Abstract
We report a new mechanism by which human mRNAs crosstalk: an Alu element in the 3'-untranslated region (3' UTR) of one mRNA can base-pair with a partially complementary Alu element in the 3' UTR of a different mRNA thereby creating a Staufen1 (STAU1)-binding site (SBS). STAU1 binding to a 3' UTR SBS was previously shown to trigger STAU1-mediated mRNA decay (SMD) by directly recruiting the ATP-dependent RNA helicase UPF1, which is also a key factor in the mechanistically related nonsense-mediated mRNA decay (NMD) pathway. In the case of a 3' UTR SBS created via mRNA–mRNA base-pairing, we show that SMD targets both mRNAs in the duplex provided that both mRNAs are translated. If only one mRNA is translated, then it alone is targeted for SMD. We demonstrate the importance of mRNA–mRNA-triggered SMD to the processes of cell migration and invasion.
INTRODUCTION
SMD downregulates hundreds of human transcripts that regulate myogenesis1, cutaneous wound-healing2, adipogenesis3, and many if not most other cellular processes4. One of the first SMD targets identified was human c-JUN mRNA5, which encodes a protein that, together with c-FOS, forms the AP-1 early-response transcription factor6,7. Since AP-1 regulates cell proliferation, differentiation, apoptosis, organogenesis and stress responses, SMD does also.
SMD is a translation-dependent process that targets mRNAs harboring a 3' UTR SBS5,8. SBSs can form intramolecularly, as best exemplified by the 19-base-pair stem and associated apex that typifies the 3' UTR of mRNA encoding human ADP ribosylation factor 1 (ARF1)8. In support of its functional significance, this structure is conserved in the 3' UTR of ARF1 mRNA in rat and mouse8. SBSs can also form intermolecularly via base-pairing between the Alu element within the 3' UTR of an SMD target and one or more partially complementary Alu elements within a long noncoding RNA (lncRNA), which we have called ½-sbsRNAs because they comprise one strand (i.e. half) of a double-stranded SBS2. As an example, the 3' UTR Alu element within mRNA encoding CUB domain-containing protein 1 (CDCP1), which functions at the cell surface as an anti-apoptotic agent that promotes tumor-cell viability during or in early states of metastatic colonization9, is targeted for SMD upon base-pairing with the lncRNA we have named ½-sbsRNA2 (i.e., the second of those ½-sbsRNAs that we have studied)2. Alu elements are a type of short interspersed element (SINE) that are unique to primates, ~300 base-pairs in length, and constitute ~13% of the human genome10. Remarkably, B and identifier (ID) SINEs within lncRNAs of mouse, which we call m½-sbsRNAs, function analogously to Alu elements within ½-sbsRNAs of primates and trigger SMD despite the independent evolution of B and ID elements relative to Alu elements11.
SMD, like the mechanistically related pathway nonsense-mediated mRNA decay (NMD), involves the UPF1 RNA helicase4,12. An SBS bound by STAU1 and/or is paralog STAU213 is thought to function during SMD analogously to how a splicing-dependent exon-junction complex (EJC) functions during NMD: when translation terminates sufficiently upstream of either complex so that the terminating ribosome fails to displace the associated proteins, then mRNA decay will occur provided that the complex has acquired UPF1. STAU1 and STAU2 bind UPF1 directly during SMD13 as does the EJC constituent UPF214,15 or, in some cases, the EJC constituent UPF3X (also called UPF3B)1,16,17 during NMD. While studies of intermolecular SBSs have been restricted to sequences containing no more than two consecutive nucleotide mismatches, SBSs are apt to be more tolerant of mismatches given that their lengths can range from ~86–300 base-pairs2. Tolerance is also likely to be engendered by the ability of STAU1 and STAU2 to form homodimers and heterodimers, if not homomultimers and heteromultimers18,19.
The finding that mRNA–lncRNA interactions occur in cells via Alu element base-pairing raised the intriguing issue of whether mRNA–mRNA interactions also occur in cells via Alu element base-pairing. Additionally, because only the mRNA strand of those mRNA–lncRNA duplexes studied to date is targeted for SMD, if mRNA–mRNA duplexes do form, would only one mRNA strand or would both mRNA strands be targeted for SMD?
In this communication, we computationally determine that 15,530,838 mRNA–mRNA duplexes potentially exist in human cells via intermolecular 3' UTR Alu element base-pairing. We predict that a single mRNA can interact with multiple mRNAs via 3' UTR Alu element base-pairing and demonstrate that mRNA–mRNA duplex formation can compete with mRNA–lncRNA duplex formation. Each mRNA of an mRNA–mRNA duplex is targeted for SMD provided that each is translated and that translation terminates sufficiently upstream of the SBS. We demonstrate the functional significance of mRNA–mRNA duplex formation in studies of mRNA encoding CUB domain-containing protein 1 (CDCP1), which functions at the cell surface as an anti-apoptotic agent that promotes tumor-cell viability during or in early states of metastatic colonization9. Our findings uncover a previously unappreciated function for mammalian-cell mRNAs. This function adds a new and unanticipated layer of complexity to the intricate network of post-transcriptional interactions that regulate human gene expression.
RESULTS
Computational predictions of human-cell mRNA–mRNA duplexes
Of the 129,138 human transcripts reported in ENSEMBL, we determined that 3,943 contain a single 3' UTR Alu element (RepeatMasker). We employed DuplexFold20 to obtain the predicted ΔG values of computationally derived mRNA–mRNA duplexes formed via base-pairing of partially complementary 3' UTR Alu elements (Supplementary Table 1). Results revealed that duplexes between bona fide SMD targets were among the 15,530,838 duplexes obtained. Two of the bona fide SMD targets, CDCP1 mRNA2 and Sosondowah ankyrin repeat domain family member C (SOWAHC) mRNA, formerly called FLJ21870 mRNA2, are predicted to duplex via their 3' UTR Alu elements with a ΔG value of −224 kcal/mol (Fig. 1a, Supplementary Fig. 1a, b and Supplementary Table 2). siRNA to SOWAHC mRNA, which encodes a protein (Supplementary Fig. 1c) of unknown function, stimulates “wound healing” after scrape injury to keratinocyte monolayers as monitored by the migration rate of human keratinocyte HaCaT cells into a denuded wound track2. Intriguingly, upregulating CDCP1 protein also increases cell motility9. We hypothesized that if CDCP1 mRNA–SOWAHC mRNA duplexes were to form, recruit STAU1 and trigger the SMD of each constituent mRNA, then downregulating SOWAHC mRNA would upregulate CDCP1 mRNA. The resulting increase in CDCP1 protein would promote cell motility, since there is no evidence that SOWAHC protein affects cell motility (Supplementary Fig. 1d). The CDCP1 mRNA 3' UTR Alu element is also predicted to duplex with the 3' UTR Alu element of another bona fide SMD target that encodes RAB11 family interacting protein 1 (RAB11FIP1) with a ΔG value of −251.2 kcal/mol (Fig. 1a, Supplementary Fig. 1a, b and Supplementary Table 2). We hypothesized that downregulating RAB11FIP1 mRNA would likewise upregulate the abundance of CDCP1 mRNA. However, since RAB11FIP1 protein has been shown to promote wound healing2, downregulating RAB11FIP1 protein may mask the promotion of wound-healing attributable to the upregulation of CDCP1 protein (Supplementary Fig. 1d).
