Abstract
Neuronal nicotinic acetylcholine receptor (nAChR) genes (CHRNA5/CHRNA3/CHRNB4) have been reproducibly associated with nicotine dependence, smoking behaviors, and lung cancer risk. Of the few reports that have focused on early smoking behaviors, association results have been mixed. This meta-analysis examines early smoking phenotypes and SNPs in the gene cluster to determine: (1) whether the most robust association signal in this region (rs16969968) for other smoking behaviors is also associated with early behaviors, and/or (2) if additional statistically independent signals are important in early smoking. We focused on two phenotypes: age of tobacco initiation (AOI) and age of first regular tobacco use (AOS). This study included 56,034 subjects (41 groups) spanning nine countries and evaluated five SNPs including rs1948, rs16969968, rs578776, rs588765, and rs684513. Each dataset was analyzed using a centrally generated script. Meta-analyses were conducted from summary statistics. AOS yielded significant associations with SNPs rs578776 (beta = 0.02, P = 0.004), rs1948 (beta = 0.023, P = 0.018), and rs684513 (beta = 0.032, P = 0.017), indicating protective effects. There were no significant associations for the AOI phenotype. Importantly, rs16969968, the most replicated signal in this region for nicotine dependence, cigarettes per day, and cotinine levels, was not associated with AOI (P = 0.59) or AOS (P = 0.92). These results provide important insight into the complexity of smoking behavior phenotypes, and suggest that association signals in the CHRNA5/A3/B4 gene cluster affecting early smoking behaviors may be different from those affecting the mature nicotine dependence phenotype.
Keywords: CHRNA5, CHRNA3, CHRNB4, meta-analysis, nicotine, smoke
Introduction
Tobacco use usually begins during adolescence; over 90% of those who smoke more than 100 cigarettes become regular smokers and one-third of those adolescents who experiment with tobacco go on to become nicotine dependent [Anthony et al., 1994; CDC, 2005; Mayhew et al., 2000]. Symptoms of dependence in these youths may appear within weeks of occasional tobacco use [DiFranza, 2008; DiFranza et al., 2000, 2002; Doubeni et al., 2010; Escobedo et al., 1993; O’Loughlin et al., 2002a, 2002b], particularly loss of control over their smoking [DiFranza et al., 2007; Scragg et al., 2008; Soteriades et al., 2011]. Early initiation of tobacco use is also associated with an increased number of daily cigarettes smoked in adulthood [Chen and Millar, 1998; D’Avanzo et al., 1994; Eisner et al., 2000; Everett et al., 1999; Fernandez et al., 1999] and a decreased rate of smoking cessation [Breslau and Peterson, 1996; Chen and Millar, 1998; Eisner et al., 2000; Lando et al., 1999]. Age of smoking onset in women is a significant predictor of cessation during pregnancy; a woman who initiates earlier is more likely to smoke while pregnant [Chen et al., 2006]. Individuals who initiate tobacco use early in life are also at greater risk for smoking-related lung cancer, in part because this tissue type continues to develop into early adulthood [Hirao et al., 2001; Wiencke and Kelsey, 2002; Wiencke et al., 1999]. In addition, smoking accelerates the rate of cervical maturation and cell proliferation, therefore resulting in an increased risk of cervical cancer [Hwang et al., 2009; Ma et al., 2011]. Given the long-term health implications of early tobacco use, many studies have worked to identify and understand the underlying genetic and environmental risk factors for early smoking behaviors.
It is well established that genetic factors contribute to risk for tobacco initiation, quantity smoked, nicotine dependence, and smoking persistence [Li et al., 2003; Rhee et al., 2003; Rose et al., 2009; Schnoll et al., 2007]. Neuronal nicotinic acetylcholine receptor (nAChR) subunit genes are among the top candidates for several smoking-related phenotypes because a cluster of nicotinic receptor genes (CHRNA5/CHRNA3/CHRNB4) on chromosome 15q25 has been repeatedly associated with nicotine dependence, smoking behaviors, and lung cancer [Amos et al., 2008; Berrettini et al., 2008; Broms et al., 2012; Furberg et al., 2010; Greenbaum and Lerer, 2009; Hung et al., 2008; Liu et al., 2010; Rose, 2007; Saccone et al., 2010a, 2007; Thorgeirsson et al., 2008; 2010]. This region also contains the nonsynonymous risk variant rs16969968 G/A, that causes a functional amino acid change in the α5 subunit protein (Asp398Asn); the Asparagine (Asn) allele decreases response to a nicotine agonist [Bierut et al., 2008]. The high-risk α5 Asn 398 is associated with reduced Ca2+ permeability and desensitizes faster than the wild type Aspartic acid (Asp) at 398 (α4β2)2 α5 [Kuryatov et al., 2011]. In addition, Alpha5 knockout mice showed increased nicotine intake [Fowler et al., 2011]. This variant accounts for 4% of the variance in the serum levels of the long-term metabolite of nicotine, cotinine [Etter et al., 2009; Keskitalo et al., 2009]. In addition to this well-characterized functional variant, there have been other robust associations within the CHRNA5/CHRNA3/CHRNB4 gene cluster to smoking quantity. These were reported in three genome-wide meta-analyses identifying SNPs in the cluster as top association signals with the outcome of cigarettes smoked per day (CPD) [Furberg et al., 2010; Liu et al., 2010; Thorgeirsson et al., 2010]. These reports support the prevailing view that smoking quantity/frequency as a proxy for nicotine dependence is most consistently associated with SNPs at the CHRNA5/CHRNA3/CHRNB4 locus. A fourth meta-analysis [Saccone et al., 2010a] focused on four statistically distinct loci in the CHRNA5/CHRNA3/CHRNB4 cluster tagged by SNPs rs16969968 (CHRNA5), rs578776 (CHRNA3), rs588765 (CHRNA5), and rs12914008 (CHRNB4) [Saccone et al., 2009a]. SNPs rs16969968 and rs588765 were each strongly associated with CPD in a joint model that adjusts for the effect of each, suggesting independent effects; rs578776 is also associated with CPD, but its effect size is smaller after adjustment for SNP rs16969968 in the association model [Saccone et al., 2010a].
Although the majority of the studies of CHRNA5/ CHRNA3/CHRNB4 variation and tobacco have targeted smoking behaviors in adulthood, a few reports specifically focused on early smoking behaviors. Weiss et al. found that a CHRNA5/CHRNA3/CHRNB4 haplotype including SNPs rs16969968 and rs578776 was significantly associated with severity of nicotine dependence among individuals who began daily smoking at or before age 16, but not in those who began smoking after age 16 [Weiss et al., 2008]. Correlated SNPs rs1948 (CHRNB4) and rs8023462 (CHRNA3/CHRNB4) (r2 = 0.8) were found to be associated with early age of tobacco initiation in two independent samples [Schlaepfer et al., 2008]. These two SNPs are not in high linkage disequilibrium with any of the four loci mentioned above (rs1696998 r2 = 0.32, rs578776 r2 = 0.09, rs588765 r2 = 0.56, and rs12914008 r2 = 0.02) (1000 Genomes Pilot) 1 CEU [Abecasis et al., 2012]. A second study targeting age of tobacco initiation in a Korean sample found that SNPs rs6495308, rs11072768, and rs951266 were significant for association [Li et al., 2010], but it is not clear whether in this sample these SNPs are in high linkage disequilibrium with the four previously described loci. Two genome-wide meta-analyses (with CPD as the primary phenotype) also included age of tobacco initiation as an outcome and did not find any association that exceeded genome-wide significance [Furberg et al., 2010; Thorgeirsson et al., 2010]. In another large meta-analysis, age at onset of regular smoking modified the association between rs16969968 and CPD, with early onset of smoking being associated with a stronger gene–phenotype association. In that analysis, no association was seen between dichotomized age of onset (at age 16) and rs16969968 (P = 0.77) among 67,128 smokers [Hartz et al., 2012]. Thus, while some reports have found evidence for association in this region with age of tobacco initiation, others have not, and it has not been clear whether separate independent loci in these genes may be associated with different phenotypes related to early smoking behavior.
The goal of the current study was to perform a meta-analysis specifically targeting early smoking phenotypes and SNPs in the CHRNA5/CHRNA3/CHRNB4 gene cluster. These analyses were undertaken to determine: (1) whether the most robust association signal in this region shown for other smoking behaviors (rs16969968) is also seen for age of smoking initiation and age of onset of regular smoking, and/or: (2) if additional statistically independent signals are important in adolescent initiation of tobacco use and progression to regular smoking.
