phase unwrapping and background phase removal can directly influence the accuracy of the result and the quantitative values of the magnetic susceptibility.
To date, a variety of phase unwrapping and background phase removal approaches have been proposed (12,14,29–31). For example, phase unwrapping can be achieved using conventional path-based methods in the spatial domain (32–34) and linear fitting methods in the temporal domain (11,30). Compared to phase unwrapping, background phase removal is more challenging, since the local frequency component of the brain phase contains both background phase and the nonlocal tissue phase contributions. Traditional high-pass spatial filtering, e.g. homodyne filtering that have been very successfully used in susceptibility weighted imaging (35), can remove, along with the background phase, a substantial contribution of low frequency components of the tissue phase (29). This will inevitably lead to inaccurate magnetic susceptibility values using QSM. Recently, the harmonic property of the background phase has been used in several methods to remove the background phase while preserving the low spatial frequency components of the tissue phase. These methods include, for example, the sophisticated harmonic artifact reduction for phase data (SHARP) method and its variants (14,16), and the projection onto dipole fields (PDF) method (13,29). The combination of these phase unwrapping and background removal methods provides a number of viable solutions to extract the tissue phase for QSM.
In addition to the aforementioned techniques, the phase can also be unwrapped using a Laplacian-based method (6), which is fast, robust, and can theoretically suppress or completely remove the background contribution to the phase signal. However, this ability of background phase removal has not been demonstrated due to the difficulty of computing the Laplacian of the phase signal outside the brain. Previous studies have primarily assumed that the phase Laplacian is zero outside the brain. In this study, we estimate the Laplacian of the phase image outside the brain using L2 norm minimization. By extending the Laplacian to the whole field-of-view (FOV), we achieve phase unwrapping and background phase removal in a single step without compromising performance. With the Laplacian-based phase processing methods, existing QSM and susceptibility tensor imaging pipelines can provide high image quality using a simple implementation, reduced computation time, and fewer or no spatial constraints. The proposed technique has been implemented with existing QSM and STI algorithms into a single Matlab toolbox, named “STI Suite”.
Materials and Methods
Integrated Phase Unwrapping and Background Phase Removal using Laplacian
The Laplacian of the phase can be derived from the sine and cosine functions of the wrapped phase directly (36):
| [1] |
The relationship between the Laplacian of the phase image and the underlying magnetic susceptibility distribution is given by (37):
| [2] |
Eq. [2] is a Poisson equation, in which the local elements of (∇2/3− ∂2/∂z2)χ can be regarded as sources that generate the tissue phase obeying the principle of superposition. Solving Eq. [1] yields the unwrapped phase that is free of contributions from sources outside the FOV, while Eq. [2] gives the susceptibility maps. Importantly, according to Eq. [2], the unwrapped phase should be free from contributions outside of the FOV, since the region outside the FOV fulfills the Laplace equation.
If the phase measurement is available everywhere within the whole imaging FOV including areas without tissue support, then both Eq. [1] and [2] can be solved in the spatial frequency domain by assuming periodic boundary conditions at the edges of the FOV. This approach is fast as it takes advantage of the Fast Fourier Transform (FFT) algorithm. Specifically, the Laplacian of the sine and cosine can be calculated using Fourier transforms (6).
Unfortunately, phase measurements are typically not available outside the tissue. Therefore, generally, Eq. [1] and [2] must be solved with boundary conditions set at the irregularly shaped tissue boundaries and the FFT algorithms can no longer be applied. In addition, although the boundary conditions are governed by Maxwell’s equations in principle, it is difficult to define them rigorously as only the z-component of B-field is measurable by MRI. Even if the boundary conditions were defined properly, solving the partial differential equations would still be computationally intensive.
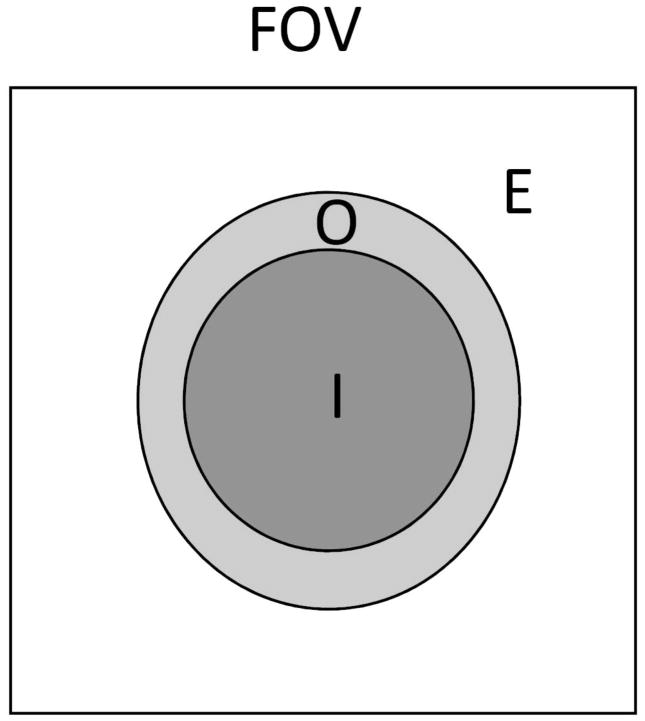
To take advantage of the simplicity of the Fourier approach and the FFT algorithm, the phase outside the tissue has to be determined. Previously, the spherical mean value property of harmonic functions has been successfully used in SHARP (14). In this study, we applied the same spherical mean value filtering to estimate the phase Laplacian outside the FOV. Let I and O be the interior and boundary regions of the tissue respectively, and E is the relative complement of I and O with respect to the FOV, i.e. I ∪ O ∪ E = FOV (Fig. 1). Region O is the set of tissue voxels next to the boundaries that are within a distance of the radius of the spherical mean value filter. Then the phase Laplacian within the region of E, ∇2ϕE, shall be the solution of the following minimization problem:
| [3] |
where “S” represents the spherical mean value operator over a sphere centered around a given spatial location, and the residual susceptibility sources d are estimated as the mean over trustable region I
Fig. 1.