Figure 1. SOWAHC mRNA reduces the abundance of mRNAs to which it is predicted to base-pair via partially complementary 3' UTR Alu elements.
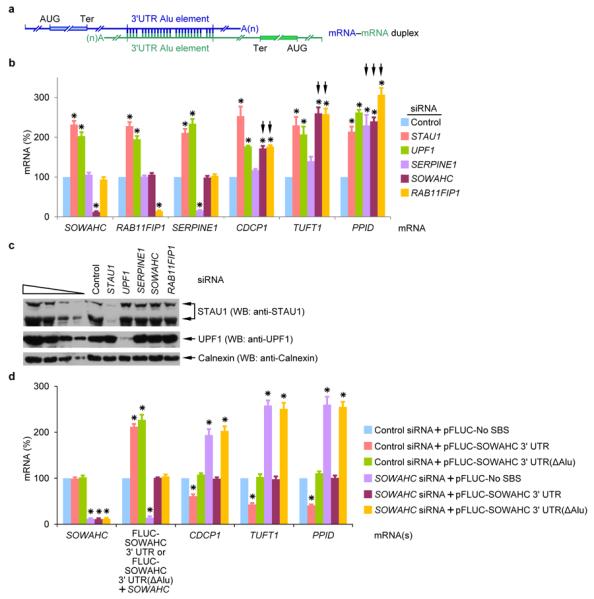
a, Diagram of computationally predicted putative mRNA–mRNA duplexes that form in human cells via base-pairing between partially complementary 3' UTR Alu elements (Supplementary Table S1). b, Histogram representation of RT-sqPCR analyses of the indicated mRNA using lysates of HeLa cells treated with the specified siRNA (see Supplementary Fig. 1o for data using alternative siRNAs). The level of each mRNA was normalized to the level of GAPDH mRNA, and normalized levels in the presence of Control siRNA are defined as 100. Black arrows denote results that implicate mRNA–mRNA duplex formation. c, Western blot (WB) using the designated antibody and lysates analyzed in c. Calnexin serves as a loading control. d, Histogram representation of RT-sqPCR analyses of lysates of HeLa cells that had been transiently transfected with the specified siRNA and pFLUC-SOWAHC 3' UTR, pFLUC-SOWAHC 3' UTR(ΔAlu) or, as a negative control, pFLUC-No SBS. The level of each mRNA was normalized to the level of MUP mRNA (from the phCMV-MUP reference plasmid), and normalized levels in the presence of Control siRNA + pFLUC-No SBS are defined as 100. Error bars, s.e.m.; # of independently performed experiments = 3; *, P < 0.05 (one-tail t-test).
SOWAHC mRNA reduces the levels of predicted mRNA partners
In tests of these hypotheses, downregulating the cellular abundance of either SOWAHC or RAB11FIP1 mRNA to ~10-fold below normal indeed increased the level of CDCP1 mRNA to ~2-fold above normal (Fig. 1b and Supplementary Fig. 1e, f). As expected since each is an SMD target, the level of SOWAHC, RAB11FIP1 or CDCP1 mRNA was increased above normal by either STAU1 or UPF1 siRNA (Fig. 1b, c and Supplementary Fig. 1e, f). The reduction in CDCP1 mRNA abundance by SOWAHC mRNA is attributable to SOWAHC 3' UTR sequences: a comparable level of an exogenous source of this 3' UTR (i.e., expressing the plasmid pFLUC-SOWAHC 3' UTR, which encodes mRNA harboring the firefly luciferase (FLUC) open translational reading frame upstream of the SOWAHC 3' UTR) decreased the level of CDCP1 mRNA (Fig. 1d and Supplementary Fig. 1g, h). This decrease was eliminated when the abundance of cellular SOWAHC mRNA was reduced (Fig. 1d and Supplementary Fig. 1g, h). In contrast, an exogenous source of SOWAHC 3' UTR that lacks the Alu element (i.e., expressing the plasmid pFLUC-SOWAHC 3' UTR(ΔAlu)) failed to decrease the level of CDCP1 mRNA (Fig. 1d and Supplementary Fig. 1g, h).
Computational analyses indicated that the 3' UTR Alu element of SOWAHC mRNA could additionally base-pair with the 3' UTR Alu element of mRNA encoding the phosphorylated glycoprotein tuftelin1 (TUFT1) as well as the 3' UTR Alu element of mRNA encoding cyclosporin-binding enzyme peptidylprolyl isomerase D (PPID, also called cyclophilin D) with predicted ΔG values of −439.4 kcal/mol and −207.3 kcal/mol, respectively. The 3' UTR Alu element of RAB11FIP1 mRNA is predicted to do likewise with ΔG values of −434.6 kcal/mol and −307.4 kcal/mol, respectively (Fig. 1a, Supplementary Fig. 1a, b and Supplementary Table 2). Remarkably, downregulation of either SOWAHC or RAB11FIP1 mRNA increased the levels of TUFT1 and PPID mRNAs to 2.5–3-fold above normal (Fig. 1b and Supplementary Fig. 1e, f). These increases were attributable to SMD because downregulating either STAU1 or UPF1 increased the levels of TUFT1 and PPID mRNAs to 2-2.5-fold above normal (Fig. 1b, c and Supplementary Fig. 1e, f). Furthermore, these increases were mediated by the SOWAHC 3' UTR Alu element (Fig. 1d and Supplementary Fig. 1g, h).
