Abstract
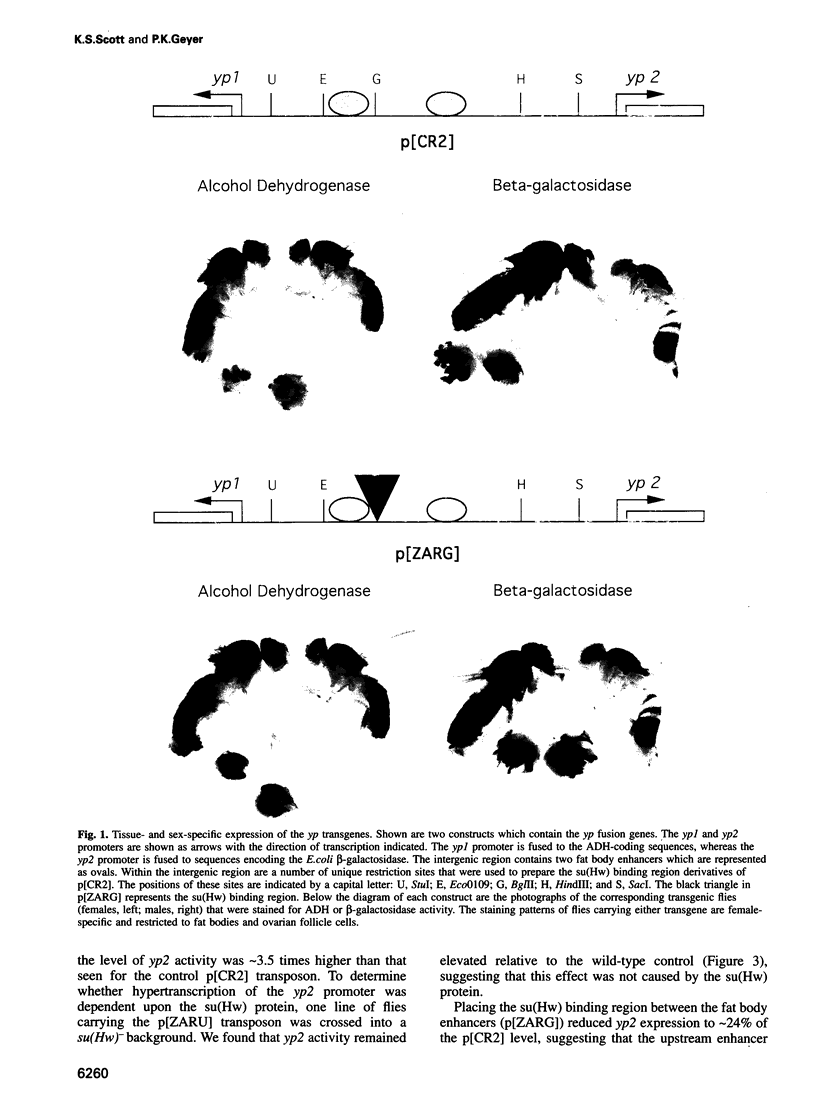
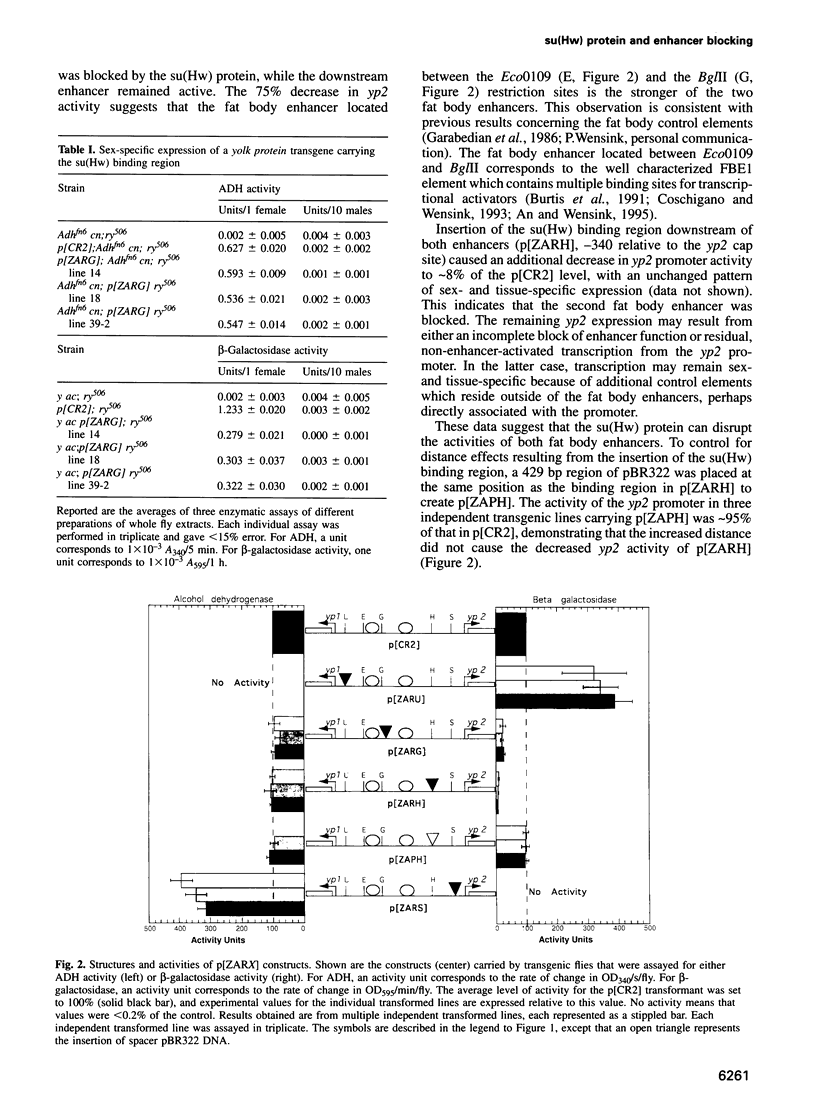
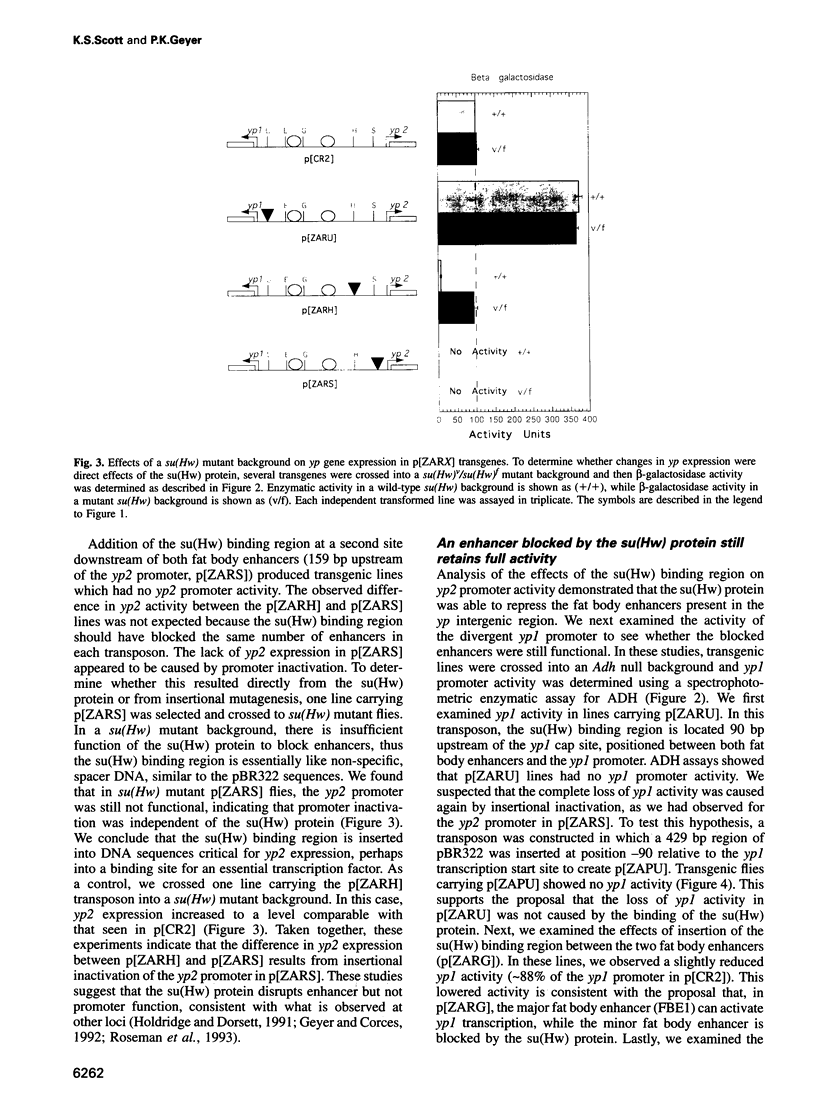
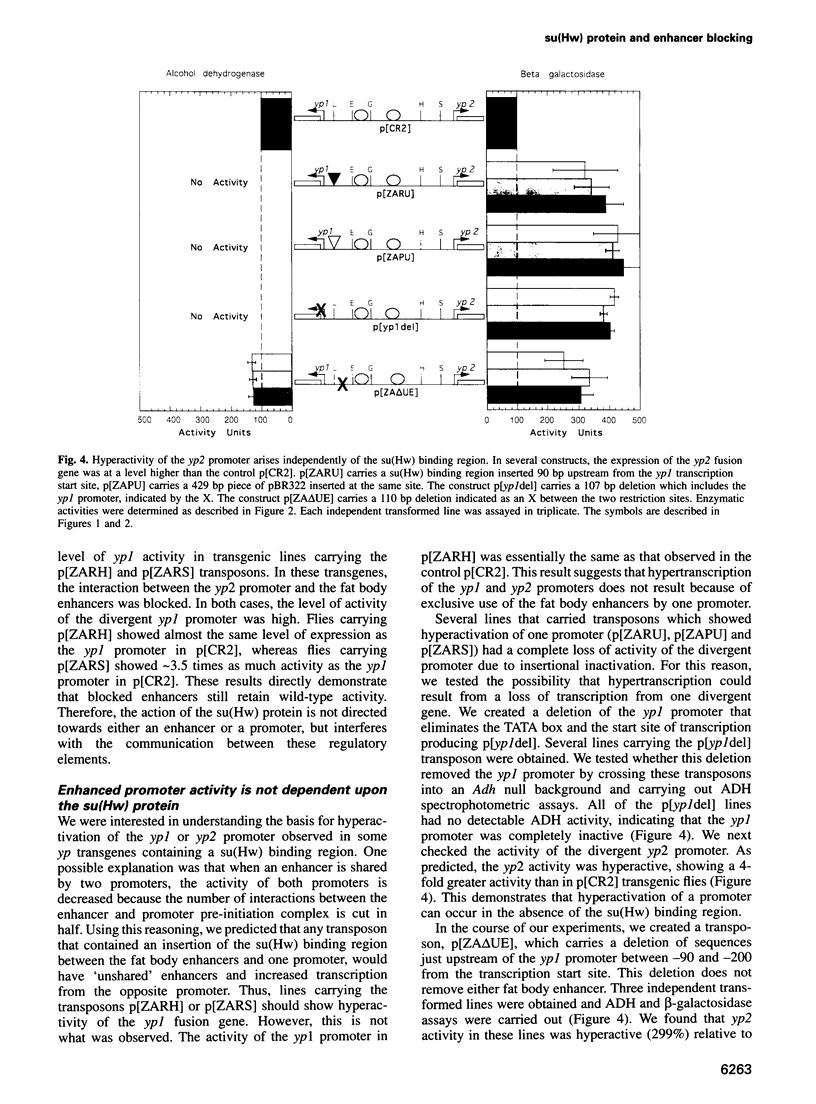
The suppressor of Hairy-wing [su(Hw)] protein mediates the mutagenic effects of the gypsy retrotransposon by blocking enhancer activity. These repressive effects are general, can occur over long distances and require that the su(Hw) protein is bound between the affected enhancer and promoter. The effects of the su(Hw) binding region on yolk protein (yp) gene expression were determined. These genes are regulated by shared enhancers in the intergenic region, which provided a method to examine whether an enhancer blocked by the su(Hw) protein remained functional. We demonstrate that a blocked enhancer is completely active, supporting the proposal that the su(Hw) protein is an insulator protein that acts by forming a new boundary in a pre-existing chromatin domain, thereby preventing the interaction of regulatory elements located upstream of the insertion site with the promoter. In addition, we found that yp promoter function is not diminished by sharing enhancers, suggesting that these enhancers are not rate limiting for transcriptional activation. Lastly, our data indicate that yp promoter activity is interdependent, such that transcription from one promoter influences the level of activity of the linked promoter.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahamsen N., Martinez A., Kjaer T., Søndergaard L., Bownes M. Cis-regulatory sequences leading to female-specific expression of yolk protein genes 1 and 2 in the fat body of Drosophila melanogaster. Mol Gen Genet. 1993 Feb;237(1-2):41–48. doi: 10.1007/BF00282782. [DOI] [PubMed] [Google Scholar]
- Adami G., Babiss L. E. Evidence that USF can interact with only a single general transcription complex at one time. Mol Cell Biol. 1992 Apr;12(4):1630–1638. doi: 10.1128/mcb.12.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W., Wensink P. C. Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. EMBO J. 1995 Mar 15;14(6):1221–1230. doi: 10.1002/j.1460-2075.1995.tb07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M. D., Weiner A. J., Goralski T. J., Mahowald A. P. The follicle cells are a major site of vitellogenin synthesis in Drosophila melanogaster. Dev Biol. 1982 Jan;89(1):225–236. doi: 10.1016/0012-1606(82)90309-8. [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Coschigano K. T., Baker B. S., Wensink P. C. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991 Sep;10(9):2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces V. G., Geyer P. K. Interactions of retrotransposons with the host genome: the case of the gypsy element of Drosophila. Trends Genet. 1991 Mar;7(3):86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics. 1993 Aug;134(4):1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M. J., Hung M. C., Wensink P. C. Independent control elements that determine yolk protein gene expression in alternative Drosophila tissues. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1396–1400. doi: 10.1073/pnas.82.5.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M. J., Shepherd B. M., Wensink P. C. A tissue-specific transcription enhancer from the Drosophila yolk protein 1 gene. Cell. 1986 Jun 20;45(6):859–867. doi: 10.1016/0092-8674(86)90560-x. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992 Oct;6(10):1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G. Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8593–8597. doi: 10.1073/pnas.85.22.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Spana C., Corces V. G. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 1986 Oct;5(10):2657–2662. doi: 10.1002/j.1460-2075.1986.tb04548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., Gdula D. A., Coyne R. S., Corces V. G. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 1993 Oct;7(10):1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- Holdridge C., Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol. 1991 Apr;11(4):1894–1900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack J., Dorsett D., Delotto Y., Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991 Nov;113(3):735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- Jackson M. E. Negative regulation of eukaryotic transcription. J Cell Sci. 1991 Sep;100(Pt 1):1–7. doi: 10.1242/jcs.100.1.1. [DOI] [PubMed] [Google Scholar]
