Abstract
The M and S forms of A. gambiae have been the subject of intense study, but are morphologically indistinguishable and can only be identified using molecular techniques. PCR-based assays to distinguish the two forms have been designed and applied widely. However, the application of these assays towards identifying hybrids between the two forms, and backcrossed hybrids in particular, has been problematic as the currently available diagnostic assays are based on single loci, and/or are located within a multi-copy gene. Here we present an alternative genotyping method for detecting hybridization and introgression between M and S molecular forms based on a multi-locus panel of single nucleotide polymorphisms (SNPs) fixed between the M and S forms. The panel of SNPs employed are located in so called “islands of divergence” leading us to describe this method as the “Divergence Island SNP” (DIS) assay. We show this multi-locus SNP genotyping approach can robustly and accurately detect F1 hybrids as well as backcrossed individuals.
Introduction
Populations of the African malaria vector, Anopheles gambiae, are thought to be undergoing speciation and have been the focus of numerous studies aimed at evaluating models of ecological speciation (Ayala& Coluzzi 2005; Manoukis 2008; White et al. 2010). Discrete subpopulations of A. gambiae have been grouped into two morphologically indistinguishable molecular forms, defined according to fixed SNP differences located within a 2.3 kb fragment at the 5’ end of the multi-copy rDNA IGS region on the X chromosome (Favia et al. 2001).
Consistent with strong reproductive barriers between the M and S forms, field surveys indicate hybrids to be rare in most of the regions where the M and S forms occur sympatrically (della Torre et al. 2005). Moreover, the molecular forms display phenotypic divergence in different locations within their geographic range (Lehmann& Diabate 2008). Most notable differences include differential insecticide resistance (Chandre et al. 1999; Tripet et al. 2007), desiccation resistance (Lee et al. 2009), larval habitat segregation (Gimonneau et al. 2012), and wing morphological differentiation (Sanford et al. 2011). It has been proposed that mechanisms responsible for divergence between M and S include pre-zygotic reproductive isolation (Diabate et al. 2007) associated with mate selection (Diabate et al. 2009; Manoukis et al. 2009)and post-zygotic isolation in the form of reduced hybrid fitness (White et al. 2010).
Although the M and S forms are thought to be largely reproductively isolated in most places where they occur in sympatry, this is not true everywhere. Hybridization between forms were reported to occur rarely (<1%) in Mali (Tripet et al. 2001) and reproductive isolation between M and S is thought to be complete in Cameroon (Wondji et al. 2005). Whereas, in The Gambia, M/S hybrids were identified from a number of sites at frequencies as high as 16.7% (Caputo et al. 2008) and in Guinea-Bissau hybrids were recovered at frequencies over 20% (Marsden et al. 2011; Oliveira et al. 2008). In addition, a cryptic subgroup of A. gambiae known as the "Goundry" population collected in Burkina Faso was recently found to be composed of 36% M/S hybrids (Riehle et al. 2011). These studies suggest that rates of hybridization and backcrossing may be higher than previously thought.
Two opposing models exist that describe the relationship between the M and S forms. The “genomic islands of speciation” model suggests that divergence between the M and S genomes is restricted to small regions (~3%) of the genome that may contain the genes responsible for reproductive isolation between forms and that ongoing gene flow is responsible for very low levels of divergence over the remaining 97% of the genome (Lanzaro et al. 1998; Turner& Hahn 2007; Turner et al. 2005; Wang-Sattler et al. 2007; Wang et al. 2001). The second model, the “incidental islands of divergence” model, suggests that divergence between the two forms is far more extensive and widely distributed over the genome, that gene flow between the two forms is nearly zero and that the M and S forms therefore represent distinct species (Lawniczak et al. 2010; Neafsey et al. 2010; White et al. 2010). This work has recently culminated in the formal recognition of the M form as a species distinct from A. gambiae and given the designation Anopheles coluzzii (Coetzee et al. 2013). We continue to use the designation “M form” for A. coluzzii throughout this paper.