It should be noted that while the 3' UTR Alu element of SERPINE1 mRNA is predicted to form a duplex with the 3' UTR Alu element of CDCP1, TUFT1 and PPID mRNAs (Supplementary Fig. 1a) with ΔG values of −137.6 kcal/mol, −100.7 kcal/mol and −74.1 kcal/mol, respectively (Supplementary Fig. 1b), only the level of PPID mRNA was increased when the level of SERPINE1 mRNA was decreased. Thus, ΔG values alone are insufficient indicators of cellular base-pairing. For example, while BAG5 mRNA and CDCP1 mRNA are predicted to duplex with the lncRNA ½-sbsRNA2 with ΔG values of −416 kcal/mol and −513.7 kcal/mol, respectively9, only CDCP1 mRNA is targeted for SMD in HeLa cells because BAG5 mRNA fails to bind ½-sbsRNA2 (and undoubtedly other ½-sbsRNAs that are computationally predicted to base-pair with BAG5 mRNA) and, thus, STAU12,21. Furthermore, each of two 30-nucleotide 3' UTR sequences that perfectly base-pair with an artificial ½-sbsRNA trigger SMD when situated 74-nucleotides downstream of a termination codon but not when situated 44-nucleotides downstream of the same termination codon (Supplementary Fig. 1i–n). By analogy to the rule establishing which termination codons trigger NMD based on their distance from a downstream exon-junction complex12,22, our results suggest that the 30-nucleotide SBSs must reside a minimum distance (> 44 nucleotides) downstream of a termination codon to trigger SMD. Thus, multiple factors determine whether a potential mRNA–mRNA duplex will target either constituent mRNA for SMD (see also below).
mRNA–mRNA duplexes create SBSs
If an SBS could be formed by the 3' UTR Alu element of SOWAHC mRNA base-pairing with the 3' UTR Alu element of CDCP1, TUFT1 or PPID mRNA, then it should be possible to co-immunoprecipitate the respective mRNA–mRNA complexes. To test this possibility, HeLa cells that transiently expressed two plasmid DNAs were crosslinked using formaldehyde so that only bona fide cellular complexes would be examined. The first plasmid encoded either FLUC-SOWAHC 3' UTR-MS2bs mRNA, which contains 12 copies of the MS2 coat-protein binding site (MS2bs) upstream of the FLUC-SOWAHC 3' UTR polyadenylation signal or, as negative controls, FLUC-MS2bs mRNA or FLUC-SOWAHC 3' UTR mRNA (Fig. 2a); the second plasmid encoded Flag-MS2-hMGFP protein, which consists of the MS2 coat-protein tagged with the Flag epitope and fused to monster green fluorescent protein (hMGFP)2. A fraction of each lysate was then immunoprecipitated using anti-Flag antibody. Prior to IP, when compared to FLUC-MS2bs mRNA expression, FLUC-SOWAHC 3' UTR mRNA or FLUC-SOWAHC 3' UTR-MS2bs mRNA expression decreased the abundance of CDCP1, TUFT1 and PPID mRNAs without affecting the level of other SMD targets such as SERPINE1 mRNA (Fig. 2b).
Figure 2. mRNA–mRNA duplexes create SBSs and trigger SMD.
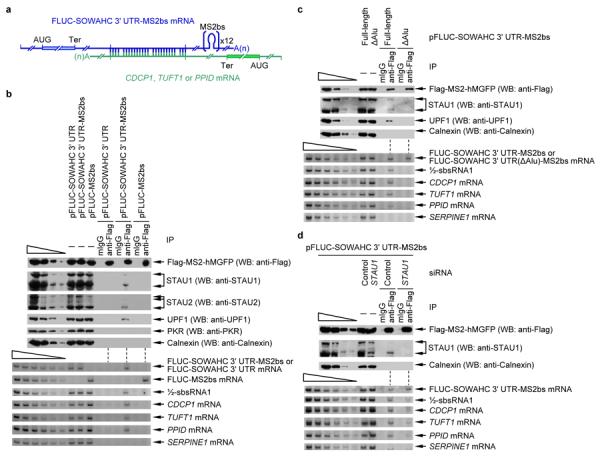
a, Diagram of FLUC-SOWAHC 3' UTR mRNA harboring 12 copies of MS2bs base-paired with CDCP1, TUFT1 or PPID mRNA via complementary 3' UTR Alu elements. b, Western blot (upper) or RT-sqPCR(lower) before (−) or after immunoprecipitation (IP) of lysates of formaldehyde-crosslinked HeLa cells that had been transiently transfected with pFlag-MS2-hMGFP and pFLUC-SOWAHC 3' UTR, pFLUC-SOWAHC 3' UTR-MS2bs or pFLUC-MS2bs. IP was performed using either anti-Flag or, as a control for nonspecific IP, mouse immunoglobulin G (mIgG). c, essentially as in b except pFLUC-SOWAHC 3' UTR-MS2bs or pFLUC-SOWAHC 3' UTR(ΔAlu)-MS2bs was used. d, essentially as in b except cells were transfected with Control or STAU1 siRNA. Error bars, s.e.m.; # of independently performed experiments = 3; P < 0.05 (one-tail t-test).
In support of the concept that the 3' UTR Alu element of SOWAHC mRNA forms an SBS through intermolecular base-pairing with the 3' UTR Alu element of CDCP1, TUFT1 or PPID mRNA, retrieval of the SOWAHC 3' UTR from lysates of cells expressing FLUC-SOWAHC 3' UTR-MS2bs mRNA using the anti-Flag antibody also immunoprecipitated cellular STAU1, STAU2 and UPF1 and cellular CDCP1, TUFT1 and PPID mRNAs (Fig. 2c). This did not occur in lysates expressing FLUC-SOWAHC 3' UTR mRNA, FLUC-MS2bs mRNA (Fig. 2b) or FLUC-SOWAHC 3' UTR(ΔAlu)-MS2bs mRNA (Fig. 2c), from which the SOWAHC 3' UTR could not be retrieved. Results obtained in a “rescue” experiment also suggested that mRNA–mRNA duplexes form via base-pairing between complementary 3' UTR Alu elements (Supplementary Fig. 2a–c): Retrieval of the FLUC-SOWAHC 3' UTR-MS2bs mRNA using the anti-Flag antibody immunoprecipitated RLUC-PPID 3' UTR mRNA but not RLUC-PPID 3' UTR(AluR) mRNA, in which exclusively the 3' UTR Alu element of PPID mRNA was in reversed orientation; however, RLUC-PPID 3' UTR(AluR) mRNA could be retrieved using FLUC-SOWAHC 3' UTR(AluR)-MS2bs mRNA, the latter of which also harbored exclusively its 3' UTR Alu element in reversed orientation. Notably, STAU2, which is a paralog of STAU1, also functions in SMD13. Calnexin, protein kinase RNA-activated (PKR) and SERPINE1 mRNA, which are irrelevant, were not detectably immunoprecipitated under any condition tested (Fig. 2b, c). Furthermore, downregulating STAU1 decreased the co-IP of CDCP1, TUFT1 and PPID mRNAs with FLUC-SOWAHC 3' UTR-MS2bs mRNA (Fig. 2d), indicating that STAU1 stabilizes the mRNA–mRNA complexes. In agreement with formaldehyde-crosslinking results, compared to cells that were untreated, mRNA–mRNA base-pairing between complementary 3' UTR Alu elements was enhanced using cells treated with psoralen (Supplementary Fig. 2d, e), which forms covalent RNA–RNA crosslinks between two adjacent pyrimidine bases that are situated on opposite strands of dsRNA23.