Methods
Subjects
A total of 56,034 subjects from 41 datasets spanning nine countries were included in a meta-analysis. Sample sizes for each SNP and the two phenotypes differ (age of tobacco initiation N ~ 12,662, age of onset of regular smoking N ~ 55,317) depending on which of the studies, listed in Table 1, had contributed information. All but 11 of these datasets consisted of unrelated ever-smokers of European descent. Details regarding individual study characteristics are provided in supplementary Table S1. Eight (Dental Caries, VA-Twin, NAG-FIN, NAG-OZALC, NTR1, NTR2, QIMR, and STR) were subsets of unrelated subjects selected from family-based datasets. The Add Health, NYS-FS, and SMOFAM datasets included family members of European descent. Informed consent was obtained from all participants and was approved by the local institutional review boards. Inclusion into this study required ever smokers to be assessed for (1) Age of tobacco initiation (AOI); and/or (2) Age of onset of regular smoking (AOS). Mean AOI and AOS values are shown in Table 1. Phrasing of AOI and AOS questions is provided in supplementary Table S1.
Table 1.
Summary of studies participating. Column 1 lists the acronym for each group, defined in the Supplementary Information (Dataset Descriptions). Column 2 indicates the number of subjects utilized for this meta-analysis. Columns 3 and 4 provide the mean age of initiation of tobacco (AOI) and onset of regular smoking (AOS) for groups that have those phenotypes. Columns 5–9 give minor allele frequencies at target SNPs for individual groups
| Datasets | N | AOI | AOS | Recruitment location | rs16969968 A | rs578776 T | rs588765 T | rs1948 A | rs684513 G |
|---|---|---|---|---|---|---|---|---|---|
| ACS-COPD | 2,791 | 19.2 | US | 0.35 | 0.27 | 0.42 | 0.34 | 0.21 | |
| ACS-LCA | 988 | 18 | US | 0.38 | 0.25 | 0.42 | 0.34 | – | |
| Add Health | 690 | 12.8 | 15.5 | US | 0.31 | 0.26 | 0.41 | 0.34 | 0.22 |
| ARIC | 5,775 | 18.5 | US | 0.34 | 0.28 | 0.42 | – | 0.21 | |
| BOMA | 828 | 18.1 | Germany | 0.36 | 0.27 | 0.41 | 0.34 | – | |
| CADD | 397 | 14 | 15.4 | US | 0.32 | 0.24 | 0.43 | 0.35 | 0.23 |
| COGA | 1,704 | 14.3 | 21.9 | US | 0.34 | 0.29 | 0.41 | – | – |
| COGEND | 2,057 | 13.3 | 15.5 | US | 0.35 | 0.27 | 0.42 | 0.33 | – |
| Dental Caries | 459 | 13.8 | US | 0.36 | 0.27 | 0.42 | 0.33 | – | |
| EAGLE-PLCO | 4,948 | 18 | Italy | 0.4 | 0.26 | 0.37 | 0.31 | – | |
| FINRISK | 7,542 | 21.6 | Finland | 0.33 | 0.32 | 0.38 | – | – | |
| GEOS | 475 | 16.9 | US | 0.37 | 0.3 | 0.39 | 0.31 | – | |
| KCI-WSU | 945 | 16.4 | US | 0.38 | – | – | – | – | |
| LHS-Utah | 1,943 | 17.4 | US | 0.39 | 0.24 | – | – | – | |
| LOLIPOP | 643 | 17.6 | UK | 0.31 | 0.31 | 0.43 | – | – | |
| MDACC LCA | 2,289 | 17.5 | US | 0.37 | 0.19 | 0.43 | 0.34 | 0.2 | |
| MDACC Melanoma | 825 | 18.6 | US | 0.33 | 0.27 | 0.44 | 0.35 | 0.23 | |
| MUC12 cases | 339 | 19.5 | Germany | 0.36 | 0.19 | 0.43 | 0.35 | – | |
| MUC12 controls | 216 | 20.1 | Germany | 0.34 | 0.21 | 0.43 | 0.32 | – | |
| MUCMD cases | 523 | 18.9 | Germany | 0.37 | 0.25 | 0.42 | 0.33 | 0.19 | |
| MUCMD controls | 989 | 20.2 | Germany | 0.35 | 0.27 | 0.42 | 0.32 | 0.21 | |
| NAG OZALC | 792 | 17 | Australia | 0.36 | 0.21 | 0.43 | 0.34 | – | |
| NAG FIN | 714 | 15.4 | 18.1 | Finland | 0.37 | 0.3 | 0.36 | 0.31 | – |
| NESDA | 1,138 | 16.3 | Netherlands | 0.31 | 0.3 | 0.38 | – | – | |
| NFBC1966 | 2,243 | 16.1 | 18.3 | Finland | 0.32 | 0.27 | 0.37 | 0.34 | – |
| NHS BRCA | 1,198 | 19.5 | US | 0.35 | 0.27 | 0.42 | 0.34 | – | |
| NHS CHD | 724 | 19.7 | US | 0.35 | 0.22 | 0.43 | – | – | |
| NHS T2D | 1,590 | 19.6 | US | 0.34 | 0.24 | 0.4 | – | – | |
| NTR1 | 464 | 16.2 | 18.7 | Netherlands | 0.31 | 0.17 | 0.9 | 0.33 | 0.28 |
| NTR2 | 593 | 15.9 | 18.1 | Netherlands | 0.31 | 0.3 | 0.46 | 0.36 | – |
| NYS-FS | 422 | 14.2 | 17 | US | 0.32 | 0.24 | 0.46 | 0.36 | 0.21 |
| QIMR | 258 | 16.4 | Australia | – | 0.29 | – | 0.34 | – | |
| SHIP | 1,871 | 18.5 | Germany | 0.34 | 0.22 | 0.43 | – | – | |
| SMOFAM | 398 | 17.8 | US | 0.32 | 0.28 | 0.46 | 0.38 | 0.18 | |
| STR | 4,096 | 18 | Sweden | 0.34 | 0.28 | – | 0.34 | – | |
| UTAH | 484 | 14.8 | 18 | US | 0.39 | 0.24 | – | – | – |
| VA-Twin | 1,960 | 14.4 | 16.5 | US | 0.33 | 0.28 | – | – | – |
| WTCCC-CHD | 1,219 | 17.7 | UK | 0.33 | 0.26 | 0.46 | – | – | |
| WTCCC-HT | 766 | 17.3 | UK | 0.31 | 0.31 | 0.44 | – | – | |
| Yale | 942 | 13.8 | 17.5 | US | 0.35 | 0.29 | – | – | 0.22 |
| YFS | 365 | 10.9 | 14.6 | Finland | 0.32 | 0.31 | 0.4 | 0.35 | – |
SNPs
SNP rs1948 was chosen due to a previously established association with AOI [Schlaepfer et al., 2008]. Based on previous results [Saccone et al., 2010a; Saccone et al., 2009a], we also included three statistically distinct nicotine dependence loci in the CHRNA5/A3/B4 gene cluster: rs16969968, rs578776, and rs588765. If a group did not have genotype data for SNPs rs16969968, rs578776, or rs588765 we chose a highly correlated (r2 > 0.8) SNP proxy. Another option would have been to use imputed data. However, in that situation, it would have been important for all imputation to be performed consistently between groups which would have required some groups to reimpute specifically for this project. The advantage of using an imputed SNP instead of a high-LD proxy for this study was not compelling because the imputation quality of the SNP would need to be taken into account as well. Thus, imputed SNPs were unlikely to add a significant amount of information to this study and/or change the take home message. SNP proxies for applicable datasets are shown in Table 2. Information on r2 was obtained using Haploview HapMap II+III CEU [Barrett et al., 2005] and the 1000 Genomes Pilot 1 CEU [Abecasis et al., 2012]. SNP rs684513 was included due to associations with nicotine dependence [Winterer et al., 2010] and lung cancer risk [Amos et al., 2010]; its r2 is less than 0.8 with rs16969968, rs578776, and rs588765 in both Hapmap [Altshuler et al., 2010] and the 1000 genomes project (http://www.1000genomes.org) CEU data [Abecasis et al., 2012]. Linkage disequilibrium (as measured by r2) for all five loci is presented in Table 3. Allele frequencies for SNP proxies and target SNPs in each dataset are given in Table 1.
