Summary
While structural symmetry is a prevailing feature of homo-oligomeric proteins, asymmetry provides unique mechanistic opportunities. We present the crystal structure of full-length TRAP1, the mitochondrial Hsp90 molecular chaperone, in a catalytically active closed state. The TRAP1 homodimer adopts a distinct, asymmetric conformation, where one protomer is reconfigured via a helix swap at the Middle:C-terminal Domain (MD:CTD) interface. Importantly, this interface plays a critical role in client binding. Solution methods validate the asymmetry and show extension to Hsp90 homologs. Point mutations that disrupt unique contacts at each MD:CTD interface reduce catalytic activity, substrate binding, and demonstrate that each protomer needs access to both conformations. Crystallographic data on a dimeric NTD:MD fragment suggests that asymmetry arises from strain induced by simultaneous NTD and CTD dimerization. The observed asymmetry provides the potential for an additional step in the ATPase cycle, allowing sequential ATP hydrolysis steps to drive both client remodeling and client release.
Introduction
Protein folding is a fundamental process and protein misfolding can dramatically impact cellular and organismal fitness. To ensure proper folding and function, cells have evolved molecular chaperones to maintain cellular homeostasis. These proteins interact with substrates at different folding stages and act in concert throughout the maturation process (Hartl et al., 2011).
Heat shock protein 90 (Hsp90) is a highly conserved ATP-dependent chaperone essential to the folding and activation of nearly 10% of the proteome (Zhao et al., 2005). Unlike other chaperones, Hsp90 preferentially interacts with partially folded substrate “client” proteins (Jakob et al., 1995) and primes them for downstream protein-protein or protein-ligand interactions (Taipale et al., 2012). Thus, beyond initial folding Hsp90 plays an important role throughout the functional lifetime of many clients. Client proteins vary widely in sequence, structure, size and function, and for this reason client specificity is not well understood (Echeverria et al., 2011). Disruption of Hsp90 activity can affect evolutionary outcomes (Lindquist, 2009) and has been linked to human diseases including cancer (Whitesell and Lindquist, 2005), vascular disease (Shah et al., 1999), and neurodegeneration (Luo et al., 2010).
Hsp90 functions as a homodimer with each protomer consisting of three major domains: the N-terminal domain (NTD), responsible for ATP binding and hydrolysis, the middle domain (MD) that aids in hydrolysis, and the C-terminal dimerization domain (CTD) (Figures 1A and 1B). Crystal structures, electron microscopy (EM) and small angle X-ray scattering (SAXS) in the presence or absence of nucleotide have revealed large conformational changes at the major domain boundaries. In the absence of nucleotide, a variety of distinct open states are populated that are thought to aid in binding a diverse set of clients (Krukenberg et al., 2008; Krukenberg et al., 2009). The crystal structure of yeast Hsp90 (yHsp90) (Ali et al., 2006) along with biochemical studies (Prodromou et al., 2000) demonstrate that NTD dimerization mediates ATP hydrolysis and is essential for function. Following ATP hydrolysis, Hsp90 transiently adopts a compact ADP-bound conformation initially observed in the bacterial Hsp90 (bHsp90) (Shiau et al., 2006) and later trapped in yHsp90 and human Hsp90 (hHsp90) (Southworth and Agard, 2008). Together, these data establish a model for the conformational cycle of Hsp90 whereby the protomer arms undergo concerted conformational changes through rounds of ATP binding, hydrolysis, and release (Krukenberg et al., 2011). Though hydrolysis is essential for in vivo function across Hsp90 homologs (Panaretou et al., 1998), it remains unclear how hydrolysis is coupled to client maturation.
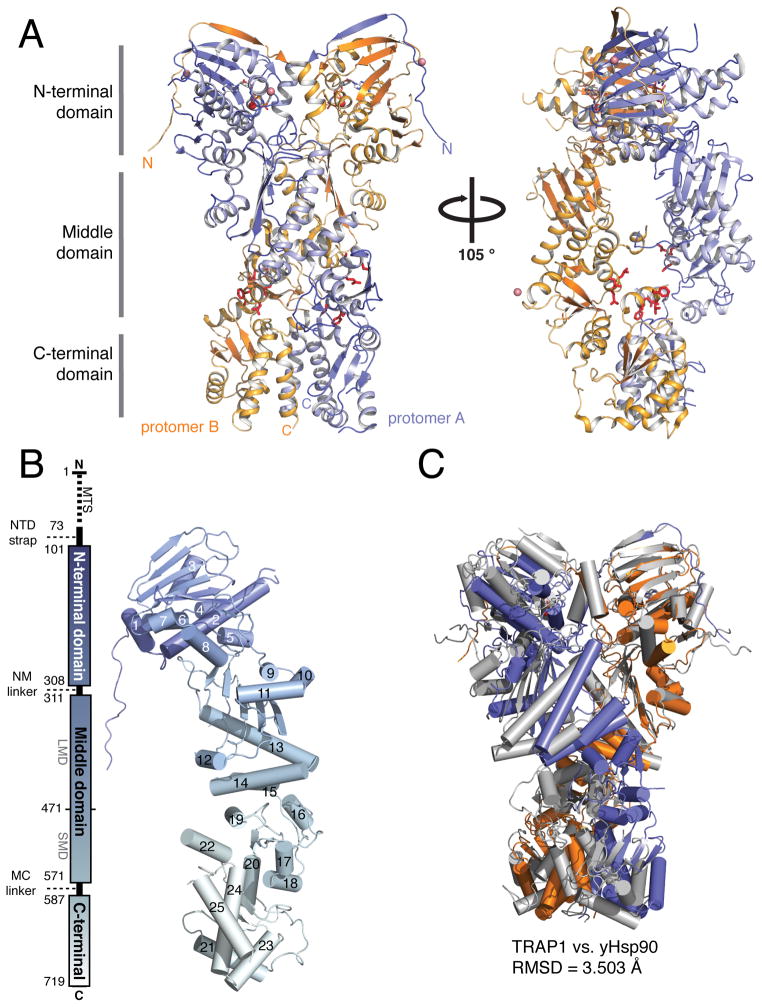
Figure 1. Crystal structure of full-length TRAP1 in an asymmetric closed state.
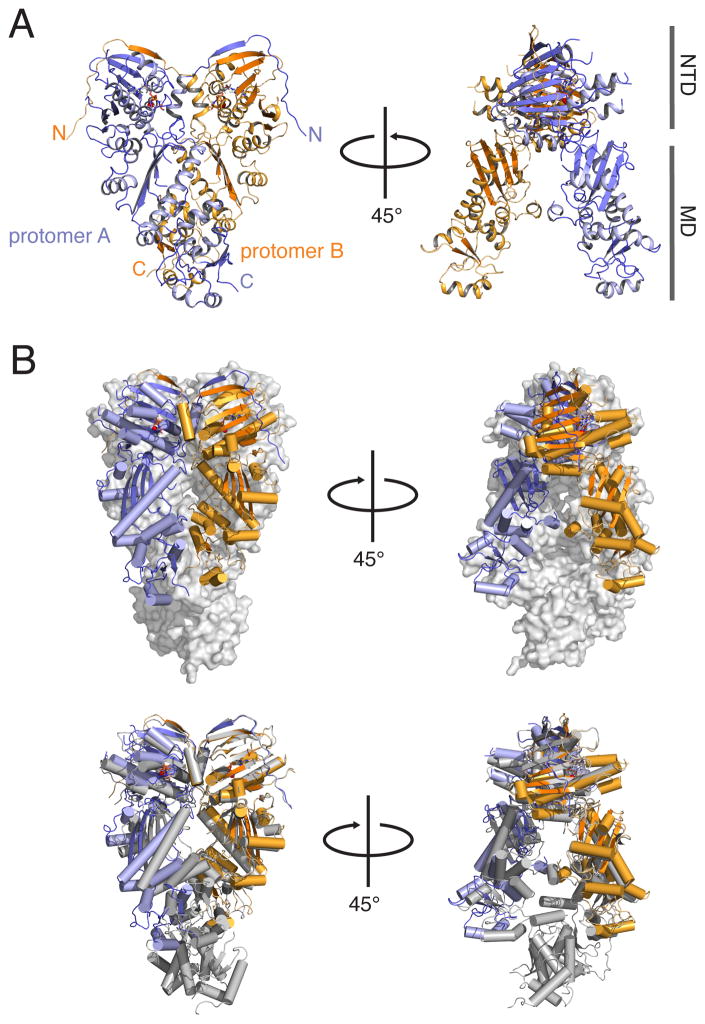
A) Full-length TRAP1 homodimer from D. rerio. Protomer B (orange) is similar to the p23-stabilized yHsp90 structure, while protomer A (blue) makes novel contacts between the MD and CTD. Residues known to bind clients in this region are shown in red. Also visible is an N-terminal extension that exchanges between protomers. Cobalt atoms (pink) help stabilize crystal contacts. B) TRAP1 domain boundaries (left), map of the helical positions within a single protomer (right). N to C-terminal progression is indicated by a transition from blue to gray. C) Comparison of TRAP1 structure to the closed state of yHsp90 (gray) (RMSD = 3.5Å). (See also Movie S1)
Most eukaryotes have four Hsp90 homologs: two in the cytosol (Hsp90α/β), and one in both the endoplasmic reticulum (GRP94) and mitochondria (TRAP1) (Johnson, 2012). In vitro experiments demonstrate that although the fundamental conformational states are well conserved, equilibria and kinetics are unique to each Hsp90 homolog (Southworth and Agard, 2008), suggesting adaptation to the specific needs of clients in each cellular compartment and species. Within the eukaryotic cytosol, Hsp90 function is aided by numerous Hsp90-specific co-chaperones, though only one Hsp90-specific co-chaperone has been detected for the prokaryotic or organellar Hsp90s (Johnson, 2012; Liu et al., 2010).