- Karch F., Galloni M., Sipos L., Gausz J., Gyurkovics H., Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994 Aug 11;22(15):3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. E., Rubin G. M. Analysis of P transposable element functions in Drosophila. Cell. 1984 Aug;38(1):135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Kellum R., Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992 May;12(5):2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991 Mar 8;64(5):941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Levine M., Manley J. L. Transcriptional repression of eukaryotic promoters. Cell. 1989 Nov 3;59(3):405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Logan S. K., Garabedian M. J., Wensink P. C. DNA regions that regulate the ovarian transcriptional specificity of Drosophila yolk protein genes. Genes Dev. 1989 Sep;3(9):1453–1461. doi: 10.1101/gad.3.9.1453. [DOI] [PubMed] [Google Scholar]
- Logan S. K., Wensink P. C. Ovarian follicle cell enhancers from the Drosophila yolk protein genes: different segments of one enhancer have different cell-type specificities that interact to give normal expression. Genes Dev. 1990 Apr;4(4):613–623. doi: 10.1101/gad.4.4.613. [DOI] [PubMed] [Google Scholar]
- Marlor R. L., Parkhurst S. M., Corces V. G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986 Apr;6(4):1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., Coyne R. S., Corces V. G. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988 Oct;2(10):1205–1215. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- Peifer M., Bender W. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9650–9654. doi: 10.1073/pnas.85.24.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Bender W. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 1986 Sep;5(9):2293–2303. doi: 10.1002/j.1460-2075.1986.tb04497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Qian S., Capovilla M., Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991 Jun;10(6):1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., Engels W. R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988 Mar;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R. R., Pirrotta V., Geyer P. K. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993 Feb;12(2):435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Rutledge B. J., Mortin M. A., Schwarz E., Thierry-Mieg D., Meselson M. Genetic interactions of modifier genes and modifiable alleles in Drosophila melanogaster. Genetics. 1988 Jun;119(2):391–397. doi: 10.1093/genetics/119.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Lis J. T. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acids Res. 1987 Apr 10;15(7):2971–2988. doi: 10.1093/nar/15.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Peifer M., Bender W., O'Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990 Dec;9(12):3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. A., Corces V. G. The suppressor of Hairy-wing binding region is required for gypsy mutagenesis. Mol Gen Genet. 1992 May;233(1-2):65–70. doi: 10.1007/BF00587562. [DOI] [PubMed] [Google Scholar]
- Smith P. A., Corces V. G. The suppressor of Hairy-wing protein regulates the tissue-specific expression of the Drosophila gypsy retrotransposon. Genetics. 1995 Jan;139(1):215–228. doi: 10.1093/genetics/139.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S. U., Gerasimova T., Kurkulos M., Boeke J. D., Corces V. G. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994 Sep 1;8(17):2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Spana C., Corces V. G. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 1990 Sep;4(9):1505–1515. doi: 10.1101/gad.4.9.1505. [DOI] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988 Nov;2(11):1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Vazquez J., Farkas G., Gaszner M., Udvardy A., Muller M., Hagstrom K., Gyurkovics H., Sipos L., Gausz J., Galloni M. Genetic and molecular analysis of chromatin domains. Cold Spring Harb Symp Quant Biol. 1993;58:45–54. doi: 10.1101/sqb.1993.058.01.008. [DOI] [PubMed] [Google Scholar]
- Vazquez J., Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994 Dec 15;13(24):5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988 Oct 30;70(2):399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]