Reconciliation of the opposing models awaits the resolution of a number of outstanding questions concerning interactions between the M and S forms. These include accurate assessment of the spatial and temporal distribution of hybridization rates, the frequency of backcross hybrids and hybrid fitness in nature. The most widely used PCR-based diagnostics to differentiate M molecular forms from S forms are based on single base pair substitutions at either the 540th or 649th nucleotide position in the 28S rDNA locus (Fanello et al. 2002; Favia et al. 2001; Santolamazza et al. 2004). However, the 28S rDNA is a multi-copy gene making it less than ideal for taxonomic differentiation. Consequently, further development of methods for differentiating M and S molecular forms based on a single locus marker continued, ultimately resulting in a new method based on polymorphism in insertion sites for a group of retrotransposons known as short interspersed elements (SINEs) (Santolamazza et al. 2008). One of the SINE insertion sites, located on the X chromosome and referred to as SINE X6.1, was found to be fixed in the M form and absent in the S form. In subsequent studies in which multiple M/S diagnostic methods were employed, some discrepancies in results were observed (Santolamazza et al. 2011) especially in populations where M/S hybridization is common (e.g. Guinea-Bissau) which were attributed to the different biases in the various methods (Santolamazza et al. 2011). In particular, diagnostics based on the 28S IGS rDNA (i.e. Favia/Fanello assays) were unreliable in geographic regions with higher levels of hybridization due to the presence of different copy numbers of 28S IGS between M and S in hybrid/introgressed individuals. However, as the SINE-X is based on a single locus, this diagnostic cannot distinguish F1 hybrids from backcrossed individuals and mis-identifies a proportion of backcrossed hybrids. For example, 50 % of the progeny of an F1 [SINE genotype MS] x parental [SINE genotype SS] would have SINE genotype MS, and the remainder would have a SINE genotype SS.
It is clear that determination of the frequency of hybrid individuals requires that individuals be identified using multi-locus genotypes located across multiple linkage groups, such as those employed by White et al. (White et al. 2010), as opposed to the widely used single locus X-linked markers. This would allow not only the recognition of F1 hybrids but backcrossed individuals as well. Determination of the frequencies of both F1 and backcrossed genotypes would provide information on the level of introgression and hybrid fitness. Moreover, a multi-locus approach would allow identification of hybrid males, which cannot be identified using current single locus X linked diagnostics, as males are hemizygous. The application of this method to populations throughout the sympatric range of M and S would allow a description of spatial heterogeneity in levels of introgression that could be related to key environmental parameters that include mating cues that sustain assortative mating within forms as well as conditions that favor the survival of hybrid genotypes.
In this paper, we introduce a new method of multi-locus SNP genotyping composed of markers known to be fixed differences between M and S molecular forms. This multi-locus SNP genotyping approach can robustly and accurately detect F1 hybrids as well as backcrossed individuals resulting in introgression.
Method
Development of Divergence Island SNP (DIS) genotyping assay
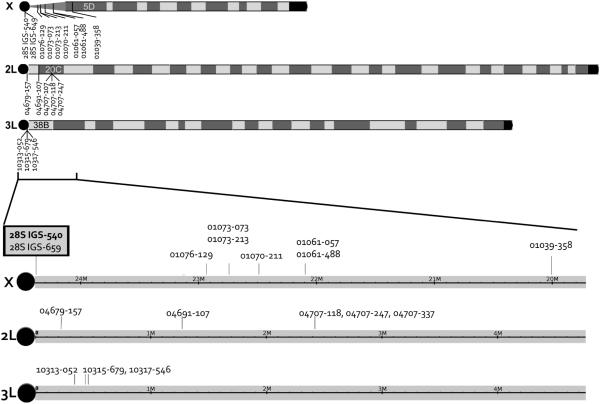
Single nucleotide polymorphisms reported to be fixed between M and S molecular forms were selected for genotyping from four different studies (Stump et al. 2005; Turner& Hahn 2007; Turner et al. 2005; White et al. 2010). We used Typer® AssayDesigner software (Sequenom, San Diego, CA) to devise a multiplex SNP genotype assay consisting of 17 SNPs that occur on all three chromosomes; 9 on the X, 5 on the 2L, 3 on the 3L (Figure 1). A list of SNPs genotyped is provided in Table 1. Full details of the DNA sequence around each SNP, iPLEX assay primer sequences, etc. are provided in Supplemental Table S1. We employed SNP IDs indicating the last 5 digits of the relevant A. gambiae gene ID starting with "AGAP0" concatenated with 3 digits of the relative nucleotide position from published DNA sequences. For example, 01039-044 indicates a SNP on the 44th nucleotide of the published fragment sequence of AGAP01039. In addition we included the molecular form diagnostic SNP utilized in conventional molecular form diagnostic methods (Fanello et al. 2002; Favia 1997) denoted 28S-IGS-540 as well as an additional linked SNP, 28S-IGS-649, also reported to be fixed between M and S molecular forms (Gentile et al. 2002; Santolamazza et al. 2004). The designation of M and S alleles were based on previously published information (Stump et al. 2005; Turner& Hahn 2007; Turner et al. 2005; White et al. 2010).
Figure 1.