TRAP1 is encoded in the nucleus and was originally identified as a binding partner of tumor necrosis factor receptor 1 (Song et al., 1995). TRAP1 has since been shown to localize primarily to the mitochondria (Felts et al., 2000) and most abundantly in the matrix (Cechetto and Gupta, 2000). Although few TRAP1 clients have been identified, there is an emerging appreciation for the role of TRAP1 in regulating mitochondrial protein homeostasis (Altieri et al., 2012). Specifically, TRAP1 has been implicated in critical pathways including mitochondrial fission/fusion (Takamura et al., 2012), mitophagy (Costa et al., 2013; Zhang et al., 2013), and the necrotic form of cell death through regulating cyclophilin D (Kang et al., 2007). TRAP1 appears closely linked to cancer progression (Sciacovelli et al., 2013; Yoshida et al., 2013), and appears relevant for both familial and idiopathic forms of Parkinson’s disease (Butler et al., 2012; Costa et al., 2013; Zhang et al., 2013). These characteristics highlight a growing understanding of TRAP1’s biological role and underscore its potential as a unique therapeutic target.
To better understand both shared and unique aspects of TRAP1 mechanism, we determined the crystal structure of TRAP1 at 2.3 Å resolution. The full-length mature protein was crystallized as a dimer in a closed conformation with AMPPNP, ADP-AlF4−, or ADP-BeF3−. In the absence of client or co-chaperones, TRAP1 adopts an unexpected asymmetric conformation with the largest deviations at the MD:CTD interface. Complementary solution-based methods (SAXS and Dipolar Electron-Electron Resonance, DEER) confirm that this asymmetric state is populated in solution and structure-based point mutations demonstrate the functional relevance of contacts at the distinct MD:CTD interfaces. Further, in vitro experiments show conservation of asymmetry between TRAP1 homologs as well as in bHsp90. Additionally, we crystallized a truncated version of a TRAP1 dimer bound to AMPPNP but lacking the CTD. This 1.7 Å NTD:MD structure is perfectly symmetric and has implications for understanding how asymmetry is formed in the full-length dimer. Both sets of structures display a previously unknown N-terminal extension that we show serves a regulatory role for ATPase activity. Finally, we propose that our TRAP1 structure depicts an important functional state with significant implications for the utilization of energy from ATP hydrolysis during client remodeling.
Results
Structure of Full-length TRAP1 in an active closed state
High-quality diffracting crystals of the mature form of TRAP1 from zebrafish- zTRAP1 (80% identical/90% similar to human TRAP1, hTRAP1), diffracted to 2.3 Å, however initial attempts at molecular replacement with previously solved Hsp90 models (full-length and NTD, MD, or CTD domains) proved unsuccessful. Instead, the structure was experimentally phased using selenomethionine and SAD to 3.0 Å and native structures of TRAP1 bound to three nucleotide analogs (AMPPNP, ADP-BeF3−/AlF4−) were refined at a maximum resolution of 2.3 Å (Table 1). No significant differences between the analogs were observed.
Table 1.
Crystallographic Data Table
| Data Set | TRAP1 AMPPNP native | TRAP1 AlF native | TRAP1 BeF native | TRAP1 (NTD:MD) AMPPNP native | TRAP1 AlF Se-methionine | TRAP1 BeF Se-methionine |
|---|---|---|---|---|---|---|
| PDB Code | 4IPE | 4IYN | 4J0B | 4IVG | ||
| Data Collection | ||||||
| Wavelength | 1.1159 | 1.1159 | 1.1159 | 1.1159 | 0.9796 | 0.9796 |
| Space group | C2 | C2 | C2 | I222 | C2 | C2 |
| Cell dimensions: | ||||||
| a,b,c (Å) | 176.0, 96.1, 124.5 | 178.9, 96.6, 125.9 | 177.8, 96.6, 125.0 | 85.1, 94.5, 155.5 | 177.9, 96.7, 125.2 | 177.4, 96.8, 125.3 |
| β (°) | 134.6 | 134.3 | 134.3 | 90.0 | 134.6 | 133.8 |
| Resolution (Å) | 50-2.3 (2.4-2.3) | 50-2.3 (2.4-2.3) | 45-2.35 (2.43- | 50-1.75(1.81-1.75) | 50-3.0 (3.1-3.0) | 45-2.7 (2.8-2.7) |
| 2.35) | ||||||
| Rsymma (%) | 5.1 (39.5) | 9.0 (43.1) | 8.7 (54.4) | 8.0 (99.9) | 10.0 (28.7) | 12.0 (95.3) |
| Rr.i.m.b (%) | 5.2 (37.6) | 6.8 (36.0) | 5.8 (50.8) | 9.1 (99.9) | 9.4 (24.3) | 12.6 (98.0) |
| Rp.i.m.c (%) | 2.6 (18.7) | 3.4 (18.3) | 3.3 (29.1) | 3.4 (45.8) | 3.4 (8.7) | 4.9 (40.1) |
| Completeness (%) | 98.6 (97.6) | 99.1 (91.1) | 99.5 (98.1) | 99.4 (94.6) | 100 (100) | 100 (99.8) |
| Redundancy | 4.1 (4.1) | 4.8 (3.8) | 3.8 (3.1) | 7.8 (6.2) | 14.3 (8.0) | 7.4 (6.7) |
| I/σ | 24.9 (3.1) | 18.6 (2.4) | 18.0 (1.9) | 24.9 (1.43) | 25.6 (7.7) | 16.8 (1.2) |
| Wilson B factor (Å2) | 48.16 | 50.2 | 59.1 | 29.8 | 69.9 | 56.84 |
| FOM 0.44 | FOM 0.40 | |||||
| Refinement | ||||||
| Resolution (Å) | 26-2.3 | 30-2.3 | 30-2.3 | 30-1.75 | ||
| Reflections | 67,272 | 66,710 | 62,239 | 63,000 | ||
| Nonhydrogen Atoms | 9,684 | 10,148 | 9,705 | 3,913 | ||
| Water Molecules | 279 | 288 | 189 | 344 | ||
| Rworkd | 18.0 | 18.5 | 20.5 | 18.86 | ||
| Rfreee | 22.5 | 23.14 | 24.8 | 20.26 | ||
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.01 | 0.01 | 0.01 | 0.008 | ||
| Bond angles (°) | 1.16 | 1.18 | 1.17 | 1.17 | ||
| B factors (Å2) | ||||||
| Protein | 73.5 | 67.8 | 82.3 | 47.8 | ||
| Water | 59.6 | 51.2 | 59.4 | 49.8 | ||
| Coordinate error (Å) | 0.25 | 0.27 | 0.33 | 0.20 | ||
| Ramachandran plotf | ||||||
| Favored (%) | 95.22 | 94.75 | 95.01 | 97.05 | ||
| Allowed (%) | 4.61 | 4.08 | 4.65 | 2.53 | ||
| Outliers (%) | 0.17 | 1.17 | 0.34 | 0.42 |
Rsymm = Σ|Ii − <Ii>|/ΣIi, where Ii is the intensity of the ith observation and <Ii> is the mean intensity of the reflection.