A schematic representation of the different regions in the FOV: I and O are interior and boundary regions of the brain, respectively, and E is the exterior of the brain.
| [4] |
The reason why ∇ϕE has to satisfy Eq. [3] is that phase contributions from sources outside the FOV have been already removed by the Laplacian operator. When sources within E are also removed, the only remaining susceptibility sources originate from the region of I ∪ O. Because of the inaccuracy at the boundary region O, these remaining sources are estimated based on region I as given by Eq. [4]. In short, Eq. [3] simply states that when all background sources are removed, the only sources of phase reside in the trustable region.
Once ϕE is determined, the Laplacian for the whole FOV, ∇2ϕFOV, can be determined as
| [5] |
Finally, the background removed and unwrapped phase can be obtained using the following FFT-based inverse Laplacian:
| [6] |
For convenience, we refer to this method as HARmonic PhasE REmoval with LapLAcian, or HARPERELLA in short. It is emphasized that HARPERELLA achieves both phase unwrapping and background phase removal in a single integrated procedure purely based on the Laplacian operator.
Numerical Simulations
A 3D 128×128×128 Shepp-Logan phantom was used to evaluate the accuracy of HARPERELLA. The phantom was zero padded to 384×384×384 for accurate simulation of the corresponding resonance frequency map. The phantom was composed of multiple ellipsoids placed in a homogenous background with zero susceptibility. The susceptibility values for the ellipsoids were 0, 0.2 and 0.3 ppm, respectively. A cubic object of 100 ppm was placed outside the multiple ellipsoids and served as the external source generating the background field. The background phase was removed using HARPERELLA, with a spherical kernel radius of 5 voxels. The result is compared with the ground truth.
Human Brain Imaging
In vivo brain imaging of 10 adult subjects was conducted on a GE MR750 3.0T scanner (GE Healthcare, Waukesha, WI) equipped with an 8-channel head coil. Phase images with whole-brain coverage were acquired using a standard flow-compensated 3D Fast spoiled-gradient-recalled (FSPGR) sequence with the following parameters: TE = 23 ms, TR = 30 ms, flip angle = 20°, field-of-view (FOV) = 256×256×176 mm3, matrix size = 256×256×176, SENSE factor = 2. All experiments were approved by the local institutional review board.
Image Analysis
The real and imaginary data from the scanner were combined to form the complex data, and then separated into magnitude and phase. The resulting magnitude images were used to obtain the mask of the brain tissue using the BET tool in FSL (FMRIB, University of Oxford) (38). The background phase was then removed using the proposed HARPERELLA method. Briefly, the Laplacian of the phase was calculated using Eqs. [1–4], with zero padding to increase the numerical accuracy. Values outside the brain were set to zero while large Laplacian values near the boundary of the brain (<3 voxels from the boundary of the brain) were also considered inaccurate and set to zero. The phase Laplacian values outside the brain were then estimated with Eq. [3], using a spherical kernel radius (R) of 10 mm. This equation is solved using the preconditioned LSQR method, an iterative linear equation solver in Matlab. The Laplacian values for the whole FOV were then obtained by combining the Laplacian values inside the brain with the calculated Laplacian values outside the brain. Eventually, the background removed phase was calculated using Eq. [6] followed by zero filling areas outside the brain.
The phase processed by HARPERELLA was compared with other state of the art phase processing methods. In contrast to the Laplacian-based phase unwrapping in HARPERELLA, we first performed 3D phase unwrapping of the total phase using the algorithm developed by Abdul-Rahman et al. (34). The first background phase removal method compared is a SHARP method with a variable radius of spherical kernel at the brain boundary (16), referred to as V-SHARP for short. The second one is the method of Projection onto the Dipole Field (PDF) developed by Liu et al (29). All susceptibility maps were computed using the LSQR method (6). The anatomical structures in the susceptibility maps were segmented manually using a Matlab-based ROI tool as described previously (39). The selected ROIs include 6 iron rich gray matter nuclei, namely putamen, globus pallidus, caudate nuclei, red nuclei, substantia nigra, dentate nuclei, and 3 subcortical white matter regions, namely internal commissure, splenium of corpus callosum and optic radiation. All algorithms were implemented in Matlab R2011b (Mathworks, Natick, MA).
STI Suite: A Software Package for Phase Processing, QSM and STI
The Laplacian-based phase processing offers excellent robustness, and can produce local tissue phase free of erroneous phase discontinuities at the tissue boundaries. As such, QSM and STI processing can be achieved using a simpler implementation with fewer or no spatial constraints. To facilitate dissemination and evaluation, all algorithms and tools described here are available in a shared Matlab-based toolbox, “STI Suite”. The toolbox includes implementations of HARPERELLA and V-SHARP (6,16) methods for phase processing, the LSQR method for QSM (6), the k-space-based method for STI (19–21), and graphical user interfaces for easy visualization and ROI editing. STI Suite is available online at http://people.duke.edu/~cl160/.