Table 2.
Summary of SNP proxies for participating groups. Column 1 lists the acronym for each group, defined in the Supplementary Information (Dataset Descriptions). Columns 2–4 show the SNP proxies utilized by each group for rs16969968, rs578776, and rs588765 respectively. If a space is blank then the group genotyped the SNP of interest directly. A “–” indicates the group neither genotyped the SNP of interest directly, nor had an available proxy
| Datasets | rs16969968 proxy | rs578776 proxy | rs588765 proxy |
|---|---|---|---|
| ACS-COPD | |||
| ACS-LCA | rs8034191 | rs6495306 | |
| Addhealth | rs11637630 | rs680244 | |
| ARIC | rs17486278 | rs6495306 | |
| BOMA | rs1051730 | rs6495306 | |
| CADD | rs11637630 | rs680244 | |
| COGA | rs6495306 | ||
| COGEND | |||
| Dental Caries | rs1051730 | rs6495306 | |
| EAGLE | rs1051730 | rs6495306 | |
| FINRISK | |||
| GEOS | rs6495306 | ||
| KCI-WSU | – | – | |
| LHS-Utah | – | ||
| LOLIPOP | |||
| MDACC LCA | rs1051730 | rs6495309 | rs6495306 |
| MDACC Melanoma | rs6495306 | ||
| MUC12 cases | rs1051730 | rs6495309 | rs6495306 |
| MUC12 controls | rs1051730 | rs6495309 | rs6495306 |
| MUCMD cases | rs680244 | ||
| MUCMD controls | rs680244 | ||
| NAG OZALC | rs1051730 | rs6495309 | rs680244 |
| NAG FIN | rs1051730 | rs6495306 | |
| NESDA | rs11637635 | ||
| NFBC1966 | rs1051730 | rs6495309 | rs6495306 |
| NHS BRCA | rs1051730 | rs6495306 | |
| NHS CHD | rs951266 | rs6495306 | rs938682 |
| NHS T2D | rs951266 | rs6495306 | rs938682 |
| NTR1 | rs1163635 | ||
| NTR2 | rs1051730 | rs680244 | |
| NYS-FS | rs11637630 | rs680244 | |
| QIMR | |||
| SHIP | rs951266 | rs938682 | rs6495306 |
| SMOFAM | rs1317286 | rs555018 | |
| STR | 680244 | ||
| UTAH | rs1051730 | – | |
| VA-Twin | – | ||
| WTCCC-CHD | |||
| WTCCC-HT | |||
| Yale | rs621849 | ||
| YFS | rs1051730 | rs6495306 | |
Table 3.
Linkage Disequilibrium values (as measured by r2) utilized for the meta-analysis and taken from the 1000 genomes project Pilot 1 http://www.1000genomes.org/
| SNP/Locus | rs16969968 | rs578776 | rs588765 | rs1948 |
|---|---|---|---|---|
| rs16969968 | ||||
| rs578776 | 0.23 | |||
| rs588765 | 0.46 | 0.04 | ||
| rs1948 | 0.32 | 0.09 | 0.56 | |
| rs684513 | 0.1 | 0.61 | 0.1 | 0.06 |
Quality Control
Quality control procedures were carried out to ensure proper coding of variables across studies. Specifically, this was carried out by examination of univariate and multivariate distributions for phenotypic variables within each study, allele frequencies for SNPs, and conformity with Hardy-Weinberg Equilibrium (HWE).
Statistical Analyses
Case/Control Data
To ensure consistent analyses across individual datasets, a unified script was created using SAS (SAS Institute, Cary, NC) and R (http://www.r-project.org/). These scripts were developed and distributed following protocols comparable to Saccone et al. [Saccone et al., 2010a]. Briefly, participating groups held several planning conference calls to decide on the analytical model and scripts were written through collaborative efforts between SHS at the University of Colorado and SMH at Washington University in St. Louis. Individual analyses were carried out by each participating group and summary results returned to the coordinating group.
AOS and AOI phenotypes were normalized within each dataset. Z-scores were calculated to correct for age and sex prior to analysis. Scripts were written to perform a linear regression using AOS or AOI as the dependent variable and each SNP as the independent variable. Each SNP was coded according to an additive model (0, 1, or 2 copies of the minor allele) and each SNP was analyzed separately.
Family Data
PLINK [Purcell et al., 2007] was used to analyze the three datasets drawn from families (Add Health, NYS-FS, and SMOFAM). The QFAM total association test (–qfam-total) was used to conduct a family-based analysis that partitions the between family variance and the within family variance [Fulker et al., 1999] and is similar to that incorporated in the QTDT package [Abecasis et al., 2000]. This linear regression approach was ideal for this study because beta values from each family study analysis could be included.
In order to maintain uniformity in individual analyses, SHS compiled detailed instructions and distributed these to the groups analyzing family data with information to Z-score AOS or AOI in SAS (SAS Institute, Cary, NC) and R (http://www.r-project.org/), to code additively with a consistent minor allele, and to perform the analyses in PLINK with the –qfam-total option. Results were returned to SHS.
Meta-Analyses
Meta-analysis was carried out using the meta-analysis procedure in PLINK [Purcell et al., 2007]. This procedure requires results files (for a quantitative trait) with beta estimates, standard errors, and an optional P-value. PLINK meta-analysis output includes a beta summary estimate, and a summary P-value for both random and fixed effects models. The R package rmeta was used to create forest plots.
In addition, the PLINK meta-analysis output provides a measure of heterogeneity with a P-value for Cochrane’s Q statistic. This heterogeneity index in a meta-analysis refers to the variation in outcomes (betas) between studies. The Q statistic is calculated as the weighted sum of the squared differences between individual study effects (individual betas) and pooled effects (summary beta) across studies. Q has a chi-squared distribution with number of studies minus one degree of freedom.
A total of five SNPs and two phenotypes were examined, so the Bonferroni correction for multiple testing would be 0.05/10 = 0.005. This threshold for significance is overly conservative because of a correlation that exists between certain of these SNPs (rs684513/rs578776 r2 = 0.6). However, the goal of this study was not to discover a novel association but to disentangle the well-established association between these genes and tobacco behaviors. Thus, the nominal P-values provided should be interpreted with this in mind.
Results
Descriptive
Allele frequencies were utilized to confirm correct coding of the minor allele for each locus and are presented in Table 1. Frequencies were similar among individual datasets and also to those of the target SNPs in the CEU population in HapMap. The mean AOI for the 15 contributing datasets was 14.4 years (range, 10.9–16.4). The mean AOS for the 39 contributing datasets was 18 years (range, 14.6–21.9).
Single SNP Analysis for AOI and AOS Phenotypes
Tables 4 and 5 show PLINK meta-analysis results for each single locus and the AOI and AOS quantitative phenotypes. Results were nearly identical for both the fixed effects and random effects models. Table 4 presents results of AOI analysis where column one shows the SNP/Locus and columns two and three are the number of contributing datasets and the number of subjects with genotypic and phenotypic information at that SNP/Locus. Columns four and five show the summary beta and P-value, respectively. Column six is the P-value for the Cochrane’s Q statistic. There were no significant associations for AOI at any of the five loci.
Table 4.
Results of single SNP analysis and the age of initiation of tobacco (AOI) outcome. Column 1 = SNP/Locus, column 2 = number of contributing datasets for each locus, column 3 = number of total subjects with genotypic and phenotypic information, column 4 = summary Beta, column 5 = summary P-value, column 6 = Cochrane’s Q statistic P-value. Random effects versus a fixed effects model produce nearly identical betas and P-values
| SNP/Locus | Number of contributing datasets (AOI) | Number of Subjects | Summary beta (SE) | Summary P-value | Cochrane’s Q Statistic P-value |
|---|---|---|---|---|---|
| Locus 1: rs16969968 | 15 | 12,662 | 0.007 (0.013) | 0.588 | 0.261 |
| Locus 2: rs578776 | 15 | 12,662 | 0.02 (0.014) | 0.233 | 0.723 |
| Locus 3: rs588765 | 12 | 10,269 | − 0.019 (0.015) | 0.198 | 0.084 |
| rs1948 | 11 | 9,278 | − 0.027 (0.016) | 0.362 | 0.138 |
| rs684513 | 5 | 2,821 | 0.023 (0.034) | 0.507 | 0.531 |
Table 5.
