Abstract
Integral membrane proteins play central roles in controlling the flow of information and molecules across membranes. Our understanding of membrane protein structure and function, however, is seriously limited, mainly due to difficulties in handling and analyzing these proteins in aqueous solution. The use of a detergent or other amphipathic agents is required to overcome the intrinsic incompatibility between the large lipophilic surfaces displayed by membrane proteins in their native forms and polar solvent molecules. Here we introduce new tripod amphiphiles displaying favourable behaviours toward several membrane protein systems, leading to enhanced protein solubilization and stabililization compared to both conventional detergents and previously-described tripod amphiphiles.
Keywords: amphiphiles, membrane proteins, stabilization, molecular design, protein structure
Introduction
Integral membrane proteins (IMPs) perform a wide range of functions including the transfer of biologically relevant molecules and information across the lipid bilayer of cells and organelles. Membrane proteins constitute about a third of open reading frames in the human genome and are the targets of more than half of current pharmaceuticals.[1] Detailed structural and functional information for IMPs is essential to provide a fundamental understanding of their mechanism of action as well as to facilitate rational design of new drug molecules. Despite extensive efforts, however, our understanding of IMP structure and function lags far behind that of soluble proteins. Only a few hundred IMP structures are known, corresponding to less than 1.0 % of the total number of membrane proteins. This low number is attributed to difficulties associated with handling these proteins.[2] A key requirement of isolation and structural studies of IMPs is that they must be maintained in solution by amphipathic additives which shield their large hydrophobic surface area from the aqueous environment. Conventional detergents such as dodecyl-β-D-maltoside (DDM), lauryldimethylamine-N-oxide (LDAO) and n-octyl-β-D-glucopyranoside (OG) are widely used to both extract IMPs from the lipid bilayer and to maintain the native state in solution.[3] However, many membrane proteins solubilized with even these popular reagents have a tendency to denature and/or aggregate, [4] excluding them from further study.
Various classes of novel amphiphiles have been developed to tackle this challenging problem.[5] Examples include amphipols,[5a,b] lipopeptide detergents (LPDs),[5c] hemifluorinated surfactants (HFSs),[5d,e] nanodiscs (NDs),[5f] short peptide designers,[5g] cholate-based facial amphiphiles[5h,i] and cholesterol-derived agents (chobimalt).[5j,k] The published studies using these agents tend to focus on membrane protein stabilization. In contrast, we have focused on the development of novel agents with favourable membrane protein solubilization properties in addition to stabilization, exemplified by tripod amphiphiles (TPAs),[5l–n] maltose-neopentyl glycol (MNG)[5o,p] and glucose-neopentyl glycol (GNG) amphiphiles[5q]. It is noteworthy that members of the NG class have facilitated the crystal structure determinations of more than 10 new membrane proteins including several independent G-protein-coupled receptors (GPCRs).[6]
Our previous research has shown the potential of tripod amphiphiles to both solubilize and stabilize IMPs.[5l–n] A TPA with an N-oxide headgroup (commercial name is TRIPAO) efficiently extracted bacteriorhodopsin (bR) and rho protein from membranes (Figure S1). TPA-solubilized bR and the potassium channel from Streptomyces lividans have been crystallized, although their structures have not yet been solved.[5n] In a recent study, glycotripod amphiphiles (TPA-2 and TPA-2-S; commercially available) displayed favourable behaviours for the solubilization and stabilization of the photosynthetic superassembly comprised of the light harvesting I (LHI) and reaction center (RC) complex from Rhodobacter capsulatus (Scheme 1).[5m] Such generally favourable behaviours of these molecules have prompted us to expand the set of tripod amphiphiles by introducing structural variation into the previously-described agents. Here we prepared two new tripod amphiphiles and evaluated their properties with several membrane protein systems including a G-protein coupled receptor (GPCR). We found that the new agents displayed superior properties in the solubilization and stabilization of membrane proteins compared to conventional detergents and previously-described TPAs.
Scheme 1.