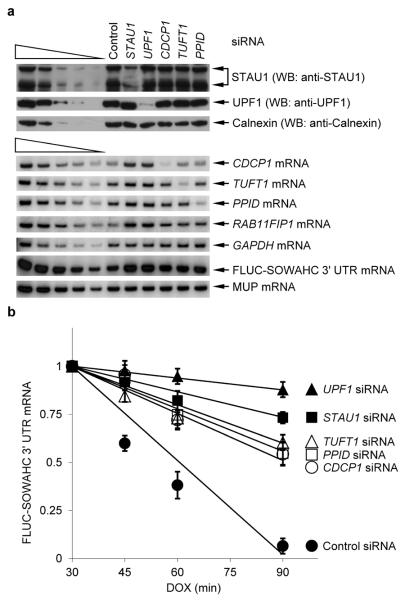
mRNA–mRNA duplexes trigger SMD
To determine if the reduction in mRNA abundance brought about by the observed mRNA–mRNA duplexes triggers mRNA decay, cells transiently expressing pTRE-FLUC-SOWAHC 3' UTR, which produces FLUC-SOWAHC 3' UTR mRNA from a doxycycline-repressible promoter, in the presence of one of several siRNAs (Fig. 3a) were harvested 30, 45, 60 or 75 min after doxycycline treatment. Compared to Control siRNA, siRNAs that downregulated STAU1 or UPF1 or CDCP1, TUFT1 or PPID mRNA significantly increased the half-life of FLUC-SOWAHC 3' UTR mRNA (Fig. 3b and Supplementary Fig. 3a, b) as well as the abundance of FLUC-SOWAHC 3' UTR mRNA and RAB11FIP1 mRNA (Fig. 3a).
Figure 3. mRNA–mRNA duplexes create SBSs and trigger SMD.
a, Western blot (upper) or RT-sqPCR(lower) of lysates of HeLa Tet-Off cells that had been transiently transfected with the specified siRNA (see Supplementary Fig. 3c for data using alternative siRNAs) in the presence of 2 μg/ml of doxycycline (DOX) and, after removing doxycycline 48-h later, with pTRE-FLUC-SOWAHC 3' UTR, which contains a doxycycline-repressible promoter, and the phCMV-MUP reference plasmid. The removal of DOX elicits a transcriptional burst in FLUC-SOWAHC 3' UTR mRNA production. A fraction of cells was harvested after an additional 4-h (time 0) and analyzed. b, Plot of RT-sqPCR data using samples shown in a that were further processed as follows. At time 0, 2 μg/ml of doxycycline were added to the remaining cells. Additional fractions of cells were harvested at the specified times thereafter. RT-sqPCR analyses were essentially as in Fig. 1b. As in Fig. 1d, the levels of exogenous FLUC-SOWAHC 3' UTR mRNAs were comparable to the level of cellular SOWAHC mRNA. Error bars, s.e.m.; # of independently performed experiments = 3; P < 0.05 (Chi-squared test).
SMD targets only translated strand(s) of mRNA–mRNA duplexes
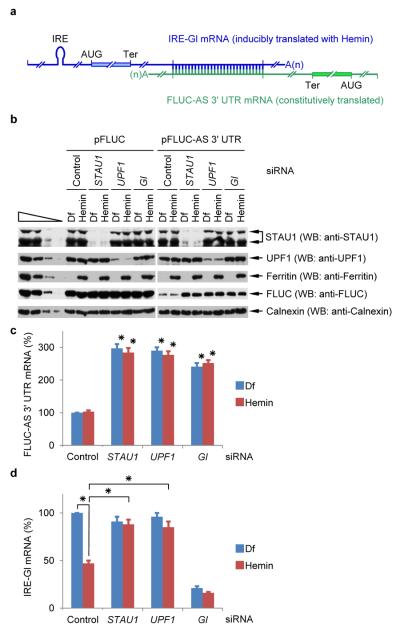
To date, only the mRNA strand of an mRNA–lncRNA (i.e. an mRNA–½-sbsRNA) duplex has been found to be targeted for SMD2, possibly, we hypothesized, because only the mRNA in the duplexes studied is translationally active2,24. However, this generalization is complicated by the finding that some transcripts that have been designated as lncRNAs footprint with ribosomes, suggesting that their short open reading frames might be translated25. Nevertheless, data suggest that the RNP structures of our lncRNAs differ in uncharacterized ways from those of mRNAs even though both are largely cytoplasmic. Nine-in-ten siRNAs chosen to downregulate an mRNA are generally successful, and the degree of downregulation is to ~10% of normal; however, only one-in-ten siRNAs chosen to downregulate an lncRNA is generally successful, and the degree of downregulation is to only ~50% of normal11 (data not shown). To determine the fate of mRNA–mRNA duplexes in which one mRNA is translationally active and the other is not, HeLa cells stably expressing IRE-Gl mRNA26 and transiently expressing (i) FLUC-AS 3' UTR mRNA or FLUC mRNA, (ii) Control, UPF1, STAU1 or Gl siRNA, and (iii) the reference MUP mRNA were cultured in the presence of either hemin or deferoxamine mesylate (Df) for 18-h prior to harvesting. IRE-Gl mRNA harbors the 35-nucleotide iron-responsive element (IRE) of ferritin mRNA in its 5' UTR26, and the 3' UTR of FLUC-AS 3' UTR mRNA contains 66 nucleotides that perfectly duplex with 3' UTR nucleotides in IRE-Gl mRNA (Fig. 4a).
Figure 4. An mRNA that duplexes with another mRNA by 3' UTR base-pairing is targeted for SMD provided it is translationally active.
a, Diagram of IRE-Gl mRNA base-paired with FLUC-AS 3' UTR mRNA, which contains 3' UTR sequences engineered to be complementary to 3' UTR sequences of Gl mRNA. b, Western blot (WB) of lysates of HeLa cells stably expressing IRE-Gl mRNA and transiently expressing FLUC-AS 3' UTR mRNA (+) or, as a negative control, FLUC mRNA (−), the specified siRNA, and the reference MUP mRNA. Cells were cultured in the presence of either hemin or deferoxamine mesylate (Df) for 18-h prior to harvesting. c, Histogram representations of RT-sqPCR analyses of samples analyzed in b. The level of FLUC-AS 3' UTR mRNA was normalized to the level of MUP mRNA (Supplementary Fig. 4b), and the normalized level in the presence of Df and Control siRNA is defined as 100. d, as in c, but the level of IRE-Gl mRNA was normalized to the level of cellular SMG7 mRNA (Supplementary Fig. 4b). Error bars, s.e.m.; # of independently performed experiments = 3; *, P <0.05 (one-tail t-test).
