Abstract
Great interest persists in useful prognostic and therapeutic targets in glioblastoma (GBM). In this study, we report the definition of miR-148a as a novel prognostic oncomiR in GBM. miR-148a expression was elevated in human GBM specimens, cell lines and stem cells (GSCs) compared to normal human brain and astrocytes. High levels were a risk indicator for GBM patient survival. Functionally, miR-148a expression increased cell growth, survival, migration, and invasion in GBM cells and GSCs and promoted GSC neurosphere formation. Two direct targets of miR-148a were identified, the EGFR regulator MIG6 and the apoptosis regulator BIM, which rescue experiments showed were essential to mediate the oncogenic activity of miR-148a. By inhibiting MIG6 expression, miR-148a reduced EGFR trafficking to Rab7-expressing compartments which includes late endosomes and lysosomes. This process coincided with reduced degradation and elevated expression and activation of EGFR. Lastly, inhibition of miR-148a strongly suppressed GSC and GBM xenograft growth in vivo. Taken together, our findings provide a comprehensive analysis of the prognostic value and oncogenic function of miR-148a in GBM, and further defining it as a potential target for GBM therapy.
Keywords: microRNA-148, MIG6, BIM, EGFR, glioblastoma
INTRODUCTION
Glioblastoma (GBM) is an extremely aggressive tumor that accounts for the majority of deaths due to primary brain neoplasms (1). Despite the most advanced treatment with combinations of surgery, radiotherapy and chemotherapy, GBM is associated with a median survival of only 14 months (2). Factors responsible for GBM malignancy and poor prognosis include rapid cell growth, resistance against apoptosis, and distant invasion of the surrounding brain (1, 3).
Receptor tyrosine kinase (RTK) pathways are deregulated in the vast majority of GBMs (4, 5). Among RTKs, the epidermal growth factor receptor (EGFR) is the most commonly altered (6). It is mutated and/or amplified in 40% and overexpressed in > 60% of tumors (7, 8). Activation of EGFR induces tumor cell growth, migration, and invasion, as well as resistance to chemotherapy and radiation (6, 9). EGFR signaling and protein half-life are tightly regulated (10). Mitogen-inducible gene 6 (MIG6) regulates EGFR signaling and turnover by binding EGFR and directly inhibiting tyrosine kinase activity, increasing clathrin-dependent EGFR endocytosis and trafficking into the lysosome, and promoting EGFR degradation (11-13). Ablation of MIG6 induces tumor formation, supporting a tumor suppressor function of MIG6 (11, 14). The MIG6 gene is located on chromosome 1p36 which is subject to focal deletions in GBM. A Cancer Genome Atlas (TCGA) data analysis showed that 15 out of 430 GBM samples contain homozygous deletions in 1p36 (14) but that MIG6 expression is downregulated in ~50% of primary tumor samples and GBM cell lines (11). Therefore MIG6 deletions only account for a small fraction of the GBM tumors with reduced MIG6 expression.
Resistance to apoptosis is a big obstacle in GBM therapy (15, 16). Apoptosis in the intrinsic pathway is regulated by the balance between pro-apoptotic (Bax, Bak, BIM and Bad) and anti-apoptotic (Bcl-2 and Bcl-xL) members of the Bcl-2 family (17). Pro-apoptotic BIM (BCL2L11) is localized to the mitochondria where it initiates the mitochondrial cell death pathway by directly activating Bax/Bak-dependent apoptosis. BIM has been shown to be an important mediator of targeted therapy-induced apoptosis in solid tumors. BIM is downregulated in 29% of GBM cases based on TCGA analysis (18, 19). However, the causes of BIM downregulation in GBM are not known.
microRNAs (miRNAs) are short noncoding RNA molecules that regulate gene expression by binding to the 3’ untranslated region (3’UTR) of target mRNA and inducing mRNA degradation and/or inhibition of protein synthesis (20, 21). Deregulation of miRNA expression has been associated with cancer formation through alterations in either oncogenic or tumor suppressor gene targets (20, 22). A number of miRNAs are deregulated in GBM and play important roles in tumor formation and growth (23-31). However, a role for miR-148a in GBM has not been described before.
We analyzed miRNA expression in > 500 patient GBMs in the TCGA database and found that miR-148a is upregulated and predicted poor patient survival. We therefore embarked on a comprehensive study of miR-148a in GBM. Our data show for the first time that miR-148a is upregulated in GBM, where it exerts oncogenic effects in vitro and in vivo by regulating BIM, MIG6, and EGFR. MiR-148a is therefore a novel oncomiR and potential therapeutic target in GBM.
MATERIALS AND METHODS
Cells and tumor specimens
GBM cell lines U87, U373, A172, T98G, SNB-19 and U251 were from ATCC, who authenticates cell lines with short tandem repeat (STR) profiling. Cells lines that were used for more than six months after purchase were re-authenticated by STR profiling in 2013 by Laragen, Inc. GBM stem cells (GSCs) 1228, 0802 and 0308 (a kind gift from Dr. Jeongwu Lee, Cleveland Clinic) were isolated from patient surgical specimens and characterized for tumorigenesis, pluripotency, self-renewal, stem cell markers, and neurosphere formation (32). GBM surgical specimens (n=18) and normal brain (n=7) were obtained from patients undergoing surgery at the University of Virginia Hospital according to protocols approved by the internal Review Board.
TCGA data analysis
The collection of data from The Cancer Genome Atlas (TCGA) was compliant with all laws and regulations for the protection of human subjects, and necessary ethical approvals were obtained. Analysis of all data was done in the R project (33). For analysis of differential expression and determination of the effects of miR-148a on patient survival, Agilent 8x15k microRNA expression for 491 glioblastoma and 10 normal unmatched brain samples was downloaded along with clinical information from the TCGA database (Level 2 (normalized) data, November 2012). Cox regression analysis of all samples with miRNA and survival data (n=482) was performed to determine whether miR-148a levels were a risk indicator for survival. The expression of miR-148a was also compared in normal brain (n=10) to GBM (n=491) using the R-based Limma package (34).