Rr.i.m. = Σhkl[N/(N − 1)]1/2 Σi |Ii(hkl) − <I(hkl) >|/Σhkl Σi Ii(hkl), where Ii(hkl) is the observed intensity and <I(hkl)> is the average intensity of multiple observations of symmetry-related reflections.
Rp.i.m. = Σhkl[1/(N − 1)]1/2 Σi |Ii(hkl) − <I(hkl) >|/Σhkl Σi Ii(hkl), where Ii(hkl) is the observed intensity and <I(hkl)> is the average intensity of multiple observations of symmetry-related reflections.
Rwork = Σ(||Fobs| − |Fcalc||/Σ|Fobs|)
Rfree = R value for a randomly selected subset (2000 reflections) of the data that were not used for minimization of the crystallographic residual. Highest resolution shell is shown in parenthesis.
Calculated with the program PROCHECK(32).
The asymmetric unit contains a single closed-state dimer, with dimerization interfaces at both the CTD and the NTD (Figure 1A). At 2.3 Å resolution our model presents the highest resolution picture of full-length Hsp90 catalytic machinery, showing that the nucleotide coordinating residues and catalytic water necessary for hydrolysis (Ali et al., 2006) are positioned in a manner consistent with this being a catalytically active state. Distinguishing TRAP1 features are also visible, including the characteristic “LXCXE” motif in the MD (Chen et al., 2006), which we observe to form a weak disulfide bond between C516 and C542. From sequence alignments and in comparison to other Hsp90 homologs, vertebrate TRAP1 proteins have an inserted 8-residue motif that is part of helix 19 (h19).
Comparing the TRAP1 structure to the previously solved yHsp90 closed state (stabilized by the co-chaperone p23) (Ali et al., 2006) it is clear that while the general architecture of the Hsp90 dimer is conserved there are striking differences (Figure 1C). This is best illustrated with a morph between the two structures starting with TRAP1 and transitioning to the yHsp90 structure (Movie S1). In addition, an ordered 14-residue N-terminal extension significantly lengthens the previously observed β-strand swap (Figures 1A, 1B and 2). In general, TRAP1 adopts a more compact conformation, shrinking the overall dimer height by ~4 Å and rotating clockwise by ~20° to constrict the cleft between protomers. Importantly, the major differences between the yHsp90 and TRAP1 structures arise from the non-equivalent conformations of protomers in TRAP1, illustrating striking asymmetry in the closed TRAP1 structure. Asymmetry is unlikely to be a consequence of crystal contacts as protomer B has more substantial contacts but is most similar in conformation to the protomers from the yHsp90 structure. Motivated by the unique features of our TRAP1 structure we investigated the functional relevance of these novel structural elements.
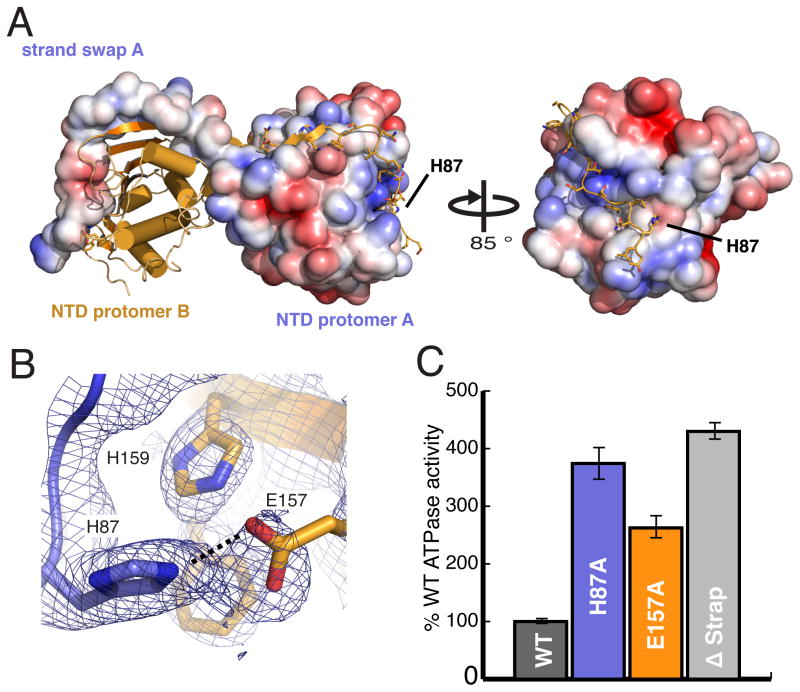
Figure 2. N-terminal strand extension regulates TRAP1 activity.
A) The surface representation of protomer A highlights the electrostatic charge distribution of the NTD and the extensive NTD-strand swap (1,484 Å2/monomer) made with each neighboring protomer. B) The salt bridge between H87 and E157 is displayed with the electron density map calculated using experimental phase restraints. C) ATPase activities of WT TRAP1, Δstrap, and H87 or E157 mutations indicate that disruption of strap contacts leads to a significant acceleration of ATPase activity. (Error bars are propagated standard deviations).
The N-terminal extension (“strap”) regulates ATP hydrolysis
Previous studies with truncation mutants have implicated the N-terminus of yHsp90 in regulating ATPase activity, with truncations accelerating activity (Richter et al., 2006; Richter et al., 2002). These residues form an initial β-strand and helix (h1), and are coupled to nucleotide binding and subsequent lid closure prior to hydrolysis. In isolated NTD structures (Li et al., 2012; Shiau et al., 2006) and an apo-NTD:MD structure (Dollins et al., 2007), the first 7 residues form β-strand interactions within the NTD of the cis-protomer, while in the yHSP90 closed state these elements cross over to form analogous β-strand contacts with the trans-protomer. TRAP1, GRP94, and cytosolic hHsp90α/β all contain an extension of the N-terminal strand between 10–50 residues long. Surprisingly this extension is not conserved in yHsp90 or in bHsp90 to which TRAP1 is most closely related (Chen et al., 2006). Our TRAP1 structure reveals that 14 ordered residues of N-terminal extended sequence (out of 26) make extensive trans-protomer interactions to fully envelope the neighboring NTD, adding an additional 771 Å2 of surface area to the N-terminal strand swap (Figure 1A and 2A). This extension seems to act as a “strap” to further stabilize the closed state. Notably, the crystal structure of Grp94 was truncated to exclude this extension (Dollins et al., 2007).
To test the relevance of our observed strap, we truncated the extension to match yHsp90 (Δstrap), and saw a 4-fold increase in ATPase activity relative to WT (Figure 2). As a more conservative test we made point mutations aimed at disrupting the H87:E157 salt-bridge located at the extreme N-terminus of the strap (Figure 2A and 2B). Mutating either H87 or E157 to alanine in zTRAP1 resulted in a ~3-fold increase in ATPase activity over WT (Figure 2C). Intriguingly, the identity of equivalent residues in the cytosolic hHsp90α are swapped, preserving the salt-bridge. These data support a role for the strap in regulating the Hsp90 ATPase cycle and suggest that elaborations at the N-terminus may serve to further tune the ATPase cycle for the particular demands of each Hsp90 homolog.
Novel asymmetry in the closed state dimer
Unexpectedly, the two protomers from the TRAP1 homodimer have significantly different conformations resulting in a distinct closed state. Although the differences are most pronounced at the domain interfaces, asymmetry pervades the entire structure (Figure 3). To accomplish the significant global changes between TRAP1 protomers, hinge points between domains allow for flexibility and movement. Figure 3A illustrates the differences in domain orientation within the TRAP1 dimer compared to yHsp90 (Ali et al., 2006). Alignments between each individual domain illustrates the large changes in domain orientation relative to the overall dimer arrangement, and highlights the pronounced rotation at a hinge point between the large-MD (LMD) and small-MD (SMD) between, h14 and h15 (Figure 1B) near residue 471. To compensate, the NTD and CTD counter-rotate to maintain functional NTD and CTD interfaces (Figure 3A); likely explaining the difficulty in using molecular replacement to phase our diffraction data. The separation of the MD into LMD/SMD sub-domains is consistent with previous studies predicting nucleotide dependent flexing (Morra et al., 2012). Interestingly, the identified break point is where Hsp90 and other GHKL family members significantly diverge, suggesting unique functions of the SMD and CTD in Hsp90 systems (Meyer et al., 2003).