Location of divergence island SNPs (DIS). Black circles indicate centromeres. The two markers highlighted in gray box (28S IGS-540 and 28S IGS-659) indicate the molecular form diagnostic SNPs used for typical molecular form determination.
Table 1.
Details of SNPs utilized for M, S and hybrid characterization. Additional sequence and primer design data provided in Supplemental Table S1.
| SNP ID | Chrom. | genome coordinate |
SNP | M genotype | S genotype | reference |
|---|---|---|---|---|---|---|
| 28S-IGS-540 | X | 24,391,149 | T/C | TT | CC | Favia et al. 1997, Fanello et al. 2002 |
| 28S-IGS-649 | X | 24,391,258 | A/T | AA | TT | Gentile et al. 2002 |
| 01076-129 | X | 22,944,682 | T/G | TT | GG | Stump et al. 2005 |
| 01073-073 | X | 22,750,572 | G/T | GG | TT | Stump et al. 2005 |
| 01073-213 | X | 22,750,432 | G/A | GG | AA | Stump et al. 2005 |
| 01070-211 | X | 22,497,157 | A/G | AA | GG | Turner et al. 2005 |
| 01061-057 | X | 22,105,860 | T/C | TT | CC | Stump et al. 2005 |
| 01061-488 | X | 22,10,5429 | A/T | AA | TT | Stump et al. 2005 |
| 01039-358 | X | 20,015,634 | C/A | CC | AA | Stump et al. 2005 |
| 04679-157 | 2L | 209,536 | C/T | CC | TT | White et al. 2010 |
| 04691-107 | 2L | 1,274,353 | A/G | AA | GG | Turner& Hahn 2007; Turner et al. 2005 |
| 04707-118 | 2L | 2,430,786 | C/T | CC | TT | Turner et al. 2005 |
| 04707-247 | 2L | 2,430,915 | A/G | AA | GG | Turner et al. 2005 |
| 04707-337 | 2L | 2,431,005 | C/T | CC | TT | Turner et al. 2005 |
| 10313-052 | 3L | 296,897 | G/A | GG | AA | White et al. 2010 |
| 10315-679 | 3L | 387,877 | G/A | GG | AA | White et al. 2010 |
| 10317-546 | 3L | 413,944 | T/C | TT | CC | White et al. 2010 |
Assessment of the DIS genotyping assay
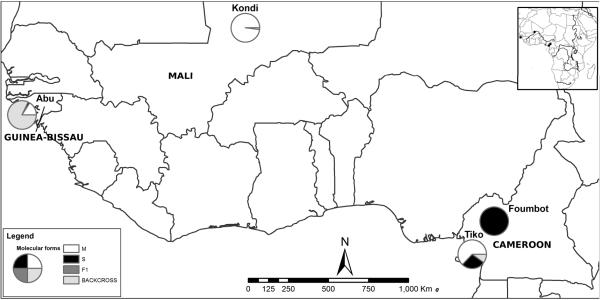
To assess the performance of our assay, we screened 92 M form samples from Kondi, Mali (allopatric M form population), 94 S form samples from Foumbot, Cameroon (allopatric S population), 81 M and S forms from Tiko, Cameroon (sympatric population), and 50 samples from Abu, Guinea-Bissau (hybridizing population) (Marsden et al. 2011). Adult females of A. gambiae s.s. were collected indoors using aspirators between 2002 and 2009. Geographic coordinates for collection sites are provided in Table 2 and geographic locations indicated on a map in Figure 2.
Table 2.
Collection site information. M, S, F1 and backcross frequencies are based on DIS assay result.
| Site | Country | Latitude | Longitude | Collection Date | Sample size |
% M |
% S |
% F1 |
% backcross |
|---|---|---|---|---|---|---|---|---|---|
| Kondi | Mali | 16.36670 | −3.38330 | October- November, 2002 |
92 | 98 | 0 | 0 | 2 |
| Foumbot | Cameroon | 5.48505 | 10.60005 | August 2006 | 94 | 0 | 99 | 0 | 1 |
| Tiko | Cameroon | 4.07860 | 9.36810 | September- October, 2003 |
81 | 63 | 27 | 0 | 10 |
| Abu | Guinea- Bissau |
11.46144 | −15.91411 | November, 2009 | 50 | 16 | 2 | 0 | 82 |
Figure 2.
Collection site map with molecular form composition based on DIS assay.
The Sequenom iPLEX®Gold assay was used for SNP genotyping following the manufacturer's protocol (Jurinke et al. 2002). This genotyping method utilizes the MALDI-TOF mass spectrometry to determine genotypes based on the mass of allele-specific fragments (Fu et al. 2008; Wright et al. 2008; Zhang et al. 2010). Mass spectrogram visualization and genotype calls were conducted using TyperAnalyzer software version 4.0 (Sequenom, San Diego, CA).
