Abstract
Colon cancer (CC) is a leading cause of cancer mortality. Novel biomarkers are needed to identify CC patients at high risk of recurrence and those who may benefit from therapeutic intervention. The aim of this study is to investigate if miR-21 expression from RNA isolated from formalin-fixed paraffin-embedded (FFPE) tissue sections is associated with prognosis and therapeutic outcome for patients with CC. The expression of miR-21 was measured by quantitative reverse transcriptase-polymerase chain reaction in a Japanese cohort (stage I–IV, n = 156) and a German cohort (stage II, n = 145). High miR-21 expression in tumors was associated with poor survival in both the stage II/III Japanese (P = 0.0008) and stage II German (P = 0.047) cohorts. These associations were independent of other clinical covariates in multivariable models. Receipt of adjuvant chemotherapy was not beneficial in patients with high miR-21 in either cohort. In the Japanese cohort, high miR-21 expression was significantly associated with poor therapeutic outcome (P = 0.0001) and adjuvant therapy was associated with improved survival in patients with low miR-21 (P = 0.001). These results suggest that miR-21 is a promising biomarker to identify patients with poor prognosis and can be accurately measured in FFPE tissues. The expression of miR-21 may also identify patients who will benefit from adjuvant chemotherapy.
Keywords: microRNA, prognosis, colorectal cancer
Introduction
Colorectal cancer is a leading cause of cancer mortality worldwide. Adjuvant chemotherapy after surgical resection decreases recurrences and improves survival in stage III colon cancer (CC) 1. Yet current adjuvant therapy does not work equally well for all patients and identifying classifiers to predict response to therapies will help guide medical decisions and result in improved patient outcomes. The role of adjuvant chemotherapy for stage II CC remains controversial. Many stage II patients will benefit from therapy. But if surgery is curative, additional therapy may harm quality of life with little therapeutic benefit. The high-risk features of stage II CC patients include T4 tumors, poor differentiation, perforation, and an inadequate number of evaluated lymph nodes 2. Yet these features cannot completely identify which patients are at low- or high-risk for disease recurrence. Therefore, it is important to develop novel biomarkers to identify high-risk patients who may be suitable for therapeutic intervention.
Cancer develops as a result of multiple genetic and epigenetic alterations 3. Better knowledge about the changes in gene expression that occur during carcinogenesis may lead to improvements in diagnosis, treatment, and prevention. Identifying novel biomarkers that can guide therapeutic decisions is a major goal. Although several RNA-based biomarkers have been reported to identify high-risk patients 4–6, measurement methods of these biomarkers usually require freshly frozen tissues. In contrast, formalin-fixed paraffin-embedded (FFPE) tissue samples have been collected through decades of routine histopathological examination and are the most widely available materials for use in clinical research. For evaluation of biomarkers in FFPE tissue, immunohistochemistry or in situ hybridization is currently the diagnostic standard. However, the degree of expression of the marker can only be described in a semiquantitative way. In contrast, quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) is a quantitative, reliable, and standardized method to investigate RNA expression. But formaldehyde-containing fixatives cause cross-linkage between nucleic acids and proteins and make subsequent extraction and quantification of RNA challenging 7. A major obstacle to RNA expression analysis of FFPE tissues has been the uncertainty about whether gene expression analyses from routinely archived tissues accurately reflect the expression before fixation, and this is likely due to high fragmentation 8. Because fragmentation does not cause further loss of quality when naturally occurring small RNAs are targeted, microRNA is more ideal for analysis of RNA extracted from FFPE samples.
MicroRNAs are 18- to 25-nucleotide, noncoding RNA molecules that regulate the translation of many genes 9. MicroRNA expression levels are altered in most types of human cancers 10–12. We have previously shown that microRNA expression is associated with prognosis in patients with lung 13, 14, colon 15, gastric 16 and esophageal cancer 17, 18. Specifically for CC, we have shown that patients with tumors expressing high levels of an oncogenic microRNA, miR-21, have a worse prognosis for stage II or stage III colon cancer, demonstrating its potential as a prognostic indicator for colon cancer 15. Consistent results have also been observed in other malignancies with increased expression of miR-21 being associated with a worse prognosis and/or therapeutic outcome in multiple cancers including lung cancer 13, 14, 19, colon cancer 15, 20–22, pancreatic cancer 23, 24, breast cancer 25, 26, head and neck cancer 27, tongue cancer 28, astrocytomas 29 and chronic lymphocytic leukemia 30. These data support the hypothesis that expression of miR-21 has potential as a prognostic indicator for a wide variety of malignancies. In most of these reports, miR-21 expression was measured from freshly-frozen tissues and the utility of measuring miR-21 expression from FFPE samples still remains unclear. For clinical application, it will be useful if miR-21 can be accurately measured from FFPE tissue and if its expression can stratify patients into risk groups. In situ hybridization of miR-21on FFPE colon cancer tissues has been used to stratify patients into low and high risk groups for survival 22, but qRT-PCR measurements was not performed and the association between miR-21 and therapeutic outcome was not examined. Therefore, we determined if miR-21 expression from FFPE tissues was associated with cancer-specific mortality and therapeutic outcome for CC.