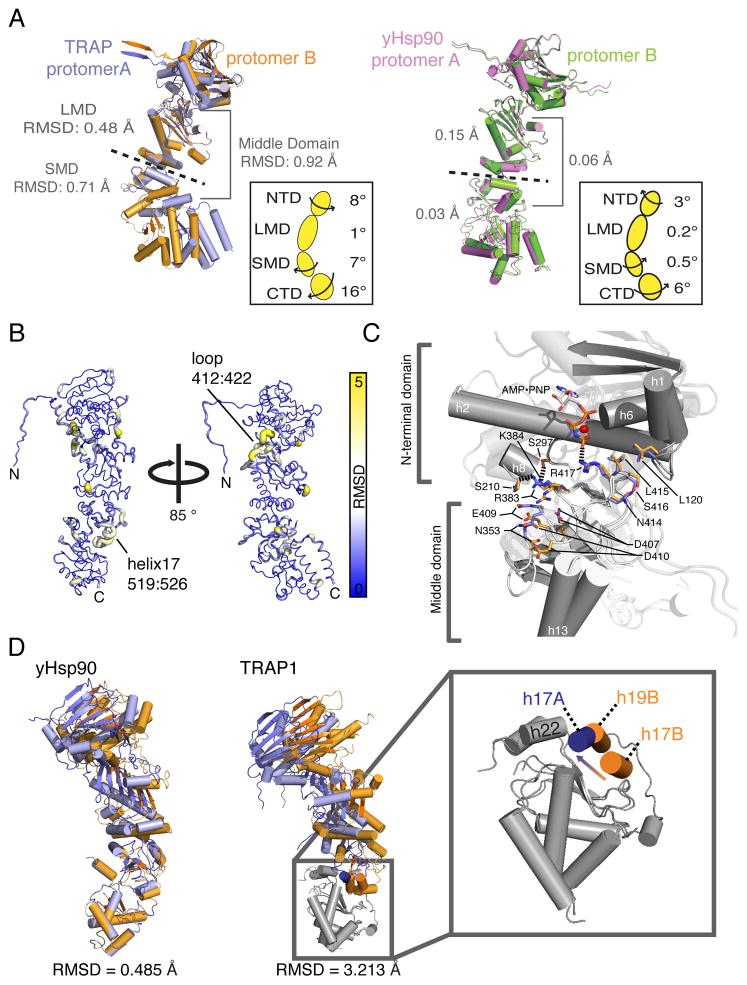
Figure 3. Novel asymmetry revealed in the TRAP1 dimer.
A) Protomers from TRAP1 and yHsp90 full-length structures are aligned at the LMD, highlighting asymmetry, between TRAP1 domains. RMSD values highlight differences between the LMD and SMD versus the entire MD; the direction and degree of rotation calculated between subdomains is indicated in the inset. B) Domain differences between Trap1 protomers are illustrated with a thicker diameter and the color yellow highlighting regions of higher variability (higher RMSD). C) View of the NTD:MD interface with protomers aligned at the NTD illustrates that asymmetry starts near R417. D) Global alignment yHsp90 and TRAP1 protomers shows significant differences in overall RMSD values. The zoomed panel shows helix swapping at the unique interface formed at the MD:CTD interface. (see also Movie S2)
By aligning domains individually and mapping inter-protomer RMSDs back to the structure of protomer B, it is evident that the individual NTDs and CTDs are similar and the largest deviations occur at the NTD:MD interface and the MD:CTD interface (Figure 3B). At the NTD:MD interface, the loss of hydrogen bonds between D407:K384 and E409:R383 in protomer B lead to a shift of 4.4 Å and a deviation from symmetry (Figure 3C). Notably, this shift occurs in a loop within the LMD (411–422), which contains residues necessary for stabilizing an active closed state (Ali et al., 2006). Here protomers A and B display significantly different B-factors (~40 and 60, respectively) suggesting differences in energetics of the ATP γ-phosphate coordination by R417 within the LMD.
The largest deviations occur at the MD:CTD interface where the CTD amphipathic helix (h22) interacts with either h17 of protomer A (h17A) or h19 of protomer B (h19B) (Figure 3D). The different contacts with h22 alter which face of h22 is exposed to the dimer cleft by inducing an 8 Å translation and 87° rotation towards the MD when exchanging from h17A to h19B (Movie S2). Most importantly, this motion significantly changes the spatial arrangement of residues directly implicated in client binding in bHsp90 and yHsp90 (Figure 1A, Movie S2) (Genest et al., 2013), also highlighted in the morph between yHsp90 and TRAP1 (Movie S1). These conformational changes suggest a potential use of asymmetry to influence client interactions.
Solution based measurements support an asymmetric closed state
To confirm the observed asymmetry we sought methods that could detect an asymmetric closed conformation in solution. SAXS has a proven ability to determine novel solution conformations for Hsp90s and to allow for deconvolution of conformational states in a mixed population (Krukenberg et al., 2008). To confirm that the observed differences could be distinguished, we overlaid theoretical P(r) curves calculated for the asymmetric TRAP1 structure, the yHsp90 crystal structure, a model of TRAP1 in the yHsp90 conformation, and two symmetric models based on our asymmetric structure (homodimers of protomers A or B; Figure S1A).
As expected, SAXS shows a compaction from a broad apo P(r) distribution upon the addition of AMPPNP or ADP-BeF3− (Figure 4A). At this resolution the structures are nearly identical across species and ATP analogs, suggesting a well-conserved closed state. To quantitatively compare the calculated P(r) curves from our models with those obtained from experimental data, the P(r) from corresponding apo data was used in a linear combination with each closed state model for fitting (Methods). Fit quality was gauged using a residual R, analogous to the crystallographic R-factor. Qualitatively and quantitatively, the asymmetric conformation is a much better fit to the experimental data of TRAP1 in solution (Figure 4B, Figure S1B and Table S1). Fits of the experimental data using symmetric models (homodimers of protomer A and protomer B) results in inferior R-values (Table S1), supporting the existence of closed asymmetric homodimers in solution.
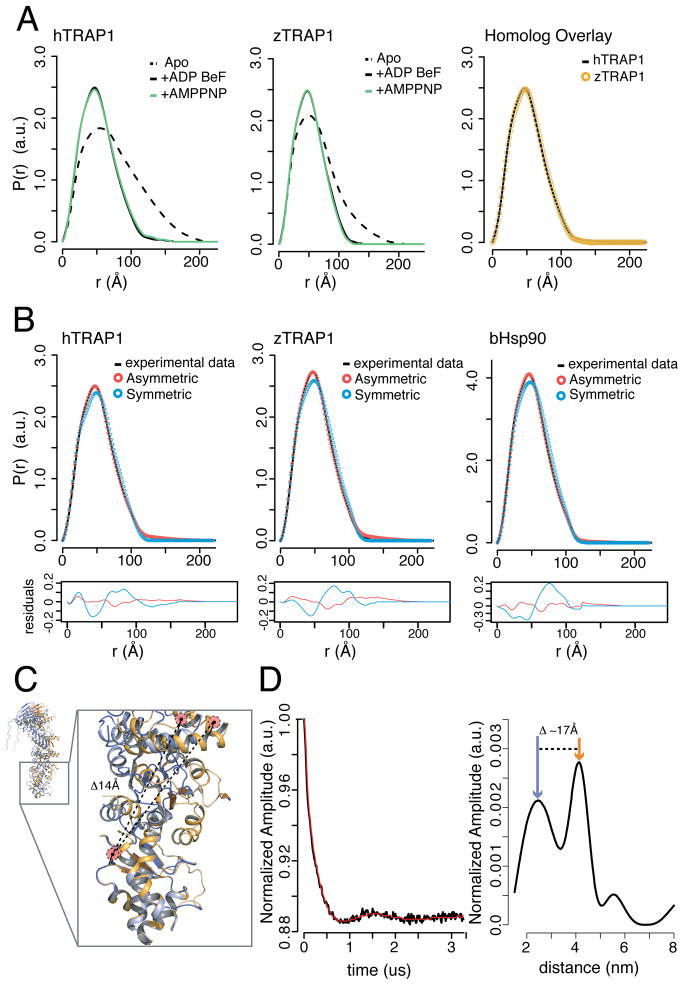
Figure 4. Solution methods support a conserved asymmetric closed state.