Quantitative RT-PCR
miScript Primer Assay Hs-miR-148a was used for measuring miR-148a. Total RNA was extracted from GBM cell lines and GSCs. RNA samples were reverse-transcribed using the miScript Reverse Transcriptase kit (QIAGEN, Valencia, CA), and quantitative real-time PCR analysis was performed using the 7500 Real-time PCR System (Applied Biosystems, Carlsbad, CA). qRT-PCR was also used to assess the mRNA levels of MIG6 and BIM. The primer sequences were: MIG6-forward: 5′-GACAATTTGAGCAACTTGACTTGG-3′, MIG6-reverse: 5′-GGTTACTTAGTTGTTGCAGGTAAG-3; BIM-forward: 5′-TGGCAAAGCAACCTTCTGATG-3′ and BIM-reverse: 5′-GCAGGCTGCAATTGTCTACCT-3′. Human U6B and GAPDH primers (QIAGEN, Valencia, CA) were used as controls.
Cell transfections
GBM cells and GSCs were transfected with 20 nM pre-miR-148a, anti-miRNA-148a or control-miR (Ambion, Carlsbad, CA), using Oligofectamine or Lipofectamine RNAimax (Invitrogen, Grand Island, NY) according to the manufacturer’s instructions. Plasmid transfections were performed with Fugene 6 (Roche, Indianapolis, IN). miR-148a expression was verified by qRT-PCR 72 hrs and 7 days post-transfection.
Generation of anti-miR-148a stable expressing GBM cells
Lentiviruses encoding the pEZX-AM04 expression cassette containing a hygromycin resistance gene as well as the antisense sequence for miR-148a and the red fluorescent protein mCherry gene under the U6 promoter (pEZX-AM04; GeneCopoeia) (Fig. S2B, S2C) were generated with pPACKH1 Lentivector packaging Plasmid mix (System Biosciences, Mountain View, CA) and concentrated using PEG-it Virus Precipitation Solution (System Biosciences.). U87 cells were infected with the lentiviruses or control viruses lacking the anti-miR-148a sequence. After culturing in selection media, mCherry was detected by fluorescence microscopy. A stable infection efficiency of ~100% was attained (Fig. S2A).
Cell growth and apoptosis assays
For growth, GBM cells and GSCs were transfected with pre-miR-148a, anti-miR-148a, or control. Three days post-transfection, the cells were counted for 5 days with a hemocytometer. For apoptosis, cells were transfected as above and Annexin V-PE/7AAD flow cytometry was used to determine the dead and apoptotic cell fractions as previously described (35).
Cell migration and invasion assays
The effects of miR-148a expression on cell migration and invasion were assessed using the wound healing and trans-well assays as previously described (36).
Neurosphere formation assay
GSCs were grown in low EGF and FGF medium (20 ng/ml each) and transfected with either anti- or pre-miR-148a or controls for 72 h. The cells were dissociated into single cells in buffer (EDTA 1mM, BSA 0.5% in PBS) and 1000 single cells were incubated for 7 days. Secondary neurospheres containing more than 30 cells were counted.
In vivo tumor formation
Tumor xenografts were generated by implantation of 1228 GSCs transfected with anti-miR-148a and U87 cells engineered to stably express anti-miR-148a. 1228 (1 × 105 cells; n=6) and U87 cells (3 × 105 cells; n=10) were stereotactically implanted into the striata of immunodeficient mice. Four weeks after tumor implantation, the animals were subjected to brain magnetic resonance imaging (MRI). To measure tumor size, 30 μl of gadopentetate dimeglumine (Magnevist, Bayer Healthcare, NJ) was intraperitoneally injected 15 minutes prior to scanning and tumor volume was quantified as previously described (37, 38).
Immunoblotting
Immunoblotting was performed as previously described using antibodies for MIG6 (Santa Cruz Biotechnologies, Santa Cruz, CA), BIM, EGFR and p-EGFR (Cell Signaling, Danvers, MA). All blots were stripped and re-probed with β-actin or GAPDH (Santa Cruz, Dallas, Texas) as control. Blots in which differences were not obvious were quantified by densitometry on film as previously described (39).
Generation of MIG6 and BIM 3’UTR constructs
The MIG6 3′-UTR reporter plasmid was constructed via insertion of the MIG6 3’-UTR (2561 bp) downstream of the Renilla luciferase stop codon in the pMIR vector (Promega, Madison, WI) generating the pMIR-MIG63’UTR plasmid. For BIM a commercially available 3’-UTR reporter plasmid, pEZX-BIM3UTR-1, was used (Genecopoeia, Madison, WI). QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to generate mutations in the 3′ UTR of MIG6 and BIM by PCR using the pMIR-MIG6 3’UTR and pEZX-BIM 3’UTR as constructs templates. Primers containing the mutation TGCACTGA (1370-1377)→CCGGGCCG in the 3′ UTR of MIG6 gene and TGCACTG (1029-1035)→GCGCGCC 3’UTR of BIM were used.
3’UTR reporter assays
GBM cells were transfected with pre-miR-148a or pre-miR control for 6 hrs. For MIG6, the cells were then transfected with either the reporter vector with 3′UTR-MIG6 or with mutant-3’UTR, in addition to a control β-galactosidase reporter plasmid. For BIM, the cells were transfected with either 3’UTR BIM or BIM mutant-3’UTR. Luciferase assays were performed 48 hrs later using the Luciferase System Kit (Promega, Madison, WI) for MIG6 or the Dual Luciferase Assay (Promega, Madison, WI) for BIM, and luminescence was measured on a Promega GloMax 20/20 luminometer. Firefly luciferase activity was double normalized by dividing each well first by β-galactosidase activity and then by average luciferase/β-galactosidase value in a parallel set done with a constitutive luciferase plasmid.