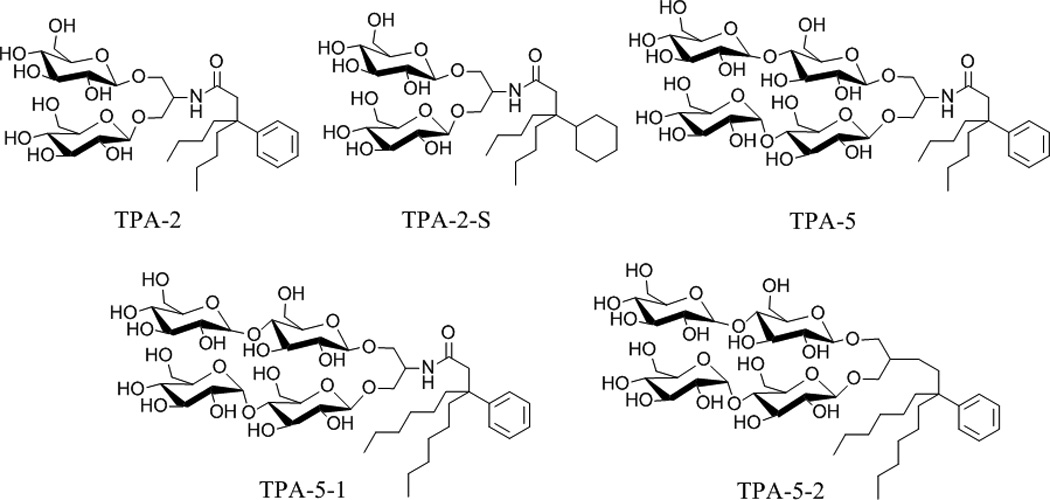
Chemical structures of previously-reported (TPA-2, TPA-2-S and TPA-5) and new tripod amphiphiles (TPA-5-1 and TPA-5-2).
Results
The chemical structures of previously reported TPAs (TPA-2, TPA-2-S and TPA-5) and newly-synthesized tripod amphiphiles (TPA-5-1 and TPA-5-2) are illustrated in Scheme 1. The new amphiphiles share their hydrophilic head group with TPA-5 but have a variable hydrophobic group; the alkyl chain of TPA-5 was extended from a butyl to a hexyl chain to give TPA-5-1 while further modification was introduced by directly attaching the hydrophilic group to the hydrophobic group without an amide linkage to produce TPA-5-2. These molecules were easily prepared on a multi-gram scale that would support biochemical research (see supporting information for details). All new TPAs were highly water-soluble (up to ~20 wt %). Critical micelle concentration (CMC) values were estimated by solubilization experiments employing the hydrophobic fluorescent dye, diphenylhexatriene,[7] and the hydrodynamic radii (Rh) of the micelles were estimated through dynamic light scattering (DLS) measurements. The data for tripod amphiphiles (TPA-2, TPA-2-S, TPA-5, TPA-5-1 and TPA-5-2) and two conventional detergents (DDM and OG[8]) are presented in Table 1. The CMC values of TPA-5-1 and TPA-5-2 are ~0.30 mM (0.031 wt %) and ~0.021 mM (0.002 wt %), respectively. These values are much smaller than that of TPA-5 (~8.0 mM; 0.78 wt %) and two branched diglucoside-bearing TPAs, TPA-2 and TPA-2-S (3.6 mM, 0.24 wt %; 1.8 mM, 0.12 wt %, respectively). This is likely due to increased hydrophobicity induced by the alkyl chain extension and removal of the polar amide functional group. The small CMC values of the new agents indicate their strong aggregation tendency, which may imply the formation of protein-detergent complexes (PDCs) with enhanced stability. All TPAs except TPA-5 form smaller micelles than DDM and form a single micelle size population as assessed by DLS (see Figure S2 for the two sets of micelle distribution of TPA-5).
Table 1.
Critical micelle concentrations (CMCs) and hydrodynamic radii (Rh) of the micelles (mean ± SD, n = 4) for tripod amphiphiles (TPA-2, TPA-2-S, TPA-5, TPA-5-1 and TPA-5-2) and conventional detergents (DDM and OG).