A) SAXS P(r) curves show that addition of ATP analogs to TRAP1 homologs results in a characteristic shift towards a more compact conformation. Overlaying the nucleotide bound SAXS curves reveals little difference in the closed conformation between homologs. B) SAXS curves of closed state hTRAP1, zTRAP1 from panel A, and bHsp90 from Krukenberg et al. fit using a linear combination of apo data and theoretical scattering for the zTRAP1 crystal structure (asymmetric), or a closed state model of zTRAP1 in the yHsp90 conformation (symmetric). Residuals below clearly show that asymmetric structure is best fit. (see also Table S1, Figure S1) C) DEER probe design shown on protomers A and B aligned at the CTD with a predicted distance change of 14 Å. D) Background corrected and normalized time-domain DEER data (black) fit by Tikhonov regularization (red) are shown on the left. Calculated distance distributions for closed state hTRAP1 (right) obtained after Tikhonov regularization. Two major peaks are observed in the presence of ADP-BeF supporting an asymmetric closed state in solution. Colored arrows show calculated distances from protomer A (blue) or B (orange). (see also Figure S2)
DEER was used as a complementary measure of asymmetry in solution, which is capable of measuring distances between two paramagnetic centers at a range of 18–60 Å to produce a distance distribution that resolves simultaneous states within the sample (Jeschke, 2012). For our experiment, we chose a pair spanning the MD:CTD interface within one protomer and having a predicted distance change of ~14 Å (Figure 4C, Methods). Native cysteine residues were removed to facilitate site-specific labeling, which did not negatively affect ATPase activity and displayed a closed state distribution by SAXS indistinguishable from WT (Figure S2). After spin-labeling, heterodimers were formed by incubating with 10-fold excess cysteine-free TRAP1. Closure was induced with ADP-BeF3−. The resulting DEER distribution clearly shows two distinct peaks confirming two conformations in the closed state (Figure 4D).
To assess the generality of the TRAP1 asymmetric state we evaluated solution scattering for the closely related bHsp90. Using previously reported closed state SAXS data for bHsp90 (Krukenberg et al., 2008) we find that our asymmetric conformation is a much better fit than the previous symmetric closed state model (Figure 4B). This analysis extends our observation of an asymmetric closed state to other Hsp90 systems. Taken together, this data supports an asymmetric closed conformation that is evolutionarily conserved between bacteria and humans.
Structure-based MD:CTD interface mutations severely impair activity
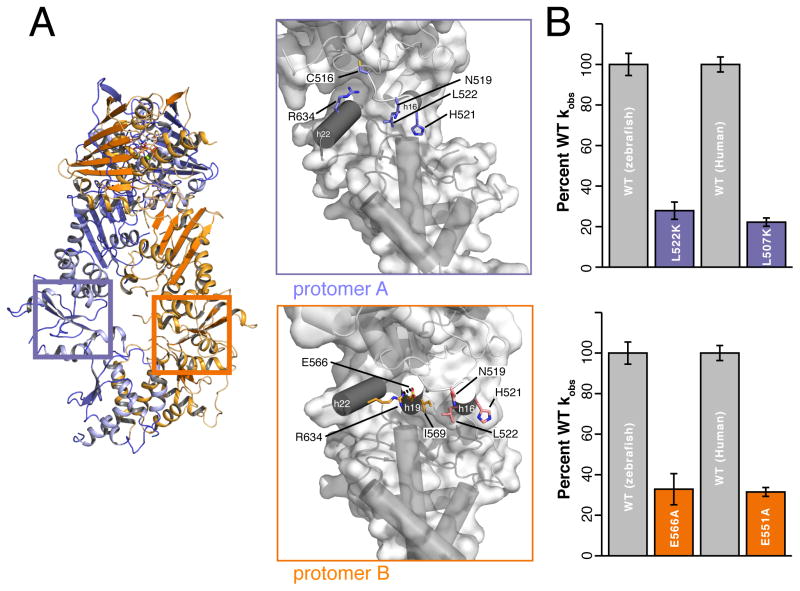
Having validated our crystallographic observations, we aimed to test the functional relevance of conformational asymmetry that either exposes or buries distinct surfaces at the MD:CTD interfaces of chemically identical protomers. These interfaces vary significantly in buried surface area (A:410 Å2, B:1057 Å2) and bonding networks. Notably, MD:CTD interface residues N519, H521, and L522 from h17 are buried, making hydrophobic and polar interactions in protomer A that are fully exposed in protomer B (calculated using PISA) (Krissinel and Henrick, 2007). Conversely, residues E566 and I569 make interactions in protomer B and are exposed in protomer A (Figure 5A). To test the functionality of each MD:CTD interface, we mutated these residues and confirmed structural integrity with circular dichroism spectroscopy of WT and mutant proteins (Figure S3A and S3B). Electron microscopy confirmed that the mutants could reach a closed state (Figure S3C). To assess function, ATPase activities were measured using an enzyme-coupled assay (Leskovar et al., 2008). The protomer A triple mutant (N519A.H521A.L522K) and protomer B double mutant (E566A.I569K) show a ~80% and 75% decrease in respective activity. Single mutations (protomer A:L522K and protomer B:E566A) were additionally tested and demonstrate similarly decreased activity (Figure 5B and Table S2).
Figure 5. Structure based MD:CTD interface mutations impair ATPase activity.
A) Crystal structure of TRAP1 rotated 75° from view in Figure 1A. Highlighted regions are the distinct MD:CTD interfaces generated by the helix swap of protomers A and B highlighted in Figure 3. B) Relative kobs of zTRAP1 and hTRAP1 with single point mutations designed to disrupt unique contacts at the MD:CTD interfaces. The equivalent drop in activity establishes conservation of asymmetric interfaces between homologs. (Error bars are propagated standard deviations).
Having confirmed the relevance of our asymmetric MD:CTD interfaces from the zTRAP1 structure, we wanted to extend these observations to hTRAP1. Mapping single point mutants from zTRAP1 to hTRAP1 (L507K and E551A), we observed an equivalent drop in ATPase activity (Figure 5B and Table S2) demonstrating conservation of both MD:CTD interfaces across homologs. Importantly, we find that the severity of mutations decrease with less disruptive substitutions (L507S) as well as when moving away from the interface (V555S) (Table S2). Not all mutations in the MD:CTD region are deleterious: C501A and C527A actually increase the ATPase rate by ~2-fold (Figure S2B and Table S2). The mutant data supports our structural observations and demonstrates that both observed MD:CTD interfaces are functional for ATPase activity.
While the mutant homodimers can form a closed state, albeit with greater difficulty, (Figure S4B), they are severely impaired in their ATPase activity. To test whether simply forming an asymmetric dimer can restore the activity we mixed the MD:CTD interface mutants at different ratios (1:3, 1:1 and 3:1) to form heterodimers (Figure S3D). Heterodimer formation failed to restore ATPase activity suggesting that each dimer must be able to access both conformations in order to efficiently hydrolyze ATP.
To probe the function of the MD:CTD interfaces for client interactions, we tested the ability of WT and MD:CTD interface mutations to interact with a previously reported model substrate (Δ131Δ) (Street et al., 2011). Consistent with these studies, the addition of WT Δ131Δ accelerates the ATPase of WT hTRAP1, however no acceleration was observed for the mutants (Figure S5A). The lack of mutant acceleration for both mutants is consistent with the requirement to access both conformations for proper progression of the ATP cycle. To measure binding we used D19C Δ131Δ (Gillespie and Shortle, 1997) labeled with IAEDANS and measured binding by fluorescence polarization anisotropy (Methods). Pre-closed WT hTRAP1 bound with a Kd of ~9 μM, while the mutant designed to disrupt the interface observed in protomer A (L507K) somewhat compromised binding (Kd ~35 μM) whereas no binding could be detected above background for the protomer B disrupting mutation (E551A) (Figure S5B). These results demonstrate that conformational asymmetry is connected to functional asymmetry for client binding, and supports a preferential interaction of Δ131Δ with the B interface in which the binding residues are in closer juxtaposition (Figure 1, Movie S2).