Rescue experiments
To determine if MIG6 and BIM mediate the effects of miR-148a, rescue experiments were conducted in which the effects of anti-miR-148a were measured in the setting of inhibited MIG6 or BIM. Cells were either transfected with anti-miR-148a for 6hrs (1228) or U87 cells stably expressing anti-miR-148a were used. The cells were then transfected with siRNA against MIG6 (Thermo Fisher Scientific, Waltham, MA) or BIM (Cell Signaling, Danvers, MA) and cell growth and death were assessed as described above. MIG6, EGFR and BIM expression changes were verified by immunoblotting.
EGFR tracking assays
Cells were plated and transfected with either pre-miR-148a or pre-miR control for 24hrs followed by transfection with Rab7-mCherry for 24 hrs (kindly provided by Marc G. Coppolino, University of Guelph). Cells were serum starved overnight, followed by stimulation with 50 ng/mL EGF for 30 minutes. Samples were then washed, fixed, and permeabilized before immunostaining using primary antibodies (EGFR, Abcam, Cambridge, MA; MIG6, Santa Cruz, Dallas, Texas). Samples were imaged using a 63X (NA 1.4) lens on a Zeiss LSM 700 with 405, 488, 543, 633 nm lasers using ZEN software (Carl Zeiss, Oberkochen, Germany). Captured images were analyzed for colocalization using ImageJ software. Briefly, images were initially thresholded, and the Colocalization Finder tool was used to determine the area and intensity of colocalizing pixels of EGFR.
Statistics
All experiments were performed at least 3 times. Two group comparisons were analyzed with t-test and p values were calculated. For rescue experiments, the anti-miR-148a-induced change in the setting of inhibited target protein was compared with the anti-miR-148a-induced change in the control setting. For TCGA data, Cox regression analysis was performed to determine the correlation between miR-148a expression and patient survival. More detailed TCGA data statistical analyses are described in the corresponding sections. For all analyses, P < 0.05 was considered significant.
RESULTS
MiR-148a expression is upregulated in GBM cells, GSCs and human tumors and inversely correlates with patient survival
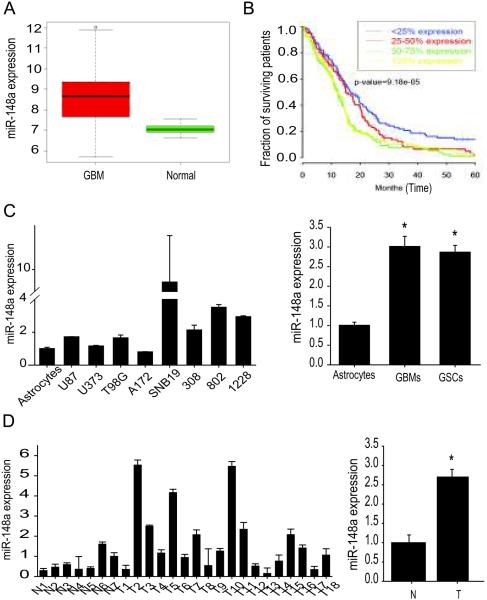
We analyzed TCGA data for miR-148a levels and for correlation with patient survival. The comparison of tumor (n=491) with normal tissue samples (n=10) showed a significant (59%) increase of miR-148a levels in the tumors as compared to normal brain (p=3×10-4) (Fig 1A). Cox regression analysis of 482 GBM samples in the TCGA dataset revealed that elevated miR-148a expression is a highly significant negative risk factor (p=9.9×10-6). The hazard ratio was 1.19 with confidence intervals 1.10-1.29. The Kaplan-Meier curve of the TCGA patient cohort is shown in Figure 1B. The lower quartile (with the lowest miR-148a expression) had longer overall survival than those with higher miR-148a expression. The median survivals of the different groups in the Kaplan Meier curve are <25% expression = 515, 25-50%= 463, 50-75%= 377, 75-100% = 382 (days). Log-rank analysis of 482 samples revealed that miR-148a was highly significant as a negative risk factor (p=9.18×10-5) (Fig. 1B). We also measured miR-148a levels in GBM cells (U87, U373, T98G, A172, and SNB19), GSCs (0308, 0822, and 1228) and human tumor specimens (n=18) as well as normal human astrocytes and normal brain (n=7). MiR-148a was significantly higher in GBM cells and GSCs than in astrocytes (p<0.05) (Fig. 1C) and significantly higher in tumors than in normal brain (p<0.05) (Fig. 1D). Altogether, these data demonstrate that miR-148a is upregulated in GBM and that high miR-148a expression predicts poor patient survival.
Figure 1. miR-148a is upregulated in GBM cells, GSCs and human tumors and inversely correlates with patient survival.