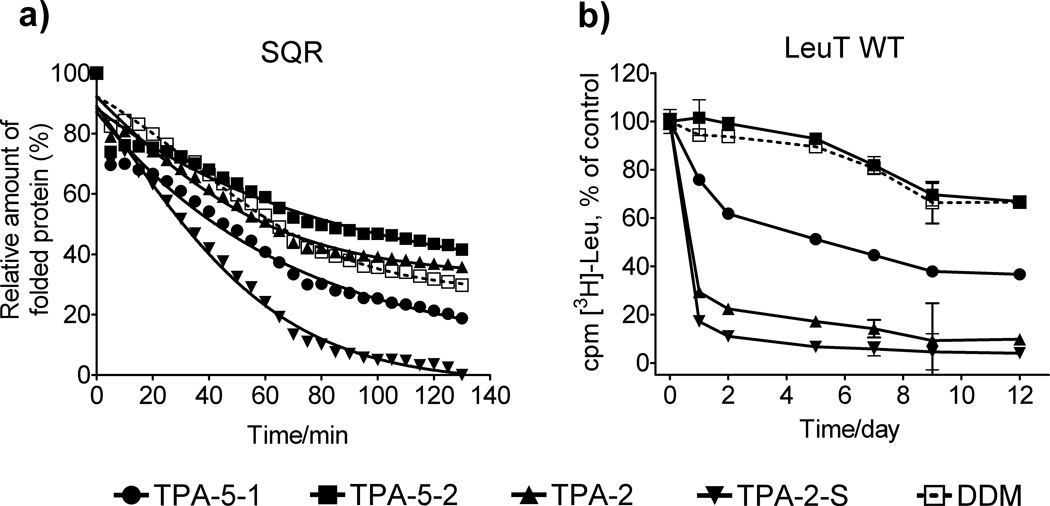
We first evaluated the newly-prepared agents (TPA-5-1 and TPA-5-2) and compared with previously-reported ones (TPA-2 and TPA-2-S) for the stability of succinate:quinone oxidoreductase (SQR) expressed in Escherichia coli; [9] a previous study showed that TPA-5 was too poor to solubilize R. capsulatus superassembly from the membrane and thus this amphiphile was not included in this experiment. The structural stability of SQR was assessed with a reactive probe, N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM).[10] In this assay, the probe molecules react with thiol groups that become solvent-accessible upon protein unfolding, leading to fluorescence emission. The assay can therefore be used to estimate the relative amount of protein unfolding by monitoring fluorescence change of assay solutions. SQR with four subunits (SdhA, SdhB, SdhC, and SdhD) was initially extracted from the native membrane with DDM. The pure protein was diluted to generate solutions containing CMC + 0.04 wt % amphipathic agents. Protein integrity was monitored at elevated temperature (30 °C) for 130 min. Under these conditions, a previously-reported tripod amphiphile, TPA-2, was comparable to DDM at maintaining the folding structure of SQR while TPA-2-S and TPA-5-1 were worse than DDM (Figure 1a). Notably, TPA-5-2 was better than DDM. When we increased detergent concentration to CMC + 0.2 wt %, all TPAs except TPA-2-S were comparable to DDM (Figure S3a). Similar results were observed for another membrane protein, the rhomboid intramembrane serine protease GlpG (Figure S3b).[11]
Figure 1.
(a) CPM assays for SQR and (b) time course activity of LeuT solubilized with tripod amphiphiles (TPA-2, TPA-2-S, TPA-5-1 and TPA-5-2) or DDM at CMC + 0.04 wt % determined by scintillation proximity assay (SPA) based on [3H]-Leu binding. The CPM assay was performed at 30 °C while the SPA was conducted on samples stored at room temperature. Results are expressed as % relative folded protein at time 0 or % activity (mean ± SEM, n = 2) relative to the day 0 measurements for the CPM assay and SPA, respectively.