Results
Numerical Simulations
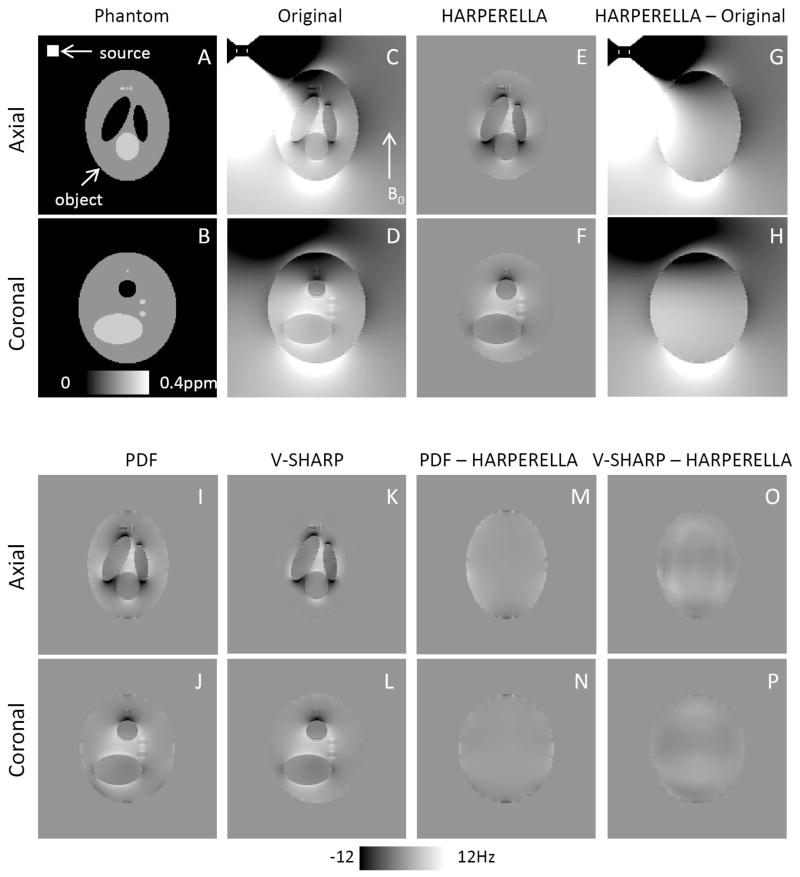
Fig. 2 shows the accuracy of the HARPERELLA for background phase removal. A very strong background phase inside the ellipsoids can be observed when the external source is included in the simulation (Fig. 2C and D). The HARPERELLA methods effectively removed the background phase (Fig. 2E and F). The difference between the HARPERELLA filtered phase and the phase generated by the object in the presence of the external source does not contain contrast of the internal ellipsoids (Fig. 2G and H), indicating the accuracy of HARPERELLA in preserving the phase contrast of internal structures. Similarly, the background phase was also removed effectively using PDF (Fig. 2I, J) and V-SHARP (Fig. 2K, L). The difference image between PDF and HARPERELLA are homogenous, indicating the similar accuracy of PDF (Fig. 2M, N). In contrast, there are some low frequency heterogeneity in the difference between HARPERELLA and V-SHARP (Fig. 2O, P).
Fig. 2.
Numerical simulations using a Shepp-Logan phantom. A, B: The phantom in two orthogonal views. C, D: Simulated phase with the external susceptibility source. E, F: Background removed phase using HARPERELLA. G, H: The difference between the background-removed phase (E, F) and the phase generated by the phantom with the external susceptibility source (C, D). I–L: The background removed phase using PDF (I, J) and V-SHARP (K, L), respectively. M, N: The phase difference between PDF and HARPERELLA. O, P: The phase difference between V-SHARP and HARPERELLA.
HARPERELLA of Human Brain in vivo
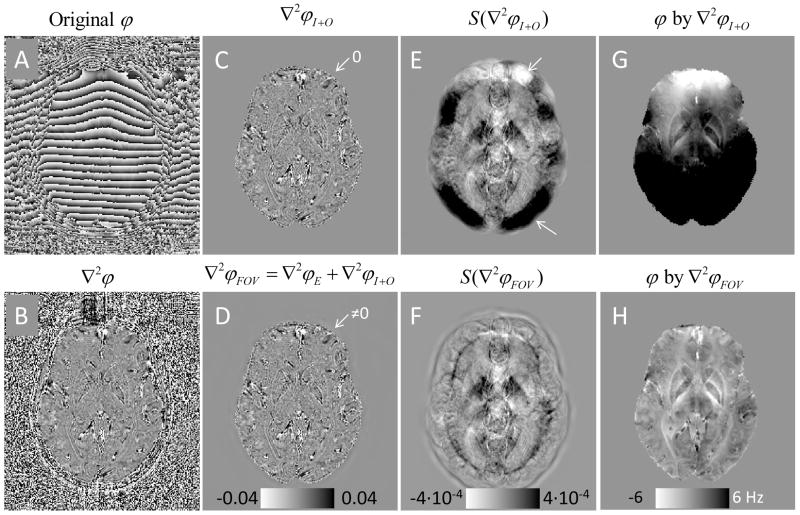
Fig. 3 illustrates the procedures and underlying principles of HARPERELLA. The phase Laplacian (Fig. 3B) calculated from the original phase (Fig. 3A) is inaccurate outside the brain. If the Laplacian outside the brain is simply masked out (Fig. 3C), the unwrapped phase calculated using Eq. 7 still contains a significant contribution of background phase (Fig. 3G). This can be understood using the spherical mean values of the Laplacian (Fig. 3E). The Laplacian of the brain tissue mainly contains edges that are the “sources” of phase contrast. Inside the brain, the “positive sources” cancel the adjacent “negative sources”, so that the magnitude of the spherical mean value of the phase Laplacian is two orders of magnitude smaller than that of the phase Laplacian values at the boundary of the brain. The inaccurate Laplacian values at the brain boundary give rise to net “positive” or net “negative” sources, which can be easily seen in the spherical mean value maps (Fig. 3E, arrows). It is these net “positive” or net “negative” sources that give rise to the background phase in Fig. 3G.
Fig. 3.
HARPERELLA phase processing. A: Original tissue phase. B: Laplacian of the raw phase. C: Laplacian of the raw phase inside the brain. D: Laplacian of the phase throughout the FOV computed using HARPERELLA. Note that the Laplacian outside the brain is non-zero in D. E, F: The spherical mean value filtered images of C and D, respectively. G, H: The unwrapped phase calculated from E and F using the FFT-based inverse Laplacian.
With HARPERELLA, the Laplacian values of the brain tissue are intact. The Laplacian values outside the brain are estimated (Fig. 3D), so that the spherical mean values is uniform throughout the FOV (Fig. 3F). More intuitively, the “positive sources” always cancel the adjacent “negative sources”, so that there is no net background source throughout the FOV. Correspondingly, the background phase contribution is effectively removed in the unwrapped phase image (Fig. 3H) using Eq. 7.