In the present study, we investigated the association of miR-21 expression, determined by qRT-PCR from FFPE samples, with prognosis of patients with CC. We also investigated the influence of hematoxylin contamination on qRT-PCR results.
Materials and Methods
Patients and tissue samples
We used archival FFPE tissues from 301 patients who had undergone surgical excision of CC. Rectal cancer patients were excuded. Of 301 patients, 156 patients were treated at the Hiroshima University Hospital (Hiroshima, Japan) between 1997 and 2003, and 145 patients were treated at the Mainz University Clinic Center and its teaching hospitals in Germany between 2005 and 2007. Detailed background information including age, sex, TNM stage, tumor location, survival times from diagnosis, receipt of adjuvant chemotherapy (for patients with stage I–III CC), and receipt of postoperative chemotherapy (for patients with stage IV CC) has been collected. The final date of follow-up was December 31, 2008 for the Japanese cohort and March 25, 2013 for the German cohort. Tumor histopathology was classified according to the World Health Organization Classification of Tumors system. Tumors were staged according to the TNM classification system. This study was approved by the Institutional Review Board of the National Institutes of Health, the Ethical Committee for Human Genome Research of Hiroshima University (Hiroshima, Japan) and Landsaerztekammer Rheinland-Pfalz (Mainz, Germany).
RNA extraction
FFPE samples were sectioned (10 μm), deparaffinized, and stained with hematoxylin and eosin to ensure that the sectioned block contained tumor cells. For the Japanese cohort, adjacent sections were stained by hematoxylin and the tumor area was marked under a light microscope. For German cohort, the tumor areas in adjacent sections were marked under a light microscope without hematoxylin staining. Tumor areas were macrodissected with sterile disposable scalpels (Cincinnati Surgical Company, Cincinnati, OH) and subjected to RNA isolation using the PureLink FFPE Total RNA Isolation Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions with small modifications. This included a 10 minute centrifugation at maximum speed in an Eppendorf 5415C benchtop centrifuge after proteinase K digestion to remove trace hematoxylin from the sample. Total RNA was quantified using the NanoDrop ND-1000 spectrometer (NanoDrop, Wilmington, DE) and both OD 260/280 and OD 260/230 ratios utilized for quality control.
qRT-PCR
Expression levels of miR-21 and RNU48 were measured using Taqman MicroRNA Assays (Applied Biosystems) while blinded to clinical outcomes. cDNA was synthesized using microRNA-specific primers and a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Austin, TX) according to the manufacturer’s instructions. Briefly, 40 ng of RNA was reverse transcribed in a 20 μL reaction with gene specific RT probes. qPCR was performed using the 7900 HT-Fast real-time PCR system (Applied Biosystems). We used small nuclear RNA (RNU48) as an endogenous normalization control for miR-21. All assays were performed in triplicate. Relative expression quantitation of miR-21 was calculated with RQ manager 1.2 (Applied Biosystems).
MSI analysis
Genomic DNA was extracted from FFPE samples (10μm) from individual patients. The tumor area was marked under a light microscope without hematoxylin staining. Tumor areas and adjacent nonneoplastic areas were macrodissected with sterile disposable scalpels (Cincinnati Surgical Company) and subjected to DNA extraction by a phenol-chloroform method with proteinase K digestion 31. The five microsatellite markers that were recommended by a National Cancer Institute (NCI) workshop on MSI (BAT25, BAT26, D2S123, D5S346, and D17S250) were used to examine paired nonneoplastic and tumor DNA for MSI status. PCR and subsequent analyses using ABI 3130XL Genetic Analyzer (Applied Biosystems) were performed, and the shift of PCR products from tumor DNA was compared to that of DNA from corresponding nonneoplastic tissue. The size of each fluorescent PCR product was calculated using GeneMapper software (Applied Biosystems). According to the guidelines of the international workshop of NCI 32, tumors were classified as MSI-H when at least 2 of the 5 markers displayed novel bands, MSI-L when additional alleles were found with 1 of the 5 markers, and MSS when all microsatellite markers showed identical patterns in both tumor and non-neoplastic tissues. MSI-H and MSI-L were considered as MSI. MSI status was scored independently by two examiners.