We next turned to bacterial leucine transporter (LeuT) from Aquifex aeolicus.[12] Protein activity was estimated by assessment of radiolabeled leucine binding using scintillation proximity assay (SPA).[13] The transporter was initially extracted with 1.0 wt % DDM and then diluted in solutions containing individual amphiphiles (DDM, TPA-5-1, TPA-5-2, TPA-2 and TPA-2-S). Fig 1b shows that at lower amphiphile concentration (i.e. CMC + 0.04 wt %) TPA-2 and TPA-2-S-solubilized LeuT showed rapid activity loss, similarly to our previous observations for LDAO and OG-solubilized samples.[5o] TPA-5-1, the amphiphile with amide linkage to the hydrophobic group, was more efficient than glucose-bearing TPA agents (TPA-2 and TPA-2-S), yet inferior to DDM, in maintaining the transporter activity during the 12-day experimental period. In contrast, TPA-5-2, the amphiphile with a direct connection to the hydrophobic group displayed favourable stabilization effect comparable to DDM. When we increased detergent concentration to CMC + 0.2 wt %, similar results were observed (Figure S4a). For a continuing evaluation of the TPAs with bacteriorhodopsin (bR), the protein was initially solubilized from the native purple membrane with 2.0 wt % octyl-β-D-thioglucoside (OTG), the most commonly used detergent for manipulation of this protein.[14] Following ultracentrifugation to remove insoluble debris, the bR solution was diluted with amphiphile-containing solutions to give the final concentration of 0.2 wt % OTG + 0.8 wt % TPA; TPA-2-S was not included in this experiment because the agent consistently showed the worst results among the current TPA set for the stability of the proteins investigated. The stability of bR was assessed by spectrophotometry measuring the strong 554 nm absorbance of the native structure. When the absorbance was monitored over a period of 20 days, two of the new agents, TPA-5-1 and TPA-5-2, were observed to be more effective than OTG and TPA-2 at maintaining the native structure of bR, with TPA-5-2 being the best agent (Figure S4b).
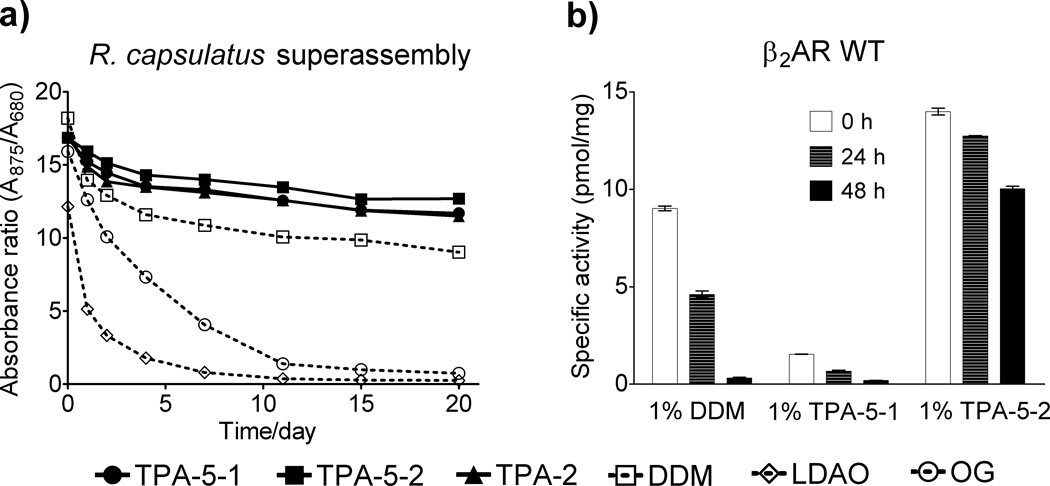
We then turned to a photosynthetic superassembly, composed of the light harvesting complex I (LHI) and the reaction center (RC), from R. capsulatus.[15] This LHI-RC superassembly contains a large number of protein subunits and multiple cofactors such as bacteriochlorophyll and carotenoids, thereby giving a highly characteristic UV-Vis absorption spectrum. Thus, the protein integrity can be unambiguously assessed by spectrophotometry. We monitored the absorbance ratio (A875/A680) of protein solutions to investigate the integrity of LHI-RC complexes (the absorbances at 875 nm and 680 nm arise from cofactors embedded in the native conformation of the complexes and the oxidation of bacteriochloropyll freed from LHI upon denaturation, respectively). The superassembly was extracted from the native membrane with 1.0 wt % DDM and purified with the same detergent at the CMC (0.009 wt %) via Ni2+-NTA affinity chromatography, utilizing a seven-membered histidine tag on the C-terminus of the M-subunit of the RC. This protein was diluted with amphiphile-containing solutions, so that the final concentration of each agent is CMC + 0.04 wt %, with residual DDM far below its CMC (0.0004 wt %). As expected from a previous study, the LHI-RC complexes in the two conventional detergents (OG and LDAO) was rapidly destroyed. In contrast, TPA-2, TPA-5-1 and TPA-5-2-solubilized complexes showed enhanced stability over 20 days as compared to DDM-solubilized sample with the best results achieved using TPA-5-2 (Figure 2a). Similar results were obtained in increasing detergent concentrations (Figure S5).