Results of single SNP analysis and the age of onset of regular smoking (AOS) outcome. Column 1 = SNP/Locus, columns 2 = number of contributing datasets for each locus, column 3 = number of total subjects with genotypic and phenotypic information, column 4 = summary Beta, column 5 = summary P-value, column 6 = Cochrane’s Q statistic P-value. Significant summary P-values are indicated with bold. Random effects versus a fixed effects model produce nearly identical betas and P-values
| SNP/Locus | Number of contributing datasets (AOS) | Number of Subjects | Summary Beta (SE) | Summary P-value | Cochrane’s Q Statistic P-value |
|---|---|---|---|---|---|
| Locus 1: rs16969968 | 39 | 55,317 | 0.0004 (0.004) | 0.917 | 0.052 |
| Locus 2: rs578776 | 38 | 54,796 | 0.02 (0.007) | 0.004 | 0.601 |
| Locus 3: rs588765 | 34 | 49,240 | − 0.009 (0.008) | 0.297 | 0.01 |
| rs1948 | 24 | 28,970 | 0.023 (0.009) | 0.018 | 0.772 |
| rs684513 | 12 | 15,950 | 0.032 (0.013) | 0.017 | 0.676 |
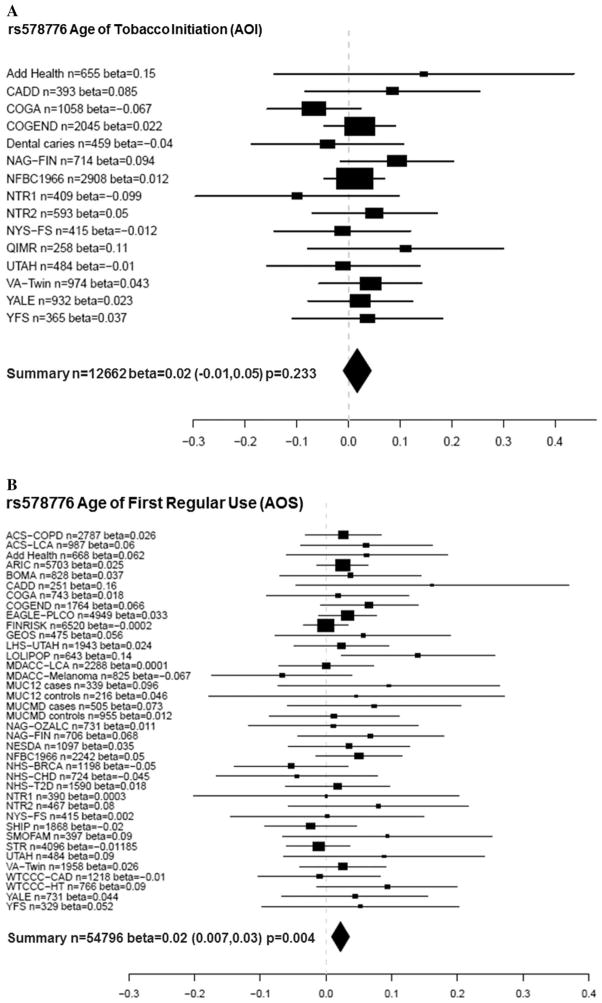
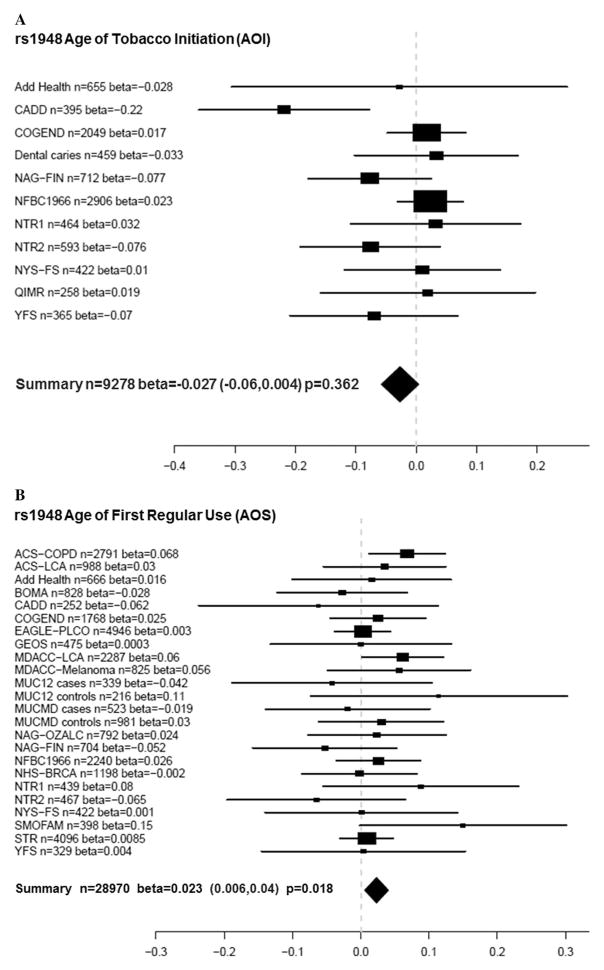
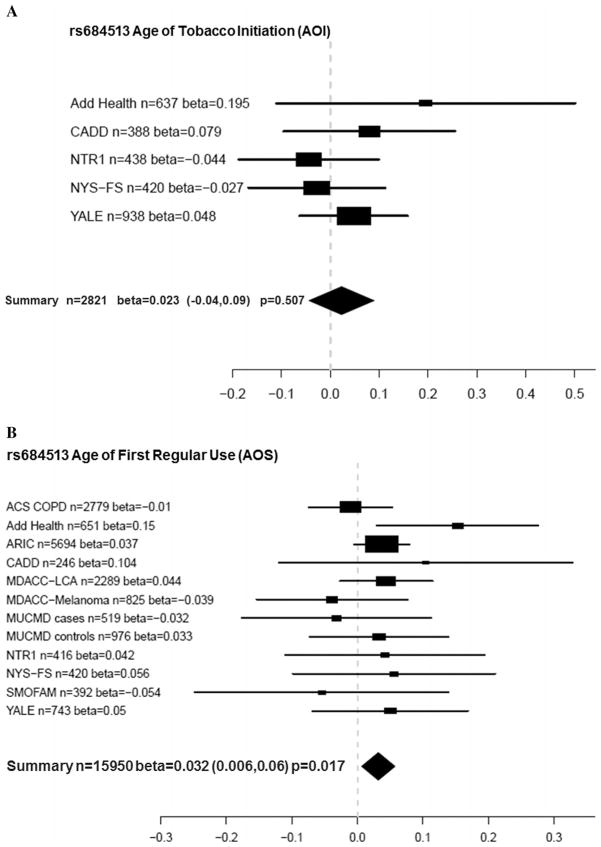
Table 5 shows the corresponding results of AOS meta-analyses. Locus rs578776 was significantly associated with AOS (beta = 0.02, nominal P = 0.004, adjusted P = 0.04). Figure 1A shows the meta-analysis forest plot for the association between rs578776 with a phenotype of AOI while Figure 1B shows these results for AOS. Although the rs578776 locus was not significantly associated with AOI, both betas were 0.02 and suggest that for AOS, the minor allele confers a protective effect because it is associated with a later age of smoking onset. In addition, SNPs rs1948 and rs684513 were associated with AOS (rs1948 beta = 0.023, P = 0.018; rs684513 beta = 0.032, P = 0.017). Figure 2A and B show meta-analysis forest plots for rs1948 with AOI and AOS, respectively. Figure 3A and B show forest plots for rs684513 AOI and AOS, respectively.
Figure 1.
(A) Meta-analysis forest plot for rs578776 age of tobacco (AOI) initiation showing the individual betas (for each study), summary beta, individual number of subjects (n), summary n, and a summary P-value. Individual and summary betas are for the rs578776 T allele. (B) Meta-analysis forest plot for rs578776 age of first regular tobacco use (AOS) showing the individual betas (for each study), summary beta, individual number of subjects (n), summary n, and a summary P-value. Individual and summary betas are for the rs578776 T allele.
Figure 2.
(A) Meta-analysis forest plot for rs1948 age of tobacco initiation (AOI) showing the individual betas (for each study), summary beta, individual number of subjects (n), summary n, and a summary P-value. Individual and summary betas are for the rs1948 A allele. (B) Meta-analysis forest plot for rs1948 age of first regular tobacco use (AOS) showing the individual betas (for each study), summary beta, individual number of subjects (n), summary n, and a summary P-value. Individual and summary betas are for the rs1948 A allele.