Statistical analysis
Univariate and multivariate Cox regression was used to evaluate the associations between clinical covariates and cancer-specific mortality in Stata 11 (College Station, TX). Hazard ratio (HR) and 95% confidence interval (CI) were estimated from Cox proportional hazard models. For all analyses, age was treated as a categorical variable (greater than or equal to 65 years old versus less than 65 years old). Multivariable Cox regression models were built using stepwise removal of variables with a threshold for removal at P < 0.10. Differences in miR-21 expression levels between 2 groups were tested by the Mann–Whitney U test using Graphpad Prism v5.0 (Graphpad Software Inc, San Diego, CA.). High miR-21 expression was defined on the basis of highest tertile for each cohort, similar to our previous publication on miR-21 in two independent patient cohorts from USA and Hong Kong15. Kaplan-Meier survival curves were constructed for high-miR-21 and low-miR-21 patients using Graphpad Prism v5.0. Differences between survival curves were tested for statistical significance by a Log-rank test. A P value of < 0.05 was considered statistically significant. Two sided P values are reported for all statistical tests.
Results
Expression of miR-21 in stages I–IV of CC (Japanese cohort)
We examined whether the expression of miR-21 in FFPE CC tissues was associated with cancer-specific mortality. While optimizing our RNA isolation techniques from FFPE tissue, we found that there were trace amounts of hematoxylin in several RNA preparations (Supplementary Fig. S1) and that hematoxylin contamination in RNA preparations interferes with qRT-PCR (Supplementary Fig. S2). An extra centrifugation step after proteinase K digestion was found to remove most of the hematoxylin (Supplementary methods and Supplementary Fig. S1). Therefore, all RNA samples were prepared in this manner.
qRT-PCR of miR-21 was performed in all samples of the Japanese cohort (n = 156, Table 1). We first investigated the association between miR-21 expression levels and clinico-pathological characteristics (Supplementary Fig. S3). Expression levels of miR-21 were significantly higher in T4 cases than T1, T2, and T3 cases (P = 0.03, 0.03, and 0.04, respectively; Mann–Whitney U-test), indicating that elevated miR-21 expression was associated with a more aggressive histology of the primary tumor. Expression of miR-21 was also significantly elevated in node-positive cases (N1) compared to node-negative cases (N0, P = 0.03; Mann–Whitney U-test). Expression levels of miR-21 were not associated with age, sex, M classification, or TNM stage.
Table 1.
Characteristics of the study populations
| Japanese cohort (n = 156) | German cohort (n = 145) | |
|---|---|---|
| Recruitment area | Hiroshima, Japan | Mainz, Germany |
| Age, mean (range), year | 63 (29–89) | 70 (39–90) |
| Sex, No. (%) | ||
| Male | 93 (60) | 75 (52) |
| Female | 63 (40) | 70 (48) |
| Follow-up time, median (range), month | 54.0 (2.8–111.8) | 51.6 (1.2–72.4) |
| Adjuvant chemotherapy* (for stage I–III), No. (%) | ||
| Received | 59 (43) | 25 (17) |
| Did not received | 77 (57) | 120 (83) |
| Postoperative chemotherapy* (for stage IV), No. (%) | ||
| Received | 17 (85) | 0 |
| Did not received | 3 (15) | 0 |
| TNM stage, No. (%) | ||
| I | 49 (31) | 0 |
| II | 40 (26) | 145 (100) |
| III | 47 (30) | 0 |
| IV | 20 (13) | 0 |
| Tumor Location, No. (%) | ||
| Proximal | 28 (18) | 63 (43) |
| Distal | 128 (82) | 73 (50) |
| Not available | 0 (0) | 9 (6) |
| MSI status, No. (%) | ||
| MSI | 7 (11) | 19 (15) |
| MSS | 54 (89) | 111 (85) |
| Not available | 95 | 15 |
Abbreviations: MSI, microsatellite instability; MSS, microsatellite stable.
Chemotherapy regimens were primarily 5-fluorouracil based regimens.
Next, we evaluated the association between miR-21 expression levels and prognosis. When analyzing all cases regardless of stage, we found that cases with high miR-21 expression had slightly worse cancer-specific mortality although this did not achieve statistical significance (P = 0.083, Log-rank test, Supplementary Fig. S4). Similar results were seen with multivariate Cox regression analyses (Supplementary Table S1).