As expected IRE-Gl mRNA was translationally active in the presence of hemin, but translationally inactive in the presence of Df (Supplementary Fig. 4a), as was ferritin mRNA (Fig. 4b). In contrast, FLUC-AS 3' UTR mRNA was constitutively translated (Fig. 4b). The level of FLUC mRNA was unaffected by hemin, Df or any siRNA (Supplementary Fig. 4b). The levels of FLUC-AS 3' UTR mRNA and its product protein were likewise constant in the presence of either hemin or Df, but elevated 2-3-fold in the presence of STAU1, UPF1 or Gl siRNA (Fig. 4b, c and Supplementary Fig. 4b, c) indicating that it is targeted for SMD regardless of the translational status of IRE-Gl mRNA. Compared to Df, hemin decreased the level of IRE-Gl mRNA in the presence of Control siRNA, suggesting that IRE-Gl mRNA translation is required for its own SMD (Fig. 4d and Supplementary Fig. 4b, c). Consistent with this interpretation, in the presence of hemin, the level of IRE-Gl mRNA after STAU1 or UPF1 siRNA treatment, compared to Control siRNA treatment, was upregulated ~2-fold (Fig. 3d and Supplementary Fig. 4b, c), i.e., was upregulated when the level of FLUC-AS 3' UTR mRNA was upregulated by either STAU1 or UPF1 siRNA treatment (Fig. 4c and Supplementary Fig. 4b, c). As expected, the level of IRE-Gl mRNA was downregulated in the presence of either hemin or Df and the presence of Gl siRNA (Fig. 4d and Supplementary Fig. 4b, c). Thus, translationally active IRE-Gl mRNA, unlike translationally inactive IRE-Gl mRNA, is an SMD target by virtue of its base-pairing with FLUC-AS 3' UTR mRNA. We conclude that when an mRNA–mRNA duplex creates an SBS via base-pairing between complementary 3' UTR sequences, each constituent mRNA is targeted for SMD provided that it is translated and the SBS resides sufficiently downstream of the termination codon.
SMD of mRNA–mRNA duplexes is functionally important
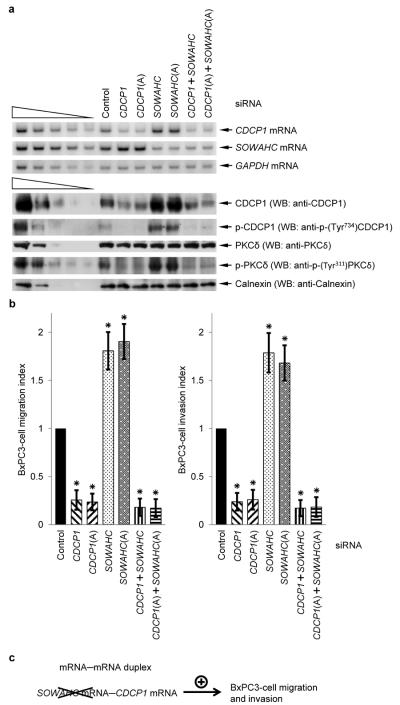
We next examined the biological relevance of the complex web of mRNA–mRNA duplexes that are SMD targets using a trans-well9 assay to measure the migration and invasion indices of human pancreatic adenocarcinoma BxPC3 cells after introducing one or more siRNAs. Additionally, cell migration rates were assessed using human keratinocyte HaCaT cells in a wound-healing, also called or scrape-injury, assay9. As expected9, downregulating CDCP1 protein inhibited cell migration and invasion (Fig. 5, Supplementary Fig. 5). Additionally, downregulating SOWAHC mRNA promoted cell migration and invasion (Fig. 5, Supplementary Fig. 5). To test the hypothesis that downregulating SOWAHC mRNA promotes cell migration and invasion by inhibiting the SMD of CDCP1 mRNA, we assessed the phosphorylation status of CDCP1 and PKCδ proteins using antibody to either phospho-(Tyr734)CDCP1 or phospho-(Tyr311)PKCδ. CDCP1 protein is known to undergo autophosphorylation, the level of which increases with an increase in CDCP1 protein cellular abundance9. Once autophosphorylated, CDCP1 protein binds PKCδ protein, resulting in an increased level of PKCδ phosphorylation that promotes cell motility9. In support of our hypothesis, downregulating SOWAHC mRNA was found to augment the degree of CDCP1 and PKCδ phosphorylation (Fig. 5a). Furthermore, downregulating SOWAHC mRNA failed to increase the level of PKCδ phosphorylation or promote cell migration and invasion in the presence of CDCP1 siRNA (Fig. 5a, b).
Figure 5. Evidence that the SMD of mRNA–mRNA duplexes contributes to pancreatic adenocarcinoma cell migration and invasion.
a, RT-sqPCR and Western blotting (WB) analyses of lysates of BxPC3 cells that had been transiently transfected with the specified siRNA(s). GAPDH mRNA and Calnexin represent loading controls. b, Migration and invasion assays of BxPC3 cells after transfection with the specified siRNA(s). c, Model for the mechanism by which siRNA to SOWAHC mRNA, which encodes a protein of unknown function, promotes BxPC3-cell migration and invasion: downregulating SOWAHC mRNA inhibits SOWAHC mRNA-CDCP1 mRNA base-pairing and, thus, the SOWAHC mRNA-mediated SMD of CDCP1 mRNA, which encodes a protein that inhibits cell migration and invasion. # of independently performed experiments = 3, *, P <0.05 (one-tail t-test).
DISCUSSION
Mammalian-cell mRNAs have long been known to constitute the intermediate between the DNA from which they are synthesized and the proteins that they encode. Recently, mRNAs have been shown to crosstalk with other mRNAs by serving as competing endogenous RNAs (ceRNAs), also called target mimics, that “sponge up” microRNAs, thus minimizing the microRNA-mediated downregulation of other target mRNAs27–31. Here we report a new and unanticipated physiologically important function for mRNAs: the SMD of one or more translationally active mRNAs that base-pair via partially complementary 3' UTR Alu elements. When both mRNAs of such an mRNA–mRNA duplex are translationally active, then each is at once regulated and regulator since both are subjected to SMD.