Test for linkage disequilibrium was done using likelihood method implemented in Arlequin version 3.5 (Excoffier& Lischer 2010) with 100,000 permutations.
Comparison of molecular form diagnostic methods
To assess the accuracy of our DIS assay in comparison to other molecular form diagnostic methods, we selected a set of 79 samples from Mali, Cameroon and Guinea-Bissau and assayed them using: (1) our DIS genotyping assay, (2) the Short Interspersed Element (SINE)-PCR (Santolamazza et al. 2008), (3) a molecular form diagnostic PCR method (Santolamazza et al. 2004) and (4) PCR-RFLP assay for two SNPs on 2L and 3L "islands of divergence" (White et al. 2010). The relative accuracy of these methods was assessed through comparison with DNA sequence data. Specifically, we sequenced the 28S-IGS fragment amplified in the species diagnostic PCR (Scott et al. 1993) which includes the 28S IGS-540 SNP targeted by the Favia PCR (Favia et al. 2001) and Fanello RFLP (Fanello et al. 2002) as well as the 28S IGS-649 SNP which is the target of the Santolamazza assay (Santolamazza et al. 2004). We also sequenced the undigested fragment of the 2L PCR-RFLP and 3L PCR-RFLP autosomal assays of White et al. (2010). All sequencing was conducted on an ABI 3730 by the UCDNA facility at the University of California Davis. The SINE-X assay screens the SINE200 insertion which is larger than the maximum amplimer size allowed for iPLEX assays. Thus this particular marker was not included in the iPLEX assay, but we assessed consistency of standard SINE-X assay with the molecular form calls based on our DIS method.
Results
A sample of multi-locus genotypes generated by the DIS assay are presented for illustration in Table 3. We excluded the multi-copy SNPs (28S IGS-540 and 28S IGS-649) because we detected heterozygotes on these markers from males collected in Abu, indicating that single X chromosome frequently carry both SNPs, indicating that these are not truly allelic and are therefore unsuitable for further analysis. By the proportion of M or S alleles, S ancestry values can be calculated for each individual. For instance, individuals from pure M parental populations will have S ancestry of 0 while individuals from pure S parental populations will have S ancestry of 1. F1 individuals will be heterozygous at all loci, resulting in S ancestry of 0.5. The entire genotype data for all individuals can be visualized using heatmap-like figures as shown in Figure 3. An Illustration of how these figures were generated and how they represents genotypes for all samples in each population are provided in Supplemental Figure S1.
Table 3.
A sample of multi-locus SNP genotyping results. Each row represents an individual mosquito sample. Each column represents a locus. Loci are grouped by linkage group (X, 2L and 3L). Homozygotes for M alleles (MM) are marked in black, homozygotes for S alleles (SS) are marked in light gray, and heterozygotes (MS) are marked in dark gray. Consensus molecular form is determined based on all 15 locus genotypes. We allowed up to 2 mismatched calls. Individuals that have more than 13 MM genotypes were called M form. Individuals carrying more than 13 SS genotypes were called S form. Individuals carrying more than 13 MS genotypes were called F1. And the rest were called as backcross (F1+n). S ancestry is the proportion of S alleles at all loci.