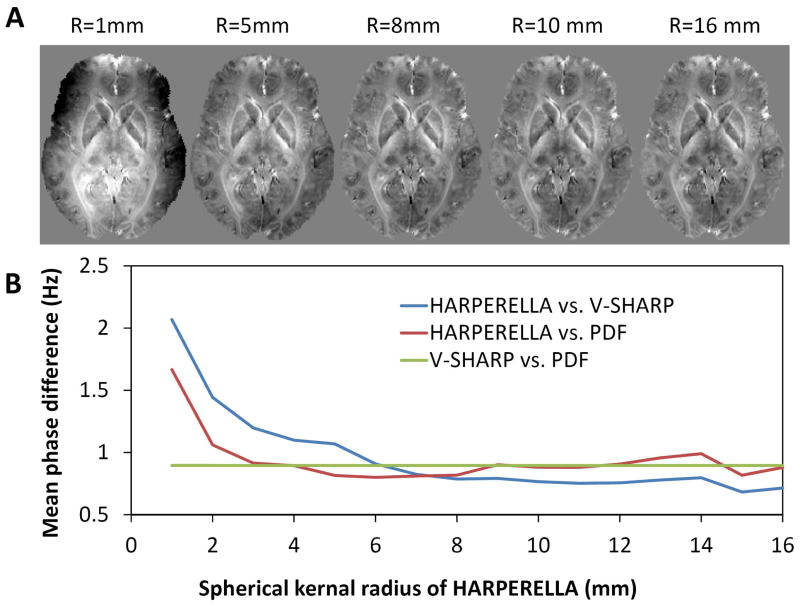
Fig. 4A shows the influence of spherical kernel size on HARPERELLA. With very small kernel radius of 1mm, the background phase cannot be effectively removed. With the kernel size increased to 4mm, the background phase removal is getting more and more effective (data not shown). When the spherical kernel radius increases to 5mm, the background phase is already effectively removed. Further increase of kernel radius up to 16 mm will not significantly change the efficacy of background removal. While the kernel radius between 5 to 16 mm yields similar results, no gold standard is available to determine which one is the best. To give a tentative measure, we calculated the mean phase difference between the HARPERELLA with that of V-SHARP and PDF (Fig. 4B). The phase differences decrease with increasing radius from 1~5mm, and nearly plateau with the kernel radius changing from 6 to 16mm.
Fig. 4.
Influence of spherical kernel size on HARPERELLA. A: Background removed phase using different spherical kernel size. B: Mean phase difference between phase images obtained using PDF, V-SHARP and HARPERELLA with different spherical kernel sizes.
Comparison with Other Phase Processing Methods for QSM
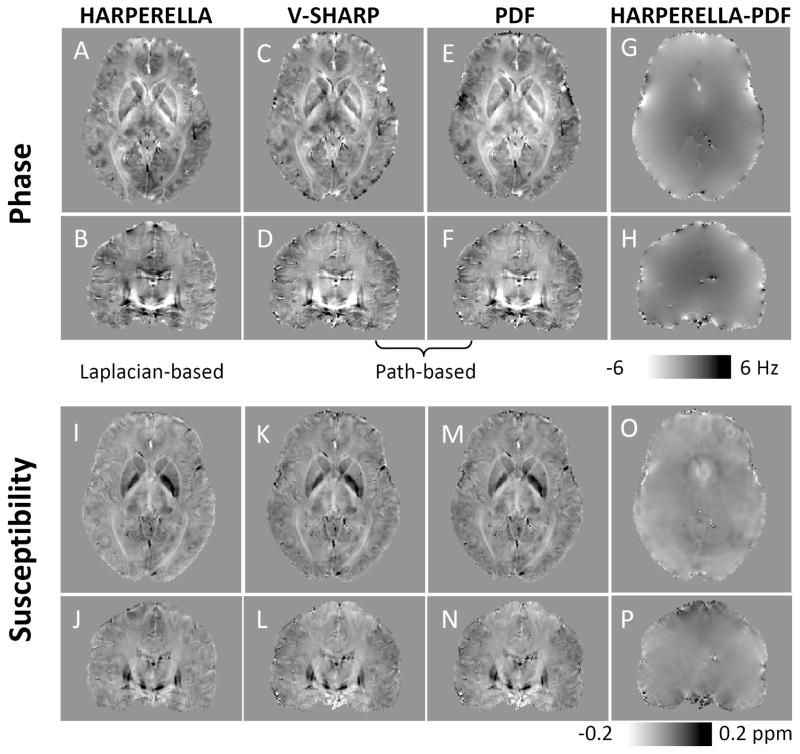
Fig. 5 compares HARPERELLA with the other two methods for phase processing and subsequent QSM. It can be observed that all three methods generate comparable phase maps with some visual differences (Fig. 4A–F). As will be discussed later, the three methods are based on very similar physical principles. The major difference is that HARPERELLA uses a Laplacian-based phase unwrapping algorithm, while the other two methods use the traditional path-based phase unwrapping technique. Therefore, we only show the difference map between the HARPERELLA and the PDF processed phase images (Fig. 5G, H). The difference map is smooth without clear tissue contrast except for a few local phase differences near blood vessels. After carefully examining the whole brain, we did not observe any local phase differences related to the contrast between gray matter, white matter and cerebrospinal fluid.
Fig. 5.
Comparison of the background-removed phase and magnetic susceptibility obtained using different phase processing methods. A, B: Tissue phase images obtained using HARPERELLA. C, D: Tissue phase images obtained using path-based phase unwrapping and V-SHARP. E, F: Tissue phase images obtained using path-based phase unwrapping and PDF. G, H: Phase difference between that results obtained with HARPERELLA and path-based phase unwrapping plus PDF. I, J: Susceptibility maps derived from HARPERELLA-processed phase images. K, L: Susceptibility maps derived from phase images obtained using path-based phase unwrapping and V-SHARP. M, N: Susceptibility maps derived from phase images obtained using path-based phase unwrapping and PDF. O, P: Difference between the susceptibility maps obtained with HARPERELLA and path-based phase unwrapping plus PDF.