Figure 3.
(A) Meta-analysis forest plot for rs684513 age of tobacco initiation (AOI) showing the individual betas (for each study), summary beta, individual number of subjects (n), summary n, and a summary P-value. Individual and summary betas are for the rs684513 G allele. (B) Meta-analysis forest plot for rs684513 age of first regular tobacco use (AOS) showing the individual betas (for each study), summary beta, individual number of subjects (n), summary n, and a summary P-value. Individual and summary betas are for the rs684513 G allele.
Cochrane’s Q statistic P-values (column six of Tables 4 and 5) indicate that there was one SNP/loci (rs588765 AOS only) showing modest heterogeneity.
Discussion
This meta-analysis was designed to examine the relationship of SNPs in the CHRNA5/CHRNA3/CHRNB4 gene cluster with early smoking behaviors including AOI and AOS. Genetic variation in this genomic region shows robust association to nicotine dependence and tobacco use behaviors in adult samples [Furberg et al., 2010; Liu et al., 2010; Saccone et al., 2010a; Thorgeirsson et al., 2010]. Although previous studies examining association with tobacco initiation [Furberg et al., 2010; Thorgeirsson et al., 2010] and first regular use have been inconclusive, most have not found association with the locus including SNP rs16969968 [Furberg et al., 2010; Hartz et al., 2012; Thorgeirsson et al., 2010]. We hypothesized that a possible explanation for conflicting reports may be related to the multiple independent loci in this region [Saccone et al., 2009a; Saccone et al., 2010b], if different loci contribute to different smoking-related phenotypes. This analysis expands on previous work by examining two additional SNPs (rs1948 and rs684513) based on previous associations with smoking-related phenotypes [Amos et al., 2010; Schlaepfer et al., 2008; Winterer et al., 2010], in addition to three statistically independent nicotine dependence loci in the CHRNA5/A3/B4 gene cluster: rs16969968, rs578776, and rs588765.
Results revealed evidence for association between AOS and three of the five target SNPs. Importantly, AOS shows no association with rs16969968, which is strongly and reproducibly associated with nicotine dependence, smoking quantity, and cotinine levels [Furberg et al., 2010; Greenbaum and Lerer 2009; Keskitalo et al., 2009; Saccone et al., 2010b; Saccone et al., 2007; Thorgeirsson et al., 2010]. This is consistent with previous reports [Broms et al., 2012; Ducci et al., 2011; Furberg et al., 2010; Thorgeirsson et al., 2010; Weiss et al., 2008]. One explanation put forward for the lack of findings with tobacco initiation and rs16969968 is a report by Hong et al where the “risk” allele (A) predicts a defect in a dorsal anterior cingulate-ventral striatum/extended amygdala circuit whose weakness predicts addiction severity [Hong et al., 2011]. One could also reason that rs16969968 is not associated with initiation and instead more strongly related to craving and ability to quit due to the action of CHRNA5 to alter output of deep cortical layers [Picciotto and Kenny, 2012].
In the current report, the strongest evidence for association with AOS was detected with SNP rs578776 (P = 0.004). There was also evidence for association between SNPs rs1948 (beta = 0.023, P = 0.018) and rs684513 (beta = 0.032, P = 0.017) and AOS. Similar to rs578776, the betas for these SNPs were positive, signifying that having more copies of the minor alleles is associated with a later age of smoking onset. Results with rs578776 and rs684513 are in agreement with a previous study [Broms et al., 2012], where there was evidence for association of age of onset of smoking (AOS) and the Locus 2 signal (rs578776) and rs684513. In addition, significance with rs1948 in this report is supported by previous findings of significant association with “smoking at age 14” and suggestive evidence with age of onset of smoking [Broms et al., 2012].
In contrast to the AOS findings, there was no evidence for association between AOI and any of the SNPs examined in the CHRNA5/A3/B4 cluster. It might be intuitively expected that AOI and AOS would be highly correlated and that the associations with risk SNPs would not vary between them greatly even though there are differences in statistical power between AOI and AOS due to sample size. However, the correlations between AOI and AOS among the samples where both measures are available are actually rather low (ranging from 0.20–0.65, data not shown). There is additional support for the hypothesis that a low correlation exists between these two phenotypes. Breslau et al. (1993) showed the lag time from first cigarette to daily smoking onset was significantly longer in those who initiated <14 years of age relative to those initiating ≥17 years of age. A social explanation was proposed for these findings in that early initiators were said to face a nonpermissive environment (e.g. availability, community norms, adult supervision, etc.) [Breslau et al., 1993]. In addition, we performed an ad hoc exploratory analysis to determine whether there were any trends with the five loci and the delta between AOI and AOS. Utilizing a subset of samples with both phenotypes available (Add Health, CADD, COGA, COGEND, NYS-FS, YFS), we found no evidence for association between the AOI–AOS time differential and any of the loci used for this study (data not shown). The similarities and differences in results across SNPs for the two phenotypes are interesting also to consider in the context of work finding an association between externalizing behaviors and rs8040868, which is a locus not well-tagged by the SNPs investigated here [Stephens et al., 2012]. It is possible that some of the differences between AOI and AOS results are related to more general underlying behaviors, such as novelty-seeking and conduct disorder. Future studies aimed at disentangling possible unique genetic contributions to these phenotypes are merited. An alternative explanation for these findings could be biological. Age at initiation is related to behavioral traits such as Attention Deficit Hyperactivity Disorder (ADHD), Conduct Disorder (CD), Oppositional Deviant Disorder (ODD), delinquency, and criminal behavior [Armstrong and Costello, 2002; Flory et al., 2003; Han et al., 1999; Krueger et al., 2002; McGue and Iacono, 2008; McGue et al., 2001; Sihvola et al., 2011; Tarter et al., 2003; Young et al., 2000], while the time from initiation to regular smoking could be related to individual differences in neuroadaptation to nicotine and other psychoactive components of tobacco. Thus, it is reasonable to propose that nicotinic receptor properties are related to the neuroadaptation process and consequently to CHRN gene polymorphisms.
These findings corroborate and strengthen certain previous reports. Rs578776 (CHRNA3) has been previously associated with nicotine dependence in subjects of European ancestry [Saccone et al., 2009a, 2009b, 2010b; Weiss et al., 2008] and heavy smoking [Stevens et al., 2008] with the minor allele (T) acting in a protective manner against nicotine dependence. Our results also support the protective effect of the minor allele where a positive beta (0.02) indicated a later age of first regular smoking among individuals with 1 or 2 copies of the T allele. However, the SNP is not known to be functional, so the basis for the association needs to be investigated further. SNP rs1948 (CHRNB4) has previously been associated with tobacco and alcohol age of initiation in the CADD sample and NYS-FS samples [Schlaepfer et al., 2008], but few other studies have focused on this locus. SNP rs1948 was recently associated with the tolerance factor of the Nicotine Dependence Syndrome Scale (NDSS) (P = 0.0071) [Broms et al., 2012] and SNP rs684513 (major allele) has been previously associated with nicotine dependence [Winterer et al., 2010] and lung cancer risk [Amos et al., 2010]. Lori et al found an association with FTND of this CHRNA5 SNP [Lori et al., 2011]. These meta-analysis results support previous findings showing a protective effect of the minor allele for AOS (P = 0.015, beta = 0.034).
The PLINK meta-option reports the Cochrane’s Q statistic as a measure of heterogeneity for each SNP. One significant heterogeneity P-value was obtained for SNP rs588765 (P = 0.01) in the AOS analysis only. It is unlikely this heterogeneity can be accounted for by differences in phenotype assessment between studies because this would be predicted to result in significant heterogeneity for all loci in the AOS analysis. Large differences in allele frequencies between groups could also produce heterogeneity. Examination of allele frequencies for all participating groups suggests one outlier (NTR1 T allele frequency is 0.9) within this SNP/locus. In order to determine whether this sample population was driving the significant Q statistic for rs588765 and AOS, the meta-analysis was performed excluding this sample. Results for AOS were similar and the Q statistic P-value was identical (beta = −0.004, P = 0.281, Q P-value = 0.01) suggesting the NTR1 rs588765 T allele frequency is not the cause of observed heterogeneity at this locus. Furthermore, results from a random effects model (automatically generated by PLINK) were nearly identical for rs588765 (fixed effects beta = −0.009, P = 0.297, random effects beta = −0.005, P = 0.402). Thus, heterogeneity across samples is unlikely to be driving the association results for AOS with these two SNPs.