| Sample | S ancestry | consensus molecular form |
X | 2L | 3L | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01076-129 | 01073-213 | 01073-073 | 01070-211 | 01061-488 | 01061-057 | 01039-358 | 04679-157 | 04691-107 | 04707-118 | 04707-247 | 04707-337 | 10313-052 | 10315-679 | 10317-546 | |||
| 02KON014 | 1 | M | GG | AA | TT | GG | TT | CC | AA | TT | GG | TT | GG | TT | AA | AA | CC |
| 02KON019 | 1 | M | GG | AA | TT | GG | TT | CC | AA | TT | GG | TT | GG | TT | AA | AA | CC |
| 06FOUM030 | 0 | S | TT | GG | GG | AA | AA | TT | CC | CC | AA | CC | AA | CC | GG | GG | TT |
| 06FOUM031 | 0 | S | TT | GG | GG | AA | AA | TT | CC | CC | AA | CC | AA | CC | GG | GG | TT |
| 06NGAL009 | 0.5 | F1 | GT | AG | TG | GA | TA | CT | AC | TC | GA | TC | GA | TC | AG | AG | CT |
| 06NGAL020 | 0.5 | F1 | GT | AG | TG | GA | TA | CT | AC | TC | GA | TC | GA | TC | AG | AG | CT |
| 03Tiko044 | 0.16 | F1+n | GG | AA | TT | GG | TT | CC | AA | TC | GA | TC | GA | TC | AA | AA | CC |
| 03Tiko039 | 0.9 | F1+n | GG | AA | TT | GG | TT | CC | AA | TT | GG | TT | GG | TT | AG | AG | CT |
| 09ABU007 | 0.53 | F1 | GT | AG | TG | GA | TA | CT | AA | TC | GA | TC | GA | TC | AG | AG | CT |
| 09ABU008 | 0.67 | F1+n | GG | AA | TT | GG | TT | CC | AA | CC | AA | CC | AA | CC | AA | AA | CC |
| 09ABU019 | 0.7 | F1+n | GG | AA | TT | GG | TT | CC | AA | TT | GG | TC | GA | TC | GG | GG | TT |
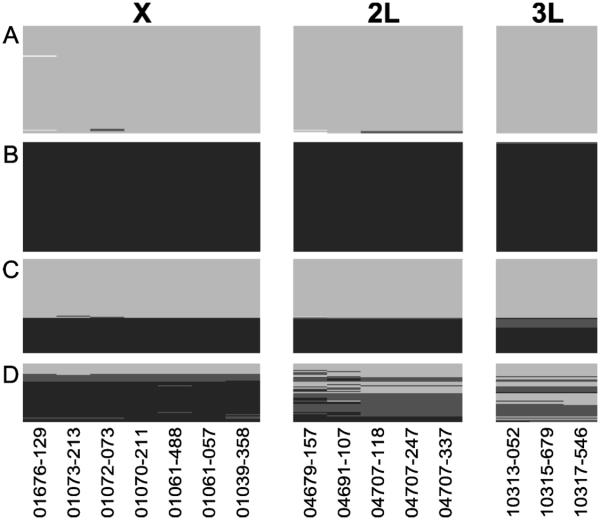
Figure 3.
Genotypes of markers used in DIS assay. Light gray color represent M genotype, black for S genotype, dark gray for heterozygotes, and white for missing data. A: Dire, Mali (M allopatric population, N=92). Each row is an individual with 15 loci genotypes. B: Foumbot, Cameroon (S allopatric population, N=94). C: Tiko, Cameroon (M and S sympatric population, N=81). D: Abu, Guinea-Bissau (hybridizing population, N=50).
Our survey of 317 samples from 4 different populations reveals not only the molecular form information but information on the history of hybridization backcrosses (Figure 3). In Dire where M is reportedly allopatric, 98% of M samples were M like across all loci, and strong linkage was found among all of the divergence island SNPs (P<0.0005). Similarly in Foumbot where S is reportedly allopatric, 99% of S samples were S like across all loci, with strong linkage among all SNPs (P<0.0005). In Abu, Guinea-Bissau, a known hybrid zone, the F1 hybrids and backcrosses are clear. Specifically, the DIS assay results show that in Guinea-Bissau M samples appear M like at all loci. However, S or hybrids based on X markers reveals extensive introgression of M alleles into S population on the autosomes. Consistent with this, significant linkage was only found between SNPs within each divergence island, but not between SNPs in different divergence islands. The weak association of 2L island with other divergence islands have also been reported Weetman et al. (2012). Together these data are consistent with extensive and asymmetric introgression between the M and S forms in Guinea-Bissau, as reported by Marsden et al. (2011). It is noteworthy that overall the X-linked SNPs exhibited lower levels of heterozygosity (H=0.0823) than autosomes (H2L = 0.354, H3L = 0.307) based on the DIS. Since these SNPs are fixed differences and therefore heterozygotes are hybrids, SNPs on the X chromosome have greater diagnostic power than those on autosomes.
To assess the accuracy for the identification of molecular forms based on DIS, we compared DNA sequence of the 28S IGS fragment used for species identification (Scott et al. 1993) with DIS genotype calls. Overall we found the DIS to have higher rates of successful amplification and genotyping (100%) than the other methods (92.3-98.7%) as shown in Table 4. Moreover, DIS genotypes were more consistent (=95.4%) with DNA sequence data compared to other methods (Table 4). Notably, 94% of the DNA sequence data of the 28S IGS-540 SNP were consistent with consensus molecular form data from 15 DIS, compared with 75% and 81% for the Fanello and Favia diagnostics. It is noteworthy that the inconsistency in the other methods is due to overestimation of heterozygotes.
Table 4.