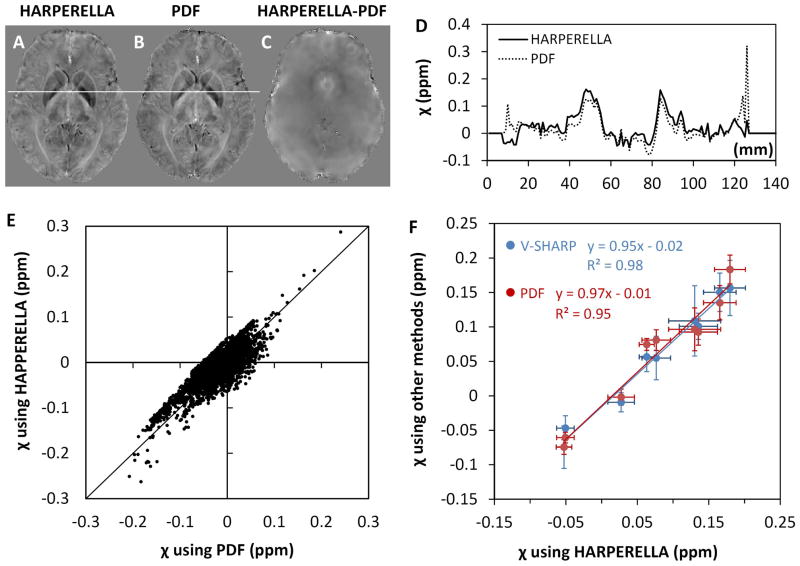
Fig. 5I–N shows magnetic susceptibility maps of the brain obtained using the background-removed phase maps in Fig. 5A–F. As expected, the magnetic susceptibility maps are also very similar. There are some global susceptibility differences unrelated to tissue contrast (Fig. 5O, P). The streaking artifacts also appeared different among V-SHARP, PDF and HARPERELLA. These streaking artifacts are related to phase errors in noisy voxels at the brain boundary and around blood vessels and can be observed in the difference maps. Throughout the brain, there are no local susceptibility differences related to the contrast between gray and white matter (Fig. 5O, P). Fig. 6 further compares a representative line profile through the magnetic susceptibility maps calculated from the HARPERELLA-processed and the PDF-processed phase images respectively (Fig. 6D). In the majority of the brain tissue, the magnetic susceptibility profiles are very similar between the two methods, despite some small differences at the brain boundary. From Fig. 6E, the susceptibility maps derived from the HARPERELLA-processed and PDF-processed phase images are linearly correlated with each other. We further investigated the quantitative relationship in the group of 10 subjects. Fig. 6F compares the mean susceptibility values in a few major brain structures measured using the three methods. It can be seen that the magnetic susceptibility maps obtained using HARPERELLA are linearly correlated with both those derived from V-SHARP (slope = 0.95, R2=0.98) and those calculated using PDF (slope = 0.97, R2=0.95).
Fig. 6.
Detailed comparison of HARPERELLA, SHARP and PDF. A–C: The susceptibility maps of the two methods and their difference. D: Difference in line profiles between the two methods. E: Scatter plot of the susceptibility values obtained using the two methods. F: Group comparison of susceptibility mapping results using HARPERELLA versus V-SHARP and PDF. The susceptibility values are obtained from the mean values of the selected ROIs in 10 subjects. The ROIs include the putamen, the globus pallidus, the caudate nuclei, the red nuclei, the substantia nigra, the dentate nuclei, the internal commissure, the splenium of the corpus callosum and the optic radiation. In this figure, path-based phase unwrapping is used for PDF and V-SHARP.
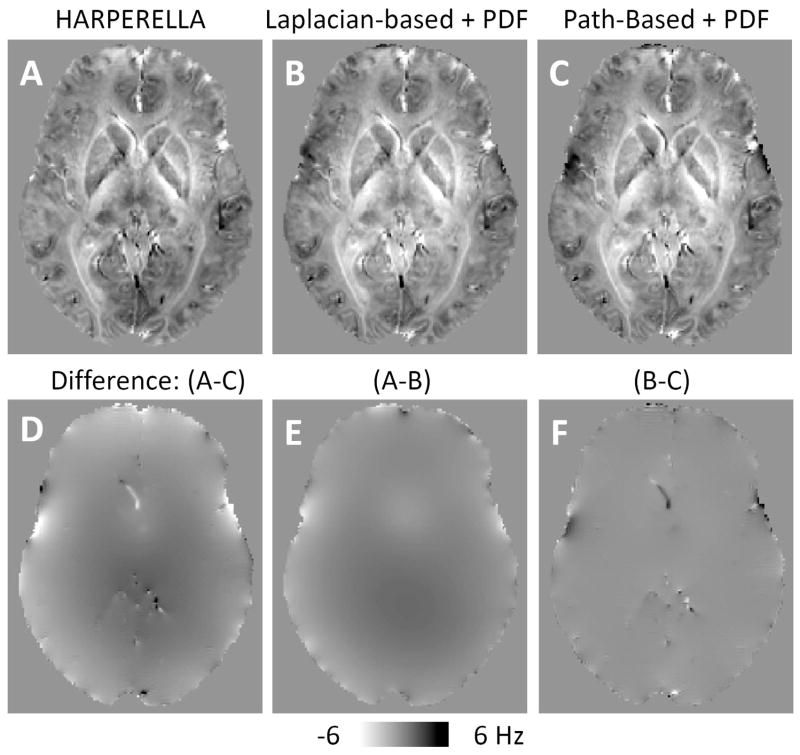
To explore the origin of the phase difference around the blood vessels (Fig. 5G, H), we compared the Laplacian-based and path-based phase unwrapping (Fig. 7). From the difference map (bottom row), there is no local phase contrast related to blood vessels between HARPERELLA and PDF, if Laplacian-based unwrapped phase is used for PDF (Fig. 7E). In contrast, if the same PDF method is applied to Laplacian-based and path-based unwrapped phase, local phase contrast related to blood vessel is seen in the difference map (Fig. 7F). This shows the fundamental difference between Laplacian-based and path-based unwrapping. While the accuracy of Laplacian-based unwrapping vs. path-based phase unwrapping is difficult to access for the complicated venous network, the Laplacian-based unwrapping offers excellent robustness, which can be seen from the continuous unwrapping results even in a blood vessel surrounded by noise (Fig. 8).