There are several limitations to this study. First, given the involvement of many groups across several countries and use of data collected for other purposes, methods for assessments were not consistent across all studies. The phrasing of questions differed across sites, and although an attempt was made to harmonize the phenotypes it would have been ideal to use the same tools and questions. Second, some groups did not have both AOI and AOS data, so direct comparison of results is incomplete because power to detect an association differs. Third, the exact same SNPs were not available for all groups, so we relied on the use of proxy SNPs based on the current HapMap data. It is possible that some of the SNPs used as proxies are not as well correlated with the target SNP signal as we believe, but given the large datasets now available, this is unlikely for most of the targets.
This study also features several strengths. First, this study was built on strong existing support for an important role of this gene cluster, enabling the efforts to be focused on characterizing the associations in the region. Second, this project began with multiple discussions by all participants about the goals for the study, after which a specific analytical plan was developed to test carefully defined hypotheses, thereby minimizing multiple testing. Third, the sample sizes are substantial and therefore have allowed us to separate the effects of unique loci in a manner not otherwise possible.
The most significant finding (rs578776) has been associated with nicotine dependence in multiple independent samples and the minor allele confers a protective effect. Results of this meta-analysis suggest the protective effect imparted by the minor allele may influence early smoking behaviors, resulting in a decreased risk of nicotine dependence later in life. Perhaps even more interesting is the fact that there was absolutely no evidence at the most highly replicated locus for smoking heaviness (rs16969968), which has been shown to confer a functional effect on receptor function [Bierut et al., 2008; Fowler et al., 2011; Kuryatov et al., 2011]. This is the third study to report nonsignificance for age of initiation or age of onset of smoking at the rs16969968 [Furberg et al., 2010; Hartz et al., 2012; Thorgeirsson et al., 2010]. This work emphasizes the fact that early smoking behaviors are related to, but different from nicotine dependence. It also demonstrates the value in pursuing multiple SNPs in a region where one SNP has been definitively associated with behavior and has concomitant evidence for molecular function. It is likely that other genes found to be associated with complex behaviors will harbor multiple functional variants, which may be involved in different aspects in the development of the disorder. For example, the most significantly associated SNP in this study (rs578776) represents a possible target for early intervention, while SNP rs16969968 may be a more effective target for later treatment. Taken as a whole, these results underscore the complexity of smoking behavior phenotypes, and suggest different genetic signals in this region may contribute to different stages of tobacco use and dependence.
Supplementary Material
Acknowledgments
Contract grant sponsor: NIH; Contract grant numbers: U01 HG004738; R01 DA12690, R01 DA12849, K01 DA24758, R01 AA11330, R01 AA017535, R01 AA07535, R01 AA07728, R01 AA13320, R01AA13321, R01 AA14041, R01 AA11998, R01 AA17688, R01 DA012854, K02 DA021237, UL1RR024992, R01 DA021913, P60 DA011015, K01 DA19498, R21 DA027070, K02 AA018755, U01 DA02830, R01HL089651-01, N01-AG-1-2109, K07 CA118412, R01 DA03706, U01 HG004436, R01 NS45012, U01 NS069208, P01 CA089392, R01 DA12854, R01 CA060691, N01-PC35146, R01 CA87895, P01 AG005842, T32 AG000186, K99R00 DA023549, R01 DA026911, KL2 RR024994, K08 DA032680, P30 DK-072488, U01 HG004738, R01 DE12101; Contract grant sponsor: Australian National Health and Research Council; Contract grant numbers: 241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 464914, 496739, 552485, 552498; Contract grant sponsor: Australian Research Council; Contract grant numbers: A7960034, A79906588, A79801419, APP1009839, DP0770096, DP0212016, DP0343921, and FT110100548; Contract grant sponsor: Germany Federal Ministry for Education and Research; Contract grant numbers: BMBF NGNF-2; BMBF NGFNplus; BMBF IG MooDS 01GS08144 and 01GS08147; Contract grant sponsor: Alfried Krupp von Bohlen und Halbach-Stiftung; Contract grant sponsor: 5th Framework Programme (FP-5); Contract grant sponsor: GenomEUtwin Project; Contract grant numbers: QLG2-CT-2002-01254; Contract grant sponsor: University of California; Contract grant number: 7PT2000-2004; Contract grant sponsor: Tobacco-Related Disease Research Program; Contract grant sponsor: Academy of Finland and The Center of Excellence in Complex Diseases; Contract grant numbers: 134309, 126925, 121584, 124282, 129378, 117787, 129494, 41071; Contract grant sponsor: University Hospital Oulu (Oulu, Finland); Contract grant sponsor: Tampere and Turku University Hospital Medical Funds; Contract grant numbers: 9M048 and 9N035; Contract grant sponsor: Juho Vainio Foundation; Finnish Foundation for Cardiovascular Diseases; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research; Finnish Cultural Foundation; Tampere Tuberculosis Foundation and Emil Aaltonen Foundation (T.L.); Social Insurance Institution of Finland; NARSAD Young Investigator Award; Welcome Trust; British Heart Foundation; National Institute for Health Research; Academy of Finland Center of Excellence in Complex Disease Genetics; Global Research Awards for Nicotine Dependence (GRAND); SALVE Program; NIHR Academic Clinical Fellowship at the Division of Mental Health, St George’s, University of London, UK; Medical Research Service and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs; The European Commission; GenomeEUtwin Project under the European Commission Programme Quality of Life and Management of the Living Resources of Fifth Framework Programme; the Swedish Research Council; the Swedish Council for Working Life and Social Research; the Swedish Foundation for Strategic Research; the Swedish Heart and Lung Foundation; the Ragnar Söderberg Foundation; the Jan Wallander and Tom Hedelius Foundation; Federal Ministry of Education and Research; Contract grant numbers: 01ZZ9603, 01ZZ0103, 01ZZ0403, and 03ZIK012; Contract grant sponsor: The Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania; joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. For the NTR study (www.tweelingregister.org) funding was obtained from the Netherlands Organization for Scientific Research (NWO); CMSB: Center for Medical Systems Biology (NWO Genomics); Spinozapremie (SPI 56-464-14192); the VU University Centre for Neurogenomics and Cognitive Research (CNCR); Genomewide analyses of European twin and population cohorts (EU/QLRT-2001-01254); the European Research Council: Genetics of Mental Illness (ERC 230374) and Beyond the Genetics of Addiction (ERC 284167). KCB was supported in part by the Mary Beryl Patch Turnbull Scholar Program.
NIH Grants R01 DA12690, R01 DA12849, K01 DA24758, R01 AA11330, R01 AA017535, R01 AA07535, R01 AA07728, R01 AA13320, R01AA13321, R01 AA14041, R01 AA11998, R01 AA17688, R01 DA012854, K02 DA021237, UL1RR024992, R01 DA021913, P60 DA011015, K01 DA19498, R21 DA027070, K02 AA018755, U01 DA020830, R01HL089651-01, N01-AG-1-2109, K07 CA118412, R01 DA03706, U01 HG004436, R01 NS45012, U01 NS069208, P01 CA089392, R01 DA12854, R01 CA060691, N01-PC35146, R01 CA87895, P01 AG005842, T32 AG000186, K99R00 DA023549, R01 DA026911, KL2 RR024994, K08 DA032680, P30 DK-072488, U01 HG004738, R01 DE12101; Australian National Health and Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 464914, 496739, 552485, 552498); Australian Research Council (A7960034, A79906588, A79801419, APP1009839, DP0770096, DP0212016, DP0343921, and FT110100548); Germany Federal Ministry for Education and Research (project grants BMBF NGNF-2; BMBF NGFNplus; BMBF IG MooDS 01GS08144 and 01GS08147); Alfried Krupp von Bohlen und Halbach-Stiftung; 5th Framework Programme (FP-5); GenomEUtwin Project (QLG2-CT-2002-01254); 7PT2000-2004, from the University of California Tobacco-Related Disease Research Program; Academy of Finland (project grants 134309, 126925, 121584, 124282, 129378, 117787, 129494, 41071 and The Center of Excellence in Complex Diseases); University Hospital Oulu (Oulu, Finland); Tampere and Turku University Hospital Medical Funds (project grants 9M048 and 9N035); Juho Vainio Foundation; Finnish Foundation for Cardiovascular Diseases; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research; Finnish Cultural Foundation; Tampere Tuberculosis Foundation and Emil Aaltonen Foundation (T.L.); Social Insurance Institution of Finland; NARSAD Young Investigator Award; Welcome Trust; British Heart Foundation; National Institute for Health Research; Academy of Finland Center of Excellence in Complex Disease Genetics; Global Research Awards for Nicotine Dependence (GRAND); SALVE Program; NIHR Academic Clinical Fellowship at the Division of Mental Health, St George’s, University of London, UK; Medical Research Service and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs; The European Commission; GenomeEUtwin Project under the European Commission Programme Quality of Life and Management of the Living Resources of Fifth Framework Programme; the Swedish Research Council; the Swedish Council for Working Life and Social Research; the Swedish Foundation for Strategic Research; the Swedish Heart and Lung Foundation; the Ragnar Söderberg Foundation; the Jan Wallander and Tom Hedelius Foundation; Federal Ministry of Education and Research (grants 01ZZ9603, 01ZZ0103, 01ZZ0403, and 03ZIK012); The Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania; joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. For the NTR study (www.tweelingregister.org) funding was obtained from the Netherlands Organization for Scientific Research (NWO); CMSB: Center for Medical Systems Biology (NWO Genomics); Spinozapremie (SPI 56-464-14192); the VU University Centre for Neurogenomics and Cognitive Research (CNCR); Genomewide analyses of European twin and population cohorts (EU/QLRT-2001-01254); the European Research Council: Genetics of Mental Illness (ERC 230374) and Beyond the Genetics of Addiction (ERC 284167). KCB was supported in part by the Mary Beryl Patch Turnbull Scholar Program.