Figure 2.
(a) The stability of R. capsulatus superassembly and (b) the activity of β2AR WT in TPAs and conventional detergents as a function of time. The superassembly was solubilized in individual detergents at CMC + 0.04 wt % and stability was assessed by measuring the absorbance ratio (A875/A680) over 20 days. β2AR WT was extracted with 1.0 wt % DDM, TPA-5-1 or TPA-5-2, and its activity was measured over time by radioligand binding assay using antagonist [3H] dihydroalprenolol during the storage of the receptor samples at 4 °C.
In the examples discussed so far, we used conventional detergents (DDM or OTG) to solubilize and isolate individual IMPs from the membranes. The purified proteins were then diluted in amphiphile-containing solutions for evaluation of the novel agents. Within this protocol, there is a possibility that the small residual conventional detergent could influence protein stability. To avoid this possibility, the newly-synthesized TPAs (TPA-5-1 and TPA-5-2) and DDM were used to extract wild-type β2 adrenergic receptor (β2AR WT[16], a representative of the human G-protein coupled receptor (GPCR) family), from the membrane and receptor stability was monitored over 48 h at 4 °C. The agents were selected based on a preliminary experiment with β2AR-T4L[17] showing that these TPAs resulted in similar Tm values for the purified receptor to that of DDM. A radioligand binding assay employing the antagonist [3H]-dihydroalprenolol was utilized to measure receptor activity. The DDM-solubilized receptor showed high initial activity but rapidly lost almost all activity (Figure 2b). TPA-5-1-solubilized receptor showed low initial activity as well as rapid reduction in activity. In contrast, the receptor solubilized in TPA-5-2 displayed almost a third higher initial activity compared to DDM and maintained two-thirds of the maximal activity after 48 h.
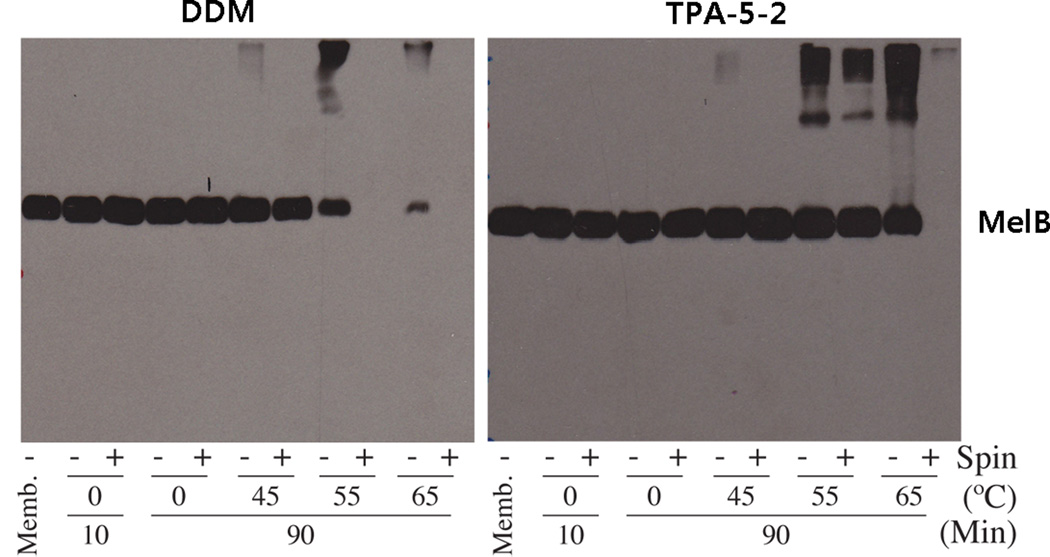
Since TPA-5-2 consistently displayed the best behaviours for several membrane protein systems, we further characterized this agent using a fusion protein of the human transporter CMP-Sia with a C-terminal GFP expressed in Saccharomyces cerevisiae. The protein solubilized by 1.0 wt % DDM, TPA-5-1, or TPA-5-2 were analyzed by fluorescent size exclusion chromatography (FSEC). TPA-5-1 and TPA-5-2 gave solubilization efficiency comparable to that displayed by DDM. In addition the TPA-solubilized protein yielded a single monodispersed peak almost identical to that obtained using DDM (Figure S6). The favourable property of TPA-5-2 at extracting β2AR WT and CMP-Sia fusion protein from the membranes led us to examine this agent with Salmonella typhimurium melibiose permease (MelB),[18] expressed in E. coli. For protein extraction, 1.5 wt % TPA-5-2 and DDM were used at 0°C for 10 min, and then aggregated material was removed via ultracentrifugation. The amount of solubilized MelB was determined after ultracentrifugation via Western blot (Figure 3). Both TPA-5-2 and DDM efficiently extracted MelB under these conditions, indicating comparably good solubilizing properties of these agents. To investigate detergent efficacy on the MelB thermostability, we elevated the temperature of solubilizing solution to 45°C, 55°C and 65°C with an increased incubation time of 90 min. DDM yields detectable MelB at 45°C, but soluble protein could not be detected at 55°C after ultracentrifugation; we assume that DDM-solubilized proteins denatured and/or aggregated at this temperature. In contrast, TPA-5-2-solubilized MelB retained its soluble form even at 55°C, further demonstrating the superior properties of the agent to conventional detergents for membrane protein stabilization. Taken together, these data indicate that the TPA-5-2 is useful for both efficient solubilization and enhanced stabilization of IMPs.