Fig. 7.
Comparison of HARPERELLA with PDF using both Laplacian-based and path-based phase unwrapping. A–C: Background removed phase using HARPERELLA (A), Laplacian-based phase unwrapping and PDF (B), and path-based phase unwrapping and PDF (C). D: Difference between images shown in panels A and C. E: Difference between images shown in panels A and B. F: Difference between images shown in panels B and C.
Fig. 8.
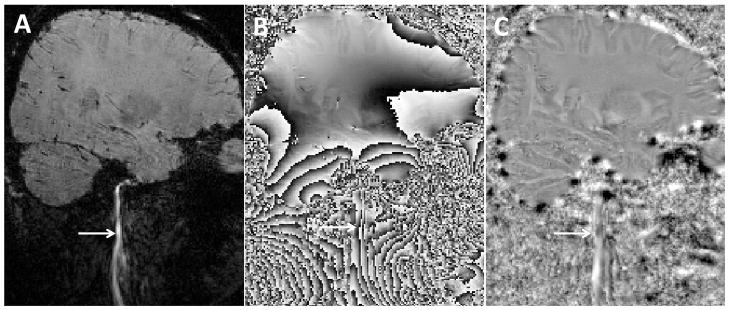
An example showing the robustness of the Laplacian-based phase unwrapping. A: magnitude. B: Raw phase. C: Unwrapped phase using the Laplacian-based phase unwrapping followed by a simple high-pass filtering. The Laplacian-based phase unwrapping can yield a continuous phase map of the blood vessel, which is surrounded by excessive amount of noise and phase wraps.
For this particular dataset, phase unwrapping using Laplacian (using Matlab) and the path-based approach (using C++) takes 6 and 20 sec, respectively. Background phase removal using HARPERELLA (including phase unwrapping), V-SHARP and PDF (excluding phase unwrapping) needs 108, 101 and 836 sec, respectively.
Discussion
In this study, we developed a novel method called HARPERELLA for integrated phase unwrapping and removal of harmonic background phase in a single step. Because the Laplacian of the phase is calculated using sine and cosine functions of the wrapped phase, our method is insensitive to phase wraps. Phase unwrapping and background phase removal is achieved using the phase Laplacian and minimizing the net susceptibility source in the FOV. We validated this technique using both numerical simulations and image analysis of human brain images acquired in vivo. HARPERELLA is fast, robust and preserves the low spatial frequency components of brain tissue phase important for QSM.
The use of the Fast Fourier Transform ensures efficiency, while the use of sine and cosine functions to calculate the Laplacian provides the robustness (Fig. 8). Despite the convenience, the accuracy of Laplacian-based phase unwrapping at the tissue interfaces must be evaluated rigorously by comparison with the conventional path-based methods. In theory, the calculation of the Laplacian using Fourier Transform requires an infinite image FOV to preserve all the spatial frequencies of the signal phase. However, in practice, a finite FOV is used for numerical practicality. Therefore, the FFT-based calculation of the Laplacian can result in phase differences around blood vessels between images processed with HARPERELLA and path-based phase unwrapping methods (Fig. 5G, H), as reported in our previous study (6). This origin of discrepancy on the edge of blood vessels is further confirmed in Fig. 7. Consequently, caution and further investigation are needed when applying HARPERELLA to evaluate the magnetic susceptibility of venous blood. Apart from the small regions around blood vessels, the difference map of the HARPERELLA- and PDF-processed phase images does not reveal any structures related to gray matter, white matter and the cerebrospinal fluid.
HARPERELLA shows very similar performance in background phase removal compared to two other background phase removal tools, i.e. V-SHARP and PDF, which are based on a very similar underlying physical principle. HARPERELLA and V-SHARP utilize the property that the background phase satisfies the Laplace equation within the brain, while PDF assumes that the background phase is generated by susceptibility sources outside the brain tissue, which also satisfies the Laplace equation (14,29). The difference is that the Laplacian-based solution may not necessarily be confined to a dipole field as in the PDF methods. As such, HARPERELLA and V-SHARP might offer more general solutions. However, PDF can remove most of the background phase, indicating the dominance of the dipole field in the background phase and the ultimate equivalency of the three methods.
With the same physical underpinnings, the main benefit of HARPERELLA is saving the step of phase unwrapping, while the accuracy of HARPERELLA is similar to that of V-SHARP and PDF, except minor differences in regions close to brain boundary. The original SHARP method eroded 5 mm tissue at the brain boundary, which cannot be recovered in the final phase images, and the deconvolution in SHARP may also add a small amount of uncertainty to the background tissue phase (16). In the V-SHARP method, the accuracy of voxels at the brain boundary is slightly lower compared to the center of the brain, due to the smaller radius of the spherical kernel used at the boundary (16). In addition, although barely noticeable in vivo, the obtained background phase using V-SHARP is not exactly harmonic likely due to the boundary issue, which can be seen from the low-frequency difference between V-SHARP and HARPERELLA in simulation (Fig. 2O, P). This is due to the use of varying spherical kernel at the brain boundary. Despite this, V-SHARP gives highly consistent results among different subjects, and has allowed successful delineation of the developmental trajectories of magnetic susceptibility over the lifespan (39). The advantage of SHARP and V-SHARP is that they solve a forward problem, which usually requires a simpler implementation and is less computationally intensive.