A) Analysis of TCGA microRNA expression data showing significantly higher expression of miR-148a in GBM tumors (n=491) than in normal brain (n=10). B) Correlation analysis of expression data and patient survival data (n=482) from TCGA showing that miR-148a levels are a risk indicator for survival. C) Quantification of miR-148a in glioblastoma (GBM) cell lines (U87, U373, T98G, A172, SNB19) and stem cell lines (GSCs) (0308, 0802, 1228) showing higher expression than in normal human astrocytes. Single cells are shown in the left panel and averages in the right panel. D) Quantification of miR-148a in human GBM tumors (T) (n=18) showing higher levels than in normal human brain (N) (n=7). Single tissues are shown in the left panel and averages in the right panel. *, p < 0.05
MiR-148a promotes GBM cell and GSC growth and survival
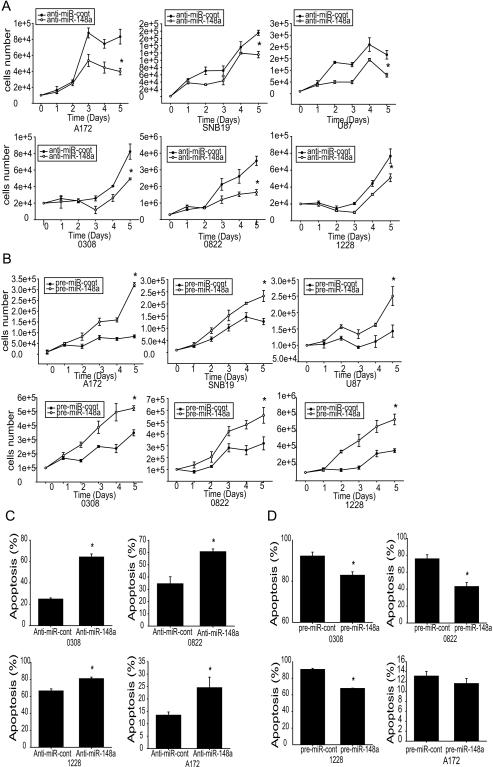
We next assessed the functional role of miR-148a in GBM (A172, SNB19, U87, and U373) and GSC (0308, 0822, and 1228) cells by determining the effects of miRNA over-expression and inhibition on cell growth and apoptosis using cell counting and Annexin V-7 AAD flow cytometry, respectively. miR-148a inhibition with antisense miRNA significantly decreased the growth rate (Fig. 2A) and overexpression of miR-148a resulted in a higher growth rate in GBM and GSC cells as compared to controls (p<0.05) (Fig. 2 B).Similarly, inhibition of miR-148a led to a significant induction of apoptosis (Fig. 2C), while overexpression of miR-148a led to a significant inhibition of apoptosis in GBM cells and GSCs (p<0.05) (Fig. 2D). MiR-148a levels were verified by qRT-PCR (Fig. S1). The above results show that miR-148a promotes cell growth and inhibits cell death in GBM.
Figure 2. miR-148a promotes GBM cell and GSC growth and survival.
GBM cell lines (A172, SNB19 and U87) and GSC (0308, 0822 and 1228) were transfected with anti-miR-148a (A), or pre-miR-148a (B) or controls. The cells were subsequently assessed for cell growth by cell counting. GBM cell line (A172) and GSCs (0308, 0822, 1228) were transfected with either anti-miR-148a (C) pre-miR-148a (D), or controls and subsequently assessed for cell death and apoptosis by AnnexinV-PE/7-AAD flow cytometry. The data show that miR-148a inhibition (A,C) inhibits and miR-148a overexpression (B,D) promotes cell growth and survival. *, p < 0.05
MiR-148a promotes GBM cell migration and invasion
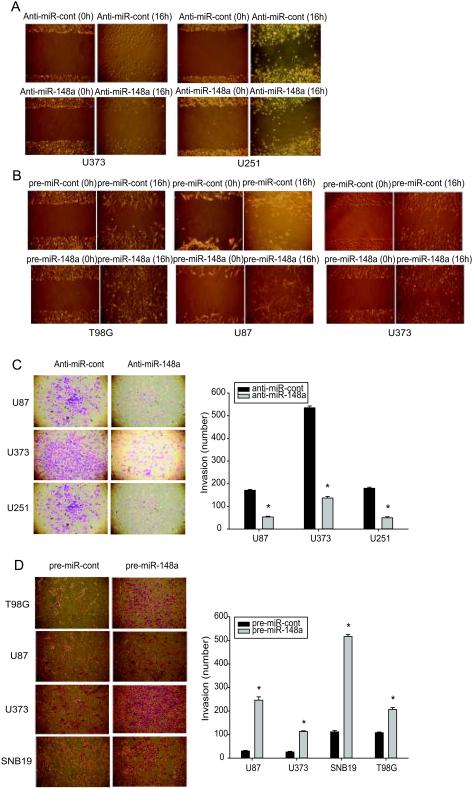
We next assessed the effects of miR-148a on GBM cell migration and invasion. GSCs were not used for these experiments because they grow as neurospheres that do not attach to tissue culture plates. Anti-miR-148a or pre-miR-148a was transfected into GBM cells followed by wound healing and invasion assays. Inhibition of miR-148a expression significantly decreased (Fig. 3A) and overexpression of miR-148a significantly increased (Fig. 3B) the migration of GBM cells. Inhibition of miR-148a expression significantly decreased (Fig. 3C) and overexpression of miR-148a significantly increased (Fig. 3D) the invasion of GBM cells. These data show that miR-148a promotes GBM cell migration and invasion.
Figure 3. MiR-148a promotes GBM cell migration and invasion.