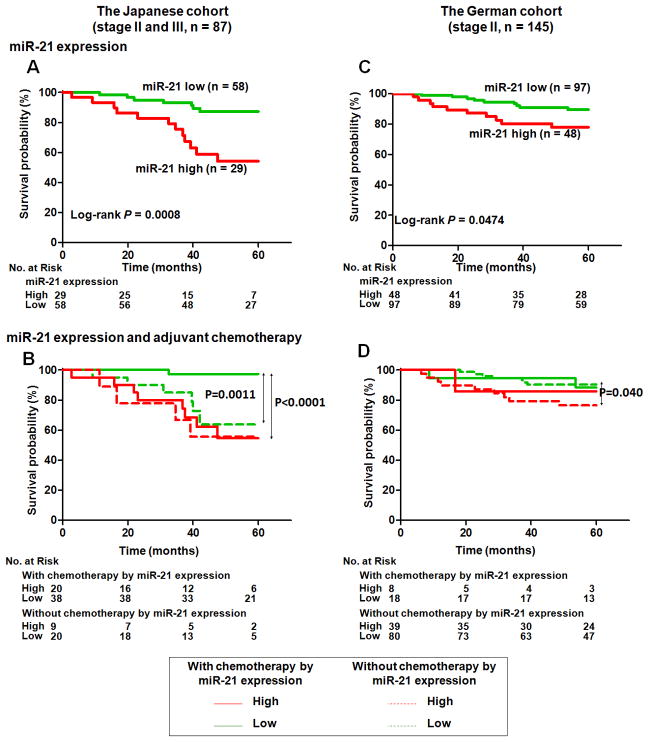
Stage I CC patients have very good survival prognosis and surgery is usually curative while stage IV CC patients have dismal prognoses with 5-year survival rates below 10%. The survival of patients with stage II or stage III CC are intermediate and it is these patients that would benefit the most from prognostic biomarkers. Therefore, we restricted our analysis of the prognostic value of miR-21 to patients with stage II and stage III CC (n = 87). We found that cases with high miR-21 expression had significantly worse cancer-specific mortality than those with low miR-21 expression (P = 0.0008, Log-rank test, Fig. 1A). Both univariate and multivariate Cox proportional hazards analysis were used to further evaluate the association of miR-21expression with cancer-specific mortality (Table 2). In univariate analysis, high expression of miR-21 (HR, 4.17; 95% CI, 1.64–10.11; P = 0.003), receipt of adjuvant chemotherapy (HR, 0.38; 95% CI, 0.16–0.94; P=0.037) and T classification (HR, 4.23; 95%CI, 1.81–10.11; P=0.001) were significantly associated with survival while TNM stage (HR, 1.59; 95% CI, 0.63–4.05; P = 0.329), age (HR, 2.05; 95% CI, 0.81–5.21; P = 0.132), tumor location (HR, 0.73; 95% CI, 0.24–2.21; P=0.582) and sex (HR, 1.12; 95% CI, 0.45–2.78; P=0.805) were not. In the final multivariate model, miR-21 expression was an independent prognostic classifier (HR, 3.13; 95% CI, 1.20–8.17; P = 0.019).
Figure 1.
Associations between miR-21 expression with cancer-specific mortality and receipt of adjuvant chemotherapy with prognosis. A and B, Kaplan–Meier plot of the cancer-specific mortality in the Japanese cohort, which includes cases classified as stages II and III. C and D, Kaplan–Meier plot of the overall survival in the German cohort, which includes only stage II cases. A and C, Patients were classified into 2 groups: patients with miR-21-high CC and patients with miR-21-low CC. B and D, Patients were classified into 4 groups: patients with miR-21-high CC who received adjuvant chemotherapy, patients with miR-21-high CC who did not receive adjuvant chemotherapy, patients with miR-21-low CC who received adjuvant chemotherapy, and patients with miR-21-low CC who did not receive adjuvant chemotherapy (B and D).
Table 2.