Together these data establish the conservation of structural, and functional importance of two distinct protomer conformations within a closed state.
Loss of CTD restores homodimer symmetry
Considering that the protomer arms are chemically identical, we questioned how the asymmetric conformation is formed. Fortuitously, crystal trials with full-length zTRAP1 mutated at the MD:CTD interface and in the presence of AMPPNP resulted in a distinct crystal form with higher symmetry and smaller unit cell (Table 1). The structure was solved by molecular replacement and refined to a resolution of 1.75 Å. The asymmetric unit contained a single protomer ending at D568, and lacking a CTD. Here, NTD dimerization is preserved by the perfect two-fold symmetry axis in the crystal lattice (Figure 6A). Given that the full-length protein was used to produce crystals, we reasoned the protein was locked in a closed and NTD-dimerized state via the straps, proteolyzed within the drop, and subsequently crystallized. This was confirmed by dissolving the crystals used to generate diffraction data (Figure S6).
Figure 6. NTD:MD crystal structure and implications for a strained closed state.
A) Crystal structure of a symmetric closed NTD:MD: AMPPNP TRAP1 dimer formed by CTD cleavage during crystallization that retains NTD dimerization. Cleavage of the CTD resulted from destabilization of the MD:CTD interface by point mutations. B) Overlay of the NTD:MD state with the full-length TRAP1 crystal structure shows that in absence of strain imposed by simultaneous NTD/CTD dimerization, TRAP1 protomers relax outward to a symmetric conformation. (see also Figure S6/S7, Movie S3).
The NTD:NTD interface in this structure is very similar to that in full-length TRAP1, and the lid remains disordered between residues 203–205 despite the lower overall B-factors and higher resolution. Importantly, by removing constraints resulting from CTD dimerization, the TRAP1 NTD:MD dimer is able to adopt a symmetric configuration and rotates outward by 10.5° pivoting from the NTD:MD interface. This movement results in a maximum change of ~10 Å measured at the SMD (Figure 6B, S7, Movie S3). This dimeric NTD:MD structure suggests that the simultaneous dimerization of NTDs and CTDs in the full length protein results in a highly strained state, which is at least partially relieved by forming the asymmetric conformation. Proteolysis and removal of the CTDs allows for relaxation to a more open, symmetric state.
Discussion
Here we present the first structure of the mitochondrial homolog of Hsp90, TRAP1. Perturbation of TRAP1 activity is linked to a number of human diseases including several cancers and neurodegeneration (Butler et al., 2012; Costa et al., 2013; Kang, 2012; Zhang et al., 2013). Though much work remains to identify particularly critical TRAP1 clients and signaling pathways, the data presented here provides a structural framework for further studies and design of TRAP1 specific therapeutic agents. Indeed, intervention with TRAP1 targeted inhibitors has already shown differential efficacy in diverse cancer models (Kang, 2012).
While it is well appreciated that Hsp90s can dynamically sample a broad range of conformations, our full-length X-ray crystal structures bound to ATP analogs show an unexpected and previously unknown asymmetric conformation. Significant asymmetry is rarely observed in homo-oligomers (Swapna et al., 2012) but notably has been reported to be a mechanistic feature of related GHKL family members (Schoeffler and Berger, 2005). Through the use of SAXS and DEER we confirm the existence of asymmetry in solution and show that it is conserved from bHsp90 to hTRAP1, suggesting that the ability to adopt unique conformations between protomers occurred early in Hsp90 evolution and persisted presumably due to functional pressure in vivo. In support of this structure-based point mutations demonstrate a functional role for asymmetry in vitro.
Our dimeric NTD:MD structure suggests that simultaneous dimerization of NTD and CTD in the ATP bound closed state results in a strained state, which is relieved by buckling at the MD:CTD interface to form the asymmetric conformation. Proteolysis and removal of the CTDs allows for relaxation to a more open, symmetric state. In support of strain being a conserved property of the closed state, single-molecule FRET experiments with yHsp90 show that binding of nucleotide in the closed state can lead to transient opening of the CTD dimer interface (Ratzke et al., 2010). As discussed below, we suggest that strain driven by ATP-induced closure and the consequent asymmetry gives rise to two conformational states having distinct functions in client maturation.
Asymmetry in the Hsp90 mechanism
Recent investigations have begun to reveal asymmetric themes in Hsp90 function. The co-chaperone Aha1 stimulates the ATPase activity of Hsp90 by binding asymmetrically at the NTD:MD interface (Retzlaff et al., 2010), while binding a single Hop co-chaperone is sufficient to block ATPase activity of both protomers (Li et al., 2011). Asymmetry in the Hsp90:Hop client loading complex has been directly observed by single particle EM (Southworth and Agard, 2011). Additionally, many client proteins have been shown to bind with a 1:1 stoichiometry (Motojima-Miyazaki et al., 2010; Street et al., 2011) and wherever examined at higher resolution, structural and mutational data directly support asymmetric binding (Street et al., 2012; Vaughan et al., 2006). Together these studies point to the Hsp90 chaperone functioning asymmetrically in concert with various co-chaperones and client proteins.
Previous models of the Hsp90 closed state
Prior to the TRAP1 structure presented here, the only available closed state structure was of yHsp90 in the presence of AMPPNP and the co-chaperone p23, required to obtain reasonable diffraction (Ali et al., 2006). Binding of p23 is important for steroid hormone receptor maturation (Hutchison et al., 1995) and has been shown to inhibit Hsp90 ATPase activity (McLaughlin et al., 2006). In the yHsp90 structure, NTDs are dimerized with both nucleotide pockets occupied by AMPPNP and two p23 monomers bound on either side of the NTD interface. Notably, if yHsp90 were to assume the TRAP1 asymmetric conformation, only one p23 could bind, as there would be a steric clash with the second. We suggest that binding of two p23 co-chaperones could be responsible for promoting the yHsp90 symmetric conformation. In support of this, NMR data has shown that binding of two p23s on hHsp90 can induce changes in the MD significantly removed from where p23 makes direct contacts (Karagoz et al., 2011). Such changes would be expected if p23 binding were to induce a rotation in the MD towards a more symmetric state.
Extension of asymmetry to other Hsp90 systems
The broader relevance of this new closed state conformation to eukaryotic cytosolic Hsp90s is also suggested by yHsp90 data that seem better explained by the asymmetric model presented here. Previous kinetic studies have shown that T22I/F (yHsp90) mutations enhanced ATP hydrolysis while seeming sterically incompatible with the yeast closed state model (Cunningham et al., 2008; Prodromou et al., 2000). By contrast, these substitutions are accommodated in our TRAP1 structure suggesting that, in the absence of p23, yHsp90 may also adopt a similar conformation during hydrolysis.
Partially buried in the yHsp90 structure, phosphorylation of S485 of yHsp90 (S526, zTRAP1) leads to a decrease in function both in vitro and in vivo (Soroka et al., 2012). Phosphomimetic mutations at this position showed profound functional defects, and substitution of alanine also compromised function. This serine is conserved among Hsp90s and fully exposed in protomer B of our TRAP1 structure. By contrast, phosphorylation of this residue in protomer A would disrupt the MD:CTD interface as it is fully buried. The degree of conservation and lack of modifying enzymes in lower species indicates this position is structurally important and perhaps was later utilized as a point of regulation. Preliminary investigations directly probing asymmetry in yHsp90 have been ambiguous thus far and thus remain a future direction.
A new model for the Hsp90 conformational cycle and utilization of ATP
While simple anti-aggregation functionality for Hsp90s is independent of nucleotide in vitro (Jakob et al., 1995), in vivo data clearly demonstrates the requirement for both ATP binding and hydrolysis in client maturation (Panaretou et al., 1998). However, it remains unclear how any Hsp90 homolog utilizes the available energy from two ATPs for client maturation.
Emerging evidence from a combination of NMR, genetic screens and mutagenesis suggests that initial client binding involves selection of a preferred conformation from an ensemble of open apo states (Genest et al., 2013; Street et al., 2012). Particularly well characterized is the binding site highlighted in Movies S1/S2 composed of variably exposed hydrophobic residues from both the SMD and CTD. An attractive hypothesis is that client maturation is driven by rearrangements of these binding site residues that occur upon transition to the closed state, the rate-limiting step to hydrolysis (Hessling et al., 2009; Street et al., 2011). ATP hydrolysis would then lead transiently through the compact ADP state in which these residues are buried within Hsp90, leading to client release. The challenge then is to understand the role of ATP in this process as affinities for Hsp90 clients are in the 1–100 μM range. Given the slow closure kinetics of Hsp90, the client would be predicted to fall off more than once during dimer closure. Therefore it is hard to understand how 7–14 kcals of energy available from hydrolysis of ATPs would benefit a release process.