The decay of a particular mRNA–mRNA duplex can in theory be fine-tuned by many variables. Variables include changes in the cytoplasmic abundance and/or translational state of one or both of the constituent mRNAs. Additionally, an increase or decrease in the number of mRNAs or lncRNAs that compete with mRNA–mRNA duplex formation, mRNA–mRNA duplex maintenance, or both could fine-tune decay. It should be noted that even though levels of the six mRNAs studied in this report are comparable to each other (Supplementary Fig. 1e), there is no need for these mRNAs to be in equal proportions to exert regulation of one another. Another possible means of regulation is alternative polyadenylation site usage, which could either maintain or remove a 3' UTR Alu element that is required for mRNA–mRNA duplex formation. Alternative polyadenylation site usage affects the transcripts of more than half of human genes and is highly regulated depending on tissue-type and/or proliferation/differentiation state32,19.
It is becoming clear that human cells have usurped 3' UTR Alu elements for a multitude of regulatory purposes. Studies of a newly discovered type of regulation by intramolecular base-pairing between inverted repeat 3' UTR Alu elements (3' UTR IRAlus) has revealed that STAU1 binding to 3' UTR IRAlus competes with binding of other dsRNA-binding proteins – p54nrb within nuclei, and protein kinase R (PKR) within the cytoplasm – so as to preclude p54nrb-mediated nuclear retention in paraspeckles, and also PKR-mediated translational repression in the cytoplasm33. Given the overlap with which dsRNA-binding proteins recognize duplexes formed by base-pairing between 3' UTR Alu elements, it will be interesting to determine how dsRNA-binding proteins other than STAU1 and STAU2 recognize and influence the metabolism of mRNA–mRNA duplexes.
ONLINE METHODS
Computational analyses
Two files, “Homo_sapien_GRCh37.62.cdna.all.fa”, which contains the sequence of each transcript, and “Homo_sapien_GRCh37.62.gft”, which contains splicing site(s) and coding region information for each human transcript, were downloaded from ENSEMBL (www.ensembl.org). The 3' UTR sequences were extracted and applied to RepeatMasker (www.repeatmasker.org) to identify 3' UTR Alu element(s). The DuplexFold function of RNAstructure (http://rna.urmc.rochesterledu/RNAstructure.html) was used to computationally predict the ΔG of derived human mRNA–mRNA duplexes formed via base-pairing of partially complementary 3' UTR Alu elements. Intermolecular duplexes between the 3' UTR Alu elements of specific mRNAs were computationally predicted using the Perl program “RNA_RNA_anneal”2.
Plasmid Constructions
To construct pFLUC-SOWAHC 3' UTR(ΔAlu), pFLUC-SOWAHC 3' UTR2, which contains the complete 3' UTR of SOWAHC mRNA, was PCR-amplified using two sets of primer pairs: 5'-GAGTCAAAGCTTATGGAAGACGCCAAAAACATAAAG-3' (sense A) and 5'-CATTCATCTTTCATCCATCTTTAAACAAATTC-3' (antisense A), and 5'-GGATGAAAGATGAATGAAAATATGAAATATATAAAC-3' (sense B) and 5'-GTCAGGGCCCTTTCACAAAACAAACTTTACAGATTTTACAATCTCCC-3' (antisense B), where underlined nucleotides specify HindII and ApaI, respectively. The resulting two PCR products were mixed and PCR-amplified using the sense A and antisense B primer pair. This PCR product was cleaved with HindIII and ApaI and subsequently inserted into HindIII- and ApaI-cleaved pFLUC.
To construct pFLUC-SOWAHC 3' UTR -MS2bs or pFLUC-SOWAHC 3' UTR(ΔAlu)-MS2bs, pFLUC-SOWAHC 3' UTR or pFLUC-SOWAHC 3' UTR(ΔAlu) was PCR-amplified using the primer pair 5'- GAGTCAAAGCTTATGGAAGACGCCAAAAACATAAAG-3' (sense) and 5'- GTCAGGATCCGAGACAGAGTCTCCGTTGCCCAGG-3' (antisense), where underlined nucleotides denote a HindIII or BamHI site, respectively. The resulting PCR products were cleaved using HindIII and BamHI and subsequently inserted into HindIII- and BamHI-cleaved pFLUC-MS2bs2.
To construct pFLUC-AS 3' UTR, pFLUC was PCR-amplified using two sets of primer pairs: 5'-GAGTCAAAGCTTATGGAAGACGCCAAAAACATAAAG-3' (sense C) and 5'-CTTCATAATATCCCCCAGTTTAGTAGTTGGACTTAGGGAACTAATAAAATGAGGAAA TTGCATCGC-3' (antisense C), and 5'-TGGGGGATATTATGAAGGGCCTTGAGCATCTGGATTCTGCCATGGCTGGCAACTAGA AGGCACAG-3' (sense D) and 5'-GTCACCTAGGCCTCCAAAAAAGCCTCCTCACTAC-3' (antisense D). The resulting two PCR products were mixed and PCR-amplified using the sense C and antisense D primer pair. This PCR product was cleaved with HindIII and AvrII and subsequently inserted into HindIII- and AvrII-cleaved pFLUC.
To construct pTRE-FLUC-SOWAHC 3' UTR, FLUC-SOWAHC 3' UTR was PCR-amplified using pFLUC-SOWAHC 3' UTR and the primer pair 5'-GAGTCAAAGCTTATGGAAGACGCCAAAAACATAAAG-3' (sense) and 5'-GTCAGAATTCGAGACAGAGTCTCCGTTGCCC-3' (antisense), where underlined nucleotides denote a SacII or EcoRI site, respectively. The resulting PCR product was digested with SacII and EcoRI and subsequently inserted into SacII- and EcoRI-cleaved pTRE (Clontech).
To construct pFLUC-SOWAHC 3' UTR(AluR)-MS2bs, in which specifically the Alu element was present in reverse orientation (R), pFLUC-SOWAHC 3' UTR-MS2bs was PCR-amplified using three sets of primer pairs to generate PCR products E, F and G, respectively: 5'-CTTAATTTTCCAAGCTTTTCTTGAC-3' (sense E) and 5'-TTTCATCCATCTTTAAACAAATTCAATGACC-3' (antisense E), 5'-CATATTTTCATTCATCGGCCAGGCATGGTGGCTCATGCC-3' (sense F) and 5'-GTTTAAAGATGGATGAAATTTTTTTTTTTTTTTTTTGAGACAGAGTCTCCGTTGCCC-3' (antisense F), and 5'-GATGAATGAAAATATGAAATATATAAAC-3' (sense G) and 5'-CAGTGGGAAAACCATGGTAAATGAC-3' (antisense G). PCR products E and F were mixed and PCR-amplified using the sense E and antisense F primer pair to generate PCR product EF. Then PCR products EF and G were mixed and PCR-amplified using the sense E and antisense G primer pair. This PCR product was cleaved with HindIII and NcoI and subsequently inserted into HindIII- and NcoI-cleaved pFLUC-SOWAHC 3' UTR-MS2bs.