Univariate and multivariate Cox regression analysis of miR-21 expression levels and overall survival in the Japanese cohort (stage II and III, n = 87)
| Characteristic | Univariate analysis
|
Multivariate analysisa
|
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| miR-21 expression | ||||
| Low | 1 (Ref.) | 0.003 | 1 (Ref.) | 0.019 |
| High | 4.17 (1.64–10.61) | 3.13 (1.20–8.17) | ||
| T classification | ||||
| T1–3 | 1 (Ref.) | 0.001 | 1 (Ref.) | 0.005 |
| T4 | 4.23 (1.81–10.11) | 3.92 (1.52–10.07) | ||
| Adjuvant chemotherapyb | ||||
| Did not receive | 1 (Ref.) | 0.037 | ||
| Received | 0.38 (0.16–0.94) | |||
| Age | ||||
| < 65 | 1 (Ref.) | 0.132 | 1 (Ref.) | 0.05 |
| 65 and > 65 | 2.05 (0.81–5.21) | 2.59 (1.00–6.70) | ||
| TNM stage | ||||
| Stage II | 1 (Ref.) | 0.329 | ||
| Stage III | 1.59 (0.63–4.05) | |||
| Tumor Location | ||||
| Distal | 1 (Ref.) | 0.582 | ||
| Proximal | 0.73 (0.24–2.21) | |||
| Sex | ||||
| Male | 1 (Ref.) | 0.81 | ||
| Female | 1.12 (0.45–2.78) | |||
Abbreviations: HR, hazard ratio; CI, confidence interval
Multivariable model was selected based on stepwise removal of variables with a significance threshold for removal set at p<0.1.
Chemotherapy regimens were primarily 5-fluorouracil based regimens.
Expression of miR-21 in an independent patients with stage II CC (German cohort)
We next evaluated the expression of miR-21 in an independent German cohort of stage II CC patients (n = 152). Based on our observation that that hematoxylin staining could affect qRT-PCR, we extracted RNA from the FFPE sections of the German cohort without hematoxylin staining. Although we did not stain FFPE samples with hematoxylin, the tumor area could be marked in the FFPE sections under a light microscope because the tumor/non-tumor borderline was clearly identified in the stage II tumors. After total RNA was extracted from all samples of the German cohort, 7 samples were excluded with OD 260/230 ratios < 1.00. For the remaining 145 samples (OD 260/230 ratio ranged from 1.07 to 2.04; mean, 1.65) qRT-PCR was performed.
The association between miR-21 expression levels and clinico-pathological characteristics was analyzed (Supplementary Fig. S3). Expression levels of miR-21 were not associated with age, sex or T classification. Next, association between miR-21 expression levels and patient survival was investigated by Kaplan-Meier analysis. We found that high miR-21 expression was significantly associated with increased cancer-specific mortality (P = 0.047, Log-rank test, Fig. 1C). Because adjuvant chemotherapy for patients with stage II CC influences patients’ survival, we analyzed individuals who did not receive adjuvant chemotherapy. We found that high miR-21 expression was associated with increased cancer-specific mortality in that group (P = 0.040, Log-rank test, Fig. 1D).
Univariate and multivariate Cox proportional hazards analysis was used to further evaluate the association of miR-21expression with survival to evaluate the potential for miR-21 expression as a prognostic biomarker (Table 3). In univariate analysis, high expression of miR-21 (HR, 2.65; 95% CI, 0.98–5.95; P = 0.055) and T classification (HR, 2.67; 95% CI, 0.96–7.41; P = 0.060) were marginally associated with survival while tumor location, age, sex, or adjuvant chemotherapy were not. In the final multivariate models, which included miR-21 expression and T classification and tumor location, high miR-21 expression was an independent prognostic indicator (HR, 2.65; 95% CI, 1.06–6.66; P = 0.037). These results demonstrate that miR-21 may be a useful biomarker to identify TNM stage II patients with high risk of recurrence.
Table 3.
Univariate and multivariate Cox regression analysis of miR-21 expression levels and overall survival in the German cohort (n = 145)
| Characteristic | Univariate analysis
|
Multivariate analysis (n=145)a
|
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| miR-21 expression | ||||
| Low | 1 (Ref.) | 0.055 | 1 (Ref.) | 0.037 |
| High | 2.42 (0.98–5.95) | 2.65 (1.06–6.66) | ||
| T classification | ||||
| T3 | 1 (Ref.) | 0.06 | 1 (Ref.) | 0.012 |
| T4 | 2.67 (0.96–7.41) | 4.08 (1.36–12.31) | ||
| Tumor Location (n=139) | ||||
| Distal | 1 (Ref.) | 0.118 | 1 (Ref.) | 0.044 |
| Proximal | 2.26 (0.81–6.27) | 3.06 (1.03–9.08) | ||
| Adjuvant chemotherapyb | ||||
| Did not received | 1 (Ref.) | 0.879 | ||
| Received | 0.91 (0.26–3.12) | |||
| Age | ||||
| < 65 | 1 (Ref.) | 0.16 | ||
| 65 and > 65 | 2.87 (0.66–12.46) | |||
| Sex | ||||
| Male | 1 (Ref.) | 0.648 | ||
| Female | 1.23 (0.50–3.04) | |||
Abbreviations: HR, hazard ratio; CI, confidence interval;
Multivariable model was selected based on stepwise removal removal of variables with a significance threshold for removal set at p<0.1.