Ulla Broms consulted for Pfizer on Nicotine dependence measurements in 2008. Jaakko Kaprio and Tellervo Korhonen consulted for Pfizer on nicotine dependence in 2011 and 2012. Laura J. Bierut served as a consultant for Pfizer in 2008 and is an inventor on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in the determination of diagnosis, prognosis, and treatment of addiction. Nancy L. Saccone is the spouse of Scott Saccone, who is also listed as an inventor on the above patent. These data were presented at the World Congress of Psychiatric Genetics. September 10, 2011. Washington, DC.
Footnotes
Supporting Information is available in the online issue at wileyonlinelibrary.com.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, Scheet P, Greisinger AJ, Mills GB, Spitz MR. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102(15):1199–1205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony JC, Warner LA, Kessler RC. Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants: basic findings from the National Comorbidity Survey. Exp Clin Psychopharmacol. 1994;2(3):244–268. [Google Scholar]
- Armstrong TD, Costello EJ. Community studies on adolescent substance use, abuse, or dependence and psychiatric comorbidity. J Consult Clin Psychol. 2002;70(6):1224–1239. doi: 10.1037//0022-006x.70.6.1224. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslau N, Fenn N, Peterson EL. Early smoking initiation and nicotine dependence in a cohort of young adults. Drug Alcohol Depend. 1993;33(2):129–137. doi: 10.1016/0376-8716(93)90054-t. [DOI] [PubMed] [Google Scholar]
- Breslau N, Peterson EL. Smoking cessation in young adults: age at initiation of cigarette smoking and other suspected influences. Am J Public Health. 1996;86(2):214–220. doi: 10.2105/ajph.86.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Wedenoja J, Largeau MR, Korhonen T, Pitkaniemi J, Keskitalo-Vuokko K, Happola A, Heikkila KH, Heikkila K, Ripatti S, et al. Analysis of detailed phenotype profiles reveals CHRNA5-CHRNA3-CHRNB4 gene cluster association with several nicotine dependence traits. Nicotine Tob Res. 2012;14(6):720–733. doi: 10.1093/ntr/ntr283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Annual smoking-attributable mortality, years of potential life lost, and productivity losses—United States, 1997–2001. MMWR. 2005;54(25):625–628. [PubMed] [Google Scholar]
- Chen J, Millar WJ. Age of smoking initiation: implications for quitting. Health Rep. 1998;9(4):39–46. 39–48. (Eng) (Fre) [PubMed] [Google Scholar]
- Chen X, Stanton B, Shankaran S, Li X. Age of smoking onset as a predictor of smoking cessation during pregnancy. Am J Health Behav. 2006;30(3):247–258. doi: 10.5555/ajhb.2006.30.3.247. [DOI] [PubMed] [Google Scholar]
- D’Avanzo B, La Vecchia C, Negri E. Age at starting smoking and number of cigarettes smoked. Ann Epidemiol. 1994;4(6):455–459. doi: 10.1016/1047-2797(94)90005-1. [DOI] [PubMed] [Google Scholar]
- DiFranza JR. Hooked from the first cigarette. Sci Am. 2008;298(5):82–87. doi: 10.1038/scientificamerican0508-82. [DOI] [PubMed] [Google Scholar]
- DiFranza JR, Rigotti NA, McNeill AD, Ockene JK, Savageau JA, St Cyr D, Coleman M. Initial symptoms of nicotine dependence in adolescents. Tob Control. 2000;9(3):313–319. doi: 10.1136/tc.9.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFranza JR, Savageau JA, Fletcher K, Pbert L, O’Loughlin J, McNeill AD, Ockene JK, Friedman K, Hazelton J, Wood C, et al. Susceptibility to nicotine dependence: the Development and Assessment of Nicotine Dependence in Youth-2 study. Pediatrics. 2007;120(4):e974–e983. doi: 10.1542/peds.2007-0027. [DOI] [PubMed] [Google Scholar]
- DiFranza JR, Savageau JA, Rigotti NA, Fletcher K, Ockene JK, McNeill AD, Coleman M, Wood C. Development of symptoms of tobacco dependence in youths: 30 month follow up data from the DANDY study. Tob Control. 2002;11(3):228–235. doi: 10.1136/tc.11.3.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doubeni CA, Reed G, Difranza JR. Early course of nicotine dependence in adolescent smokers. Pediatrics. 2010;125(6):1127–1133. doi: 10.1542/peds.2009-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducci F, Kaakinen M, Pouta A, Hartikainen AL, Veijola J, Isohanni M, Charoen P, Coin L, Hoggart C, Ekelund J, et al. TTC12-ANKK1-DRD2 and CHRNA5-CHRNA3-CHRNB4 influence different pathways leading to smoking behavior from adolescence to mid-adulthood. Biol Psychiatry. 2011;69(7):650–660. doi: 10.1016/j.biopsych.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner MD, Yelin EH, Katz PP, Shiboski SC, Henke J, Blanc PD. Predictors of cigarette smoking and smoking cessation among adults with asthma. Am J Public Health. 2000;90(8):1307–1311. doi: 10.2105/ajph.90.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobedo LG, Marcus SE, Holtzman D, Giovino GA. Sports participation, age at smoking initiation, and the risk of smoking among US high school students. JAMA. 1993;269(11):1391–1395. [PubMed] [Google Scholar]
- Etter JF, Hoda JC, Perroud N, Munafò M, Buresi C, Duret C, Neidhart E, Malafosse A, Bertrand D. Association of genes coding for the alpha-4, alpha-5, beta-2 and beta-3 subunits of nicotinic receptors with cigarette smoking and nicotine dependence. Addict Behav. 2009;34(9):772–775. doi: 10.1016/j.addbeh.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Everett SA, Warren CW, Sharp D, Kann L, Husten CG, Crossett LS. Initiation of cigarette smoking and subsequent smoking behavior among U.S. high school students. Prev Med. 1999;29(5):327–333. doi: 10.1006/pmed.1999.0560. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Schiaffino A, La Vecchia C, Borras JM, Nebot M, Salto E, Tresserras R, Rajmil L, Villalbi JR, Segura A. Age at starting smoking and number of cigarettes smoked in Catalonia, Spain. Prev Med. 1999;28(4):361–366. doi: 10.1006/pmed.1998.0433. [DOI] [PubMed] [Google Scholar]
- Flory K, Milich R, Lynam DR, Leukefeld C, Clayton R. Relation between childhood disruptive behavior disorders and substance use and dependence symptoms in young adulthood: individuals with symptoms of attention-deficit/hyperactivity disorder and conduct disorder are uniquely at risk. Psychol Addict Behav. 2003;17(2):151–158. doi: 10.1037/0893-164x.17.2.151. [DOI] [PubMed] [Google Scholar]
- Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471(7340):597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64(1):259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceshini N, Ardissino D, Bernardinelli L, Mannucci PM, Mauri F, Merlini PA, et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum L, Lerer B. Differential contribution of genetic variation in multiple brain nicotinic cholinergic receptors to nicotine dependence: recent progress and emerging open questions. Mol Psychiatry. 2009;14(10):912–945. doi: 10.1038/mp.2009.59. [DOI] [PubMed] [Google Scholar]
- Han C, McGue MK, Iacono WG. Lifetime tobacco, alcohol and other substance use in adolescent Minnesota twins: univariate and multivariate behavioral genetic analyses. Addiction. 1999;94(7):981–993. doi: 10.1046/j.1360-0443.1999.9479814.x. [DOI] [PubMed] [Google Scholar]