Figure 3.
Western blot analysis of MelB. Samples (10 µg) were separated by SDS-12% PAGE, and MelB was detected using an anti-histidine tag antibody. The solubilization extracts in TPA-5-2 and DDM were divided into two samples; one after ultracentrifugation (+) and one prior to ultracentrifugation (−). As a control, an untreated membrane sample (“Memb”; no ultracentrifugation) was included in each gel.
Discussion
Detergents with an appropriate hydrophile-lipophile balance (HLB) are known to be useful for membrane protein manipulation.[19] Conceivably, the strength of the interaction between the detergent molecules and the hydrophobic surfaces of membrane proteins could be optimized to effectively stabilize encapsulated protein structures. Specifically, a relatively hydrophilic detergent would weakly bind to membrane proteins, leading to protein aggregation, while a rather hydrophobic detergent will interact too strongly with proteins, resulting in destruction of forces stabilizing the protein structure. In a previous study, we found that the TPA architecture with branched head and tail groups has an advantage over conventional detergent architectures, i.e., linear structures.[5] However, TPA-5 was previously shown to be unsuccessful at solubilizing R. capsulatus superassembly from the native membrane.[5m] We reasoned that the poor behaviour of this agent could be attributed to an imbalance between the hydrophobic and hydrophilic domains. For example, TPA-5 has a large hydrophilic group and a comparatively small hydrophobic group. Therefore, we increased the length of the alkyl chain and, in addition, removed the polar amide group to give TPA-5-1 and TPA-5-2, respectively. Thus, these new TPA agents were rationally designed to attain an optimum HLB, without disruption of the favourable architectural features. The current results show that TPA-5-2, with the largest hydrophobic region was the best, supporting this design principle.
Detergents with a good track record for membrane protein crystallization such as OG and DDM appear to have some common attributes: good solubilization and stabilization efficacy. Similar properties have been demonstrated for novel classes of amphiphiles including the TPA, GNG and MNG amphiphiles; all of these classes have produced high quality membrane protein crystals.[5n,6] Novel agents with good stabilization efficacy but unfavourable solubilization efficiency, on the other hand, have failed in providing new crystal structures of membrane proteins. On the basis of this analysis, we reached a conclusion that detergent solubilization efficiency is a critical factor, in addition to protein stabilization efficacy, for the successful use of a specific detergent or amphiphile for membrane protein crystallization. In the case of the tripod amphiphiles the dual solubilization and stabilization character could be attributed to the fact that the three hydrophobic groups of the amphiphiles are available to interact with the hydrophobic surface of membrane proteins. This multiple-point interaction is likely to result in high affinity of these agents for membrane proteins, and accordingly enhanced solubilization efficiency from the membrane. Because of its favourable solubilization and stabilization characteristics, TPA-5-2 holds significant potential for the crystallization of membrane proteins.