Chemotherapy regimens were primarily 5-fluorouracil based regimens.
Expression levels of miR-21 and therapeutic outcomes
Biomarkers that can predict therapeutic outcomes may provide tools to allow physicians to better stratify patients to more effective treatments. We analyzed association with miR-21 expression and therapeutic outcomes in stage II/III CC patients from the Japanese cohort (n = 87) and that in stage II CC patients from the German cohort (n = 145). Information on the administration of adjuvant chemotherapy was available for all patients in both the Japanese cohort and the German cohorts. Chemotherapy regimens were primarily 5-fluorouracil (5-FU)-based regimens. In the Japanese cohort, receipt of adjuvant chemotherapy (n = 58) was beneficial for patients with stage II and stage III CC (P = 0.031, Log-rank test), whereas receipt of adjuvant chemotherapy (n = 31) was not beneficial for patients with stage II CC in the German cohort (P = 0.879, Log-rank test). We then analyzed individuals who received adjuvant chemotherapy. In the Japanese cohort, high miR-21 expression was significantly associated with poor therapeutic outcomes in patients with stage II and stage III CC (P < 0.0001, Log-rank test, Fig. 1B). In the German cohort, there were only 3 cancer-related deaths in patients that received chemotherapy, therefore there was not sufficient statistical power to address if miR-21 expression predicted therapeutic outcome in this cohort.
Efficacy of adjuvant chemotherapy in low- and high-miR-21 expressing patients
We have demonstrated that measuring miR-21 expression from FFPE samples has the potential to identify patients at high risk of recurrence. Furthermore, our current and previous data 15 have demonstrated that high miR-21 expression predicts worse overall rates of survival for patients who received adjuvant chemotherapy. These results raise the question whether adjuvant chemotherapy is beneficial for patients with miR-21-high CC. Therefore, we examined whether miR-21 expression can identify patients for whom adjuvant chemotherapy is beneficial in both the Japanese (stage II and stage III) and German (stage II) cohorts. As expected, in patients with high miR-21 expression levels, receipt of adjuvant chemotherapy was not beneficial (P = 0.741 for the Japanese cohort, Fig. 1B and P = 0.713 for the German cohort, Fig. 1D, respectively, Log-rank test). In contrast, in patients with miR-21-low CC, receipt of adjuvant chemotherapy was beneficial for the Japanese (P = 0.001, Log-rank test, Fig. 1B) and in the German cohort (P = 0.040, Log-rank test, Fig. 1D). These results demonstrate the potential for miR-21 expression to be used to identify patients who can benefit from adjuvant chemotherapy.
Microsatellite Instability and miR-21 expression
Microsatellite instability (MSI) describes a subgroup for colon tumors that are defective in DNA mismatch repair. MSI tumors have been regarded has having generally a better overall prognosis compared to microsatellite stable (MSS) tumors.33 This is largely due to the fact that MSI tumors have fewer metastases. MSI tumors, while having a more favorable survival outcome in general, are also more resistant to 5FU based chemotherapies. Due to the fact that MSI status can affect both prognosis and therapeutic outcomes, we evaluated if MSI status would confound the associations with miR-21 with prognosis and therapeutic outcomes. MSI analysis was carried out for 66 Japanese and 130 German tumors. Patients with MSI tumors had a slightly more favorable survival outcome in the Japanese cohort (P = 0.208, Supplementary Figure S5) but a worse prognosis in the German cohort (P = 0.027, Supplementary Figure S5). No difference in miR-21 expression was found between MSI and MSS tumors (Supplementary figure S3). While there is limited power to address the interaction of MSI status and miR-21 expression, we did not find evidence that MSI status confounds the association of miR-21 and prognosis (Supplementary Figure S5, Supplementary Table S2).