Different from SHARP or V-SHARP, HARPERELLA does not have limitations near the brain boundary, which is the same as PDF. HARPERELLA does not alter the local tissue phase contrast, because it keeps the Laplacian inside the brain, and only alters the Laplacian outside to compensate the inaccurate Laplacian values at the boundary. This compensation is achieved using the spherical mean kernel in Eq. [3] to ensure a consistent distribution of Laplacian throughout the FOV. Despite its importance, the kernel size can be selected over a wide range of values with satisfactory results (Fig. 4). This finding suggests that the outside Laplacian closer to the brain boundary plays more important roles. As such, once a critical threshold of kernel size is reached (R=6), a moderate change of the kernel size will not affect performance of HARPERELA significantly. An advantage of HARPERELLA is that it can provide a convenient way to combine all the phase data from multiple coils and/or multiple echoes, since the Laplacian of the tissue phase does not contain any contribution from the background and coil phase, and thus can be combined straightforwardly through weighted linear combination.
In conclusion, we developed a Laplacian-based method called HARPERELLA for 3D phase unwrapping and removal of harmonic background phase in a single step. HARPERELLA can effectively remove the background contribution, while preserving the local tissue phase throughout the whole brain. The local tissue phase contrast between gray matter, white matter and CSF remains the same between HARPERELLA, PDF and V-SHARP, regardless of the phase unwrapping methods. This similarity is attributed to the same underlying physical principles. In contrast to the similar contrast between gray matter, white matter and CSF, the phase contrast around the blood vessels is dependent on the choice of Laplacian-based or path-based unwrapping. As such, the accuracy of HARPERELLA for quantifying the magnetic susceptibility of the venous blood needs to be further investigated.
Acknowledgments
The study is supported in part by the National Institutes of Health (NIH) through grant R01 MH096979 and by the National Multiple Sclerosis Society through grant RG4723 to C. L.
List of abbreviations
- FFT
Fast Fourier transform
- FOV
field-of-view
- GRE
gradient echo
- HARPERELLA
(3D phase unwrapping and) harmonic (background) phase removal using Laplacian
projection onto dipole fields
- QSM
quantitative susceptibility mapping
- SHARP
sophisticated harmonic artifact reduction for phase data
- STI
susceptibility tensor imaging
- V-SHARP
SHARP with varying spherical kernel sizes
References
- 1.Rauscher A, Sedlacik J, Barth M, Mentzel HJ, Reichenbach JR. Magnetic susceptibility-weighted MR phase imaging of the human brain. Am J Neuroradiol. 2005;26(4):736–742. [PMC free article] [PubMed] [Google Scholar]
- 2.Duyn JH, van Gelderen P, Li TQ, de Zwart JA, Koretsky AP, Fukunaga M. High-field MRI of brain cortical substructure based on signal phase. Proc Natl Acad Sci USA. 2007;104(28):11796–11801. doi: 10.1073/pnas.0610821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haacke EM, Chengb NYC, House MJ, Liu Q, Neelavalli J, Ogg RJ, Khan A, Ayaz M, Kirsch W, Obenaus A. Imaging iron stores in the brain using magnetic resonance imaging. Magn Reson Imaging. 2005;23(1):1–25. doi: 10.1016/j.mri.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Duyn JH. Study of brain anatomy with high-field MRI: recent progress. Magn Reson Imaging. 2010;28(8):1210–1215. doi: 10.1016/j.mri.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C, Li W, Johnson GA, Wu B. High-field (9. 4 T) MRI of brain dysmyelination by quantitative mapping of magnetic susceptibility. Neuroimage. 2011;56(3):930–938. doi: 10.1016/j.neuroimage.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Wu B, Liu C. Quantitative susceptibility mapping of human brain reflects spatial variation in tissue composition. Neuroimage. 2011;55:1645–1656. doi: 10.1016/j.neuroimage.2010.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinman M. Multiple sclerosis: a coordinated immunological attack against myelin in the central nervous system. Cell. 1996;85(3):299–302. doi: 10.1016/s0092-8674(00)81107-1. [DOI] [PubMed] [Google Scholar]
- 8.Dexter D, Wells F, Lee A, Agid F, Agid Y, Jenner P, Marsden C. Increased nigral iron content and alterations in other metal ions occurring in brain in Parkinson’s disease. J Neurochem. 1989;52(6):1830–1836. doi: 10.1111/j.1471-4159.1989.tb07264.x. [DOI] [PubMed] [Google Scholar]
- 9.Bartzokis G, Lu PH, Tishler TA, Fong SM, Oluwadara B, Finn JP, Huang D, Bordelon Y, Mintz J, Perlman S. Myelin breakdown and iron changes in Huntington’s disease: pathogenesis and treatment implications. Neurochemical Res. 2007;32(10):1655–1664. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- 10.Yablonskiy DA, Luo J, Sukstanskii AL, Iyer A, Cross AH. Biophysical mechanisms of MRI signal frequency contrast in multiple sclerosis. Proc Natl Acad Sci. 2012;109(35):14212–14217. doi: 10.1073/pnas.1206037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu T, Spincemaille P, de Rochefort L, Kressler B, Wang Y. Calculation of susceptibility through multiple orientation sampling (COSMOS): A method for conditioning the inverse problem from measured magnetic field map to susceptibility source image in MRI. Magn Reson Med. 2009;61(1):196–204. doi: 10.1002/mrm.21828. [DOI] [PubMed] [Google Scholar]
- 12.Shmueli K, de Zwart JA, van Gelderen P, Li TQ, Dodd SJ, Duyn JH. Magnetic susceptibility mapping of brain tissue in vivo using MRI phase data. Magn Reson Med. 2009;62(6):1510–1522. doi: 10.1002/mrm.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Rochefort L, Liu T, Kressler B, Liu J, Spincemaille P, Lebon V, Wu JL, Wang Y. Quantitative susceptibility map reconstruction from MR phase data using Bayesian regularization: validation and application to brain imaging. Magn Reson Med. 2010;63(1):194–206. doi: 10.1002/mrm.22187. [DOI] [PubMed] [Google Scholar]