GBM cell lines were transfected with either pre-miR-148a, anti-miR-148a or controls and assessed for migration with the wound healing assay (A, B), and invasion with the transwell invasion assay (C, D); Left panels of (C) and (D) show representative invasion assays, right panels show the quantification of invasion. The data show that miR-148a overexpression increases and miR-148a inhibition inhibits GBM cell migration and invasion. *, p < 0.05
MiR-148a induces GSC neurosphere formation and promotes the in vivo growth of GSC- and GBM-derived xenografts
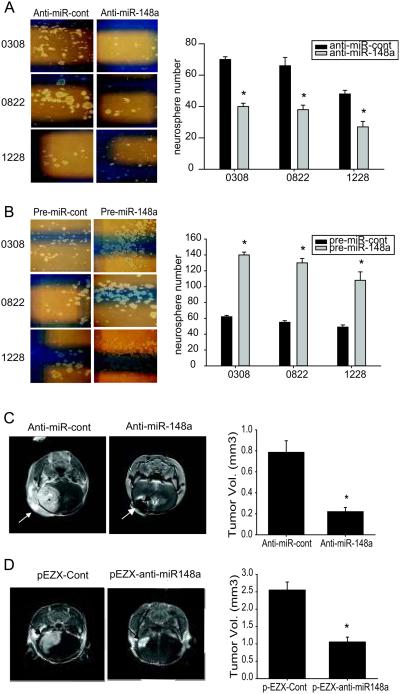
We analyzed the effects of miR-148a on GSC self-renewal using a neurosphere formation assay. Anti-miR-148a or pre-miR-148a were transfected into GSCs and neurosphere formation was assessed for one week. MiR-148a inhibition significantly reduced neurosphere size and number and miR-148a overexpression increased neurosphere size and number (p<0.05) (Fig. 4A, B). These data suggest that miR-148a promotes the self-renewal ability of GSCs. To determine if miR-148a affects GSC tumorigenesis, we assessed the effects of anti-miR-148a on orthotopic GSC xenograft formation. GSC 1228 cells were transfected with anti-miR-148a or anti-miR-control and stereotactic implanted into the striata of immunodeficient mice (n=6). Tumor sizes were measured with MRI four weeks after implantation. Anti-miR-148a significantly inhibited tumor formation by GSCs (P<0.05) (Fig. 4C). We also assessed the effects of stable anti-miR-148a expression on GBM xenograft growth. U87 cells stably expressing anti-miR-148a were orthotopically injected into NOD/SCID immunodeficient mice brains (n=10) and tumor size was measured by magnetic resonance imaging (MRI) after 3 weeks. The result shows significantly reduced tumor volume in anti-miR-148a expressing xenografts as compared to controls (p<0.05) (Fig. 4D). These data show that miR-148a promotes GSC and GBM tumor formation and growth.
Figure 4. miR-148a induces GSC neurosphere formation and antisense miR-148a inhibits the growth of GSC-derived and GBM cell-derived orthotropic xenografts.
A and B) GSCs were transfected with either anti-miR-148a, pre-miR-148a or controls and assessed for self-renewal with the neurosphere formation assay. The data show that Inhibition of miR-148a significantly inhibits neurosphere formation (A), and that overexpression of miR-148a significantly increases neurosphere formation (B). Left panels of (A) and (B) show representative assays, right panels show quantification of neurosphere formation. C) GSCs (1228) were transfected with anti-miR-148a or control and orthotopically implanted in immunodeficient mice (n=6). After 4 weeks, tumor volumes were measured by MRI. D) anti-miR-148a expressing U87 stable cells were orthotopically implanted in immunodeficient mice (n=10). After 3 weeks, tumor volumes were measured by MRI. The data from (C) and (D) show that miR-148a inhibition leads to inhibition of GSC-derived and GBM cell derived xenograft growth. Arrows point to tumors. *, p < 0.05
MiR-148a inhibits MIG6 and BIM expression and indirectly enhances EGFR expression and activation
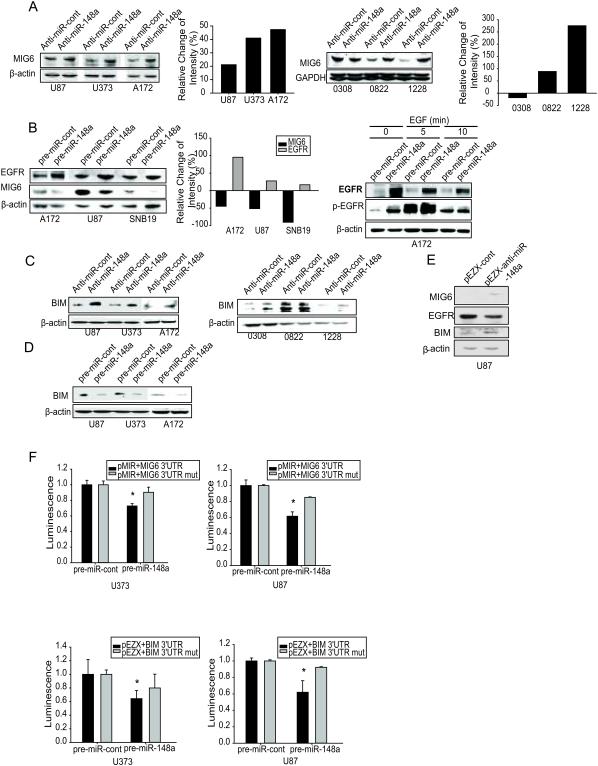
To uncover mRNA targets of miR-148a in GBM, we used bioinformatics databases (Targetscan, Pictar, RNAhybrid) to identify potential tumor suppressor targets. The following genes contained predicted binding sites for miR-148a: ERRFI1 (MIG6, NM_018948), BCL2L11 (BIM, NM_001204106), PTEN (NM_000314), SOCS3 (NM_003955), DNMT1 (NM_001130823) and JMY (NM_152405). To experimentally verify these potential targets, cells were transfected with miR-148a and assessed protein and mRNA target levels by immunoblotting and qRT-PCR, respectively. Two of the candidates were confirmed: MIG6 (ERRFI1) and BIM (BCL2L11). As MIG6 is a critical regulator of EGFR trafficking, degradation and activation, we also determined the effects of miR-148a on EGFR expression and activation. MiR-148a inhibition increased (Fig. 5A) and miR-148a overexpression reduced (Fig. 5B) the expression of MIG6 in GBM cells and GSCs. MiR-148a inhibition increased (Fig. 5C) and miR-148a overexpression reduced (Fig. 5D) the expression of BIM extra-long (most abundant form of BIM) in GBM cells and GSCs. Moreover, the effects of miR-148a on EGFR expression and activation were opposite to those on MIG6, as miR-148a overexpression led to increased EGFR and phospho-EGFR (Fig 5B). We confirmed the above results in U87 cells stably expressing anti-miR-148 (Fig. 5E). MiR-148a also inhibited MIG6 and BIM mRNA levels, suggesting that its effects are via translation inhibition as well as via mRNA degradation (Fig. S4). To determine if MIG6 and BIM 3’-UTRs are direct targets of miR-148a, MIG6 or BIM 3’-UTR reporter constructs or 3’UTR mutant controls were transfected into GBM cells prior to transfection with miR-148a and luciferase activity was measured. Overexpression of miR-148a significantly reduced luciferase activity for both MIG6 and BIM (Fig. 5F). The above data show that miR-148a directly inhibits MIG6 and BIM and indirectly up-regulates EGFR protein expression and promotes EGFR activation.