Discussion
Several lines of evidence have suggested that microRNAs have utility as both biomarkers and therapeutic targets for cancer. MicroRNAs have been found to be altered in most every malignancy examined 12 and different microRNAs have been shown to be oncogenic or tumor suppressors depending on cellular context 34–36. We have previously shown that increased expression of miR-21 in CC tissues was associated with worse cancer-specific mortality in two independent cohorts using RNA isolated from frozen specimens 15. Because biomarkers developed from FFPE samples can be more readily translated into clinical application, we measured microRNA expression in FFPE samples and examined their utility as a prognostic classifier. In the present study, we prepared RNA from FFPE CC tissue with minimal hematoxylin contamination from two independent cohorts. Univariate and multivariate analyses revealed that high miR-21 expression in CC is a prognostic classifier in both the Japanese and German cohorts. We previously reported that patients with tumors expressing high levels of miR-21 have a worse prognosis for stage II or stage III colon cancer in a cohort from Maryland, USA (the University of Maryland Medical Center) and a cohort from Hong Kong (Queen Mary Hospital in Hong Kong) 15. Additional reports have also found a link between high miR-21 expression and poor prognosis in Japanese37, Czech20 and Danish21, 22 cohorts of CC patients. Taken together, these results indicate that measurement of miR-21 expression has potential as a clinically useful biomarker for CC. Because these cohorts are from different geographical regions of the world, the findings should be representative of the majority of CC cases.
CC patients would benefit from prognostic markers that can identify those individuals that are more likely to recur by selecting patients that are suitable for adjuvant therapy. In the present study, we demonstrated that miR-21 expression was associated with the prognosis of patients from Japan and Germany. This indicates that measuring miR-21 expression in FFPE samples may help identify patients with a high risk of disease recurrence. However, 5-FU-based adjuvant chemotherapy was not advantageous for patients with miR-21-high CC, consistent with our previous study15. Therefore, patients with high miR-21 expression are at high-risk for disease recurrence, but such patients may not benefit from 5-FU-based adjuvant chemotherapy alone. Therefore, alternative therapeutic strategies, including combination therapies with 5-FU or other single agent therapies, may be more effective than 5-FU alone. In contrast, patients with low miR-21 expression should respond well to 5-FU based adjuvant chemotherapy this treatment strategy is likely to be beneficial for such patients.
High levels of miR-21 are may be partly responsible for the poor response to 5-FU. Increased levels of miR-21 leads to increased cell proliferation and decreased apoptosis both in vitro and in animal models38. Additionally, increased miR-21 expression reduces apoptosis and G2/M arrest due to damage by 5-FU in colon cancer cell lines39. Taken together, this indicates that while high levels of miR-21 predict poor response to 5-FU therapy, reducing miR-21 therapeutically could sensitize patients to enable greater effectiveness of 5-FU therapy. Recent progress has been made inhibiting specific microRNAs and anti-miRNA based therapies are already being tested in humans to treat chronic hepatitis C infection 40. If progress continues similar strategies may be found to use anti-miR-21 based therapies to treat colon cancer. For example, the combination of a miR-21 inhibitor with 5-FU based therapies may be more effective than 5-FU alone.
It is important to assess the quality of RNA isolated from FFPE tissue before measuring the expression of either microRNAs or mRNAs. In the present study, we found that microRNA expression levels determined by qRT-PCR from samples with hematoxylin contamination do not accurately reflect those without such contamination. We reviewed several manuscripts that described the use of qRT-PCR or microarray analysis of microRNA from FFPE tissues 41–49; however, descriptions of RNA purity, such as the OD 260/230 ratio, could not be found in the manuscripts. Further investigation is required to improve RNA purity from FFPE samples. For the Japanese cohort, all RNA samples were extracted from FFPE sections stained by hematoxylin. While we removed most of the hematoxylin from the RNA preparations of the Japanese cohort, we cannot exclude the possibility that the measurements of miR-21 expression levels were slightly affected by trace hematoxylin contamination. For future qRT-PCR analysis, it may be better to stain FFPE sections with regents that are dissolved in alcohol, such as toluidine blue, instead of hematoxylin.
In summary, we showed that high miR-21 expression is an independent prognostic classifier in two independent cohorts. We also demonstrated that receipt of adjuvant chemotherapy is beneficial for patients with miR-21-low CC. Therefore, measurement of miR-21 may help identify high-risk patients and also patients who benefit from adjuvant chemotherapy. In the present study, expression of miR-21 was measured from FFPE samples. Therefore, measurement of miR-21 can be readily translated into clinical applications. However, miR-21 expression was simply dichotomized as either high or low in the present study. For clinical application, determination of expression levels of miR-21 with an absolute quantification method is needed.
Supplementary Material
Novelty & Impact Statements.