Detergent properties can be significantly improved by introducing small variations in their hydrophilic and/or hydrophobic regions. For instance, glucose-containing conventional detergents display significantly different behaviour from maltose-containing detergents. The widely used detergents OG and DDM are structurally similar, but it is well-known that these agents display quite different properties in terms of solubilization, stabilization and crystallization of membrane proteins. A similar trend was observed for the GNG and MNG cases.[5q] GNG amphiphiles tend to form smaller PDCs but are less stabilizing, while MNG amphiphiles were previously shown to be more stabilizing but tend to form large PDCs. Another instance can be seen in our very recent report on carbohydrate analogues of Triton X-100, designated as CGTs; by the replacement of oxyethylene glycol units of Triton X-100 with glucose or maltose, the stabilization propensity was significantly enhanced (Figure S7).[20] In addition to the effect of hydrophilic group variation on detergent properties discussed above, hydrophobic variation may also result in a big change in detergent properties. Examples include the HF-MNGs (vs. MNGs; Figure S8).[5e] The current study provides further supportive data; the small structural variation in the TPA architecture, exemplified by the small differences among TPA-5, TPA-5-1 and TPA-5-2, leads to significant differences in detergent behaviour toward diverse membrane proteins.
There are a large number of membrane proteins yet to be structurally characterized. This is particularly true of many classes of eukaryotic IMPs, which are much less stable in solution than are the prokaryotic homologues. Thus, it is necessary to develop a number of alternative tools, including the TPAs and MNGs, for membrane protein studies. In addition, it is essential to assess these tools using multiple membrane protein systems as described here in order to establish utility across membrane protein groups.
Conclusion
We have conveniently prepared new tripod amphiphiles by taking advantage of a modular synthetic approach. These new agents, especially TPA-5-2, confer favourable solubilization efficiency and enhanced stabilization efficacy for several membrane protein systems, relative to previously reported examples. Additional questions should be addressed to maximize the utility of these new agents. For instance, the physicochemical properties of the amphiphiles, such as aggregation number and aqueous surface tension, need to be determined. A concerted effort also needs to be made to evaluate TPA-5-2 for membrane protein crystallization. However, the current study clearly shows that TPA-5-2 has significant potential for membrane protein manipulation. More importantly, it is significant that this promising TPA was obtained via rational design based on the general principle that the Hydrophile-Lipophile Balance plays a critical role in membrane protein solubilization and stabilization properties. This design principle should be useful for future development of novel amphiphiles for biochemical applications.
Experimental Section
Synthesis and characterization of amphiphiles, and membrane protein solubilisation: Details may be found in the Supporting Information
Supplementary Material
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIP) (grant number 2008-0061891 and 2012R1A1A1040964 to P.S.C., K.H.C., H.E.B.), NIH grant P01 GM75913 (S.H.G.), NS28471 (B.K.), the European Community’s Seventh Framework Programme FP7/2007–2013 under grant agreement number HEALTH-F4–2007–201924, EDICT Consortium (B.B., K.G., U.G.), the Danish National Research Council (C.J.L., U.G.), the Lundbeck Foundation (S.G.F.R., C.J.L., U.G.), and NIH grants R01 GM95538 and R21 HL087895 (to L.G.). R.R. was funded by the Defence, Science and Technology Laboratories (DSTL), Porton Down, UK. We thank Philip Laible (Argonne National Laboratory) and, Elodie Point and Jean-Luc Popot (Université Paris-7) for supplying membrane preparations from R. capsulatus and purple membrane, respectively. We also thank Chiara Lee and David Drew (Imperial College London) and, Jonathan Ruprecht (Medical Research Council-Mitochondrial Biology Unit, Cambridge) and Gary Cecchini (University of California, San Francisco) for providing purified GlpG and SQR samples, respectively. We also thank Shalika Nurva for her technical assistance in MelB assay.
Contributor Information
Prof. Pil Seok Chae, Email: pchae@hanyang.ac.kr.
Prof. Brian K. Kobilka, Email: kobilka@stanford.edu.
Prof. Claus J. Loland, Email: cllo@sund.ku.dk.
Dr. Bernadette Byrne, Email: b.byrne@imperial.ac.uk.