To construct pRLUC-PPID 3' UTR, the complete 3' UTR of PPID mRNA was amplified using HeLa-cell genomic DNA and primer pair: 5'-GATGTCTAGAGTGAAAAGGATTCAGTTTTGCTTATTGTGTG-3' (sense) and 5'-GTCAGGGCCCCAATAAGATGTGTCTTTACAAGTTTAAAAAGACTGGACACC-3' (antisense). The resulting PCR product was cleaved with XbaI and ApaI and subsequently inserted into XbaI- and ApaI-cleaved pRLUC19.
To construct pRLUC-PPID 3' UTR(AluR), pRLUC-PPID 3' UTR was PCR-amplified using three sets of primer pairs: 5'-GATGTCTAGAGTGAAAAGGATTCAGTTTTGCTTATTGTGTG-3' (sense H) and 5'-CCACTTTACTTACTGTATTGTGAC-3' (antisense H), 5'-CCATTTTAAAAAACATTTTACTTTTTTGTTTGTTTCTTTGAGATGGAG-3' (sense I) and 5'- ACAGTAAGTAAAGTGGGGCCGGGCACGGTGGCTCACACC-3' (antisense I), and 5'-AAGTAAAATGTTTTTTAAAATGG-3' (sense J) and 5'-GTCAGGGCCCCAATAAGATGTGTCTTTACAAGTTTAAAAAGACTGGACACC-3' (antisense J). PCR products H and I were mixed and PCR-amplified using the sense H and antisense I primer pair to get PCR product HI. Then HI and J were mixed and PCR-amplified using the sense H and antisense J primer pair. This PCR product was cleaved with XbaI and ApaI and subsequently inserted into XbaI- and ApaI-cleaved pRLUC-PPID 3' UTR.
To construct pFLUC-artificial 3' UTR (shuffle), pFLUC was PCR-amplified using two sets of primer pairs: 5'-GAGTCAAAGCTTATGGAAGACGCCAAAAACATAAAG-3' (sense K) and 5'-GATGGCTGAAGGAAGGAGGACAGTGGGAGTGGCACCTTCCAGGGTCGCAACTAGAA GGCACAGTCGAGGC-3' (antisense K), and 5'-ACTGTCCTCCTTCCTTCAGCCATCTGTTGTTTGCCCCTCCCCCGTGTCCTAATAAAAT GAGGAAATTGCATCGC-3' (sense L) and 5'-GTCACCTAGGCCTCCAAAAAAGCCTCCTCACTAC-3' (antisense L). The resulting two PCR products were mixed and PCR-amplified using the sense K and antisense L primer pair. This PCR product was cleaved with HindIII and AvrII and subsequently inserted into HindIII-and AvrII-cleaved pFLUC.
To construct (i) pAS lncRNA 1–68, (ii) pAS lncRNA 1–30 or (iii) pAS lncRNA39–68, p½-sbsRNA1(S)2 was PCR-amplified using two sets of primer pairs: (i) 5'-GAGTCAAAGCTTAAAGGAGAGACAGTCTCACTCTG-3' (sense M) and 5'-CCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCACTCCCACTGTCCTACAAAGAAAAT TAAATTAGCAGGG-3' (antisense N), and 5'-CCAGGGTCAAGGAAGGCACGGGGGAGGGGCAAACAACAGATGGCTGTTAGGATTA CAGGAGTGAGCC-3' (sense N) and 5'-GTCAGCGGCCGCCAGTTGTAAGCATATTTGGGTTAC-3' (antisense M), (ii) 5'-GAGTCAAAGCTTAAAGGAGAGACAGTCTCACTCTG-3' (sense M) and 5'-GAAGGTGCCACTCCCACTGTCCTACAAAGAAAATTAAATTAGCAGGG-3' (antisense O), and 5'-TGGGAGTGGCACCTTCCAGGGTCTTAGGATTACAGGAGTGAGCC-3' (sense O) and 5'-GTCAGCGGCCGCCAGTTGTAAGCATATTTGGGTTAC-3' (antisense M), or (iii) 5'-GAGTCAAAGCTTAAAGGAGAGACAGTCTCACTCTG-3' (sense M) and 5'-CTGTTGTTTGCCCCTCCCCCGTGACAAAGAAAATTAAATTAGCAGGG-3' (antisense P), and 5'-GAGGGGCAAACAACAGATGGCTGTTAGGATTACAGGAGTGAGCC-3' (sense P) and 5'-GTCAGCGGCCGCCAGTTGTAAGCATATTTGGGTTAC-3' (antisense M). The resulting two PCR products were mixed and PCR-amplified using the corresponding sense M and antisense M primer pairs. Each PCR product was subsequently cleaved with HindIII and NotI and inserted into HindIII- and NotI-cleaved p½-sbsRNA1(S).
Cell culture, transient transfections, formaldehyde crosslinking, and mRNA half-life determinations
Human HeLa cells, HeLa Tet-off cells (Clontech), HeLa cells stably expressing IRE-Gl Norm mRNA26, or HaCaT cells were grown in DMEM (Gibco-BRL) containing 10% fetal bovine serum (Gibco-BRL) at 37°C and in 5% CO2. Human BxPC3 cells (Sigma) were grown in RPMI 1640 (Gibco-BRL) containing 10% fetal bovine serum (Gibco-BRL). After reaching a density of 2 × 106/60-mm dish or 7.5 × 107/150-mm dish, cells were transiently transfected with the specified plasmids using Lipofectamine 2000 (Invitrogen) or the specified siRNA using Oligofectamine (Invitrogen) as described2. siRNAs consist of the nonspecific Silencer Negative Control #1 (Ambion), STAU1 siRNA2, STAU1(A) siRNA2, UPF1 siRNA2, UPF1(A) siRNA2, SERPINE1 siRNA (Ambion, cat#: s10013), SERPINE1(A) siRNA (Ambion, cat#: s10014), SOWAHC siRNA (Ambion, cat#: s35230), SOWAHC(A) siRNA (Ambion, cat#: s35231), RAB11FIP1 siRNA (Ambion, cat#: s37090), RAB11FIP1 (A) siRNA (Ambion, cat#: s37092), CDCP1 siRNA (Ambion, cat#: s35061), CDCP1(A) siRNA (Ambion, cat#: s35062), TUFT1 siRNA (Ambion, cat#: s14508), TUFT1(A) siRNA (Ambion, cat#: s14509), PPID siRNA (Ambion, cat#: s10913), PPID(A) siRNA (Ambion, cat#: s10915) and ½-sbsRNA1 siRNA2 (Supplementary Table 4). For all immunoprecipitations, cells were crosslinked using 1% formaldehyde for 10 min at room temperature and subsequently quenched with 0.25 M glycine for 5 min at room temperature prior to lysis using sonication and subsequently reversing the crosslinks by heating for 45 min at 65°C2. Alternatively, cells were crosslinked using psoralen23. Cells were washed with phosphate-buffered saline (PBS) twice and then incubated with 5μM psoralen (Sigma) in PBS for 10 min. A first round of psoralen crosslinking was achieved using 365nm UV light (1.2kJ/m2/min; UVP) for 5 min at room temperature, after which excess psoralen was removed by two sequential incubations in PBS at 37°C in 5% CO2 for 10 min. A second round of psoralen crosslinking was achieved in the presence of 365 nm UV light (1.2kJ/m2/min) for 15 min at room temperature. Cells were harvested as usual. For half-life measurements2, HeLa Tet-Off cells were transfected with the specified siRNA in the presence of 2 μg/ml of doxycycline and, after removing doxycycline 48-h later, were transfected with the pTRE-FLUC-SOWAHC 3' UTR reporter plasmid, which contains a doxycycline-repressible promoter, and the phCMV-MUP reference plasmid. A fraction of cells was harvested after an additional 4-h (time 0), at which point 2 μg/ml of doxycycline was added to the remaining cells to block FLUC-SOWAHC 3' UTR mRNA synthesis. Aliquots of cells were harvested at various times thereafter. HeLa cells stably expressing IRE-Gl Norm mRNA or IRE-Gl Ter mRNA were transfected with specified siRNA and, subsequently, the specified plasmids and cultured for one day after which 100 μM of deferoxamine mesylate (Sigma) was added. After 24 h, cells were washed with phosphate-buffered saline and subsequently cultured in fresh medium containing 100 μM of either deferoxamine mesylate or hemin (Sigma) for the specified time prior to lysis.