We find that high miR-21 expression is associated with poor survival in two independent cohorts of colon cancer patients from Japan and Germany. For the first time, we find that miR-21 expression from FFPE tissues is associated with poor therapeutic outcome for colon cancer. These results suggest that miR-21 is a promising prognostic and predictive biomarker for colon cancer and that FFPE tissue may be suitable starting material for this biomarker.
Acknowledgments
Financial support: This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, a Department of Defense Congressionally Directed Medical Research Program Grant PR093793, a Grant-in-Aid from the Ministry of Health, Labour and Welfare for the 3rd-term Comprehensive 10-year Strategy for Cancer Control, Japan, supported by the grant “Clinical Trial Centers, funding number FKN 01KN1103, IZKS Mainz” of German Federal Ministry of Education and Research
The authors are grateful to all patients who participated in the trial. The authors would like to thank Arzu Budak, Bjoern Sacher, Tuerkan Coskun and Stephan Biesterfeld for additional data management of the German cohort. Some of this data are parts of their MD theses.
Footnotes
Conflicts of interest: None
Reference List
- 1.Moertel CG, Fleming TR, Macdonald JS, et al. Levamisole and fluorouracil for adjuvant therapy of resected colon carcinoma. N Engl J Med. 1990;322:352–8. doi: 10.1056/NEJM199002083220602. [DOI] [PubMed] [Google Scholar]
- 2.Benson AB, 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–19. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 3.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–99. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther A, Johnstone E, Swanton C, et al. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–99. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 5.Barrier A, Boelle PY, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–91. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 6.Schetter AJ, Nguyen GH, Bowman ED, et al. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–87. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Specht K, Richter T, Muller U, et al. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–29. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Saito M, Schetter AJ, Mollerup S, et al. The Association of MicroRNA Expression with Prognosis and Progression in Early-Stage, Non-Small Cell Lung Adenocarcinoma: A Retrospective Analysis of Three Cohorts. Clin Cancer Res. 2011;17:1875–82. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathe EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Schetter AJ, Yang GB, et al. microRNA and inflammatory gene expression as prognostic marker for overall survival in esophageal squamous cell carcinoma. Int J Cancer. 2012 doi: 10.1002/ijc.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao W, Yu Y, Cao H, et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64:399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154–60. doi: 10.1016/j.cancergencyto.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen BS, Jorgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjaer-Frifeldt S, Hansen TF, Nielsen BS, et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107:1169–74. doi: 10.1038/bjc.2012.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giovannetti E, Funel N, Peters GJ, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–38. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 24.Hwang JH, Voortman J, Giovannetti E, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radojicic J, Zaravinos A, Vrekoussis T, et al. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507–17. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 27.Avissar M, McClean MD, Kelsey KT, et al. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;30:2059–63. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Huang H, Sun L, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 29.Zhi F, Chen X, Wang S, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer. 2010;46:1640–9. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Rossi S, Shimizu M, Barbarotto E, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–52. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss WM. Preparation of genomic DNA from mammalian tissue. Curr Protoc Mol Biol. 2001;Chapter 2(Unit 2) doi: 10.1002/0471142727.mb0202s42. [DOI] [PubMed] [Google Scholar]
- 32.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 33.Laghi L, Malesci A. Microsatellite instability and therapeutic consequences in colorectal cancer. Dig Dis. 2012;30:304–9. doi: 10.1159/000337003. [DOI] [PubMed] [Google Scholar]
- 34.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schimanski CC, Frerichs K, Rahman F, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–96. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibuya H, Iinuma H, Shimada R, et al. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–20. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 38.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Smyth P, Flavin R, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–74. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doleshal M, Magotra AA, Choudhury B, et al. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10:203–11. doi: 10.2353/jmoldx.2008.070153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Chen J, Radcliffe T, et al. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10:513–9. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 46.Liu A, Tetzlaff MT, Vanbelle P, et al. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2:519–27. [PMC free article] [PubMed] [Google Scholar]
- 47.Siebolts U, Varnholt H, Drebber U, et al. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62:84–8. doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreasen D, Fog JU, Biggs W, et al. Improved microRNA quantification in total RNA from clinical samples. Methods. 2010;50:S6–9. doi: 10.1016/j.ymeth.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Goswami RS, Waldron L, Machado J, et al. Optimization and analysis of a quantitative real-time PCR-based technique to determine microRNA expression in formalin-fixed paraffin-embedded samples. BMC Biotechnol. 2010;10:47. doi: 10.1186/1472-6750-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.