Figure 5. miR-148a directly targets and inhibits MIG6 and BIM and indirectly enhances EGFR expression and activation.
Human GBM cell lines and GSCs were transfected with anti-miR-148a (A, C), pre-miR-148a (B, D) or controls. The cells were assessed for MIG6 and EGFR (A, B) and BIM (C, D) expression/activation by immunoblotting. The data show that miR-148a overexpression inhibits MIG6 and BIM and enhances EGFR/p-EGFR, while miR-148a inhibition has the opposite effects. Immunoblots are from representative experiments and bar graphs show the quantification of the immunoblots. E) Immunoblots showing the regulation of MIG6, EGFR and BIM proteins in stable anti-miR-148a expressing U87 cells. F) 3'UTR luciferase assays for MIG6 and BIM showing the inhibition of luciferase activity by miR-148a in GBM cells relative to mutant (mut) controls. *, p < 0.05
MIG6 and BIM mediate the effects of miR-148a on GBM cell growth and survival
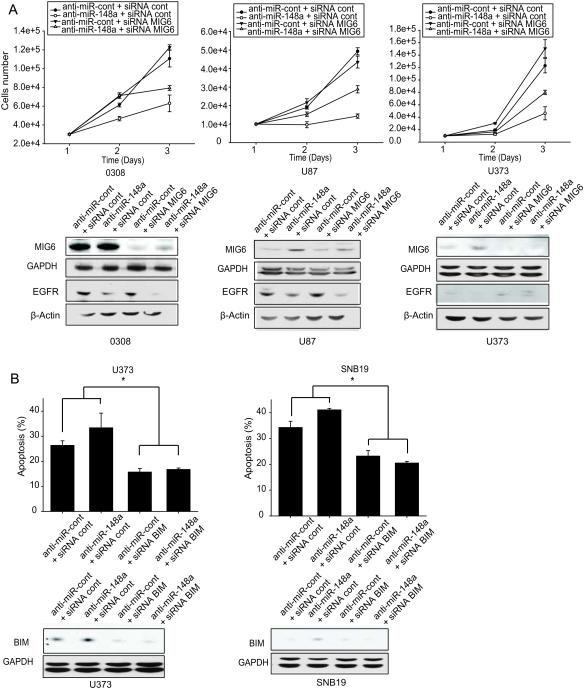
To determine if the oncogenic effects of miR-148a are mediated by MIG6 and BIM, MIG6 or BIM upregulation by anti-miR-148a was prevented using siRNAs prior to assessment of cell growth (MIG6) or apoptosis (BIM). GBM cells were transfected with MIG6, BIM or control siRNAs prior to transfection with anti-miR-148a followed by assessment of cell growth or apoptosis by cell counting and Annexin V-7AAD flow cytometry, respectively. Inhibition of miR-148a significantly inhibited GBM and GSC cell growth. MIG6 knockdown partially prevented the effects of miR-148a inhibition on cell growth (Fig. 6A). Similar to earlier results, inhibition of miR-148a increased GBM and GSC cell-line apoptosis; however, BIM knockdown prevented the increased apoptosis induced by anti-miR148a expression (Fig. 6B). MIG6 and BIM knockdown with siRNA was confirmed by immunoblotting (Fig. 6A, B). Similar rescue to the above was obtained in U87 cells stably expressing anti-miR-148a (Fig. S5, S6). The above data show that the oncogenic effects of miR-148a are partially mediated by MIG6 and BIM.
Figure 6. MIG6 and BIM mediate the effects of miR-148a on GBM cell growth and survival.