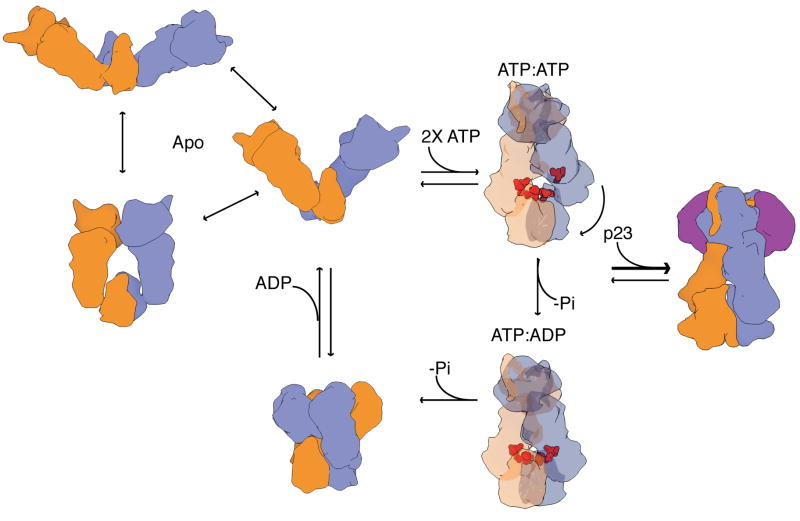
As an alternative, we propose an updated model for the chaperone cycle of Hsp90 that integrates a broad array of available information and incorporates the novel asymmetry reported here into the ATP-driven cycle of client binding, maturation and release (Figure 7). Newly added are two closed states that sequentially exploit differential ATP hydrolysis by the two protomers. Upon ATP binding by both protomers the population shifts to a closed high-energy asymmetric state as revealed in our full-length TRAP1 structure. Unique to this model is the suggestion that hydrolysis of the first ATP leads to a change in symmetry; perhaps a transition to a symmetric closed state like the yHsp90 crystal structure. As a consequence, rearrangement of the client binding site residues is directly coupled to ATP hydrolysis and would then be coupled to structural changes in the client. As our model implies, hydrolysis of the second ATP would cause the chaperone to transiently populate a compact ADP state, releasing clients, before releasing ADP and returning to the apo state.
Figure 7. New model for the conformational cycle of Hsp90.
In the absence of nucleotide, Hsp90 exists in an equilibrium of states with varying open conformations. Upon ATP binding the chaperone shifts to an asymmetric closed conformation that is significantly strained leading to buckling of the MD:CTD interface (client binding residues in red, transparency for visualization). Upon hydrolysis of one ATP, strain is relieved and the MD:CTD interface is re-arranged perhaps forming a symmetric state reminiscent of the yHsp90 conformation. This conformation can be stabilized by dual binding of co-chaperone p23 (purple) at the NTD stalling the progression of the cycle. Upon hydrolysis of the second ATP, the ADP state is transiently formed and ADP release resets the cycle to the apo state equilibrium. (Protomer arms are colored as in Figure 1).
While we tentatively favor a model where the first ATP hydrolysis leads to a closed state reminiscent of the previous yHsp90 structure, it is also possible that the protomer arms could flip-flop between conformations, or that both would assume the conformation of either protomer A or protomer B. Importantly, for each of these possibilities the MD:CTD interface must re-arrange to progress through the cycle, preserving the possibility of utilizing ATP hydrolysis for doing work on a client bound to this region.
With the introduction of inherent structural asymmetry to the Hsp90 model comes a combinatorial expansion of conformations possible for interaction with clients and co-chaperones. Modulating the asymmetry or the rate of forming the asymmetric state thus provides a convenient mode of regulation for overall Hsp90 function in various pathways. In support of this, several post-translational modifications that dramatically perturb Hsp90 function are located in or near the MD:CTD interface including the previously discussed S485 in yHsp90 (Martinez-Ruiz et al., 2005; Soroka et al., 2012).
Despite significant sequence variation there is strong structural conservation of Hsp90 from bacteria to humans. Thus, the ability of Hsp90 to sample an asymmetric conformation in a closed state is likely an intrinsic feature of several and potentially all Hsp90 homologs. Though future experiments are required to precisely define the progression of conformational states and how the structural asymmetry shown here is utilized for diverse Hsp90 functions, our findings provide a new hypothesis for how ATP hydrolysis could be directly coupled to client remodeling.
Experimental Procedures
See Extended Experimental Procedures for further details.
Protein Production and Purification
H. sapiens and D. rerio TRAP1 were cloned into the pET151/D-TOPO expression plasmid as TEV cleavable N-terminal 6x-His-tagged fusion proteins. Expression was carried out in E. coli BL21(DE3)-RIL. Cells were grown at 30°C in LB with 0.4% glucose to OD600=0.8 and induced with 0.4 mM IPTG at 16 °C for 18 hours. TRAP1 proteins were purified by Ni-affinity chromatography, anion exchange and size-exclusion chromatography.
Crystallization, structure determination
Crystals were grown in 18% (w/v) PEG3350, 0.2 M sodium malonate pH 6.6–7, 20–42 mM hexamine cobalt mixed 1:1 with TRAP1 protein at 7–10 mg/mL. The initial NTD:MD TRAP1 crystallization hit was refined to a final condition of 0.1M Sodium Phosphate monobasic pH 6.5, and 12% PEG 8000. The initial full-length TRAP1 structures were determined with SAD phasing and the NTD:MD structure was solved by molecular replacement.
Steady-state Hydrolysis Measurements
Steady-state hydrolysis rates were measured using the previously described ATP enzyme-coupled assay (Leskovar et al., 2008). Assay components were mixed to a final concentration of 200 μM NADH, 400 μM PEP, 50 U/mL PK, 50 U/mL LDH, and ATP at least 10-fold over the measured Km (Table S2). Homologs were inhibited with the Hsp90 specific inhibitors (Radicicol, data not shown).
SAXS Reaction, Data Collection and Analysis
WT TRAP1 homologs and mutant proteins were exchanged into 20 mM Hepes pH 7.5, 50 mM KCl, 2 mM MgCl2, 1mM DTT immediately prior to the experiment. Data was collected with 75 μM protein at the ALS SIBYLS beamline 12.3.1. Data was buffer subtracted and time points averaged using scripts at 12.3.1 (ogreNew) and in-house software. The scattering data was transformed to P(r) using GNOM. Further methods were used to quantitatively compare experimental SAXS data with theoretical data.
DEER Spectroscopy
Cysteine-free hTRAP1 allowed for site specific labeling with maleimide functionalized MSL and purified as above. Post labeling, heterodimers were formed with 10:1 (Cysteine-free WT:probe) and 2 mM ADP-BeF was added to induce closure as with SAXS measurements. Four-pulse DEER data were collected using a Bruker ELEXSYS E580 spectrometer at 34 GHz equipped with a SuperQ-FT pulse Q-band system with a 10 W amplifier and a 5 mm EN5107D2 resonator located at the Ohio Advanced EPR laboratory.
Supplementary Material
Highlights.
Crystal structure of the TRAP1 homodimer reveals an asymmetric closed state
SAXS and DEER validate asymmetry in solution and conservation across Hsp90 homologs
MD:CTD interfaces are functional for chaperone activity and stabilize asymmetry
Changes in asymmetry result in re-arrangement of client binding residues
Acknowledgments
We thank J. Holton, G. Meigs and ALS staff of beamline 8.3.1 for data collection and helpful discussions. We thank G. Hura, K. Dyer, J. Tanamachi and staff of beamline 12.3.1 for SAXS data collection and helpful discussions. We thank L. Peng of the M. Jacobson lab for building symmetric models of our TRAP1 structure and N. Naber of the R. Cooke lab for assistance with labeling and CW EPR. Special thanks to K. Krukenberg for bHtpG SAXS data and helpful discussions. Finally we thank members of the Agard lab for helpful discussions. Support for this work was provided by PSI-Biology grants: U01 GM098254 (DAA) and U54-GM094597 (MAK), the Larry L. Hillblom Center for the Biology of Aging (LAL), and HHMI.