Protein purification, IP and western blotting
HeLa cells were lysed, and protein was isolated using hypotonic buffer that consists of 10mM Tris-Cl (pH 7.4), 150mM NaCl, 2mM EDTA, 0.5% Triton X-100, 2mM benzamidine, 1mM PMSF and 1 tablet of protease inhibitor cocktail (Roche). If crosslinked, cells were sonicated six times for 30 sec to facilitate lysis. IP was performed as described2. Western blotting was performed as outlined2. Antibodies consisted of anti-SOWAHC (Bethyl Laboratories, cat#: A301-412A), anti-STAU134, anti-UPF111, anti-calnexin (StressGene, cat#: SPA-860), anti-Flag (Sigma, cat#: A8592), anti-ferritin (AbCam, cat#: ab7332), anti-CDCP1 (Cell Signaling Technology, cat#: 4115), anti-p(Tyr734)CDCP1 (Cell Signaling Technology, cat#: 9050), anti-PKCδ (Cell Signaling Technology, cat#: 2058), and anti-p(Tyr311)PKCδ (Cell Signaling Technology, cat#: 2058). Western blot analyses situated under wedges in the leftmost lanes of figures utilized 3-fold dilutions of protein/cells lysate and show that data fall in the linear range.
RNA purification and RT coupled to either semiquantitative (sq) or realtime (q)PCR
RNA was purified from total-cell lysates or immunoprecipitated-cell lysates using TRIzol (Invitrogen) as reported2. RT-sqPCR and RT-qPCR were performed2 using the designated primer pairs (Supplementary Table 3). RT-sqPCR analyses situated under wedges in the leftmost lanes of figures utilized 2-fold dilutions of RNA and show that data fall in the linear range. RT-PCR values include standard error of measurements obtained in the specified number of independently performed experiments.
Scrape-injury and cell migration and invasion assays
Scrape-injury repair assays were essentially as published2. Human BxPC3 cells (7.5 × 107/150-mm dish) were treated with specified siRNA for 72 h, serum-starved for 6 h, and detached using trypsin-EDTA. Then, 1 × 105 cells in 100 μl of RPMI 1640 were seeded onto either Falcon Cell Culture Inserts (BD Biosciences) for the migration assay or the upper part of a BD BioCoat Matrigel Invasion Chamber (BD Biosciences) for the invasion assay. After incubations of 6 h and 16 h, respectively, membrane-associated cells were fixed using methanol and stained using Toluidine blue (Sigma). The total number of migrated or invaded cells was determined by counting the cells that had moved from each of three wells through the membrane. Nine fields per membrane were evaluated using an Axiovert 40C inverted microscope (Zeiss).
Supplementary Material
Acknowledgements
We thank D. Mathews for computational advice, K. Nehrke for access to his inverted fluorescence microscope, S. de Lucas and J. Ortín for anti-STAU1 antibody, C. Beckham for help with the trans-well assay, and M. W.-L. Popp for comments on the manuscript. This work was supported by the US National Institutes of Health (GM074593 to L.E.M) and a University of Rochester Messersmith Graduate Student Fellowship (C.G).
Footnotes
Author Contributions C.G. wrote the Perl programs and undertook the bioinformatics analyses and performed, with assistance from Y.T., wet-bench experiments. C.G. and L.E.M. designed the experiments, analyzed experimental data, and wrote the manuscript.
References
- 1.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho H, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2013 doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 7.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 8.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 9.Miyazawa Y, et al. CUB domain-containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res. 2010;70:5136–5146. doi: 10.1158/0008-5472.CAN-10-0220. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz J. SINEs as driving forces in genome evolution. Genome Dyn. 2012;7:92–107. doi: 10.1159/000337117. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. U S A. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 15.Melero R, et al. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3' end. Nat. Struct. Mol. Biol. 2012;19:498–505. S491–492. doi: 10.1038/nsmb.2287. [DOI] [PubMed] [Google Scholar]
- 16.Gehring NH, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat. Struct. Mol. Biol. 2013;20:515–524. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martel C, et al. Multimerization of Staufen1 in live cells. RNA. 2010;16:585–597. doi: 10.1261/rna.1664210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong C, Popp MW, Maquat LE. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 2012 doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 23.Lipson SE, Hearst JE. Psoralen cross-linking of ribosomal RNA. Methods Enzymol. 1988;164:330–341. doi: 10.1016/s0076-6879(88)64053-5. [DOI] [PubMed] [Google Scholar]
- 24.Gong C, Maquat LE. “Alu”strious long ncRNAs and their role in shortening mRNA half-lives. Cell Cycle. 2011;10:1882–1883. doi: 10.4161/cc.10.12.15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009;23:2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubio-Somoza I, Weigel D, Franco-Zorilla JM, García JA, Paz-Ares J. ceRNAs: miRNA target mimic mimics. Cell. 2011;147:1431–1432. doi: 10.1016/j.cell.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elbarbary R, Li W, Tian B, Maquat LE. STAU1 binding to 3' UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27:1495–1510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marion RM, Fortes P, Beloso A, Dotti C, Ortín J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.