GBM cells and GSCs were transfected with anti-miR-148a prior to transfection with either MIG6 siRNA (A) or BIM siRNA (B). A) Growth assay showing that MIG6 inhibition partially rescues the proliferative effects of miR-148a inhibition (upper panel). Immunoblots showing the rescue of anti-miR-148a-induced upregulation of MIG6 and downregulatiopn of EGFR by the corresponding siRNA (lower panel). B) Apoptosis/cell death assay showing that BIM inhibition partially rescues the apoptotic effects of miR-148a inhibition (upper panel). Immunoblots showing the rescue of anti-miR-148a-induced upregulation of BIM by the corresponding siRNA (lower panel). *, p < 0.05
MiR-148a inhibits EGFR trafficking and degradation
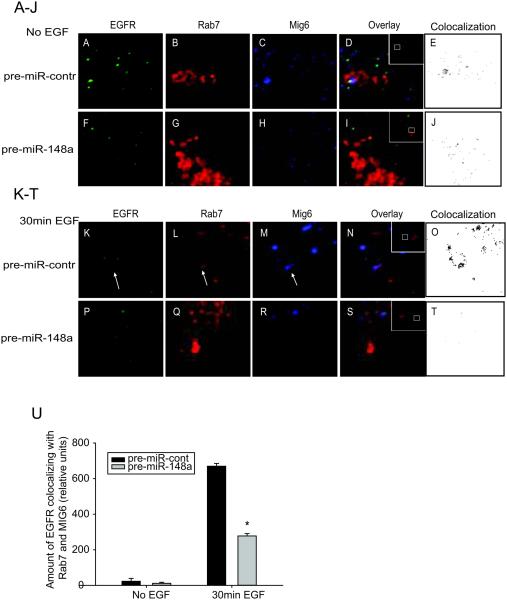
Previous research has shown that MIG6 regulates EGFR trafficking into the late endosome/lysosomes promoting EGFR degradation (11). We used confocal microscopy to determine whether miR-148a affects EGFR trafficking into a Rab7-positive late endosome/lysosomal compartment in GBM cells. Rab7 has been shown to localize to late endosomes and to be important in the maintenance of the late endosomal compartment. Rab7 also controls the fusion of late endosome with lysosomes where EGFR degradation occurs (40). First, the GBM cells were transfected with miR-148a or control before transfection with fluorescently labeled Rab7. We found reduced levels of MIG6 protein in miR-148a over-expressing cells as compared to control (data not shown). In control cells, MIG6 and EGFR colocalized in relatively large Rab7-labeled structures, likely multivesicular bodies (MVB)/late endosomes (Fig. 7A-O). This colocalization occurred at all time points, but was particularly evident 30 min after EGF stimulation in control cells (arrows, Fig. 7K-O). Importantly, in miR-148a-expressing cells colocalization between EGFR, MIG6, and Rab7 was rarely seen and never found in the large Rab7-labelled structures (MVBs) (grey circle, Fig. 7P-T). Co-localization is also shown in black and white for a clearer alternative image (Fig. 7E, J, O, and T). Quantification of the percentage of EGFR that co-localized with Rab7 and MIG6 showed a significant reduction in colocalization in miR-148a over-expressing cells compared with control cells (Fig 7U). These data demonstrate that miR-148a reduces EGFR trafficking and degradation in GBM cells.
Figure 7. miR-148a inhibits EGFR trafficking and degradation.
GBM cell lines were transfected with control (A-E and K-O) or pre-miR-148a (F-J and P-T) for 24 h and then transfected with Rab7-mCherry (red; B, G, L and Q) for 24 h. The cells were serum-starved for the last hour before being treated with EGF (50 ng/ml) for the indicated times (0 min; A-J, 30 min; K-T). Cells were fixed and stained with anti-EGFR (green; A, F, K and P) and anti-MIG6 (blue; C, H, M and R). Arrows point to the EGFR and MIG6-containing Rab7 compartment in control-transfected and EGF-treated cells (K, L and M). Note the increased amount of EGFR colocalizing with Rab7 and Mig6 in control cells (N and O) as compared with miR-148a overexpressing cells (S and T). In miR-148a-expressing cells, light gray circles point to Rab7 compartment structures, but colocalization between EGFR, MIG6 and Rab7 is rarely seen and not in large Rab7-labeled structures (MVBs) (P, Q and R). Colocalization of MIG6, EGFR and Rab7 are shown in black and white in E, J, O and T. U). Colocalization of EGFR with MIG6 and Rab7-labeled structures was quantified on the confocal images. The results are the means ± SEM of > 30 cells scored from 2 separate experiments. *, p < 0.05
DISCUSSION
MiR-148a has been investigated in some cancers but not in brain tumors (41-43). In this study, we investigated the expression, function and mechanisms of action of miR-148a in GBM. We found that miR-148a is a risk factor in GBM where its acts as an oncogene by regulating BIM, MIG6 and EFGR stability and activation.
EGFR is one of the most frequently altered genes in GBM. It is overexpressed in more than 60% of tumors but mutated and amplified in only about 40% (44, 45). Therefore, EGFR gene amplification only partially accounts for EGFR overexpression in GBM (44) suggesting that additional mechanisms may be involved. Our study suggests that miR-148a overexpression is an important mechanism of EGFR overexpression via downregulation of MIG6. Consistent with our results, others have found that MIG6 expression is downregulated in ~50% of GBM tumors without indications of MIG6 genomic deletions in the majority of samples (11). Our study also provides a new mechanism of MIG6 downregulation in GBM.
We also identified the pro-apoptotic molecule BIM as a target of miR-148a, which is downregulated in 29% of GBM cases based on TCGA analysis. Interestingly, a recent study demonstrated that elevated BIM expression levels in cancers strongly increased the anti-tumor activity of EGFR and other RTK inhibitors (46). These findings suggest that combined upregulation of BIM and inhibition of EGFR is likely to achieve synergistic anti-tumor effects. Our study shows that such combined targeting of BIM and EGFR can be achieved by inhibition of miR-148a, providing a rationale for the therapeutic targeting of miR-148a.
Previous research described miR-148a as a tumor suppressor in hepatocellular carcinoma, pancreatic cancer, gastric cancer and colorectal cancer (42, 47-49). Our study demonstrates for the first time that miR-148a is oncogenic in GBM. We show that miR-148a enhances GBM and GSC growth, survival, migration and invasion as well as GSC self-renewal and in vivo tumor growth. We also show that inhibiting miR-148a inhibits the above oncogenic endpoints. Importantly, based on our TCGA data analysis, we find that miR-148a expression displays a significant inverse correlation with GBM patient survival. A recent study identifying a ten-microRNA prognostic expression signature in GBM showed that miR-148a was among the 7 microRNAs that were associated with high risk (50). Our TCGA data analysis expanded on this finding, analyzing 482 samples to further demonstrate elevated miR-148a expression in human GBM specimens.