Footnotes
Accession Numbers
Atomic coordinates have been deposited in the Protein Data Bank (PDB) under the accession codes: 4IVG, 4IPE, 4IYN, and 4J0B.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri DC, Stein GS, Lian JB, Languino LR. TRAP-1, the mitochondrial Hsp90. Biochim Biophys Acta. 2012;1823:767–773. doi: 10.1016/j.bbamcr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EK, Voigt A, Lutz AK, Toegel JP, Gerhardt E, Karsten P, Falkenburger B, Reinartz A, Winklhofer KF, Schulz JB. The Mitochondrial Chaperone Protein TRAP1 Mitigates alpha-Synuclein Toxicity. PLoS genetics. 2012;8:e1002488. doi: 10.1371/journal.pgen.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cechetto JD, Gupta RS. Immunoelectron microscopy provides evidence that tumor necrosis factor receptor-associated protein 1 (TRAP-1) is a mitochondrial protein which also localizes at specific extramitochondrial sites. Exp Cell Res. 2000;260:30–39. doi: 10.1006/excr.2000.4983. [DOI] [PubMed] [Google Scholar]
- Chen B, Zhong D, Monteiro A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics. 2006;7:156. doi: 10.1186/1471-2164-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa AC, Loh SH, Martins LM. Drosophila Trap1 protects against mitochondrial dysfunction in a PINK1/parkin model of Parkinson’s disease. Cell Death Dis. 2013;4:e467. doi: 10.1038/cddis.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CN, Krukenberg KA, Agard DA. Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J Biol Chem. 2008;283:21170–21178. doi: 10.1074/jbc.M800046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria PC, Bernthaler A, Dupuis P, Mayer B, Picard D. An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One. 2011;6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts SJ, Owen BA, Nguyen P, Trepel J, Donner DB, Toft DO. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J Biol Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, Masison DC, Wickner S. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol Cell. 2013;49:464–473. doi: 10.1016/j.molcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels. J Mol Biol. 1997;268:158–169. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- Hutchison KA, Stancato LF, Owens-Grillo JK, Johnson JL, Krishna P, Toft DO, Pratt WB. The 23-kDa acidic protein in reticulocyte lysate is the weakly bound component of the hsp foldosome that is required for assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. The Journal of biological chemistry. 1995;270:18841–18847. doi: 10.1074/jbc.270.32.18841. [DOI] [PubMed] [Google Scholar]
- Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim Biophys Acta. 2012;1823:607–613. doi: 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Kang BH. TRAP1 regulation of mitochondrial life or death decision in cancer cells and mitochondria-targeted TRAP1 inhibitors. BMB Rep. 2012;45:1–6. doi: 10.5483/bmbrep.2012.45.1.1. [DOI] [PubMed] [Google Scholar]
- Kang BH, Plescia J, Dohi T, Rosa J, Doxsey SJ, Altieri DC. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131:257–270. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Karagoz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rudiger SG. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci U S A. 2011;108:580–585. doi: 10.1073/pnas.1011867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Southworth DR, Street TO, Agard DA. pH-dependent conformational changes in bacterial Hsp90 reveal a Grp94-like conformation at pH 6 that is highly active in suppression of citrate synthase aggregation. J Mol Biol. 2009;390:278–291. doi: 10.1016/j.jmb.2009.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Quarterly reviews of biophysics. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskovar A, Wegele H, Werbeck ND, Buchner J, Reinstein J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J Biol Chem. 2008;283:11677–11688. doi: 10.1074/jbc.M709516200. [DOI] [PubMed] [Google Scholar]
- Li J, Richter K, Buchner J. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat Struct Mol Biol. 2011;18:61–66. doi: 10.1038/nsmb.1965. [DOI] [PubMed] [Google Scholar]
- Li J, Sun L, Xu C, Yu F, Zhou H, Zhao Y, Zhang J, Cai J, Mao C, Tang L, et al. Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90. Acta Biochim Biophys Sin (Shanghai) 2012;44:300–306. doi: 10.1093/abbs/gms001. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Protein folding sculpting evolutionary change. Cold Spring Harb Symp Quant Biol. 2009;74:103–108. doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y, Wu S, Hao B, Bona R, Han D, et al. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 2010;1:79. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Sun W, Taldone T, Rodina A, Chiosis G. Heat shock protein 90 in neurodegenerative diseases. Mol Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SH, Sobott F, Yao ZP, Zhang W, Nielsen PR, Grossmann JG, Laue ED, Robinson CV, Jackson SE. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Morra G, Potestio R, Micheletti C, Colombo G. Corresponding functional dynamics across the Hsp90 Chaperone family: insights from a multiscale analysis of MD simulations. PLoS Comput Biol. 2012;8:e1002433. doi: 10.1371/journal.pcbi.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem Biophys Res Commun. 2010;400:241–245. doi: 10.1016/j.bbrc.2010.08.047. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. Embo J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. Embo J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzke C, Mickler M, Hellenkamp B, Buchner J, Hugel T. Dynamics of heat shock protein 90 C-terminal dimerization is an important part of its conformational cycle. Proc Natl Acad Sci U S A. 2010;107:16101–16106. doi: 10.1073/pnas.1000916107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, Richter K, Buchner J. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Richter K, Moser S, Hagn F, Friedrich R, Hainzl O, Heller M, Schlee S, Kessler H, Reinstein J, Buchner J. Intrinsic inhibition of the Hsp90 ATPase activity. J Biol Chem. 2006;281:11301–11311. doi: 10.1074/jbc.M510142200. [DOI] [PubMed] [Google Scholar]
- Richter K, Reinstein J, Buchner J. N-terminal residues regulate the catalytic efficiency of the Hsp90 ATPase cycle. J Biol Chem. 2002;277:44905–44910. doi: 10.1074/jbc.M208457200. [DOI] [PubMed] [Google Scholar]
- Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem Soc Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- Sciacovelli M, Guzzo G, Morello V, Frezza C, Zheng L, Nannini N, Calabrese F, Laudiero G, Esposito F, Landriscina M, et al. The Mitochondrial Chaperone TRAP1 Promotes Neoplastic Growth by Inhibiting Succinate Dehydrogenase. Cell Metab. 2013;17:988–999. doi: 10.1016/j.cmet.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V, Wiest R, Garcia-Cardena G, Cadelina G, Groszmann RJ, Sessa WC. Hsp90 regulation of endothelial nitric oxide synthase contributes to vascular control in portal hypertension. Am J Physiol. 1999;277:G463–468. doi: 10.1152/ajpgi.1999.277.2.G463. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Song HY, Dunbar JD, Zhang YX, Guo D, Donner DB. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J Biol Chem. 1995;270:3574–3581. [PubMed] [Google Scholar]
- Soroka J, Wandinger SK, Mausbacher N, Schreiber T, Richter K, Daub H, Buchner J. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth DR, Agard DA. Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol Cell. 2011;42:771–781. doi: 10.1016/j.molcel.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Molecular cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Verba KA, Lee CT, Mayer MP, Agard DA. Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. Journal of molecular biology. 2012;415:3–15. doi: 10.1016/j.jmb.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swapna LS, Srikeerthana K, Srinivasan N. Extent of structural asymmetry in homodimeric proteins: prevalence and relevance. PLoS One. 2012;7:e36688. doi: 10.1371/journal.pone.0036688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura H, Koyama Y, Matsuzaki S, Yamada K, Hattori T, Miyata S, Takemoto K, Tohyama M, Katayama T. TRAP1 controls mitochondrial fusion/fission balance through Drp1 and Mff expression. PLoS One. 2012;7:e51912. doi: 10.1371/journal.pone.0051912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Tsutsumi S, Muhlebach G, Sourbier C, Lee MJ, Lee S, Vartholomaiou E, Tatokoro M, Beebe K, Miyajima N, et al. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc Natl Acad Sci U S A. 2013;110:E1604–1612. doi: 10.1073/pnas.1220659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Karsten P, Hamm S, Pogson JH, Lutz AK, Exner N, Haass C, Whitworth A, Winklhofer K, Schulz JB, et al. TRAP1 rescues PINK1 loss-of-function phenotypes. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.