Evaluation of various genotyping platforms for molecular form diagnostic SNPs. The number of mismatches are calculated based on conventional DNA sequencing results for amplimers of the 28S IGS 540 and 649 SNPs, SINEX and the 2L and 3L SNPs described by White et al. (2010). All 79 individual samples were sequenced for each amplimer except SINEX (Santolamazza et al. 2008) method.
| Chromosome | X | 2L | 3L | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker sequence | 28S IGS-540 | SINEX | 28S IGS-649 | 04679-157 | 10313-052 | |||||
| Method | iPLEX | Favia PCR |
Fanello RFLP |
SINEX PCR |
iPLEX | White RFLP |
iPLEX | White RFLP |
iPLEX | White RFLP |
| failed amplification | 0 | 1 | 6 | 1 | 0 | 1 | 0 | 3 | 0 | 6 |
| DNA sequence mismatch |
3 | 15 | 20 | 12* | 4 | 9 | 2 | 12 | 4 | 19 |
| mismatch % | 3.8 | 19.0 | 25.3 | 13.9* | 5.1 | 11.4 | 2.5 | 15.2 | 5.1 | 24.1 |
SINEX PCR results were compared with molecular form diagnostic locus on 28S IGS for reasons provided in the Method - Comparison of molecular form diagnostic methods section. The percentage of mismatch on SINEX PCR was based on the comparison with consensus molecular form data.
Discussion
We designed a new multi-locus genotyping method, the Divergence Island SNP (DIS) genotyping assay, to improve our ability to detect and distinguish hybrids and backcrosses between the M and S forms of A. gambiae. Based on comparison with DNA sequence data, we show this approach to be more accurate than the widely used single locus Favia PCR and Fanello RFLP assays based on polymorphism in the 28S locus. The discrepancy between DNA sequence and these assays was mainly due to the overestimation of hybrids. This discrepancy may have been overlooked when reading gels due to expectations that M and S hybrids are rare in nature. In populations where hybridization is low, this bias would result in few “questionable” bands on gels and consequently few mis-identifications. However, the bias is clearly more problematic for populations with high gene flow (Caputo et al. 2011; Marsden et al. 2011; Santolamazza et al. 2011). The RFLP method for genotyping DIS in autosomes developed by White et al. (White et al. 2010) also shows a similar tendency to overestimate hybrids (Table 4). This was shown to be the result of difficulty in differentiating true hybrids from incomplete restriction enzyme cutting in these RFLP methods (Hahn et al. 2012).
In addition to being more accurate than currently available methods, we have shown that the DIS assay can reliably identify M and S forms, as well as F1 hybrids and backcrossed individuals, thus providing additional information regarding hybridization and introgression among these populations. One important consideration for the DIS relates to the question of how many mismatched loci should be allowed before classifying an M or S sample as backcrossed (i.e. a samples that have M SNPs at all loci except one; is it classified as M or backcross?). In this study, we allowed up to 2 mismatched calls to give a conservative estimate of hybridization.
The DIS method can be applied to accurately identify M and S individuals, including from sites in coastal West Africa, where conventional methods fail (Caputo et al. 2011, Santolamazza et al. 2011). Therefore, the DIS method is the only currently available method for the analysis of these populations, which includes sites in Guinea-Bissau and The Gambia. The current methods have the disadvantage of incorrectly scoring backcross individuals that are homozygous for the X-linked markers currently used, therefore the DIS method is the only available method for accurately assessing patterns of introgression between M and S. A disadvantage to the DIS method, as described here, is that it requires a multi-SNP genotyping platform, such as the Sequenom iPLEX Gold platform, that is likely to be unavailable in many labs in West Africa. However there are genotyping core facilities and commercial labs that do provide SNP genotyping services, for example we utilized the UC Davis Veterinary Genetics Laboratory facility for this study. Our cost was $4.75 to genotype all 15 SNPs per sample. This is considerably more expensive than the single SNP genotype, agarose gel-based assay currently in use, which is about $1.50 per sample and provides limited and possibly erroneous information. An obvious improvement to the single SNP assay would be to include a 3 SNP assay, with one SNP per chromosome. If this were done on an agarose gel platform the cost would be $4.50, not much less than the DIS method. So, although somewhat more expensive the DIS method allows investigators to explore problems related to hybridization and introgression of the M and S forms at the population level that were previously difficult to achieve.
The method introduced in this paper shows promise for studies of hybridization and introgression in A. gambiae. This method can differentiate F1 hybrids from backcrossed individuals and can be used to identify hybrid males. Having multiple markers within chromosomes also helps in assessing the degree of linkage and rates of recombination. For instance, linkage between divergence islands have disappeared in the Abu population (Figure 3D). Moreover, with a multiple locus assay it will be possible to investigate epistatic interactions among unlinked loci and fitness of hybrid genotypes. Overall the DIS method will allow a more in depth understanding of gene flow in this system, which has been the subject of intense debate for many years and has important implication for malaria prevention by means of mosquito population control using chemical or biological elements (Marsden et al. 2011; Marsden et al. 2012). To this end, further DIS genotyping efforts are now underway across a greater geographic and temporal scale. A similar approach has been utilized in comparing two different geographic transects across the hybrid zone of the European house mouse. In this case, the species boundary and potential mechanism of species isolation was successfully illuminated (Teeter et al. 2010).