- Hartz SM, Short SE, Saccone NL, Culverhouse R, Chen L, Schwantes-An TH, Coon H, Han Y, Stephens SH, Sun J, et al. Increased genetic vulnerability to smoking at CHRNA5 in early-onset smokers. Arch Gen Psychiatry. 2012;69(8):854–860. doi: 10.1001/archgenpsychiatry.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao T, Nelson HH, Ashok TD, Wain JC, Mark EJ, Christiani DC, Wiencke JK, Kelsey KT. Tobacco smoke-induced DNA damage and an early age of smoking initiation induce chromosome loss at 3p21 in lung cancer. Cancer Res. 2001;61(2):612–615. [PubMed] [Google Scholar]
- Hong LE, Yang X, Wonodi I, Hodgkinson CA, Goldman D, Stine OC, Stein ES, Thaker GK. A CHRNA5 allele related to nicotine addiction and schizophrenia. Genes Brain Behav. 2011;10(5):530–535. doi: 10.1111/j.1601-183X.2011.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Hwang LY, Ma Y, Benningfield SM, Clayton L, Hanson EN, Jay J, Jonte J, Godwin de Medina C, Moscicki AB. Factors that influence the rate of epithelial maturation in the cervix in healthy young women. J Adolesc Health. 2009;44(2):103–110. doi: 10.1016/j.jadohealth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, Pitkaniemi J, Peltonen L, Aromaa A, Kaprio J. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18(20):4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. J Abnorm Psychol. 2002;111(3):411–424. [PubMed] [Google Scholar]
- Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Mol Pharmacol. 2011;79(1):119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando HA, Thai DT, Murray DM, Robinson LA, Jeffery RW, Sherwood NE, Hennrikus DJ. Age of initiation, smoking patterns, and risk in a population of working adults. Prev Med. 1999;29(6 Pt 1):590–598. doi: 10.1006/pmed.1999.0590. [DOI] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98(1):23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Li MD, Yoon D, Lee JY, Han BG, Niu T, Payne TJ, Ma JZ, Park T. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS One. 2010;5(8):e12183. doi: 10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori A, Tang Y, O’Malley S, Picciotto MR, Wu R, Conneely KN, Cubells JF. The galanin receptor 1 gene associates with tobacco craving in smokers seeking cessation treatment. Neuropsychopharmacology. 2011;36(7):1412–1420. doi: 10.1038/npp.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YT, Collins SI, Young LS, Murray PG, Woodman CB. Smoking initiation is followed by the early acquisition of epigenetic change in cervical epithelium: a longitudinal study. Br J Cancer. 2011;104(9):1500–1504. doi: 10.1038/bjc.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew KP, Flay BR, Mott JA. Stages in the development of adolescent smoking. Drug Alcohol Depend. 2000;59(Suppl 1):S61–S81. doi: 10.1016/s0376-8716(99)00165-9. [DOI] [PubMed] [Google Scholar]
- McGue M, Iacono WG. The adolescent origins of substance use disorders. Int J Methods Psychiatr Res. 2008;17(Suppl 1):S30–S38. doi: 10.1002/mpr.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Iacono WG, Legrand LN, Malone S, Elkins I. Origins and consequences of age at first drink. I. Associations with substance-use disorders, disinhibitory behavior and psychopathology, and P3 amplitude. Alcohol Clin Exp Res. 2001;25(8):1156–1165. [PubMed] [Google Scholar]
- O’Loughlin J, DiFranza J, Tarasuk J, Meshefedjian G, McMillan-Davey E, Paradis G, Tyndale RF, Clarke P, Hanley J. Assessment of nicotine dependence symptoms in adolescents: a comparison of five indicators. Tob Control. 2002a;11(4):354–360. doi: 10.1136/tc.11.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin J, Kishchuk N, DiFranza J, Tremblay M, Paradis G. The hardest thing is the habit: a qualitative investigation of adolescent smokers’ experience of nicotine dependence. Nicotine Tob Res. 2002b;4(2):201–209. doi: 10.1080/14622200210123000. [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Kenny PJ. Molecular mechanisms underlying behaviors related to nicotine addiction. Cold Spring Harb Perspect Med. 2012 doi: 10.1101/cshperspect.a012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60(12):1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- Rose JE. Multiple brain pathways and receptors underlying tobacco addiction. Biochem Pharmacol. 2007;74(8):1263–1270. doi: 10.1016/j.bcp.2007.07.039. [DOI] [PubMed] [Google Scholar]
- Rose RJ, Broms U, Korhonen T, Dick DM, Kaprio J, Kim Y-K. Genetics of Smoking Behavior Handbook of Behavior Genetics. New York, NY: Springer; 2009. pp. 411–432. [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010a;6(8):e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009a;150B(4):453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D, Johnson EO, Rice JP, Goate AM, Bierut LJ. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav. 2010b;9(7):741–750. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009b;69(17):6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63(11):1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoll RA, Johnson TA, Lerman C. Genetics and smoking behavior. Curr Psychiatry Rep. 2007;9(5):349–357. doi: 10.1007/s11920-007-0045-3. [DOI] [PubMed] [Google Scholar]
- Scragg R, Wellman RJ, Laugesen M, DiFranza JR. Diminished autonomy over tobacco can appear with the first cigarettes. Addict Behav. 2008;33(5):689–698. doi: 10.1016/j.addbeh.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Sihvola E, Rose RJ, Dick DM, Korhonen T, Pulkkinen L, Raevuori A, Marttunen M, Kaprio J. Prospective relationships of ADHD symptoms with developing substance use in a population-derived sample. Psychol Med. 2011;41(12):2615–2623. doi: 10.1017/S0033291711000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soteriades ES, Spanoudis G, Talias MA, Warren CW, DiFranza JR. Children’s loss of autonomy over smoking: the Global Youth Tobacco Survey. Tob Control. 2011;20(3):201–206. doi: 10.1136/tc.2010.036848. [DOI] [PubMed] [Google Scholar]
- Stephens SH, Hoft NR, Schlaepfer IR, Young SE, Corley RC, McQueen MB, Hopfer C, Crowley T, Stallings M, Hewitt J, et al. Externalizing Behaviors are Associated with SNPs in the CHRNA5/CHRNA3/CHRNB4 Gene Cluster. Behav Genet. 2012;42(3):402–414. doi: 10.1007/s10519-011-9514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, Thun MJ, Goate A, Calle EE. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarter RE, Kirisci L, Mezzich A, Cornelius JR, Pajer K, Vanyukov M, Gardner W, Blackson T, Clark D. Neurobehavioral disinhibition in childhood predicts early age at onset of substance use disorder. Am J Psychiatry. 2003;160(6):1078–1085. doi: 10.1176/appi.ajp.160.6.1078. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiencke JK, Kelsey KT. Teen smoking, field cancerization, and a “critical period” hypothesis for lung cancer susceptibility. Environ Health Perspect. 2002;110(6):555–558. doi: 10.1289/ehp.02110555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiencke JK, Thurston SW, Kelsey KT, Varkonyi A, Wain JC, Mark EJ, Christiani DC. Early age at smoking initiation and tobacco carcinogen DNA damage in the lung. J Natl Cancer Inst. 1999;91(7):614–619. doi: 10.1093/jnci/91.7.614. [DOI] [PubMed] [Google Scholar]
- Winterer G, Mittelstrass K, Giegling I, Lamina C, Fehr C, Brenner H, Breitling LP, Nitz B, Raum E, Muller H, et al. Risk gene variants for nicotine dependence in the CHRNA5-CHRNA3-CHRNB4 cluster are associated with cognitive performance. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(8):1448–1458. doi: 10.1002/ajmg.b.31126. [DOI] [PubMed] [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96(5):684–695. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.