In summary, the present study shows that miR-148a is elevated in GBM, where it predicts poor patient survival. It demonstrates that miR-148a has powerful oncogenic and cancer stem cell regulatory effects that are mediated by BIM, MIG6 and EGFR. The study therefore represents a first characterization of miR-148a as an oncogene and promising therapeutic target in GBM.
Acknowledgement
Dedicated to the memory of Michael Skalski who passed away at a young age before seeing the fruits of his contribution to this work
Grant support: Supported by NIH grants NS045209 (R.A.) and CA134843 (R.A.)
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
REFERENCES
- 1.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–93. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–42. doi: 10.1007/978-1-60327-492-0_14. [DOI] [PubMed] [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12:675–84. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 8.Friedman HS, Bigner DD. Glioblastoma multiforme and the epidermal growth factor receptor. N Engl J Med. 2005;353:1997–9. doi: 10.1056/NEJMp058186. [DOI] [PubMed] [Google Scholar]
- 9.Sebastian S, Settleman J, Reshkin SJ, Azzariti A, Bellizzi A, Paradiso A. The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta. 2006;1766:120–39. doi: 10.1016/j.bbcan.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Fry WH, Kotelawala L, Sweeney C, Carraway KL. Mechanisms of ErbB receptor negative regulation and relevance in cancer. Exp Cell Res. (3rd) 2009;315:697–706. doi: 10.1016/j.yexcr.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci U S A. 2010;107:6912–7. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–4. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, et al. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol. 2000;20:7735–50. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan CG, Killela PJ, Payne CA, Lampson B, Chen WC, Liu J, et al. Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget. 2010;1:265–77. doi: 10.18632/oncotarget.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 16.Gunther W, Pawlak E, Damasceno R, Arnold H, Terzis AJ. Temozolomide induces apoptosis and senescence in glioma cells cultured as multicellular spheroids. Br J Cancer. 2003;88:463–9. doi: 10.1038/sj.bjc.6600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath-Engel HM, Shore GC. Regulated targeting of Bax and Bak to intracellular membranes during apoptosis. Cell Death Differ. 2006;13:1277–80. doi: 10.1038/sj.cdd.4401961. [DOI] [PubMed] [Google Scholar]
- 18.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–79. doi: 10.1371/journal.pmed.0040315. discussion 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–75. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci. 2012;17:700–12. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–76. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–72. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A. Targeting of TGFbeta signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA. 2013;19:177–90. doi: 10.1261/rna.036467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–8. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–63. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kefas B, Floyd DH, Comeau L, Frisbee A, Dominguez C, Dipierro CG, et al. A miR-297/hypoxia/DGK-alpha axis regulating glioblastoma survival. Neuro Oncol. 2013;15:1652–63. doi: 10.1093/neuonc/not118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–32. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kofman AV, Kim J, Park SY, Dupart E, Letson C, Bao Y, et al. microRNA-34a promotes DNA damage and mitotic catastrophe. Cell Cycle. 2013;12:3500–11. doi: 10.4161/cc.26459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 33.R-project.org [Internet] Vienna, Austria: Institute for Statistics and Mathematics of WU (Wirtschaftsuniversität Wien) [updated 2013 Sep 25; cited 2013 May 14]. Available from: http://www.r-project.org/
- 34.Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 35.Li Y, Lal B, Kwon S, Fan X, Saldanha U, Reznik TE, et al. The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res. 2005;65:9355–62. doi: 10.1158/0008-5472.CAN-05-1946. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Guessous F, Johnson EB, Eberhart CG, Li XN, Shu Q, et al. Functional and molecular interactions between the HGF/c-Met pathway and c-Myc in large-cell medulloblastoma. Lab Invest. 2008;88:98–111. doi: 10.1038/labinvest.3700702. [DOI] [PubMed] [Google Scholar]
- 37.Amos S, Mut M, diPierro CG, Carpenter JE, Xiao A, Kohutek ZA, et al. Protein kinase C-alpha-mediated regulation of low-density lipoprotein receptor related protein and urokinase increases astrocytoma invasion. Cancer Res. 2007;67:10241–51. doi: 10.1158/0008-5472.CAN-07-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Zhang Y, Tang Y, Butler N, Kim J, Guessous F, et al. A novel PTEN/mutant p53/c-Myc/Bcl-XL axis mediates context-dependent oncogenic effects of PTEN with implications for cancer prognosis and therapy. Neoplasia. 2013;15:952–65. doi: 10.1593/neo.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Farenholtz KE, Yang Y, Guessous F, Dipierro CG, Calvert VS, et al. Hepatocyte growth factor sensitizes brain tumors to c-MET kinase inhibition. Clin Cancer Res. 2013;19:1433–44. doi: 10.1158/1078-0432.CCR-12-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–80. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu A, Xia J, Zuo J, Jin S, Zhou H, Yao L, et al. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med Oncol. 2012;29:2701–9. doi: 10.1007/s12032-011-0134-3. [DOI] [PubMed] [Google Scholar]
- 42.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–83. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 43.Liffers ST, Munding JB, Vogt M, Kuhlmann JD, Verdoodt B, Nambiar S, et al. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest. 2011;91:1472–9. doi: 10.1038/labinvest.2011.99. [DOI] [PubMed] [Google Scholar]
- 44.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–65. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delpu Y, Lulka H, Sicard F, Saint-Laurent N, Lopez F, Hanoun N, et al. The rescue of miR-148a expression in pancreatic cancer: an inappropriate therapeutic tool. PLoS One. 2013;8:e55513. doi: 10.1371/journal.pone.0055513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng H, et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011;18:1702–10. doi: 10.1038/cdd.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JP, Zeng C, Xu L, Gong J, Fang JH, Zhuang SM. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene. 2013 doi: 10.1038/onc.2013.369. Epub 2013 Sep 9. [DOI] [PubMed] [Google Scholar]
- 50.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]