Supplementary Material
Illustration of how genotype heatmap as shown in Figure 3 were generated.
Full details on SNPs used on DIS assay.
Acknowledgments
We thank Julia Malvick and Lee V. Millon at the Veterinary Genetics Laboratory for processing iPLEX assays and advising on the Sequenom iPLEX SNP genotyping assay design. We also thank Laura C. Norris, Alison Weakley, Anthony J. Cornel at UC Davis and Michelle R. Sanford at Harris County Institute of California for the constructive comments and advise to improve the assay. This study was supported with funds from the National Institutes of Health grant R01AI40308, R21AI062929 and R01AI078183.
Footnotes
Data Accessibility
All genetic data and field collection meta data regarding samples utilized in this study is accessible from Dryad doi:10.5061/dryad.nk4m0 as well as an online database, PopI (https://grassi2.ucdavis.edu/) OpenProjects “AgDIS” page.
Author Contributions
YL devised assay, performed data analysis and wrote manuscript. CDM performed comparative diagnostic assays and wrote manuscript. CN performed the new diagnostic assay as well as comparative assays for method validation. GCL conceived experiment and wrote manuscript.
References
- Ayala FJ, Coluzzi M. Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci U S A. 2005;102(Suppl 1):6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B, Nwakanma D, Jawara M, et al. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar J. 2008;7:182. doi: 10.1186/1475-2875-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B, Santolamazza F, Vicente JL, et al. The "far-west" of Anopheles gambiae molecular forms. PLoS One. 2011;6:e16415. doi: 10.1371/journal.pone.0016415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandre F, Manguin S, Brengues C, et al. Current distribution of a pyrethroid resistance gene (kdr) in Anopheles gambiae complex from west Africa and further evidence for reproductive isolation of the Mopti form. Parassitologia. 1999;41:319–322. [PubMed] [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson RC, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Millogo N, Lehmann T. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae) J Med Entomol. 2007;44:60–64. doi: 10.1603/0022-2585(2007)44[60:eteopi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dao A, Yaro AS, et al. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae. Proc Biol Sci. 2009;276:4215–4222. doi: 10.1098/rspb.2009.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Favia G, della Torre A, Bagayoko M, Lanfrancotti A, Sagnon N'F, Touré YT, Coluzzi M. Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Mol Biol. 1997;6:377–383. doi: 10.1046/j.1365-2583.1997.00189.x. [DOI] [PubMed] [Google Scholar]
- Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol Biol. 2001;10:19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- Fu JF, Shi JY, Zhao WL, et al. MassARRAY assay: a more accurate method for JAK2V617F mutation detection in Chinese patients with myeloproliferative disorders. Leukemia. 2008;22:660–663. doi: 10.1038/sj.leu.2404931. [DOI] [PubMed] [Google Scholar]
- Gentile G, Della Torre A, Maegga B, Powell JR, Caccone A. Genetic differentiation in the African malaria vector, Anopheles gambiae s.s., and the problem of taxonomic status. Genetics. 2002;161:1561–1578. doi: 10.1093/genetics/161.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, Pombi M, Choisy M, et al. Larval habitat segregation between the molecular forms of the mosquito Anopheles gambiae in a rice field area of Burkina Faso, West Africa. Med Vet Entomol. 2012;26:9–17. doi: 10.1111/j.1365-2915.2011.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, White BJ, Muir CD, Besansky NJ. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Philos Trans R Soc Lond B Biol Sci. 2012;367:374–384. doi: 10.1098/rstb.2011.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurinke C, van den Boom D, Cantor CR, Koster H. Automated genotyping using the DNA MassArray technology. Methods Mol Biol. 2002;187:179–192. doi: 10.1385/1-59259-273-2:179. [DOI] [PubMed] [Google Scholar]
- Lanzaro GC, Toure YT, Carnahan J, et al. Complexities in the genetic structure of Anopheles gambiae populations in west Africa as revealed by microsatellite DNA analysis. Proc Natl Acad Sci U S A. 1998;95:14260–14265. doi: 10.1073/pnas.95.24.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MK, Emrich SJ, Holloway AK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Meneses CR, Fofana A, Lanzaro GC. Desiccation resistance among subpopulations of Anopheles gambiae s.s. from Selinkenyi, Mali. J Med Entomol. 2009;46:316–320. doi: 10.1603/033.046.0216. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8:737–746. doi: 10.1016/j.meegid.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukis NC, Diabate A, Abdoulaye A, et al. Structure and dynamics of male swarms of Anopheles gambiae. J Med Entomol. 