Prof. Samuel H. Gellman, Email: gellman@wisc.edu.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.a) White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; b) Moller JV, le Maire J. J. Biol. Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 4.a) Garavito RM, Ferguson-Miller S. J. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; b) Bowie JU. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; c) Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc. Natl. Acad. Sci. U. S. A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Tribet C, Audebert R, Popot J-L. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Popot J-L, Althoff T, Bagnard D, Banères J-L, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Crémel G, Dahmane T, dela Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH, Kühlbrandt W, Le Bon C, Martinez KL, Picard M, Pucci B, Rappaport F, Sachs JN, Tribet C, van Heijenoort C, Wien F, Zito F, Zoonens M. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]; c) McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat. Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; d) Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot J-L. FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]; e) Cho KH, Byrne B, Chae PS. ChemBioChem. 2013;14:452–455. doi: 10.1002/cbic.201200759. [DOI] [PubMed] [Google Scholar]; f) Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; g) Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Zhang Q, Ma X, Ward A, Hong W-X, Jaakola V-P, Stevens RC, Finn MG, Chang G. Angew. Chem. Int. Ed. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]; i) Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Howell SC, Mittal R, Huang L, Travis B, Breyer RM, Sanders CR. Biochemistry. 2010;49:9572–9583. doi: 10.1021/bi101334j. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Kruse AC, Nurva S, Gether U, Guan L, Loland CJ, Byrne B, Kobilka BK, Gellman SH. Chem.-Eur. J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]; l) McQuade, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew. Chem. Int. Ed. 2000;39:758–761. [PubMed] [Google Scholar]; m) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; n) Chae PS, Laible PD, Gellman SH. Mol. BioSyst. 2010;6:86–94. doi: 10.1039/b915162c. [DOI] [PMC free article] [PubMed] [Google Scholar]; o) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot J-L, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; p) Selao TT, Branca R, Chae PS, Lehtiö J, Gellman SH, Rasmussen SGF, Nordlund S, Norén A. J. Proteome Res. 2011;10:2703–2714. doi: 10.1021/pr100838x. [DOI] [PMC free article] [PubMed] [Google Scholar]; q) Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Capaldi S, Carlsso E, Kobilka B, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem. Comm. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rosenbaum DM, Zhang C, Lyons J, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H-J, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Haga K, Kruse AC, Asada H, Kobayashi TY, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka Brian K. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Kellosalo J, Kajander T, Kogan K, Pokharel K, Goldman A. Science. 2012;337:473–476. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]; j) Rollauer SE, Tarry MJ, Graham JE, Jääskeläinen M, Jäger F, Johnson S, Krehenbrink M, Liu SM, Lukey MJ, Marcoux J, McDowell MA, Rodriguez F, Roversi P, Stansfeld PJ, Robinson CV, Sansom MS, Palmer T, Högbom M, Berks BC, Lea SM. Nature. 2012;492:210–214. doi: 10.1038/nature11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay A, London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 8.Lorber B, Bishop JB, Delucas L. J. Biochim. Biophys. Acta. 1990;1023:254–265. doi: 10.1016/0005-2736(90)90421-j. [DOI] [PubMed] [Google Scholar]
- 9.Horsefield R, Iwata S, Byrne B. Curr. Protein Pept. Sci. 2004;5:107–118. doi: 10.2174/1389203043486847. [DOI] [PubMed] [Google Scholar]
- 10.Alexandrov A, Mileni M, Chien EY, Hanson MA, Stevens RC. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Urban S. Biochem. J. 2010;425:501–521. doi: 10.1042/BJ20090861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.Quick M, Javitch JA. Proc. Natl. Acad. Sci. USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazzacco P, Sharma KS, Durand G, Giusti F, Ebel C, Popot J-L, Pucci B. Biomacromolecules. 2009;10:3317–3326. doi: 10.1021/bm900938w. [DOI] [PubMed] [Google Scholar]
- 15.Laible PD, Kirmaier C, Udawatte CS, Hofman SJ, Holten D, Hanson DK. Biochemistry. 2003;42:1718–1730. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]
- 16.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 17.Kobilka BK. Trends Pharmacol. Sci. 2011;32:213–218. doi: 10.1016/j.tips.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan L, Nurva S, Ankeshwarapu SP. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger BW, Garcia RY, Lenhoff AM, Kaler EW, Robinson CR. Biophys. J. 2005;89:452–464. doi: 10.1529/biophysj.104.051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chae PS, Wander MJ, Cho KH, Laible PD, Gellman SH. Mol. BioSyst. 2013;9:626–629. doi: 10.1039/c3mb25584k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caffrey M, Li D, Dukkipati A. Biochemistry. 2012;51:6266–6288. doi: 10.1021/bi300010w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.