- 14.Schweser F, Deistung A, Lehr BW, Reichenbach JR. Quantitative imaging of intrinsic magnetic tissue properties using MRI signal phase: An approach to in vivo brain iron metabolism? Neuroimage. 2011;54(4):2789–2807. doi: 10.1016/j.neuroimage.2010.10.070. [DOI] [PubMed] [Google Scholar]
- 15.Wharton S, Schafer A, Bowtell R. Susceptibility mapping in the human brain using threshold-based k-space division. Magn Reson Med. 2010;63(5):1292–1304. doi: 10.1002/mrm.22334. [DOI] [PubMed] [Google Scholar]
- 16.Wu B, Li W, Guidon A, Liu C. Whole brain susceptibility mapping using compressed sensing. Magn Reson Med. 2012;67(1):137–147. doi: 10.1002/mrm.23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T, Liu J, de Rochefort L, Spincemaille P, Khalidov I, Ledoux JR, Wang Y. Morphology enabled dipole inversion (MEDI) from a single-angle acquisition: Comparison with COSMOS in human brain imaging. Magn Reson Med. 2011;66(3):777–783. doi: 10.1002/mrm.22816. [DOI] [PubMed] [Google Scholar]
- 18.Haacke E, Tang J, Neelavalli J, Cheng Y. Susceptibility mapping as a means to visualize veins and quantify oxygen saturation. J Magn Reson Imaging. 2010;32(3):663–676. doi: 10.1002/jmri.22276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C. Susceptibility tensor imaging. Magn Reson Med. 2010;63(6):1471–1477. doi: 10.1002/mrm.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Wu B, Avram AV, Liu C. Magnetic susceptibility anisotropy of human brain in vivo and its molecular underpinnings. Neuroimage. 2012;59(3):2088–2097. doi: 10.1016/j.neuroimage.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Li W, Wu B, Jiang Y, Johnson GA. 3D Fiber tractography with susceptibility tensor imaging. Neuroimage. 2012;59(2):1290–1298. doi: 10.1016/j.neuroimage.2011.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Vikram DS, Lim IAL, Jones CK, Farrell JAD, van Zijl PCM. Mapping magnetic susceptibility anisotropies of white matter in vivo in the human brain at 7T. NeuroImage. 2012;62(1):314–330. doi: 10.1016/j.neuroimage.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisnieff C, Liu T, Spincemaille P, Wang S, Zhou D, Wang Y. Magnetic susceptibility anisotropy: Cylindrical symmetry from macroscopically ordered anisotropic molecules and accuracy of MRI measurements using few orientations. NeuroImage. 2013;70:363–376. doi: 10.1016/j.neuroimage.2012.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wharton S, Bowtell R. Fiber orientation-dependent white matter contrast in gradient echo MRI. Proc Natl Acad Sci USA. 2012;109(45):18559–18564. doi: 10.1073/pnas.1211075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sukstanskii AL, Yablonskiy DA. On the role of neuronal magnetic susceptibility and structure symmetry on gradient echo MR signal formation. Magn Reson Med. 2013 doi: 10.1002/mrm.24629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duyn J. MR susceptibility imaging. J Magn Reson. 2013;229:198–207. doi: 10.1016/j.jmr.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Li W. Imaging neural architecture of the brain based on its multipole magnetic response. NeuroImage. 2013;67:193–202. doi: 10.1016/j.neuroimage.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Murphy NE, Li W. Probing white-matter microstructure with higher-order diffusion tensors and susceptibility tensor MRI. Front Int Neurosci. 2013;7 doi: 10.3389/fnint.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T, Khalidov I, de Rochefort L, Spincemaille P, Liu J, Tsiouris AJ, Wang Y. A novel background field removal method for MRI using projection onto dipole fields (PDF) NMR Biomed. 2011;24(9):1129–1136. doi: 10.1002/nbm.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kressler B, de Rochefort L, Liu T, Spincemaille P, Jiang Q, Wang Y. Nonlinear Regularization for Per Voxel Estimation of Magnetic Susceptibility Distributions From MRI Field Maps. IEEE Trans Med Imaging. 2010;29(2):273–281. doi: 10.1109/TMI.2009.2023787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T, Wisnieff C, Lou M, Chen W, Spincemaille P, Wang Y. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn Reson Med. 2013;69(2):467–476. doi: 10.1002/mrm.24272. [DOI] [PubMed] [Google Scholar]
- 32.Dias JMB, Leitao JMN. The ZpiM algorithm: a method for interferometric image reconstruction in SAR/SAS. IEEE Trans Image Processing. 2002;11(4):408–422. doi: 10.1109/TIP.2002.999675. [DOI] [PubMed] [Google Scholar]
- 33.Jenkinson M. Fast, automated, N-dimensional phase-unwrapping algorithm. Magn Reson Med. 2003;49(1):193–197. doi: 10.1002/mrm.10354. [DOI] [PubMed] [Google Scholar]
- 34.Abdul-Rahman H, Gdeisat M, Burton D, Lalor M. Fast three-dimensional phase-unwrapping algorithm based on sorting by reliability following a non-continuous path. Intl Soc Opt Photonics. 2005:32–40. [Google Scholar]
- 35.Haacke EM, Mittal S, Wu Z, Neelavalli J, Cheng YCN. Susceptibility-Weighted Imaging: Technical Aspects and Clinical Applications, Part 1. Am J Neuroradiol. 2009;30(1):19–30. doi: 10.3174/ajnr.A1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schofield MA, Zhu Y. Fast phase unwrapping algorithm for interferometric applications. Opt Lett. 2003;28(14):1194–1196. doi: 10.1364/ol.28.001194. [DOI] [PubMed] [Google Scholar]
- 37.Salomir R, De Senneville BD, Moonen CTW. A fast calculation method for magnetic field inhomogeneity due to an arbitrary distribution of bulk susceptibility. Concepts Magn Reson B Magn Reson Eng. 2003;19B(1):26–34. [Google Scholar]
- 38.Smith SM. Fast robust automated brain extraction. Human Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Wu B, Batrachenko A, Bancroft-Wu V, Morey RA, Shashi V, Langkammer C, Bellis MDD, Ropele S, Song AW, Liu C. Differential developmental trajectories of magnetic susceptibility in human brain gray and white matter over the lifespan. Human Brain Mapp. 2013 doi: 10.1002/hbm.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]