2009;46:227–235. doi: 10.1603/033.046.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukis NC, Powell JR, Touré MB, Sacko A, Edillo FE, Coulibaly MB, Traoré SF, Taylor CE, Besansky NJ. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2008;105:2940–2945. doi: 10.1073/pnas.0709806105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Cornel AJ, Lee Y, et al. An analysis of two island groups as potential sites for trials of transgenic mosquitoes for malaria control. Evol. Appl. 2012 doi: 10.1111/eva.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Lee Y, Nieman CC, et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol Ecol. 2011;20:4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Lawniczak MK, Park DJ, et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science. 2010;330:514–517. doi: 10.1126/science.1193036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E, Salgueiro P, Palsson K, et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45:1057–1063. doi: 10.1603/0022-2585(2008)45[1057:hlohbm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Riehle MM, Guelbeogo WM, Gneme A, et al. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science. 2011;331:596–598. doi: 10.1126/science.1196759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford MR, Demirci B, Marsden CD, et al. Morphological differentiation may mediate mate-choice between incipient species of Anopheles gambiae s.s. PLoS One. 2011;6:e27920. doi: 10.1371/journal.pone.0027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Caputo B, Calzetta M, et al. Comparative analyses reveal discrepancies among results of commonly used methods for Anopheles gambiae molecular form identification. Malar J. 2011;10:215. doi: 10.1186/1475-2875-10-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Della Torre A, Caccone A. Short report: A new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am J Trop Med Hyg. 2004;70:604–606. [PubMed] [Google Scholar]
- Santolamazza F, Mancini E, Simard F, et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163. doi: 10.1186/1475-2875-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, et al. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, et al. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution. 2010;64:472–485. doi: 10.1111/j.1558-5646.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Tripet F, Toure YT, Taylor CE, et al. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol Ecol. 2001;10:1725–1732. doi: 10.1046/j.0962-1083.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- Tripet F, Wright J, Cornel A, et al. Longitudinal survey of knockdown resistance to pyrethroid (kdr) in Mali, West Africa, and evidence of its emergence in the Bamako form of Anopheles gambiae s.s. Am J Trop Med Hyg. 2007;76:81–87. [PubMed] [Google Scholar]
- Turner TL, Hahn MW. Locus- and population-specific selection and differentiation between incipient species of Anopheles gambiae. Mol Biol Evol. 2007;24:2132–2138. doi: 10.1093/molbev/msm143. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Sattler R, Blandin S, Ning Y, et al. Mosaic genome architecture of the Anopheles gambiae species complex. PLoS One. 2007;2:e1249. doi: 10.1371/journal.pone.0001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng L, Toure YT, Dandekar T, Kafatos FC. When genetic distance matters: measuring genetic differentiation at microsatellite loci in whole-genome scans of recent and incipient mosquito species. Proc Natl Acad Sci U S A. 2001;98:10769–10774. doi: 10.1073/pnas.191003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Wilding CS, Steen K, Pinto J, Donnelly MJ. Gene flow-dependent genomic divergence between Anopheles gambiae M and S forms. Mol Biol Evol. 2012;29:279–291. doi: 10.1093/molbev/msr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010;19:925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C, Frederic S, Petrarca V, et al. Species and populations of the Anopheles gambiae complex in Cameroon with special emphasis on chromosomal and molecular forms of Anopheles gambiae s.s. J Med Entomol. 2005;42:998–1005. doi: 10.1093/jmedent/42.6.998. [DOI] [PubMed] [Google Scholar]
- Wright WT, Heggarty SV, Young IS, et al. Multiplex MassARRAY spectrometry (iPLEX) produces a fast and economical test for 56 familial hypercholesterolaemia-causing mutations. Clin Genet. 2008;74:463–468. doi: 10.1111/j.1399-0004.2008.01071.x. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Qiu HX, Li JY, Shi JY, Xu W. The analysis of JAK2 and MPL mutations and JAK2 single nucleotide polymorphisms in MPN patients by MassARRAY assay. Int J Lab Hematol. 2010;32:381–386. doi: 10.1111/j.1751-553X.2009.01208.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Illustration of how genotype heatmap as shown in Figure 3 were generated.
Full details on SNPs used on DIS assay.